Significance
Primary effusion lymphoma (PEL) is an AIDS-defining cancer. It is associated with Kaposi sarcoma-associated herpesvirus. To date, no sequencing studies have been conducted for this cancer. We used X chromosome-targeted next-generation sequencing to identify 33 genes with coding region mutations in 100% of cases, including in interleukin 1 receptor-associated kinase 1 (IRAK1). IRAK1 kinase modulates toll-like receptor signaling-mediated immune signaling. It binds to MyD88 adapter protein, which is mutated in a subset of diffuse large B-cell lymphomas. IRAK1, however, had not been linked to cancer. This IRAK1 mutant is constitutively active and essential for PEL survival. This highlights the importance of innate immunity signaling as drivers for cancer, particularly those caused by viruses. It also suggests IRAK1 kinase may be a potential target for therapy.
Keywords: herpesviruses, Kaposi sarcoma, IRAK, myd88, primary effusion lymphoma
Abstract
Primary effusion lymphoma (PEL) is an AIDS-defining cancer. All PELs carry Kaposi sarcoma-associated herpesvirus (KSHV). X chromosome-targeted sequencing of PEL identified 34 common missense mutations in 100% of cases. This included a Phe196Ser change in the interleukin 1 receptor-associated kinase 1 (IRAK1). The mutation was verified in primary PEL exudates. IRAK1 is the binding partner of MyD88, which is mutated in a fraction of Waldenström macroglobulinemia. Together, these two mediate toll-like receptor (TLR) signaling. IRAK1 was constitutively phosphorylated in PEL and required for survival, implicating IRAK1 and TLR signaling as a driver pathway in PEL and as a new drug development target.
Primary effusion lymphoma (PEL) is a highly aggressive subtype of diffuse large B-cell lymphoma (DLBCL) (1). With few exceptions, PEL develops in HIV-positive patients and, if left untreated, causes death within a few months. All PELs express the CD138/syndecan-1 marker and carry the Kaposi sarcoma-associated herpesvirus (KSHV). Approximately 60–90% of PELs also harbor EBV (reviewed in ref. 2). KSHV is necessary for PEL growth, but unlike EBV, it is not able to transform mature B cells in culture. PELs are fully transformed. The cells readily grow in culture and form tumors in nude mice (3, 4). This suggests that in addition to viral infection, changes in the host genome also contribute to tumor development. Thus far, no PEL-specific driver mutations have been identified.
Genomic approaches for PEL lag behind those for other cancers because of the rarity of the disease. We reasoned that the rarity was a result of a very defined tumor-development pathway. Unlike DLBCL, which has many subtypes, histological subtypes for PEL have not been described, and we hypothesized that any driver mutation should be present in 100% of samples. This hypothesis has been born out at the level of chromosome copy number variations (5) and transcriptional profiling (6). A signature translocation such as myc in Burkitt lymphoma has not been described for PEL. Over the years, we assembled a large collection of functionally characterized PEL cells (Table 1), which for this project were augmented with nine primary patient exudates.
Table 1.
Sample origin, coverage, and specificity
| Samples* | All reads | On target | Specificity | Coverage | Purpose |
| BC1 | 23,121,252 | 21,691,580 | 0.94 | 443 | Discovery |
| BC2 | 24,444,387 | 22,385,092 | 0.92 | 464 | Discovery |
| BC5 | 19,576,378 | 18,062,589 | 0.93 | 371 | Discovery |
| BCBL1 | 25,674,276 | 24,135,660 | 0.94 | 497 | Discovery |
| BCLM | 19,605,945 | 18,085,981 | 0.92 | 370 | Discovery |
| BCP1 | 16,616,184 | 13,967,038 | 0.84 | 289 | Discovery |
| JSC1 | 10,290,737 | 9,285,549 | 0.90 | 173 | Discovery |
| VG1 | 23,332,697 | 21,419,478 | 0.92 | 439 | Discovery |
| Mean ± SD | 380 ± 107 | ||||
| TY1 | 4,791,393 | 4,535,046 | 0.95 | 58 | Validation |
| AP2 | 4,096,685 | 3,330,357 | 0.81 | 42 | Validation |
| AP3 | 3,017,097 | 2,445,597 | 0.81 | 33 | Validation |
| AP5 | 3,703,516 | 3,385,599 | 0.91 | 45 | Validation |
| BC3 | 2,349,785 | 1,800,776 | 0.77 | 24 | Validation |
| Patient 1 | 4,535,193 | 4,377,692 | 0.97 | 59 | Validation |
| HHB2 | 3,057,278 | 2,698,232 | 0.88 | 31 | Validation |
| ISI1 | 1,830,663 | 1,496,222 | 0.82 | 20 | Validation |
| Mean ± SD | 39 ± 15 |
Sample origin is described in Roy et al. (5).
The PEL samples to which we had access developed in male patients. All PEL cell lines that are available for functional studies are of male origin. We reasoned that any mutation on the X chromosome would automatically be hemizygous, and thus define the phenotype. Any mutation discovered by DNA sequencing in a protein-coding region would result in a mutant protein expressed from that locus, and all mutations are linked in cis (same haplotype). In contrast, mutations in autosomal regions could be in the nonexpressed copy of the gene, and many would be heterozygous. Furthermore, ambivalent nucleotide assignments on chromosome X are most likely the result of sequencing errors, as prior studies did not find evidence of gene amplification of chromosome X. Focusing on chromosome X thus reduced the coverage requirement to ascertain single nucleotide variants (SNVs). Therefore, we conducted targeted profiling of chromosome X, using the RainDance XSeq panel and technology. This panel provides for >98% coverage of 800 known genes on chromosome X. Coverage spans all exons as well as the 5′ promoter region and 3′ UTR for each gene (7). Using this approach, we identified, among others, a new mutant in interleukin 1 receptor-associated kinase 1 (IRAK1) as being conserved in 100% of PEL. The mutation introduced a new phosphorylation site. Experimentally, we showed that constitutive IRAK-1 kinase activity was essential for PEL survival.
Results
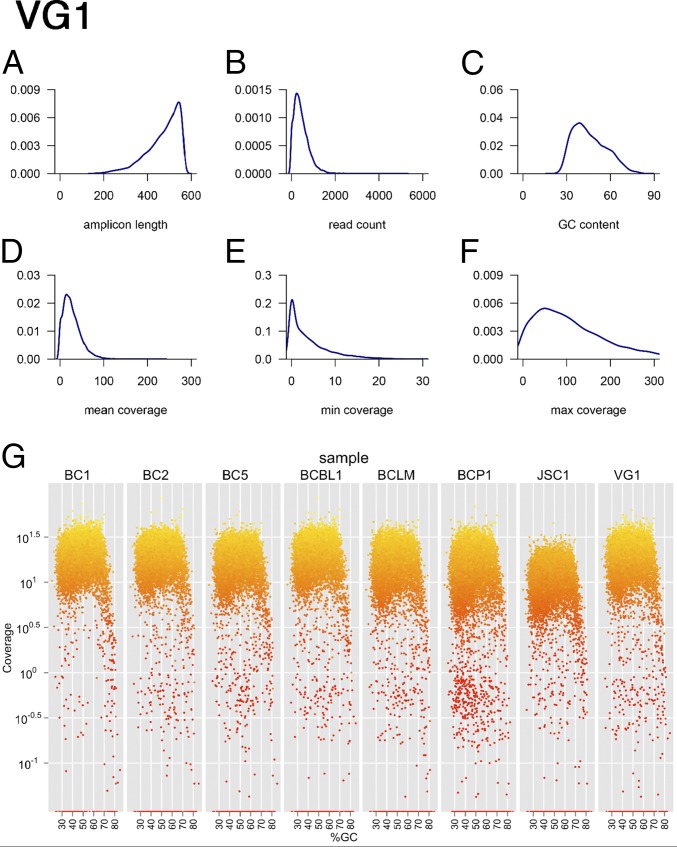
To identify single nucleotide changes on the X chromosome in PEL, we used targeted resequencing. Targets were enriched by droplet-PCR (RainDance platform) and then subjected to next-generation sequencing. We used a discovery set of eight samples, which were sequenced to a mean coverage depth of 380-fold ± 107-fold, and a second validation set of eight samples (including primary PEL exudate cells), which were sequenced to a mean coverage depth of 39-fold ± 15-fold (Table 1). Overall, we generated 76 Mb of 36-bp single-end reads using an Illumina genome analyzer and 209 Mb of 100-bp paired-end reads using an Illumina HiSeq instrument. More than 90% of the regions of interest were covered with a depth of 20× or greater, with no apparent bias for GC-content (Fig. 1).
Fig. 1.
Individual quality control (QC) of droplet-PCR enrichment and sequencing. Shown are six density curves representing the distribution of (A) the predicted amplicon length in the PCR, (B) the read-count per amplicon, (C) the GC-content across all PCR amplicons, (D) the mean coverage, (E) the minimal coverage, and (F) the maximal coverage on each position using the read data for the VG1 PEL as an example. (G) Coverage by GC-content. Shown are n = 8 samples and how coverage changes with amplicon GC-content. Data were from the discovery, single-lane multiplexed QC run, not the final data set, which includes additional sequencing runs. The final coverage is reported in Table 1. The vertical axis shows coverage on a log10 scale; the horizontal axis shows the %GC-content of each amplicon. Colors indicate coverage of each amplicon, with bright yellow indicative of higher and dark red of lower coverage. Note the drooping tail at >70% GC-content in all samples, which represents the maximal GC-content compatible with this type of PCR.
In all (n = 16; 100%) samples, 1,915 SNVs were present; of those, 512 SNVs were located in coding regions. Including 8,304 variants in the coding region, leading to 55 missense mutations, 29,313 SNVs had ≥50% frequency. We did not perform copy number analysis on the sequencing data, as we previously published the copy number profiles of these samples (5).
SNV detection remains challenging because of the high sequence error rates. We examined several factors when reporting SNVs and indels. Coverage of 30× or higher was required for each SNV call in accordance with prior work (8). If there were two variants at the same genomic location, the true variant reported had to appear at 60% frequency or higher. We aligned all of the targeted region primer sequences designed to the reference genome and excluded any variants within the primer capture sequences. We confirmed the concordance of known SNVs, using data from Affymetrix single-nucleotide polymorphism (SNP) arrays (5). As our aim was to find novel tumor-specific SNVs, we compared our data with those from Hapmap data release 28, from August 2010, on National Center for Biotechnology Information B36 assembly, dbSNP b126, and filtered out common variants. Finally, we validated individual SNVs by Sanger sequencing. This let to our consensus list of PEL-specific SNVs in chromosome X (Table 2).
Table 2.
SNVs on chromosome X that change the protein coding region in PEL
| Gene | AA | Change | Frequency* | mRNA† | Name |
| DHRSX | 292, 247 | His > Arg, Val > Leu | 8/8 | nd | Dehydrogenase/reductase (SDR family) |
| ARHGAP6 | 202, 200 | Asn > Lys, Thr > Pro | 8/8 | No | Rho GTPase-activating protein 6 |
| RAI2 | 202 | Met > Val | 8/8 | No | Retinoic acid-induced protein 2 |
| MAGEB2 | 61 | Glu > Lys | 8/8 | Yes | Melanoma-associated antigen B2 |
| TAB3 | 394 | Trp > Arg | 8/8 | nd | TGF-β-activated kinase 1 |
| DMD | 477 | Arg > Gln | 8/8 | Yes | Dystrophin Dp71ab isoform |
| RPGR | 889,887,878,880 | Glu > Gly | 8/8 | Yes | Retinitis pigmentosa GTPase regulator |
| SSX5 | 19 | Glu > Gln | 8/8 | Yes | Synovial sarcoma, X breakpoint 5 |
| WDR13 | 325 | His > Arg | 8/8 | Yes | WD repeat-containing protein 13 |
| CCDC120 | 571 | Ala > Gly | 8/8 | nd | Coiled-coil domain containing protein 120 |
| MAGIX | 112,45‡ | His > Arg | 8/8 | No | PDZ domain-containing protein MAGIX |
| 173,97 | Phe > Leu | ||||
| 323,247,242 | Val > Leu | ||||
| FAM120C | 1083, 1082 | Gln > Pro, Ile > Thr¶ | 8/8 | Yes | Family with sequence similarity 120C |
| MTRNR2L10 | 3,2 | Thr > Pro, Ala | 8/8 | nd | Humanin-like protein 10 |
| ZXDB | 764 | Asp > Asn | 8/8 | nd | Zinc finger X-linked protein ZXDB |
| EDA2R | 129 | Thr > Ala | 8/8 | No | Ectodysplasin A2 receptor |
| ATP7A | 1350 | Glu > Lys | 8/8 | Yes | Copper-transporting ATPase |
| POU3F4 | 237 | Gly > Ala | 16/16 | Yes | POU domain, class 3, factor 4 |
| RNF128 | 135 | Phe > Tyr | 8/8 | No | E3 ubiquitin-protein ligase RNF128 |
| GUCY2F | 296 | Arg > Gln | 8/8 | No | Retinal guanylyl cyclase 2 |
| LRCH2 | 398 | Asp > Asn | 8/8 | nd | Leucine-rich repeat and calponin homology domain-containing protein 2 |
| 723 | Arg > Cys | ||||
| BCORL1 | 111, 660 | Phe > Leu, Val > Gly | 16/16 | Yes | BCL6 corepressor-like protein 1 |
| RAB33A | 35 | Arg > Gly | 8/8 | Yes | Ras-related protein Rab-33A |
| DDX26B | 843 | Ile > Gly | 8/8 | nd | DEAD/H (Asp-Glu-Ala-Asp/His) box |
| ATP11C | 114 | Cys > Trp | 8/8 | nd | ATP11C ATPase, class VI, type 11C |
| MAGEC1 | 102, 103 | Ser > Pro/Cys, P > S | 8/8 | Yes | Melanoma-associated antigen C1 |
| CXorf40A | 70 | Thr > Ala | 8/8 | Yes | Chromosome X orf40 A |
| MAGEA10 | 234, 166 | Val > Ile, Arg > Lys | 8/8 | Yes | Melanoma-associated antigen 10 |
| SLC6A8 | 431,421,316 | Asp > Asn | 16/16 | No | Sodium- and chloride-dependent creatine transporter 1 |
| 454,444,339 | Cys > Arg | ||||
| 504,494,389 | Met > Thr | ||||
| 590,580,475 | Arg > Cys | ||||
| 592,582,477 | Gln > Lys | ||||
| 636,626,521 | STOP > Gly | ||||
| ABCD1 | 606 | Ser > Pro | 16/16 | Yes | ATP-binding cassette subfamily D member 1 |
| 635, 712 | Val > Met, Arg > His | ||||
| PLXNB3 | 1156, 1535 | Glu > Asp, Met > Thr | 8/8 | No | Plexin-B3 |
| IRAK1 | 196 | Phe > Ser | 8/8 | Yes | Interleukin 1 receptor kinase 1 |
| VBP1 | 123 | Met > Val | 8/8 | Yes | Prefoldin subunit 3 |
| TCEAL6 | 175 insC | Gln > fs | 8/8 | nd | Transcription elongation factor A-like 6 |
| TEX13A | 201 ins,199 ins | Trp > fs, Gly > fs | 8/8 | No | Testis-expressed 13A |
nd, not determined.
Number of mutant samples/number of samples with coverage ≥30× (frequency), additional samples had <30× coverage, and even though they often had the same mutation on confirmatory sequencing, we did not include them here.
Based on GEO data sets GSE2149 and GDS1063.
More than one amino acid position resulting from differential splicing, which generates isoforms.
Low coverage.
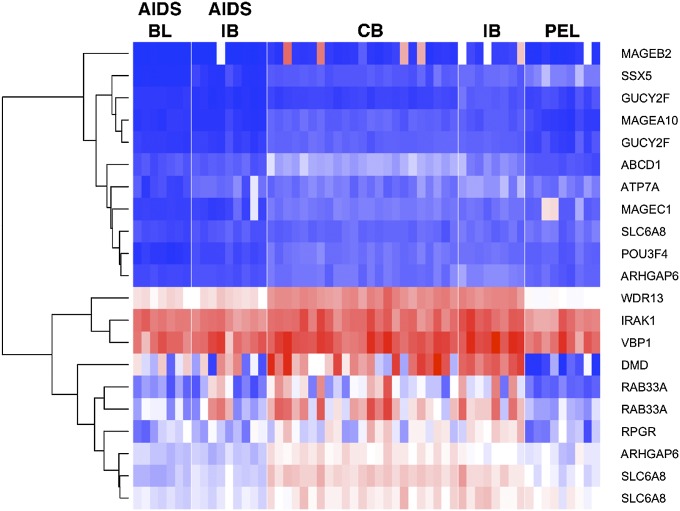
To further narrow our results and to identify the most biologically relevant alterations, we used previously published PEL mRNA profiling data (6, 9) to establish which of our target genes were transcribed in PEL (Fig. 2). The three genes IRAK1 Phe196Ser, prefoldin subunit 3 (VBP1) Met123Val, and WD repeat-containing protein 13 (WDR13) 201/199fs were transcribed at high levels in PEL, and each had a novel, PEL-defining SNV in 100% of samples (P ≤ 0.05, Bonferroni-adjusted based on a comparison of 800 targeted genes across 16 samples, assuming a 12% frequency of a new SNV being present in any one sample by chance). All three genes represent candidate driver mutations in PEL.
Fig. 2.
Heat map of transcription for highly mutated genes in PEL and other types of DLBCL. The expression values are derived from Affymetrix CEL data (GEO data set GDS1063). Red color represents high, white intermediate, and blue low expression levels. The genes shown on the vertical axis are the subset of the highly mutated genes reported in Table 2. Individual sample identifiers are shown on the bottom and classes on the top. BL, Burkitt lymphoma; CB, centroblastic; IB, immunoblastic.
We prioritized further studies on the IRAK1 Phe196Ser (C587T > C, at nt 153,284,192) mutation, which would introduce a novel phosphorylation site. The mutated residue is part of a phenylalanine triad and is 100% conserved across species. The IRAK1 Phe196Ser mutation was present in 13 of the 16 PELs subjected to targeted next-generation sequencing. For the remaining three samples, the coverage was too low to ascertain the mutation. For two of these samples (CRO-AP3 and CRO-AP5), the mutation was subsequently confirmed by PCR and Sanger sequencing. We obtained DNA from eight additional primary PEL cases and sequenced IRAK1. The IRAK1 Phe196Ser mutation was present in seven (86%) of these eight primary PEL cases (Table 3). We also obtained normal tissue from two of the patients, and neither had the IRAK Phe196Ser mutation. For the PEL cell lines, we had complete SNV data. Other SNVs in the IRAK1 gene, which do not change the amino acid sequence, were not conserved among the PEL samples (Fig. 3). This suggests that this mutation was selected for and that each case used in this collection represents an independent sample.
Table 3.
IRAK coverage and confirmation
| Phe196Ser, c587T > C (nt 153,284,192)* | |||
| Sample | Frequency (%)† | Coverage (fold) | Confirmed‡ |
| BC1 | 100 | 154 | nd |
| BC2 | 100 | 237 | Yes |
| BC5 | 100 | 362 | ndd |
| BCBL1 | 100 | 313 | Yes |
| BCP1 | 100 | 111 | Yes |
| BCLM | 100 | 141 | Yes |
| JSC1 | 99.1 | 225 | nd |
| VG1 | 100 | 116 | Yes |
| HHB2 | 100 | 13 | nd |
| BC3 | 100 | 13 | Yes |
| TY1 | 100 | 10 | nd |
| Patient 1 | 100 | 11 | nd |
| CRO-AP5 | Low¶ | 0 | Yes |
| ISI1 | Low¶ | 0 | nd |
| CRO-AP2 | 6.25 | 32 | nd |
| CRO-AP3 | Low | 0 | Yes |
| Patient 2 | —§ | — | Yes |
| Patient 3 | — | — | Yes |
| PEL625# | — | — | Yes |
| PEL533# | — | — | Yes |
| PEL534# | — | — | No |
| PEL535# | — | — | Yes |
| PEL536# | — | — | Yes |
| PEL537# | — | — | Yes |
| Skin# | — | — | No |
| Spleen# | — | — | No |
nd, not determined.
Human Genome built hg19_2011_07_29.
Frequency refers to the mutation as percentage of total reads.
Confirmed by PCR amplification and direct Sanger sequencing (Phe196Ser) or PCR amplification, subcloning and Sanger sequencing.
Too low coverage to determine.
PEL DNA from patients was not subjected to next-generation sequencing.
DNA from HIV+ samples at the AIDS Cancer Specimen Resource.
Fig. 3.
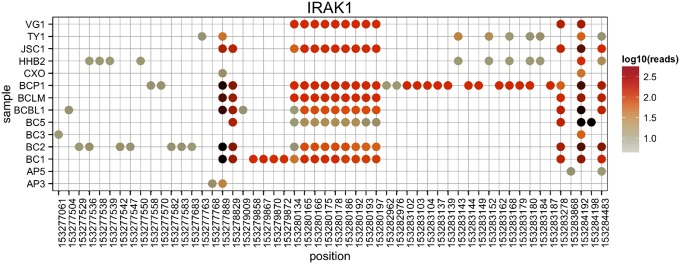
IRAK1 haplotypes. In addition to the common SNV, we also uncovered multiple sample-specific SNVs across the IRAK1 coding sequence (cds). These allowed us to establish SVN haplotypes in IRAK1 for each of the samples. Shown are the positions of all SNVs across the IRAK1 targeted regions on the x axis and the sample name on the y axis. Dots indicate a nucleotide difference from the hg19 reference genome, and the color of the dots indicates the number of reads covering that specific SNV on a log10 scale. Note that all nucleotide positions were covered in all cell lines. Note that each cell line has a different SNV pattern, demonstrating the unique origin of each of our samples.
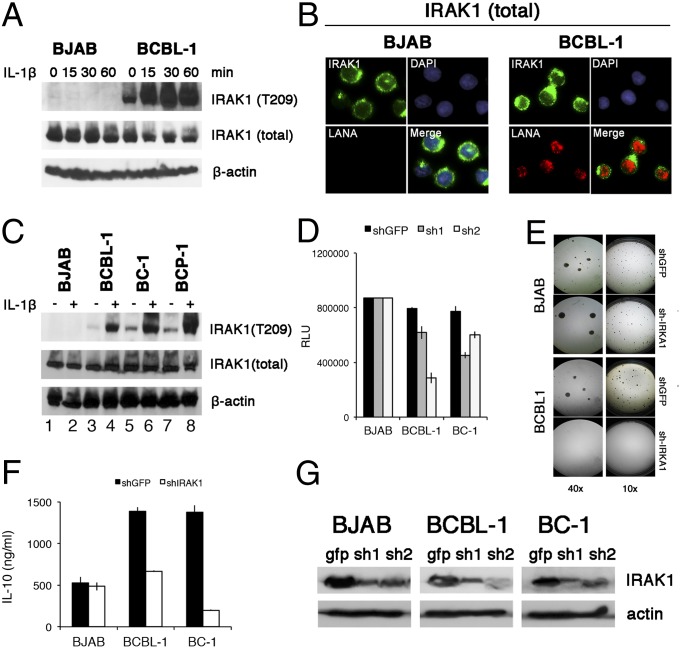
To establish a functional role for these findings, we focused on IRAK1. IRAK1 mediates toll-like receptor (TLR) and IL-1 receptor signaling (10). IRAK1 expression in PEL was confirmed by Western blot (Fig. 4A). IRAK2 and IRAK4 were also expressed, but not the inhibitory IRAK M isoform. Unlike in BJAB cells, in PEL, IRAK1 was constitutively phosphorylated at T209, an activation marker residue (11), in the absence of receptor stimulation (Fig. 4A). Immune fluorescence analysis demonstrated IRAK1 T209 phosphorylation in about 20% of BC1 cells. BJAB cells did not show evidence of IRAK1 T209 phosphorylation (Fig. 4B). This experiment was conducted in the absence of IL-1β. Even though in PEL, IRAK1 was already phosphorylated at T209 before stimulation (Fig. 5A, lane 1 compared with lanes 3, 5, and 7), upon IL-1β stimulation, T209 became more phosphorylated. IL-1β-induced IRAK1 T209 phosphorylation increased over time and was observed in multiple independent PEL cell lines (Fig. 5 A and C, lanes 4, 6, and 8). This suggests that the IL-1β-signaling pathway in PEL was functional, but that the activation threshold was lower or that IL-1β-independent signaling events drive IRAK1 activation in KSHV-infected cells.
Fig. 4.
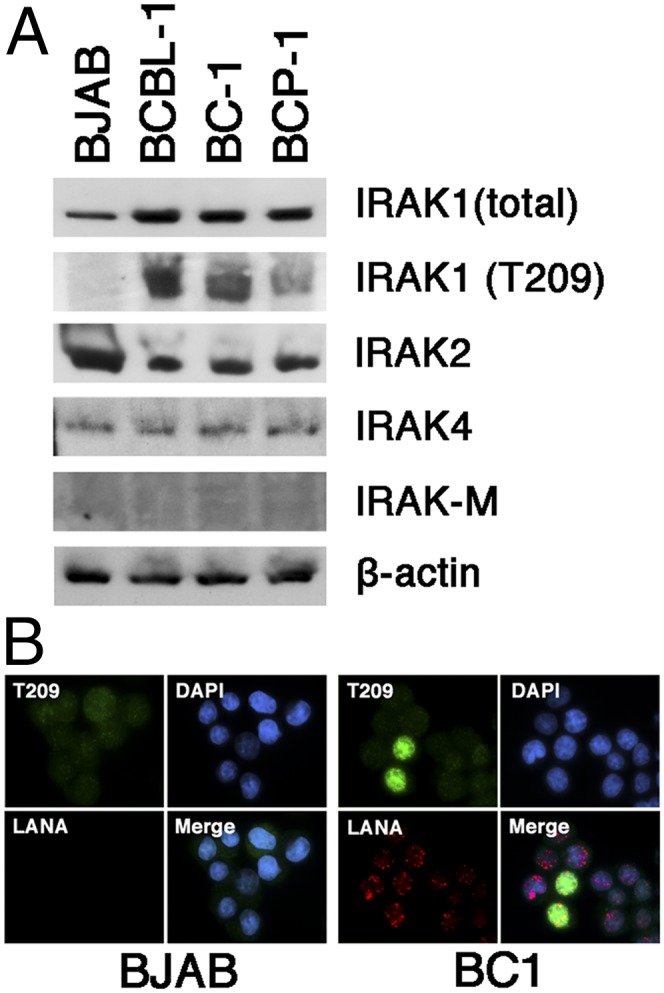
Expression of IRAK isoforms in PEL. (A) Shown is a Western blot of multiple PEL samples (BCBL-1, BC-1, BCP-1) and the BJAB Burkitt lymphoma cell line for total IRAK1, phosphoIRAK1T209, IRAK2, IRAK4, IRAK-M, and beta actin. (B) Immunofluorescence of IRAK1 phosphorylation in BJAB and BC1 cells, using antibodies against the T209 phosphorylation site in IRAK1 (green) and LANA (red). Nuclear DNA is counterstained with DAPI (blue).
Fig. 5.
Function of mutant IRAK1 in PEL. (A) BJAB and PEL cells were stimulated with 10 ng/mL IL-1β for indicated amounts of time and protein extracts were analyzed by Western blotting, using rabbit anti-IRAK1, anti-phospho IRAK1 T209, and mouse anti-β-actin, followed by secondary HRP-conjugated antibodies. (B) BJAB and PEL cells were fixed, permeabilized, and stained with rabbit anti-IRAK1 and mouse anti-LANA monoclonal antibodies, followed by fluorescence-conjugated secondary antibody. Nucleus was stained blue with DAPI. (Magnification: 600×.). (C) A panel of PEL cell lines and the BJAB Burkitt lymphoma cell line were stimulated with 10 ng/mL IL-1β for 30 min and analyzed as in A. (D) Cells viability was analyzed with a CellTiter-Glo kit. BJAB and PEL cells were infected with two independent shRNA-lentiviruses against IRAK1 for 4 d, and luminescence was measured. GFP shRNA lentiviruses were used as negative controls. (E) Result of colony formation assay at 10× and 40× magnification of BJAB and BCBL-1 cells infected with IRAK1 shRNA lentivirus vector and cultured under 5 µg/mL puromycin selection for 3 weeks. (F) Reduction in secreted IL-10 accumulated for 4 d after infection with anti-IRAK1 or anti-GFP shRNA for indicated cell lines. (G) Western blot of IRAK1 levels at 4 d after infection with anti-IRAK1 or anti-GFP shRNA for indicated cell lines.
To test the hypothesis that IRAK1 was required for PEL survival, we diminished IRAK1 expression by shRNAs. Abrogation of IRAK1 expression by multiple independent shRNAs abolished PEL growth in culture (Fig. 5D) and in soft agar (Fig. 5E) and decreased IL-10 secretion (Fig. 5F). IL-10 is expressed in most PELs and is required for survival (12). BJAB cells were oblivious to IRAK1 depletion in these assays. Fig. 5G shows the efficacy of IRAK1 knockdown for two different shRNA constructs. These data establish IRAK1Phe196Ser as a novel driver mutation in PEL.
Discussion
PEL is a rare viral lymphoma. Although transcriptional profiling has been done (6, 13), sequencing studies have not, as it has been exceedingly difficult to obtain enough cases. We collected all published cell lines as well as nine primary cases for this study. The use of cell lines allowed us to study the functional consequences of our analysis, which would not have been possible using only archival specimens.
We and others previously reported karyotyping and comparative genome hybridization (CGH) profiles for some PEL (5, 14–19). Our prior data were array-based and included a survey of known SNVs, which was used as a quality control for this study. Here, we used targeted next-generation sequencing to identify tumor-specific SNVs. We obtained a large number of significantly enriched SNVs in PEL. Each can be considered part of the mutational signature of PEL and has the potential to contribute to the PEL phenotype. Detailed validation studies on all candidates are in progress. A limitation of this study is the small number of cases, although it represents the largest number of PELs analyzed to date, and even though in two cases, nontumor tissue did not have the tumor-specific SNV. In an attempt to address this limitation, we adopted a very stringent threshold and only report SNVs that were present in >90% of cases. In comparison, ID3 was identified as a novel driver mutation in Burkitt lymphoma based on a 30–60% mutation frequency and five times higher number of cases (20–22).
From a statistical point of view, all the PEL-specific SNVs represent candidate driver mutations and deserve experimental follow-up, most notably VBP1 and WDR13, which are abundantly transcribed in PEL. We prioritized IRAK1Phe196Ser because IRAK inhibitors have entered phase I human clinical trials. Fifty-five previously identified, noncoding SNVs in the IRAK1 locus were not conserved; only the single new SNV introducing the IRAK1Phe196Ser mutation was. The IRAK1Phe196Ser mutation is not a common polymorphism, as recorded in various SNV databases. This suggests the IRAK1 locus as a whole was not selected for or fixed by chance in this set of samples; only the specific single nucleotide substitution was.
The IRAK1Phe196Ser mutation was also present in seven of eight primary PEL exudate samples, which were never cultured and thus this mutation potentially represents a driver for growth in the patient. The IRAK1Phe196Ser variant was constitutively phosphorylated at T209 in PEL without exogenous stimuli but could be further phosphorylated in response to IL-1β. At this point, we do not know the complete phosphorylation pattern of IRAK1Phe196Ser in PEL.
IRAK1 signaling was required for PEL survival in culture. The most exciting interpretation of these data would be that IRAK1Phe196Ser is a gain-of-function mutation and that activated IRAK1Phe196Ser kinase functions as a novel cell autonomous oncogene. Indeed, IRAK1Phe196Ser has different biochemical properties than the wild-type protein. IRAK1 activation is critically dependent on T209 phosphorylation (11), and T209 was phosphorylated in PEL in the absence of exogenous stimuli. The scenario is more complex, however, as IRAK1 is made in at least three isoforms and is part of a multicomponent signaling complex including MyD88, IRAK4, and various receptors, adaptors, and possibly even viral proteins. At this point, we do not know the exact composition of this complex in PEL or whether MyD88 is mutated as well. It may be that the IRAK1Phe196Ser allele is better at interacting with MyD88, IRAK4, or KSHV transmembrane oncogenes (K1, K15) than wild-type IRAK1. In EBV lymphoma, IRAK1 is essential for mediating LMP1 signaling (23). In the latter case, any phenotype of IRAK1Phe196Ser would only be visible in the context of viral infection.
In addition to PEL, KSHV also causes KS. Case- and family-based studies in pediatric KS had identified candidate susceptibility genes with immune signaling phenotypes, such as STIM1 and IFN-γR1 (24, 25). Genome-wide association studies for KS have not been reported to date. As a result of targeted sequencing of the X chromosome, we identified 55 novel, common, and by definition homozygous mutations in protein-coding regions, including IRAK1Phe196Ser. It would be interesting to see whether these same SNVs are also enriched in KS.
IRAK1 is activated by IRAK4/MyD88 in response to TLR (except TLR3) and IL-1R signaling. All TLRs are expressed in PEL and control KSHV reactivation (26). B cells from KSHV transgenic mice are hyperresponsive to TLR signaling (27). KSHV encodes a micro-RNA, mir-K12-9, which targets the IRAK1 3′ UTR (28). The mir-K12-9 is expressed in most PEL, and as shown here, so is IRAK1. If IRAK1 mRNA is a target for miR-K12-9, then its relevance must be during the initial phases of infection in primary cells before transformation into PEL, or perhaps in other cell types (endothelial cells, macrophages).
IRAK1 also contributes to the risk for systemic lupus erythematosus (29). It has been implicated in EBV LMP1 signaling (23), but mutations in IRAK1 have not previously been associated with human lymphoma. A specific (Leu265Phe) mutation in IRAK1's binding partner MyD88 was seen in 29% of activated B-cell–like DLBCL (30) and in a subset of Waldenström macroglobulinemia (31, 32). Finally, an IRAK1/4 inhibitor showed activity against Waldenström macroglobulinemia and myelodysplastic syndrome (33, 34). This work provides a rationale for testing the same inhibitor in PEL. In a larger sense, it suggests that inhibiting IRAK1/IRAK4/MyD88 immune signaling may yield new treatments for a number of viral cancers.
Materials and Methods
Sample Collection and Processing.
Fifteen PEL cell lines and one primary exudate were used. We previously reported the CGH analysis for these samples (5). We submitted two batches of samples for targeted region enrichment, using the RainDance platform at Expression Analysis and then the enriched DNA to Illumina sequencing at the University of North Carolina High Throughput Sequencing Facility. The first batch contained eight tumor cell lines, 2.5 µg DNA (BC1, BC2, BC5, BCBL1, BCLM, BCP1, JSC1, and VG1). The second batch contained eight tumor cell lines with limited DNA amounts, less than 2.5 µg (CRO-AP2, CRO-AP3, CRO-AP5, BC3, patient 1, HHB2, ISI1, and TY1). After library preparation, we validated the samples, using Quant-iT PicoGreen dsDNA (Invitrogen).
Enrichment and Sequencing.
We used RainDance technology for sequence enrichment (35) and TruSeq DNA Sample preparation Kit V2 (Illumina, catalog number FC-121–2001) for library preparation. The genomic DNA was sheared into 2–4-kb fragments, purified, and mixed with all of the PCR reagents except the primers. The mixture then formed a template droplet (∼18 pL) within the microfluidic chip. It was then combined with the primer pair droplets at a 1:1 ratio and formed a single PCR droplet (∼26 pL). The collection of the PCR droplets was amplified using Illumina paired-end PCR primers, and then the PCR amplicons were purified and normalized to 10 nM for next-generation sequencing.
For sequencing, we first used an Illumina Genome Analyzer for single reads with a target read length of 36 bp to verify the targeted regions were covered. Our evaluation showed the enrichment was successful (Table 1). Because this alone was not enough to make high confidence variant calls, we resequenced the libraries by paired-end sequencing with a mean read length of 100 bp, using the Illumina HiSEq.2000 platform.
Alignment and Variants Calling.
Read mapper on CLC genomic workbench version 5.5 (CLC Bio) was used to align all of the reads to the human reference genome build 19 Homo_sapiens_chromosome_hg19_2011_07_29 (155,270,560 bp, NC_000023). CLC read mapper is a fast and accurate high-throughput sequencing alignment tool that combines the Smith-Waterman algorithm for accuracy and a heuristic algorithm for speed. Mapping reads to a reference genome and contains three steps: preprocessing the input data, the actual mapping, and postprocessing the mapping results. Those steps become one integrated workflow in CLC genomic workbench software.
Mapping was done with a local alignment of reads to reference, in which an aligned read must have at least 50% of the length mapped with at least 80% of similarity to the reference. Mismatch, insertion, and deletion were allowed with a penalty score of 2 (a match is scored 1); any nonspecific matches (reads matched to multiple places) were discarded.
Variant calling was done with quality-based variant detection in CLC workbench. This tool is based on the Neighborhood Quality Standard algorithm (36) and finds all variants that passed the quality filters and user-specified thresholds. The minimum coverage was set to 4, and every position in the radius of 5 from a variant had to be accessed on the basis of the qualities; up to 2 mismatches and gaps were allowed for a variant call with minimum of variant frequency set to 35%. CLC variant detection also annotates where the mutation lies and the type of mutations (eg, amino acid change, insertions, and deletions). We then used the R software environment to further filter out the mutations by coverage, frequency, location, and type.
The specific parameter settings were, for short reads, mismatch cost = 2 and limit = 8; and for long reads, similarity = 0.8, length fraction = 0.5, insertion cost = 3, deletion cost = 2, and mismatch cost = 2. For paired reads, minimal distance = 80 and maximal distance = 2,500. Common to all were conflict resolution = vote, ignore nonspecific matches, no masking of reference, and local alignment. For SVN call, we used window length = 11, maximum gap and mismatch count = 2, minimum central quality = 20, minimum average quality = 15, minimum coverage = 4, minimum variant frequency = 35.0%, and maximum expected variations (ploidy) = 2.
Sanger Sequencing.
We validated missenses and frame shifts with a high mutation rate using primer sets designed to cover the specific mutation points. Amplicons were amplified by conventional PCR, using 2× GoTaq Green Master Mix (Promega, catalog number M7122), and either directly sent for sequencing or first cloned into pGEM-T Easy Vector System I (Promega, catalog number A1360) and then subjected to Sanger sequencing, using T7 promoter and Sp6 reverse primers on three or more insert positive clones (Eton Bioscience Inc or GENEWIZ Inc).
Affymetrix SNP Array Data Integration.
Since we had previously analyzed known SNPs on all cell lines (5), we had SNP calls available for each cell line from Gene Expression Omnibus (GEO) entry GSE25839. The Affymetrix Genome-Wide Human SNP 6.0 Array is a SNP-based microarray that features 1.8 million genetic markers, including 906,600 SNPs and 946,000 probes for the detection of copy number variation. We downloaded and extracted the ID_Ref for all SNPs from Affymetrix 6.0 dataset GPL6801. Then we extracted the SNPs on chromosome X and obtained the forced SNP calls for each of the samples. Next, we built an ID_Ref × PELline matrix and subjected it to unsupervised clustering by cell line. Finally, we identified SNPs that were also covered by the targeted amplification and sequencing to verify the sequencing recovered the SNP.
Expression Data Analysis for Mutates Genes.
We downloaded the expression profiling data by Affymetrix Array from the GEO database GSE2350 for the following cell lines: AIDS-Burkitt lymphoma contains 7 samples, AIDS-DLBCL-immunoblastic contains 9 samples, DLBCL-CB contains 23 samples, DLBCL-immunoblastic contains 10 samples, and PEL contains 9 samples. All of the original data files are in CEL format, so we first applied transformation to the expression value by the standard Affymetrix normalization method to eliminate bench effect, using the R packages “aroma.affymetrix” and “affy” to preprocess the CEL files and export a text file containing a table of estimated gene-level expression values. Then we applied a hierarchical clustering algorithm to cluster the genes. Here we have a matrix of expression values, in which rows are genes and columns are cell lines. We applied hierarchical clustering coupled with Euclidean distance to measure (dis)similarities and Wald linkage function to group rows.
Cell Viability Assay.
To check cell viability, BJAB and PEL (BCBL-1 and BC-1) cells were cultured in 96-well plates (10,000 cells per well) with RPMI (Gibco, 11875, 100 µL/mL), containing 2 mM l-glutamine, 10% FBS, penicillin G (100 U/mL), and streptomycin sulfate (100 µg/mL) and supplemented with 0.05 mM 2-mercaptoethanol (Invitrogen, 21985–023), 0.075% sodium bicarbonate (Life Technologies, 25080–094), and 1 U/mL human IL-6 (Peprotech, 200–06). Cells were harvested for viability analysis on day 3, the substrate of CellTiter-Glo (Promega, G7571) was added, and luminescent signals were measured by FLUOstar Optima 96-well luminometer (BMG Labtech). Three independent biological assays were performed for each experiment, and each measurement was conducted in three technical replicates.
Il-6 and Il-10 ELISA.
In a 12-well plate, 5 × 105/mL cells were seeded and treated as indicated, and supernatants were collected. The levels of secreted IL-10 were measured using commercial ELISA according to the manufacturer's protocol (eBioscience).
Knockout of IRAK1 with shRNA Lentivirus.
Two pLKO.1 lentiviral vectors (TRCN000000121137 and TRCN000000121138) targeted for IRAK1 (GeneID 3654) were obtained from Open Biosystems/Thermo Inc. The reconstructed lentiviruses were produced by the Lenti-shRNA Core Facility at the University of North Carolina. BJAB and PEL cells (5 × 105) were grown in multiwell plates, infected with 1 mL or 106 pfu lentiviruses (multiplicity of infection of 2) separately by adding Polybrene at a final concentration of 10 µg/mL, and incubated at 37 °C for 6 h. After infection, medium was replaced with fresh RPMI 1640 supplemented with 10% FBS and containing 2 mM l-glutamine, penicillin G (100 U/mL), and streptomycin sulfate (100 µg/mL) and supplemented with 0.05 mM 2-mercaptoethanol (Invitrogen Inc.), 0.075% sodium bicarbonate (Life Technologies, Inc.), and 1 U/mL human IL-6 (Peprotech, Inc.). On day 2, puromycin (5 µg/mL) was added to the medium. GFP lentivirus (shRNA-GFP) was used as control.
Immunofluorescence.
BJAB and BCBL-1 cells were fixed with 3% (wt/vol) buffered paraformaldehyde for 20 min at room temperature (RT), permeabilized with 0.2% Triton X-100 in PBS for 15 min at 37 °C. Samples were then immune-stained with rabbit anti-IRAK1 (Cell Signaling, catalog number 4504, at 1:200), anti-phosphoIRAK1 (T209, Cell Signaling, catalog number 12756, at 1:200), and mouse anti-LANA (NCL-HHV8-LNA, Leica, catalog number NCL-HHV8-LNA at 1:500), for 2 h at RT, followed by incubation with secondary antibody: Texas-Red anti-mouse IgG (Vector Laboratories, catalog number TI-2000, at 1:1,000) or fluorescein anti-rabbit IgG (Vector Laboratories, catalog number FI-1000, at 1:1,000) for 60 min at RT in the dark. Cells were washed three times with Tris-buffered saline and Tween 20, and nuclei were stained with DAPI (Sigma, catalog number D9542 at 1:1,000). Samples were observed with a fluorescence microscope type Leica DM4000B and imaged using a 100 ×/1.40–0.7 oil numerical aperture objective and a Q-Imaging Retiga 2000RV camera.
IL-1β Stimulation.
Cells were treated with 10 ng/mL IL-1β (R&Ds System, catalog number 210-LB) for 0, 15, 30, and 60 min, followed by Western blotting assay with anti-IRAK1 (Cell Signaling catalog number 4504, at 1:1,000) and anti-phoshoIRAK1(T209) (Cell Signaling catalog number 12756, at 1:1,000) antibodies. β-actin (Cell Signaling, catalog number A5441, at 1:3,000) was used as loading control. Signal was developed using Pierce ECL Western blotting substrate (Thermo Scientific, catalog number 32209) and recorded on film (Genesee Scientific, catalog number 30–101).
Colony Formation.
BJAB and PEL cells (5 × 105) were infected with shIRAK1 lentiviruses as described earlier. After 24 h, cells were mixed with Methocult M3134 medium (Stemcell Technologies, catalog number 03134) (1:10) according to manufacturer’s recommendation, puromycin (5 µg/mL) was added to the medium, and the mixture was dispensed into a 35-mm culture dish (catalog number 27100) by using a 3-mL syringe attached to a 16-gauge blunt-end needle (catalog number 28110). After 3 weeks, colonies were observed under dissecting microscope type Leica MZ6 (KL1500 LCD) with 1× objective and 10× eyepieces and were imaged using a Leica DC 150 camera with 3,072 × 2,304 pixel resolution.
Acknowledgments
The authors thank Blossom Damania and members of the D.P.D. laboratory for critical reading and Pauline Chugh for providing additional sample DNAs. Additional samples were obtained from the AIDS Cancer Specimen Resource. This work was supported by public health service Grants DE018304, CA019014, CA177315, and AI107810 (to D.P.D.) and funding through the AIDS Malignancies Clinical Trials consortium (CA121947 to D.P.D. and D.H.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data used for this analysis are available at National Center for Biotechnology Information short reads archive (accession no. SRP032975).
References
- 1.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 2.Carbone A, Cesarman E, Spina M, Gloghini A, Schulz TF. HIV-associated lymphomas and gamma-herpesviruses. Blood. 2009;113(6):1213–1224. doi: 10.1182/blood-2008-09-180315. [DOI] [PubMed] [Google Scholar]
- 3.Staudt MR, et al. The tumor microenvironment controls primary effusion lymphoma growth in vivo. Cancer Res. 2004;64(14):4790–4799. doi: 10.1158/0008-5472.CAN-03-3835. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff C, et al. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91(5):1671–1679. [PubMed] [Google Scholar]
- 5.Roy D, Sin SH, Damania B, Dittmer DP. Tumor suppressor genes FHIT and WWOX are deleted in primary effusion lymphoma (PEL) cell lines. Blood. 2011;118(7):e32–e39. doi: 10.1182/blood-2010-12-323659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein U, et al. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood. 2003;101(10):4115–4121. doi: 10.1182/blood-2002-10-3090. [DOI] [PubMed] [Google Scholar]
- 7.Tarpey PS, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41(5):535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan W, et al. Distinct subsets of primary effusion lymphoma can be identified based on their cellular gene expression profile and viral association. J Virol. 2005;79(2):1244–1251. doi: 10.1128/JVI.79.2.1244-1251.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Z, Henzel WJ, Gao X. IRAK: A kinase associated with the interleukin-1 receptor. Science. 1996;271(5252):1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 11.Kollewe C, et al. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem. 2004;279(7):5227–5236. doi: 10.1074/jbc.M309251200. [DOI] [PubMed] [Google Scholar]
- 12.Jones KD, et al. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi’s sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood. 1999;94(8):2871–2879. [PubMed] [Google Scholar]
- 13.O’Hara AJ, Vahrson W, Dittmer DP. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood. 2008;111(4):2347–2353. doi: 10.1182/blood-2007-08-104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaidano G, et al. Genetic characterization of HHV-8/KSHV-positive primary effusion lymphoma reveals frequent mutations of BCL6: Implications for disease pathogenesis and histogenesis. Genes Chromosomes Cancer. 1999;24(1):16–23. doi: 10.1002/(sici)1098-2264(199901)24:1<16::aid-gcc3>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Boulanger E, et al. A clinical, molecular and cytogenetic study of 12 cases of human herpesvirus 8 associated primary effusion lymphoma in HIV-infected patients. Hematol J. 2001;2(3):172–179. doi: 10.1038/sj.thj.6200096. [DOI] [PubMed] [Google Scholar]
- 16.Mullaney BP, Ng VL, Herndier BG, McGrath MS, Pallavicini MG. Comparative genomic analyses of primary effusion lymphoma. Arch Pathol Lab Med. 2000;124(6):824–826. doi: 10.5858/2000-124-0824-CGAOPE. [DOI] [PubMed] [Google Scholar]
- 17.Ohshima K, et al. Chromosomal and comparative genomic analyses of HHV-8-negative primary effusion lymphoma in five HIV-negative Japanese patients. Leuk Lymphoma. 2002;43(3):595–601. doi: 10.1080/10428190290012100. [DOI] [PubMed] [Google Scholar]
- 18.Nair P, Pan H, Stallings RL, Gao SJ. Recurrent genomic imbalances in primary effusion lymphomas. Cancer Genet Cytogenet. 2006;171(2):119–121. doi: 10.1016/j.cancergencyto.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan SL, et al. Primary effusion lymphoma: Genomic profiling revealed amplification of SELPLG and CORO1C encoding for proteins important for cell migration. J Pathol. 2010;222(2):166–179. doi: 10.1002/path.2752. [DOI] [PubMed] [Google Scholar]
- 20.Richter J, et al. ICGC MMML-Seq Project Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luftig M, et al. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci USA. 2003;100(26):15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camcioglu Y, et al. HHV-8-associated Kaposi sarcoma in a child with IFNgammaR1 deficiency. J Pediatr. 2004;144(4):519–523. doi: 10.1016/j.jpeds.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Byun M, et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J Exp Med. 2010;207(11):2307–2312. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory SM, et al. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc Natl Acad Sci USA. 2009;106(28):11725–11730. doi: 10.1073/pnas.0905316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sin SH, Dittmer DP. Viral latency locus augments B-cell response in vivo to induce chronic marginal zone enlargement, plasma cell hyperplasia, and lymphoma. Blood. 2013;121(15):2952–2963. doi: 10.1182/blood-2012-03-415620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abend JR, et al. Kaposi’s sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J Virol. 2012;86(21):11663–11674. doi: 10.1128/JVI.01147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob CO, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci USA. 2009;106(15):6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treon SP, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 32.Landgren O, Staudt L. MYD88 L265P somatic mutation in IgM MGUS. N Engl J Med. 2012;367(23):2255–2256, author reply 2256–2257. doi: 10.1056/NEJMc1211959. [DOI] [PubMed] [Google Scholar]
- 33.Yang G, et al. A mutation in MYD88 (L265P) supports the survival of lymphoplasmacytic cells by activation of Bruton tyrosine kinase in Waldenström macroglobulinemia. Blood. 2013;122(7):1222–1232. doi: 10.1182/blood-2012-12-475111. [DOI] [PubMed] [Google Scholar]
- 34.Rhyasen GW, et al. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 2013;24(1):90–104. doi: 10.1016/j.ccr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tewhey R, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27(11):1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altshuler D, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407(6803):513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]