Abstract
Synthetic hydrogels with engineered, cell-mediated degradation sites are an important category of biomimetic materials. Here, hydrogels are synthesized by a step-growth reaction mechanism via a radically mediated thiol-norbornene (thiol-ene) photopolymerization. This reaction combines the advantages of ideal, homogeneous polymer network formation, facile incorporation of peptides without post-synthetic modification, and spatial and temporal control over the network evolution into a single system to produce proteolytically degradable poly(ethylene glycol) (PEG) peptide hydrogels. Using a thiol-ene photopolymerization, rapid gelation times are achieved, while maintaining high cell viability for cell encapsulation. The enzyme- and cellresponsive characteristics are demonstrated by tailoring the rate of spreading of human mesenchymal stem cells (hMSCs) through both the selection of proteolytically degradable crosslinkers and the density of the adhesion peptide RGDS. Furthermore, cellular function is manipulated spatially within the thiol-ene hydrogels through biochemical photopatterning. The high degree of spatial and temporal control over gelation, combined with robust material properties, makes thiol-ene hydrogels an excellent tool for a variety of medical and biological applications.
Synthetic, covalently crosslinked hydrogels are an increasingly important class of biomaterials with applications including, but not limited to, drug delivery, contact lenses, wound dressing, and bioadhesives.[1–8] Hydrogels are of particular interest for the incorporation of cellular components into tissue-engineered materials and the study of cellular function in three dimensions.[9–11] Living cells are encapsulated and sustained by polymerizing in the presence of large, multifunctional macromolecular monomers or macromers under physiological conditions, making hydrogels an attractive platform to both evaluate and manipulate tissue development. Significant attention has recently been devoted to hydrogels comprising PEG, a so-called “blank slate” material.[12] Due to an extremely low level of protein and cellular adsorption,[13,14] PEG hydrogels enable researchers to elicit specific cellular interactions through the incorporation of biologically functional components.[15,16]
PEG hydrogels are commonly prepared by photo-crosslinking linear PEG chains that have been modified on either end with acrylate or methacrylate moieties.[8,17] This photopolymerization strategy provides excellent temporal and spatial control of network development with low cytotoxicity.[18,19] Adhesion and proteolytic degradation sites are incorporated by co-polymerization of acrylate-functionalized peptides,[20] permitting cells to remodel their microenvironments. While such hydrogel preparation has significant demonstrable benefits and has been instrumental in the development of the field of tissue engineering, hydrogels, formed fromtraditional radical-chain-growth polymerization of di(meth)acrylatemonomers, possess heterogeneous network structures with dense poly(meth)acrylate chains and longPEGcrosslinks[21,22] (for an illustration, see Supporting Information, Fig. S1).
Step-growth polymerization of co-monomer solutions with complementary reactive groups forms homogeneous network structures[23] (Fig. S1) that have been shown to possess superior strength and strain tolerance when compared to chain-growth networks of similar crosslink density.[24] Recently, base-catalyzed Michael-type addition reactions between thiols and conjugated, unsaturated functional groups have been used to create hydrogels under mild, physiologically relevant conditions.[25–28] The Michael-type addition polymerization methodology provides a simple strategy for incorporating proteolytically degradable crosslinking peptides without post-synthetic modification through the inclusion of cysteine on either end of the peptide sequence. Hydrogels that promote cell spreading and migration in a fashion similar to natural biomaterials have been synthesized with this approach.[27,29] However, the alkaline conditions required for Michael-type reactions promote disulfide formation that has been implicated in the off-stoichiometric reaction of monomers during hydrogel formation.[26] Moreover, as the Michael addition is spontaneous, all spatial and temporal control intrinsic in photopolymerization is forfeited.
The materials developed herein synergistically combine the advantages of step growth and photoinititated polymerization to synthesize bioresponsive synthetic co-peptide hydrogels. Specifically, the radical-mediated step-growth reaction between thiol and norbornene moeities[30–32] is utilized to create uniform PEG co-peptide networks, structurally similar to those resulting from Michael-type reactions. The thiol-ene photopolymerization is cytocompatible, controllable both spatially and temporally, and provides a facile means to tune biochemical and mechanical properties, making this method a versatile tool for the manipulation and study of cellular activity in three dimensions.
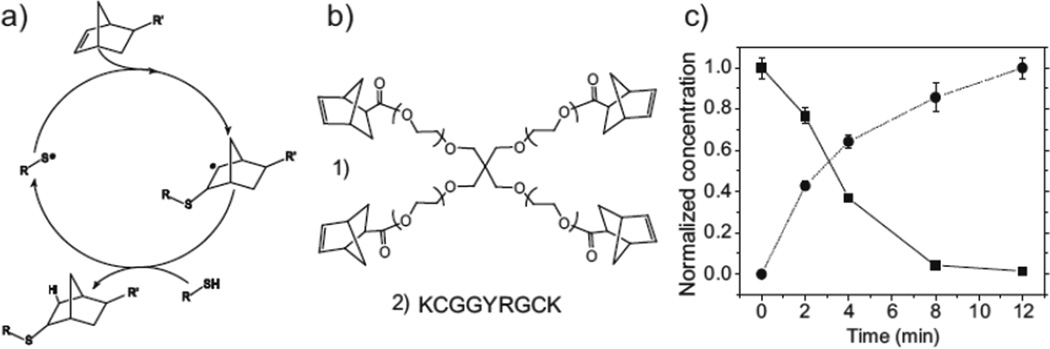
While the step-growth mechanism of the thiol-ene polymerization is well established for solvent-free reactions,[33,34] the reaction has neither been investigated in hydrated, dilute systems nor in the presence of charged peptides. Normally, step-growth thiol-ene photopolymerization proceeds via the mechanism shown in Figure 1a.[30,35] The light-activated initiating species abstracts a hydrogen atom from a thiol, forming a thiyl radical.
Figure 1.
a) General mechanism for radical, step-growth thiol-ene polymerization. An initiator abstracts a hydrogen atom from a thiol. The resulting thiyl radical propagates across the norbornene carbon–carbon double bond. The resulting norbornane radical abstracts a hydrogen atom from a thiol, completing the thioether bond formation, and regenerating a thiyl radical. b) Monomers used in this study are 1) PEG4norb (Mn 20 kDa) and 2) chymotrypsin-degradable peptide. c) Concentration of norbornene groups (■), as calculated by the disappearance of alkene proton peaks, and of thioethers (●), as calculated from the emergence of thioether α-proton peaks. Polymerization was performed at 10mW cm−2, 365-nm light with 0.05 wt% I2959.
This radical then propagates across the alkene group. Subsequently, the resulting carbon-centered radical abstracts a hydrogen atom from another thiol, forming the thioether linkage and regenerating the thiyl radical. Termination occurs through the coupling of any two radical species. The ideal step-growth mechanism presumes the alternation of propagation and chain-transfer reactions and the absence of any alkene homopolymerization.
High-resolution magic-angle spinning (HRMAS) 1H-NMR spectroscopy was used to demonstrate quantitatively the stepgrowth nature of the thiol-ene reaction under hydrated conditions. The conversion of norbornenes and thiols to thioethers via the disappearance of the alkene proton peaks of norbornene and the simultaneous appearance of the α-proton peak of the thioether were monitored during a stoichiometrically balanced photopolymerization of 20 kDa PEG–tetranorbornene (PEG4-norb, Fig. 1b) and chymotrypsin degradable peptide (Fig. 1c, see Fig. S2 for spectra). Additionally, a series of photopolymerizations was performed, in which the stoichiometric ratio of thiols to norbornenes was systematically varied. Gelation occurred only at stoichiometric ratios that satisfied the conditions of the Flory–Stockmayer equation[23,36] (Table S1). Both results indicate a 1:1 reaction between thiol and norbornene functional groups, precluding any significant chain addition of the alkene.
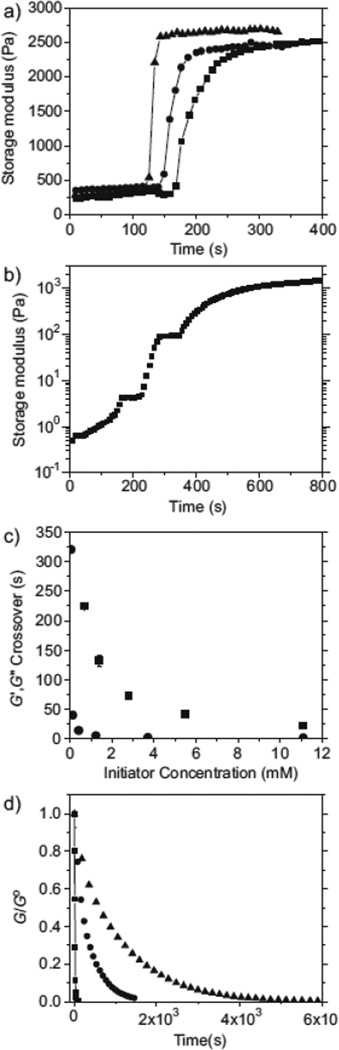
The primary objective of this work was to develop a polymerization strategy for mimicking complex biological environments, utilizing simple chemical and physical modifications for tailoring the material properties. For the thiol-ene hydrogels investigated herein, mechanical properties of the swollen state are dependent on the concentration of monomers used during polymerization, with shear elastic moduli ranging from 300±20 to 1700±360 Pa (Young’s modulus of 900–5100 Pa) for 3–10 wt% gels (Fig. S3). Importantly, the modulus values fall within the range of a variety of physiologically relevant tissues[37] and, while not investigated here, PEG monomers of lower molecular weight yield much higher modulus materials. The modulus values reported here are markedly higher than those reported by Lutholf and Hubbell for hydrogels formed via the Michael addition from monomers of similar molecular weights;[26] the average Young’s modulus of such gels formed under stoichiometric conditions and 10 wt% monomers is reported as ~1700 Pa. This disparity may be caused by a combination of potentially higher functional group conversions in the radical polymerization, as well as disulphide formation under the Michael addition conditions where increasing the relative biscysteine peptide mole fraction could result in higher-modulus materials.
Figure 2a demonstrates that elasticity is controlled within a preformed material. In this example, a preformed 5 wt% thiol-ene gel was placed in a 10 wt% solution. When exposed to light, a higher-crosslinking-density interpenetrating network (IPN) formed within the softer gel. Similar methods have been described for photopolymerized PEG diacrylate hydrogels.[38]
Figure 2.
In situ dynamic photorheometry measurements for a) shear modulus of network formed at 5 wt% and swollen in 10 wt% monomer solution with 2.2mM (0.05 wt% I2959). The gel is exposed to 5 (■), 10 (●), or 30 (▲) mW cm−2 at 120 s, creating an IPN of higher elastic modulus. b) Co-polymerization of PEG4Norb and chymotrypsin-degradable peptide at 0.05 wt% I2959 and 10mW cm−2, 365nm light, interrupted for 60 s at 165 s and again at 280 s. The plot includes a 30 s delay prior to light exposure. c) The time to gelation for a 10 wt% monomer solution plotted against the initiator concentration for I2959 (■) and LAP (●). The intensity is 10mW cm−2 at 365 nm. d) Elastic-modulus decline due to enzymatic cleavage of hydrogel crosslinks with 0.10 (▲), 0.20 (●), or 1.0mg mL−1 (■) chymotrypsin. All gels represented in this figure were formed by the co-polymerization of PEG4Norb and chymotrypsin-sensitive peptide.
The time within which the IPN is formed is controlled by varying the light intensity or initiator concentration. The ability to control hydrogel elasticity within the material has many applications in fundamental studies of cell behavior and when forming complex, engineered tissues.
A significant advantage of utilizing a light-dependent polymerization strategy is that monomer solutions can be premixed and appropriately positioned (e.g., in a tissue defect) prior to polymerization or stored until ready for use, which minimizes material use, decreases measurement errors, and provides researchers with more precise control of when and where the reaction occurs. Demonstrating that the reaction occurs exclusively during light exposure and thereby is at the discretion of the user, the polymerization in Figure 2b is interrupted at 165 and 280 s of exposure for 60 s simply by shuttering the light source. As seen from these results, little or no modulus development is observed during the periods in which there is no UV irradiation. The thiol-ene polymerization rate is also precisely controlled by changes in the initiator concentration (Fig. 2c). In the presence of either I2959, a commercial initiator commonly used for photoencapsulation of cells, or lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), a highly water-soluble initiator with strong absorbance at 365 nm,[39] gelation occurs within several minutes at even the lowest initiator concentrations studied. LAP initiates a much more rapid polymerization than I2959 for the thiol-ene polymerization, with gelation occurring in one second at 11mM LAP concentration. Initiation at even the lowest LAP concentration studied here (50µM or 0.0015 wt%), results in gelation of thiol-ene hydrogels within six minutes.With such high reactivity, the thiol-ene photopolymerization provides researchers with control over both the point at which the reaction is initiated and the rate of hydrogel formation.
While the high degree of control exercised over the evolving polymer network is an important feature of these thiol-ene co-peptide hydrogels, a particular objective of this work was the design of materials sensitive to cell-secreted proteases. As a model system, the biochemical response of the thiol-ene hydrogel to enzymatic degradation by chymotrypsin is shown in Figure 2d. Cleavage of the chympotrypsin-sensitive peptide crosslinks (sequence shown in Fig. 1b) leads to an enzymedependent decrease in the elastic modulus over time, indicating that crosslinks are being cleaved in a dose-dependent fashion.
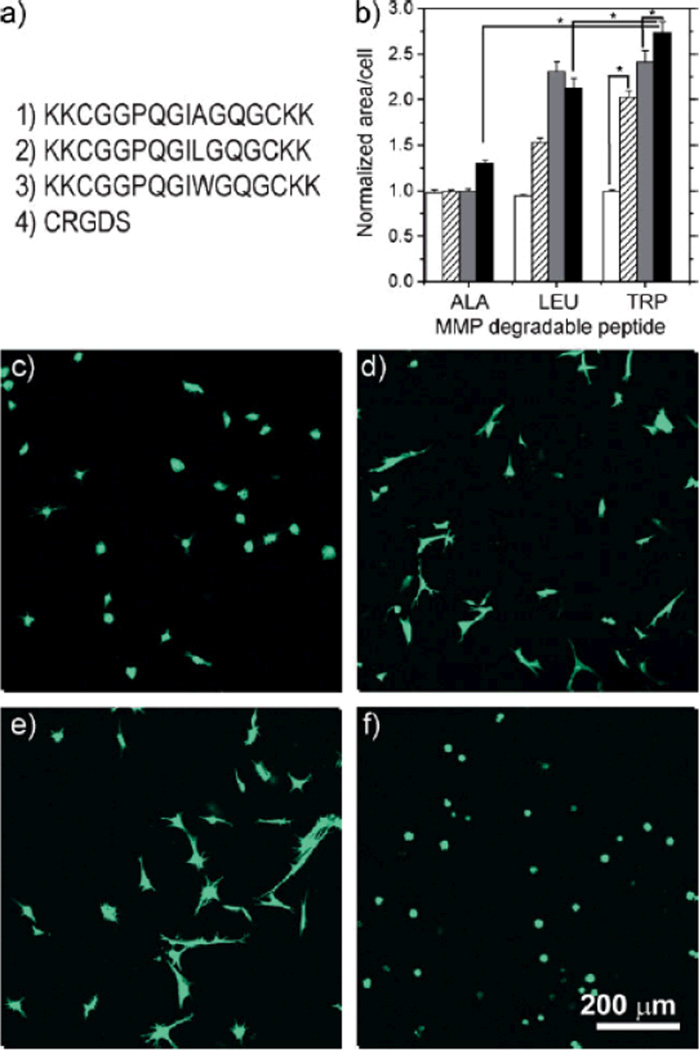
hMSCs were encapsulated in thiol-ene hydrogels to demonstrate cytocompatibility, as well as biochemical control of cell–material interactions. Figure 3 demonstrates two methods by which the response of the thiol-ene hydrogel to biological cues is biochemically modulated through choice of proteolytically degradable crosslinker and through the adhesion ligand concentration. For all studies reported here, cell viability 24 hours post polymerization was in excess of 95% and maintained for the duration of these experiments, indicating that this thiol-ene photopolymerization is cytocompatible. Figure 3b–e demonstrates that the degree of cell spreading is influenced by changing the proteolytically degradable crosslinker sequence. The matrix metalloproteinase (MMP)-degradable crosslinkers used in this study were derived from an α1(I)-collagen amino acid sequence[40–42] and are similar to amino acid sequences used in previous MMP-degradable hydrogels.[26,27] The native amino acid sequence GPQGIAGQ is susceptible to cleavage by a broad range of MMPs, including MMPs 1, 2, 3, 7, 8, and 9.[40–42] Susceptibility toMMP cleavage by all of these proteases is increased by replacing the alanine residue in the P′2 position with leucine or tryptophan.[40–42] The degree of spreading at constant adhesion ligand density is dependent on the crosslinker composition, as illustrated in Figure 3b–f, with total spreading being greatest within the MMP–Trp crosslinked gel and least in the MMP–Ala crosslinked gel, which is consistent with the comparative rates of cleavage of these peptides in solution.[40–42] The crosslinker sequence choice provides a powerful tool for controlling the degradation rate in a wide variety of environments and for potentially investigating the role of specific proteases in migration and other cellular processes.
Figure 3.
a) Peptides used in these studies 1) MMP–Ala, 2) MMP–Leu, 3) MMP–Trp, and 4) the monocysteine integrin binding motif, CRGDS. b) Quantification of the cell area for MMP–Ala, MMP–Leu, and MMP–Trp gels at day 6 relative to day 1. The degree of spreading depends on both the crosslinker and the CRGDS concentration: 0 µM (white bars), 250mM (striped), 500µM (gray), and 1mM (black). Statistical significance, indicated by (*), was determined with a one-way ANOVA followed by Tukey pairwise comparisons, α=0.05. While significant differences were found between many pairs, for clarity only a few are denoted as such here. For a complete chart of pairwise comparisons see Table S2. All hydrogels were prepared at 5 wt% concentration of PEG4Norb and MMP-degradable peptides co-polymerized with 0.05 wt% I2959 at 10mW cm−2, 365nm light. The hMSC were encapsulated using c) MMP–Ala crosslinking peptide and 1000µM RGD, d) MMP–Leu peptide and 1000µM RGD, e) MMP–Trp peptide and 1000µM RGD, f) MMP–Trp peptide and 0mM RGD.
Additionally, Figure 3 demonstrates that an adhesion ligand (in this case, RGDS integrin binding peptide[43,44]) is necessary to allow cells to spread within the matrix, and that the degree of spreading is concentration dependent for each of the peptides investigated. Without RGDS (Fig. 3b and f) cells remain rounded, without evidence of spreading, for each of the peptides investigated. It should be noted that as the cell-secreted enzymes degrade these hydrogels, the gel volumes increase and the concentration of cell-adhesion ligand necessarily decreases.
The ability to control how cells adhere to a synthetic material would be particularly important when such materials are used to induce migration in wound-healing environments such as bone defects or skin wounds.[27] These results demonstrate that the degradation rate of the material and the ability of cells to spread within it are tailored by the simple choice of the peptides that are incorporated into the hydrogel.
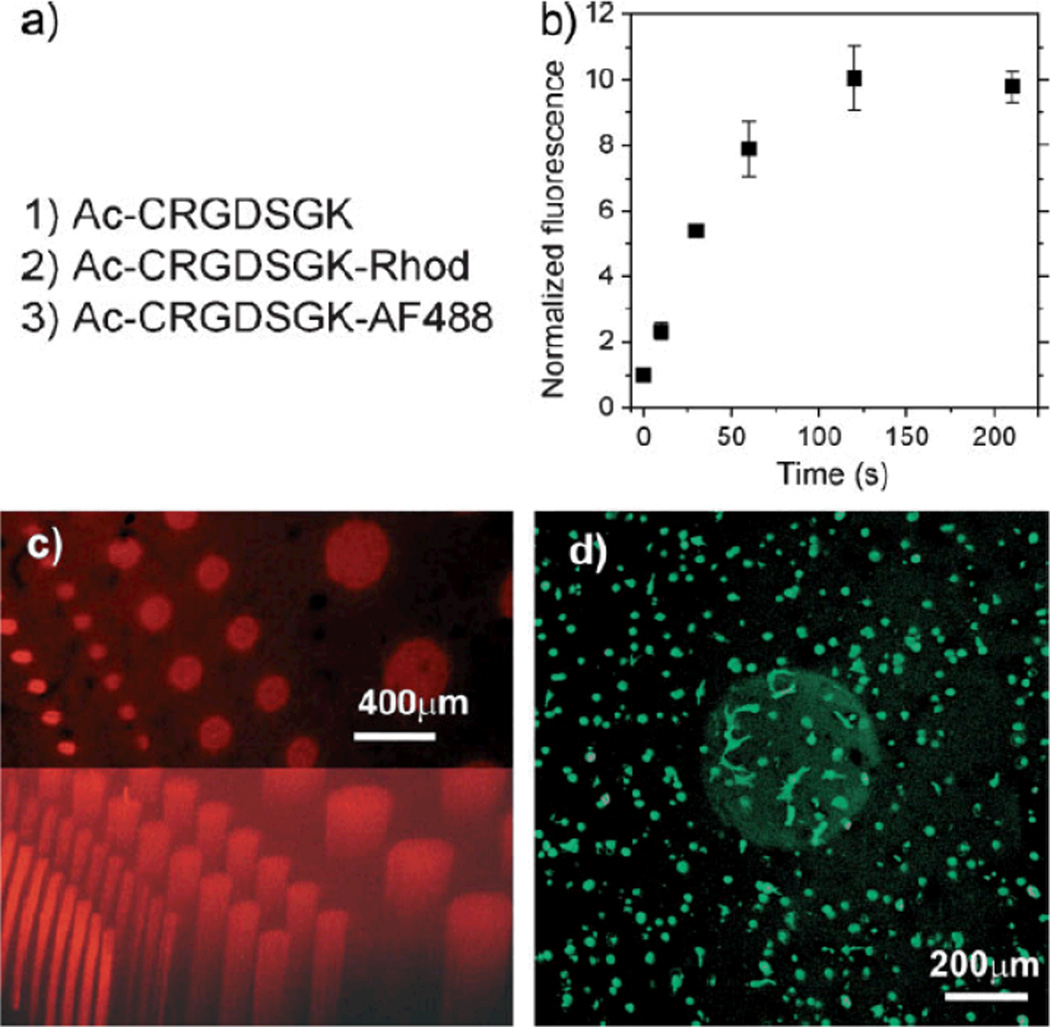
As tissues are biomechanically, biochemically, and cellularly heterogeneous structures, spatial control of cell–material interactions is a general goal for the development of biomimetic materials. Figure 4 illustrates how the thiol-ene polymerization strategy enables spatial control over gel biochemical properties. In Figure 4, gels were formed such that PEG4norb was in excess relative to the MMP-degradable crosslinker. Immediately following polymerization, a solution of CRGDS adhesion peptide, labeled adhesion peptide, and initiator were allowed to diffuse into the gel. Then, the adhesion ligand was covalently coupled within predetermined 3D regions via photopatterning techniques in a manner similar to that previously reported for PEG-acrylate hydrogels.[45–47] Following the photoconjugation, unbound ligand was allowed to diffuse into fresh media. After two minutes of exposure at these conditions, gels with photoconjugated rhodamine-labeled peptide exhibit no further increase in fluorescence, indicating that all available norbornene reactive sites are consumed (Fig. 4b). The cell-adhesion sequence was coupled to the hydrogel network only when exposed to light, as seen by the bright cylindrical regions in the confocal microscopy images in Figure 4c and d.
Figure 4.
a) Peptides used in photopatterning experiments. b) Normalized fluorescence (relative to gels with no exposure) of 1-mm-thick hydrogels saturated in a solution of 2.2mM LAP, 1mM Ac-CRGDSGK-rhodamine, and 4mM of CRGDS and illuminated for times indicated under identical conditions as for patterning experiments. c) 3D projection of an 800-µm-thick section of gel, in which Ac-CRGDSGK-rhodamine cylinders were photopatterned and angled view of the same (bottom). d) 3D projection of a 400-µm gel section. hMSCs are encapsulated in MMPdegradable hydrogel with RGDS displayed only where photopatterned (day 6). The image does not include gel/media interphases. Cells spread mostly within the volume of the photopatterned cylinder. Gels were formed at 5 wt% monomers PEG4Norb and MMP–Trp with a 1mM excess of norbornene functional groups. Adhesion ligand CRGDS and its fluorescent counterpart were diffused into the gels at a concentration of 5mM with 2.2mM LAP initiator and patterning was performed at 10 mW cm−2 for 2 min.
When hMSCs were encapsulated prior to incorporation of adhesion peptide in similar MMP-degradable hydrogels, spreading was greatest within the patterned cylinder (Fig. 4d); the average area for those cells within the photopatterned cylinders was 2.2 times the area of those outside. This value is clearly in the higher range of areas observed for cellular spreading within these hydrogels, as shown in Figure 3. It is, however, significantly lower than that of the MMP–Trp crosslinked gel with 1mM CRGDS, with which the patterned column should have a similar composition. This small discrepancy is due to greater-than-expected spreading outside of the cylinder, presumably the result of non-specifically bound CRGDS and fluorescently labeled RGDS. Nevertheless, it is demonstrated that the degree to which cells interact with the hydrogels is controlled spatially, giving researchers the flexibility to design more complex, improved strategies for inducing tissue formation or to study specific biological processes.
The unique chemistry and method for fabrication of biomimetic hydrogels utilizing thiol-ene polymerization provides excellent versatility with respect to creating and manipulating cell-encapsulation environments. The radical photoinitiation imparts rapid, yet cytocompatible, network development that is under almost complete control of the user while the step-growth mechanism imparts network homogeneity. The utility of such an approach is augmented by the ease of the monomer synthesis; as thiols on cysteine residues are used, peptide crosslinks require no post-synthetic modification and PEG is modified with norbornene via a common bioconjugation reaction. While photopatterning of an adhesion peptide is demonstrated, the strategy should broadly apply to any thiolated molecule of sufficient size and quality to diffuse within the hydrogel network. In a single gel one could imagine the creation of numerous spatially isolated and distinct mechanical and biochemical environments to better mimic the interfaces of natural tissues. The results presented in this paper suggest that the thiol-ene photopolymerization can be applied generally to the demands for a versatile, tailorable, bioresponsive hydrogel in the fields of cell biology, drug delivery, and regenerative medicine.
Experimental
Synthesis of 4-Arm PEG4norb
Norbornene-functionalized PEG was prepared by the addition of norbornene acid via the symmetric anhydride N,N’-dicyclohexylcarbodiimid (DCC; Sigma) coupling. The 4-arm PEG, MW 20000 (JenKemUSA, Allen, TX), was dissolved in dichloromethane (DCM) with 5× (with respect to hydroxyls) pyridine and 0.5×4-(dimethylamino)pyridine (DMAP; Sigma). In a separate reaction vessel, DCC 5× with respect to PEG hydroxyls, was reacted at room temperature with 10×5-norbornene-2-carboxylic acid (Sigma). A few seconds after addition of the acid, a white byproduct precipitate formed (dicycolhexylurea), indicating the formation of dinorbornene carboxylic acid anhydride. The anhydride was allowed to stir for 30 min, following which the 4-arm PEG, pyridine, and DMAP solution were added. The reaction was stirred overnight, after which the mixture was filtered. The filtrate was washed with 5% sodium bicarbonate solution and the product was precipitated in ice-cold diethyl ether.
Peptide Synthesis
All peptides were synthesized on solid Rink-amide resin using fluorenylmethoxycarbonyl (Fmoc) chemistry on a model 433A peptide synthesizer (Applied Biosystems) or a Tribute peptide synthesizer (Protein Technologies). Lysines were added to the terminal positions of the peptide sequences to encourage aqueous solubility. The peptide CRGDSGK was synthesized as the others, but with the orthogonal protecting group 1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl (Dde) on the C-terminal lysine. On resin, the N-terminus was capped with acetic anhydride and Dde was selectively removed with 2% hydrazine allowing labeling with 5(6)-carboxy rhodamine (Anaspec) by HATU coupling or with AlexaFluor 488 terafluorophenyl ester (AlexaFluor 488 5-TFP Invitrogen). Peptides were analyzed by reverse-phase high-performance liquid chromatography (HPLC) and matrix-assisted laser desorption ionisation (MALDI) mass spectrometry and purified by reverse-phase HPLC.
HRMAS 1H-NMR: HRMAS 1H-NMR spectroscopy was performed on a Varian Inova at 400mHz with a 4-mm GHX-Nano probe. Monomer solutions of 10 wt% in phosphate-buffered D2O and 0.05 wt% I2959 inside 40mL capacity nanotubes were exposed to 365-nm light at 10mW cm−2 and periodic NMR measurements were taken. It should be noted that the curvature of the nanotubes, in which polymerization occurs and spectra are taken, interferes with the light exposure, making this a poor technique to measure kinetics. It is, however, an appropriate means to determine the conversion of alkenes relative to the generation of thioethers, confirming the reaction mechanism.
Rheometry
In situ dynamic rheometry was performed with parallelplate geometry on an Ares 4400 rheometer (TA Instruments; setup shown in Fig. S4). Light at 365nm and 10mW cm−2 was directed through a flat quartz plate through the sample, while dynamic stress measurements were made at a 100-µm gap, 300% strain, and 100 rad s−1. Strain sweeps were performed prior to and post polymerization to verify the linear response regime. The gel point was taken as the storage/loss moduli crossover point, which indicates a conversion in the vicinity of the precise gel point [48]. For rheometric measurements of degradation, a 10 wt% PEG4Norb/Chymotrypsin-degradable peptide and 20mM I2959 in phosphate-buffered saline (PBS) solution was mixed and placed on ice. Concentrated and chilled chymotrypsin was mixed with the monomers to appropriate concentrations and the solution was placed on the thermal-control Peltier plate of the rheometer, set to 2°C. The solutions were polymerized in situ at 20mW cm−2 and the temperature of the plate was raised to 37°C (requiring roughly 20 s) for the monitoring of enzymatic degradation.
Cell Culture and Encapsulation
hMSCs, provided by the Tulane Center for Gene Therapy through a grant from NCRR of the NIH (grant P40 RR0 17 447) were used at passage 3 for all experiments. hMSCs were expanded using growth media (low-glucose Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen). hMSCs were encapsulated by resuspension in monomer solution containing a 5 wt% PEG and peptide in a stoichiometrically balanced ratio and 2.2mM (0.05 wt%) I2959 in PBS at a density of 300000 cells mL−1. Suspensions, in 6mm × 1mm circular molds, were exposed to 7–10 mW cm−2 352-nm centered light (40W black-light blue lamp from Sankyo Deiki) for 5 min. Following polymerization, hydrogel discs were removed and placed into growth media. For morphology and viability experiments, gels were incubated for 30 min in PBS with 2µM calcein 4mM ethidium homodimer (Live/Dead cytotoxicity kit from Invitrogen). Confocal images were taken using a Zeiss 510 laser scanning confocal microscope. Image analysis and cell-area calculations were performed using MetaMorph software for 3D stacks flattened to 2D images. A minimum of three spots on three different hydrogels was used and standard error is reported relative to individual cells. A one-way ANOVA and Tukey’s test with α=0.05 were used to determine statistical differences among the data sets.
Photopatterning
For column images, 4mM of the peptide with the sequence CRGDS and 1mM of the labeled peptide Ac-CRGDSGKRhodamine in PBS with 2.2mM LAP were allowed to diffuse for 20 min into preformed 1-mm-thick gels with 1mM unreacted norbornenes. A transparency photomask was placed directly on the upper surface of the gel, which was then exposed to collimated light at 365nm and an intensity of 10mW cm−2 intensity (Omnicure 1000, EXFO, Mississaugua, Ont., Canada) for 2 min. Gels were then removed to fresh FBS-supplemented media, which was changed every 2 h for 6 h. For patterned columns with encapsulated cells the procedure was identical, except that the RGDS peptide solution that was allowed to diffuse in was a 5mM, 50:1 mixture of Ac-CRGDSGK and Ac-CRGDSGK-AlexaFluor 488.
Supplementary Material
Acknowledgements
This work was supported the National Institute of Health (grant DE016523) and NSF (grant CBET 0626023).
Footnotes
Supporting Information is available online from Wiley InterScience or from the author.
Contributor Information
Benjamin D. Fairbanks, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80309-0424 (USA)
Michael P. Schwartz, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80309-0424 (USA)
Alexandra E. Halevi, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80309-0424 (USA)
Charles R. Nuttelman, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80309-0424 (USA)
Christopher N. Bowman, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80309-0424 (USA)
Kristi S. Anseth, Email: kristi.anseth@colorado.edu, Department of Chemical and Biological Engineering and the Howard Hughes Medical Institute, University of Colorado, Boulder, CO 80309-0424 (USA).
References
- 1.Nguyen KT, West JL. Biomaterials. 2002;23:4307. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 2.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 3.Bryant SJ, Anseth KS. J. Biomed. Mater. Res. 2002;59:63. doi: 10.1002/jbm.1217. [DOI] [PubMed] [Google Scholar]
- 4.Voldrich Z, Tomanek Z, Kopecek J. J. Biomed. Mater. Res. 1975;9:675. doi: 10.1002/jbm.820090612. [DOI] [PubMed] [Google Scholar]
- 5.Ramaswamy S, Wang DA, Fishbein KW, Elisseeff JH, Spencer RG. J. Biomed. Mater. Res. B. 2006;77B:144. doi: 10.1002/jbm.b.30404. [DOI] [PubMed] [Google Scholar]
- 6.Nicolson PC. J. Vogt, Biomaterials. 2001;22:3273. doi: 10.1016/s0142-9612(01)00165-x. [DOI] [PubMed] [Google Scholar]
- 7.Mandy SH. J. Dermatol. Surg. Oncol. 1983;9:153. doi: 10.1111/j.1524-4725.1983.tb00778.x. [DOI] [PubMed] [Google Scholar]
- 8.Peppas NA. Hydrogels in Medicine and Pharmacy. Vol. 1. Boca Raton, FL: CRC Press; 1986. [Google Scholar]
- 9.Nuttelman CR, Tripodi MC, Anseth KS. Matrix Biol. 2005;24:208. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Hwang NS, Kim MS, Sampattavanich S, Baek JH, Zhang ZJ, Elisseeff J. Stem Cells. 2006;24:284. doi: 10.1634/stemcells.2005-0024. [DOI] [PubMed] [Google Scholar]
- 11.Miot S, Woodfield T, Daniels AU, Suetterlin R, Peterschmitt I, Heberer M, van Blitterswijk CA, Riesle J, Martin I. Biomaterials. 2005;26:2479. doi: 10.1016/j.biomaterials.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 12.Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. Biomaterials. 2006;27:4881. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Xu Z, Marchant RE. Biomaterials. 2000;21:1075. doi: 10.1016/s0142-9612(00)00004-1. [DOI] [PubMed] [Google Scholar]
- 14.Yang ZH, Galloway JA, Yu HU. Langmuir. 1999;15:8405. [Google Scholar]
- 15.Hern DL, Hubbell JA. J. Biomed. Mater. Res. 1998;39:266. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 16.Drumheller PD, Elbert DL, Hubbell JA. Biotechnol. Bioeng. 1994;43:772. doi: 10.1002/bit.260430812. [DOI] [PubMed] [Google Scholar]
- 17.Sawhney AS, Pathak CP, Hubbell JA. Macromolecules. 1993;26:581. [Google Scholar]
- 18.Bryant SJ, Nuttelman CR, Anseth KS. J. Biomater. Sci. Polym. Ed. 2000;11:439. doi: 10.1163/156856200743805. [DOI] [PubMed] [Google Scholar]
- 19.Williams CG, Malik AN, Kim TK, Manson PN, Elisseeff JH. Biomaterials. 2005;26:1211. doi: 10.1016/j.biomaterials.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 20.West JL, Hubbell JA. Macromolecules. 1999;32:241. [Google Scholar]
- 21.Lin-Gibson S, Jones RL, Washburn NR, Horkay F. Macromolecules. 2005;38:2897. [Google Scholar]
- 22.Bowman CN, Kloxin CJ. AIChE J. 2008;54:2775. [Google Scholar]
- 23.Odian G. Principles of Polymerization. New York: Wiley; 1991. [Google Scholar]
- 24.Malkoch M, Vestberg R, Gupta N, Mespouille L, Dubois P, Mason AF, Hedrick JL, Liao Q, Frank CW, Kingsbury K, Hawker CJ. Chem. Commun. 2006:2774. doi: 10.1039/b603438a. [DOI] [PubMed] [Google Scholar]
- 25.Lutolf MP, Tirelli N, Cerritelli S, Cavalli L, Hubbell JA. Bioconjugate Chem. 2001;12:1051. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 26.Lutolf MP, Hubbell JA. Biomacromolecules. 2003;4:713. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 27.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Proc. Natl. Acad. Sci. USA. 2003;100:5413. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutolf MP, Raeber GP, Zisch AH, Tirelli N, Hubbell JA. Adv. Mater. 2003;15:888. [Google Scholar]
- 29.Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N, Schmokel H, Bezuidenhout D, Djonov V, Zilla P, Hubbell JA. FASEB J. 2003;17:2260. doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- 30.Morgan CR, Magnotta F, Ketley AD. J. Polym. Sci. A. 1977;15:627. [Google Scholar]
- 31.Hoyle CE, Lee TY, Roper T. J. Polym. Sci, Part A: Polym. Chem. 2004;42:5301. [Google Scholar]
- 32.Cramer NB, Bowman CN. J. Polym. Sci. A. 2001;39:3311. [Google Scholar]
- 33.Jacobine AF, Glaser DM, Grabek PJ, Mancini D, Masterson M, Nakos ST, Rakas MA, Woods JG. J. Appl. Polym. Sci. 1992;45:471. [Google Scholar]
- 34.Carioscia J, Lu H, Stansbury J, Bowman C. Dent. Mater. 2005;21:1137. doi: 10.1016/j.dental.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Cramer NB, Reddy SK, O’Brien AK, Bowman CN. Macromolecules. 2003;36:7964. [Google Scholar]
- 36.Stockmayer WH. J. Chem. Phys. 1944;12:125. [Google Scholar]
- 37.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 38.Hahn MS, Miller JS, West JL. Adv. Mater. 2006;18:2679. [Google Scholar]
- 39.Majima T, Schnabel W, Weber W. Makromolekulare Chemie–Macromol. Chem. Phys. 1991;192:2307. [Google Scholar]
- 40.Nagase H, Fields GB. Biopolymers (Peptide Science) 1996;40:399. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 41.Fields GB, Vanwart HE, Birkedalhansen H. J. Biol. Chem. 1987;262:6221. [PubMed] [Google Scholar]
- 42.Netzelarnett S, Fields G, Birkedalhansen H, Vanwart HE. J. Biol. Chem. 1991;266:6747. [Google Scholar]
- 43.Pierschbacher MD, Ruoslahti E. Nature. 1984;309:30. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 44.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 45.Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 46.Haraldsson KT, Hutchison JB, Sebra RP, Good BT, Anseth KS, Bowman CN. Sens. Actuators B. 2006;113:454. [Google Scholar]
- 47.Polizzotti BD, Fairbanks BD, Anseth KS. Biomacromolecules. 2008;9:1084. doi: 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- 48.Winter HH. Polym. Eng. Sci. 1987;27:1698. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.