Abstract
Epigenetic reprogramming of myeloid cells by infection or vaccination, termed trained immunity, confers non-specific protection from secondary infections. We characterized genome-wide transcriptome and histone modification profiles of human monocytes trained with β-glucan and identified induced expression of genes involved in glucose metabolism. Trained monocytes display high glucose consumption, lactate production, and NAD+/NADH ratio, reflecting a shift in the metabolism of trained monocytes with an increase in glycolysis dependent on the activation of mammalian target of rapamycin (mTOR) through a dectin-1/Akt/HIF1α pathway. Inhibition of Akt, mTOR, or HIF1α blocked monocyte induction of trained immunity, whereas the AMPK activator metformin inhibited the innate immune response to fungal infection. Finally, mice with a myeloid cell-specific defect in HIF1α were unable to mount trained immunity against bacterial sepsis. In conclusion, Akt/mTOR/HIF1α-dependent induction of aerobic glycolysis represents the metabolic basis of trained immunity.
Introduction
Classical descriptions of host defense mechanisms distinguish innate immune responses that are rapid, non-specific, and lack memory, from the specific T/B cell-dependent immune responses that are highly specific and have the capacity to build immunological memory. However, the dogma of an innate immune system incapable of mounting adaptive responses is contradicted by earlier studies showing that organisms lacking a specific immune system, such as plants or insects, are able to respond adaptively to infection (1, 2), and that classical innate immune cells such as macrophages have adaptive characteristics (3). In line with the concept suggesting the presence of non-specific adaptive responses in the innate immune system, recent studies have demonstrated T/B cell-independent protective effects of monocytes and natural killer (NK) cells in models of bacterial and viral infections, respectively (4, 5). Furthermore, epigenetic reprogramming at the level of histone H3 methylation has been proposed as the molecular mechanism responsible for the long-term memory of innate immunity (4, 6) and this process has been termed trained immunity.
Initiation of innate immune memory through trained immunity is the likely mechanism behind the non-specific protective effects of certain vaccines (7), while an increased inflammatory responsiveness of monocytes and macrophages due to trained immunity could also play a central role in inflammatory diseases (8). From this perspective, the capacity of innate immunity to mount adaptive responses both redefines the normal host defense and identifies a novel potential therapeutic target in human diseases. It thus becomes essential to understand the cellular and molecular mechanisms that mediate trained immunity, to ultimately harness their therapeutic potential. While epigenetic modifications are the fundamental process responsible for information storage during innate immune memory in both plants (9) and mammals (6), much less is known regarding the molecular pathways and downstream mechanisms that lead to trained immunity.
Transcriptome and epigenetics of monocytes
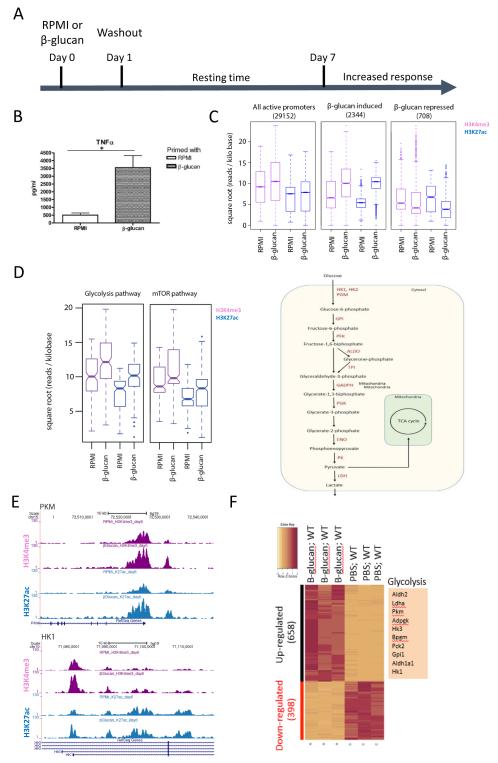
Previously it has been demonstrated that Candida albicans and its main cell wall constituent, β-glucan, induced trained innate immune memory both in vitro and in vivo (6). We performed an unbiased assessment of whole-genome mRNA expression, histone methylation and acetylation patterns after training human primary monocytes with β-glucan, the major Candida cell wall structure that mediates trained immunity, and that has been previously shown to induce non-specific protection against both infections and malignancies (10). A model of trained immunity in vitro (6) (Fig. 1A), led to a strongly potentiated cytokine production upon restimulation with lipopolysaccharide (LPS) seven days later (Fig. 1B). A similar enhanced response was observed after stimulation with the TLR2 ligand Pam3Cys or with non-related Gram-negative and Gram-positive bacteria (Fig. S1). Assessment of H3K4me3 and H3K27Ac identified a large number of promoters that were specifically induced by β–glucan training (Fig. 1C). Pathway analysis of the promoters potentiated by β–glucan identified several innate immune and signaling pathways up-regulated in trained cells that are responsible for the induction of trained immunity (ref.(6) and Saeed et al, submitted).
Figure 1. Monocyte Trained Immunity.
(A) Schematic of in vitro trained immunity experimental setup. (B) TNFα levels after 7 days in β-glucan treated cells. (C) Genome-wide H3K4me3 (blue) and H3K27me3 (purple) epigenetic modification analysis after 7 days post β-glucan treatment. (D) The epigenetic modifications in the promoter regions of the genes involved in glycolysis and mTOR (left panels) pathways and the schematic representation of the upregulated enzymes (red) in the glycolysis pathway. (E) Representative screenshots of the H3K4me3 (blue) and H3K27Ac (purple) modification in the promoter region of pyruvate kinase (PKM) and hexokinase. (F)The heat-map analysis of the total RNA expression determined by RNA sequencing between β-glucan treated group and control group was presented. Genes in the glycolysis pathway that are upregulated by the β–glucan training are highlighted in the box. Three mice per group were used for the RNA sequencing.
In addition to the immune signaling pathways, epigenetic profiling of trained monocytes based on both methylation and acetylation patterns identified a signature associated with central metabolism (Fig. S2), most interestingly an increase in the promoters of genes encoding enzymes involved in glycolysis and its master regulator mTOR (Fig. 1D). Furthermore, genes involved in glycolysis, such as hexokinase and pyruvate kinase, were epigenetically up-regulated (Fig. 1E and S3). The gene expressing mTOR and the glycolytic genes that are targets of the transcription factor HIF1α were also enhanced by β-glucan (Fig. S4). In line with this, the HIF1α activation was increased in β–glucan-trained monocytes (Fig. S5). In addition to these ex-vivo experiments, glycolysis genes were also up-regulated in vivo in mice challenged with β-glucan, as revealed by total RNA sequencing analysis in splenocytes of these mice (Fig. 1F and Fig. S6).
Glycolysis and Monocytes
Monocytes from peritoneal exudates rely on glycolysis as a main energy source (11). The role of glucose as an energy substrate for monocytes is demonstrated by the blockade of monocyte stimulation and trained immunity by incubation of cells with 2-deoxy-D-glucose (Fig. S7). Moreover, a switch from oxidative phosphorylation to aerobic glycolysis is an important feature of activated macrophages, dendritic cells, and Th1/Th17 lymphocytes (12).
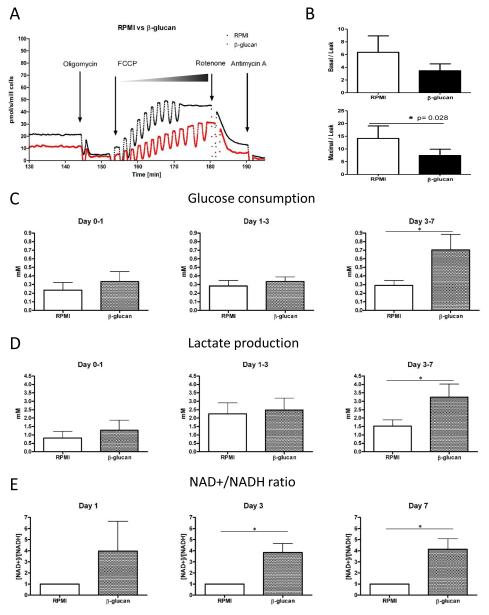
Consistent with these findings, monocytes trained with β–glucan showed a reduced baseline oxygen consumption on day 7 compared with naïve cells, compatible with a shift from oxidative metabolism towards glycolysis. Moreover, trained cells showed a decreased maximal rate of oxygen consumption after complete uncoupling with FCCP (Fig. 2A, B), while the rate of proton leak-dependent oxygen consumption was not altered (Table S1). The latter result indicates a reduction of the capacity of the mitochondrial electron transport chain (ETC); as observed after a period of hypoxia (13). Hypoxia decreases the activity of the ETC complexes I and IV through a postulated role of HIF-1α. This hypothesis was reinforced by observations of increased glucose consumption (Fig. 2C), lactate production (Fig. 2D), and NAD+/NADH ratio (Fig. 2E) in trained monocytes.
Figure 2. Post β-glucan Treatment Physiology.
(A) The representative oxygen consumption rate of RPMI and β-glucan trained monocytes determined by high-resolution respirometry (Oxygraph) (B) Baseline (upper panel) and maximum oxygen consumption rate (lower panel)of untrained (open bar) and β-glucan-trained (closed bar) monocytes determined by respirometry and normalized to the leak oxygen consumption. Kinetic changes of (C) glucose consumption and (D) lactate production from day 1, day 3 and day 7 of the RPMI and β-glucan trained monocytes. (E) The kinetics of NAD+/NADH determined at day 1, day 3 and day 7.
The glucose concentrations remained comparable among the groups during the culture, as the differences in glucose consumption did not offset the high glucose concentrations in the RPMI medium, suggesting that glucose availability is not the limiting factor for the observed training phenotype (Fig. S8). In addition, the training effect induced by β-glucan was not determined by pyruvate, an intermediate metabolite in glycolysis, which is present in most of the culture medium, as in its absence the training process was undisturbed (Fig. S9).
A high cellular NAD+/NADH ratio acts through sirtuin-1 to decrease the mitochondrial content (14). This mechanism can explain the observed β–glucan-induced reduction in ETC capacity. In contrast, LPS stimulation leads to a strong but transient increase in the glycolytic process in monocytes (Fig. S10), supporting studies suggesting that while the acute response of monocytes to LPS is characterized by glycolysis (15), the response at later time points switches to oxidative phosphorylation - a process that subsequently induces immune tolerance by activation of sirtuin-1 and sirtuin-6 histone deacetylases (16). In contrast to LPS-induced tolerance, β-glucan training inhibited the expression of Sirtuin1 (Fig. S11). Moreover, addition of resveratrol, a sirtuin-1 activator, during the first 24 hours of β-glucan training partially inhibited the enhanced IL-6 production (Fig. S11). This suggests that sirtuin deacetylases do play a role in modulated monocyte functional phenotype and highlights the intertwined complex interaction between the intermediate metabolites and subsequent immune responses through the chromatin modifying enzymes (17).
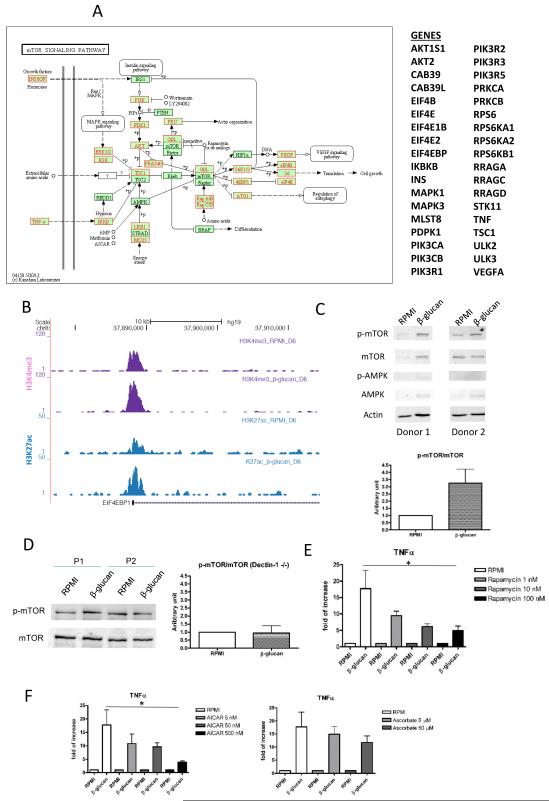
mTOR acts as a sensor of the metabolic environment (18) and functions as a master regulator of glucose metabolism in activated lymphocytes (19). Epigenetic signal at promoters of genes in mTOR pathway is significantly higher in β-glucan trained monocytes (paired t-test, p-value<0.001) compared to the cells exposed to culture medium (Fig. 3A). Target genes of mTOR, such as EIF4EBP1, displayed a similar pattern (Fig. 3B). In line with this finding, Western blot assessment of mTOR phosphorylation demonstrated significant activation (paired t-test, p-value=0.04) in trained monocytes (Fig. 3C). mTOR phosphorylation is dependent on the dectin-1 C-type lectin receptor, supported by the fact that monocytes isolated from patients with a complete deficiency in dectin-1 (20) failed to activate mTOR upon stimulation with β–glucan (Fig. 3D), and failed to enhance TNF production upon LPS restimulation (Fig. S12).
Figure 3. mTOR Signaling in β-glucan Treated Monocytes.
(A) Schematic representation of the upregulated enzymes (red) in mTOR signaling pathway. (B) Screenshot of the H3K4me3 (blue) and H3K27Ac (purple) modification in the promoter region of EIF4eBP, the main target of mTOR. (C) Western blot from cell lysate harvest at day 7 for endogenous phospho-mTOR, total mTOR, phospho-AMPK, AMPK and actin. p-mTOR/mTOR ratio is shown as a bar chart. (D) The endogenous p-mTOR status of Dectin-1 deficient patients and healthy control by Western blot from cell lysate harvest at day 7. p-mTOR/mTOR ratio is shown as a bar chart. The relative cytokine production was determined from the cells incubated with (E) rapamycin (mTOR inhibitor), (F) AICAR (AMPK inhibitor) and ascorbate (HIF1α inhibitor) in a dose-dependent manner.
Glycolysis in Trained Immunity
As the data presented above demonstrate activation of mTOR and glycolysis in trained monocytes, we next investigated the causality between these two processes by blocking glycolysis during β–glucan training. Inhibition of mTOR using rapamycin during the first day of stimulation resulted in a dose-dependent inhibition of the training effect induced by β-glucan (Fig. 3E). Indirect inhibition of mTOR with AICAR, an AMPK activator, had similar effects (Fig. 3F). As mTOR induction of glycolysis is mediated through activation of HIF1α and stimulation of glycolytic enzymes (21) and the rapamycin has been shown to inhibit HIF-1α expression (22), we assessed the effect of a HIF1α inhibitor on monocyte training, finding that the HIF1α inhibitor ascorbate also blocked trained immunity in a dose-dependent manner (Fig. 3F).
Furthermore, the link between metabolic effects to epigenetic changes was investigated by assessing the effects of the epigenetic inhibitors, MTA (methylthioadenosine, methyltransferase inhibitor) and ITF (ITF2357, histonedeacetylase inhibitor) during the training setup on the lactate measurements. As expected, the epigenetic inhibitors had no effect on lactate production in the acute phase (24 hours post β-glucan stimulation, Fig. S13). However, the lactate production was significantly reduced in the trained monocytes on day 7 when MTA was added to the monocytes with β-glucan during the first 24h in the incubation period (Fig. S13), suggesting that histone methylation also partially modifies the induction of glycolysis in the trained monocytes.
Monocyte mTOR Activation
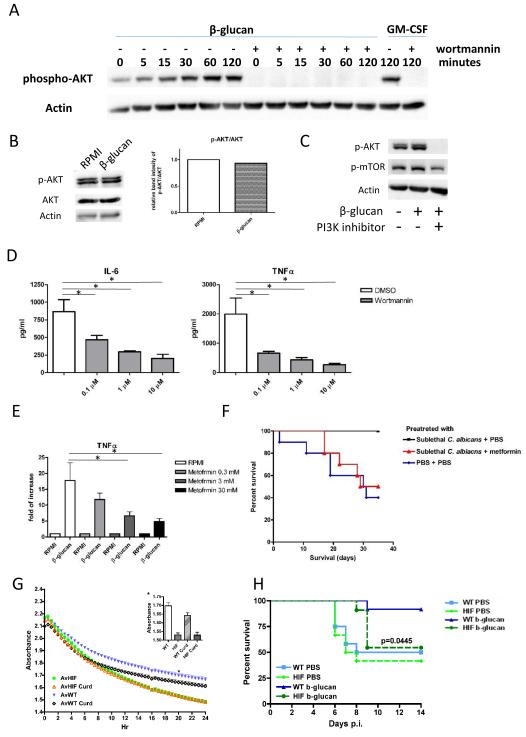
Activation of mTOR by insulin or colony-stimulating factors such as GM-CSF is mediated by intermediary activation of the Akt/PI3K pathway (23). A similar signal route is induced in monocytes by β-glucan as stimulation with β–glucan induced a strong phosphorylation of Akt (Fig. 4A). This effect was again dectin-1-dependent, as it was absent in monocytes isolated from dectin-1-deficient patients (Fig. 4B). Inhibition of Akt phosphorylation also resulted in down-regulation of mTOR activation (Fig. 4C), demonstrating the relationship between Akt and mTOR activation. Finally, the Akt inhibitor wortmannin inhibited monocyte training by β–glucan in a dose-dependent manner (Fig. 4D).
Figure 4. AKT/mTOR/HIF1α Pathway downstream of β-glucan Stimulation.
(A) AKT phosphorylation induced by β-glucan was determined in a kinetic manner by Western blot in monocytes in the presence or absence of wortmannin. (B) AKT phosphorylation and p-AKT/AKT ratio induced by β-glucan from the dectin-1 were determined by Western blot. The effect of PI3 kinase inhibitors on (C) AKT and mTOR phosphorylation were determined by Western blot or (D) by cytokine production upon LPS restimulation. (E) The relative cytokine production was determined from the cells incubated with metformin (AMPK inhibitor) in a dose-dependent manner. (F) Survival of wild type C57BL/6J mice infected with live C. albicans and AMPK-mTOR pathway training, Metformin or PBS were given from one day before the first non-lethal dose live C. albicans challenge till 3 days post challenge on a daily basis. (G) HIF-KO alveolar macrophages have increased metabolic activity. *Inset shows 20hr time point absorbance values. (F) Survival curve of wild type or mHIF-1α KO mice primed with β-glucan and challenge with a lethal dose of Staphylococcus aureus infection.
Epigenetic reprogramming of monocytes by trained immunity has been reported as a mechanism of nonspecific protection in different models. Mice were protected from lethal disseminated candidiasis after an initial non-lethal Candida albicans infection (6). Similarly, β–glucan also induced protection against infection with a lethal Staphylococcus aureus inoculum (24). Bacillus Calmette-Guérin (BCG) vaccination protected mice from systemic candidiasis (4). We assessed whether the inhibition of glycolysis abrogates the protective effects in these experimental models with the drug metformin, which acts through AMPK activation and subsequently mTOR inhibition (25) and is commonly used for the treatment of type 2 diabetes (26). In vitro, metformin suppressed trained immunity induced by β-glucan (Fig. 4E), and administration of metformin to mice during and after primary infection with a low-inoculum C. albicans completely inhibited the protective effects leading to increased survival during disseminated candidiasis induced by a primary C. albicans injection (Fig. 4F), demonstrating the crucial role of mTOR-mediated effects for mounting protective trained immunity in vivo.
In a following set of experiments, we assessed whether these effects were mediated at the level of innate immunity, and not at the level of adaptive T/B cell immunity elicited during vaccination. An experimental model of β–glucan-induced protection against S. aureus sepsis can be observed in myeloid cell-specific HIF1α conditional knock-out mice (mHIF1α-KO) (27). These mice are unable to mount glycolysis specifically in cells of the myeloid lineage. We assessed metabolic activity of wild-type and mHIF1α-KO macrophages when stimulated with β-glucan. mHIF1α-KO macrophages showed increased chemical reduction of the metabolic indicator resazurin (Fig. 4G), consistent with the hypothesis that HIF1α is critical to induce the switch to aerobic glycolysis in response to β-glucan. In this model, the cells do not undergo the switch in the absence of HIF1α and are “metabolically” dysregulated. While β–glucan increased the survival of wild-type mice infected with S. aureus from 40% to 90%, the induction of trained immunity was completely abrogated in mHIF1α-KO mice (Fig. 4H). To further dissect which pathways are modulated in the mHIF1α-KO mice, we performed RNA sequencing and compared the differential RNA expression profile between wild-type and mHIF1α-KO mice. Several interesting genes were specifically up-regulated in wild-type but not in mHIF1α-KO (Fig. S14 and Table S2.), including beclin-1 (autophagy related gene), STK11 (an AMPK related serine/threonine kinase), JHDM1D ((jumonji C domain containing histone demethylase), or FOXO4 transcription factor that is involved in PI3K/Akt stimulation. These data will form the basis of future studies to further characterize the function of these target genes for trained immunity. To sum up, these results demonstrate that stimulation of HIF1α-mediated glycolysis in myeloid cells is crucial for mounting trained immunity in vivo.
Discussion
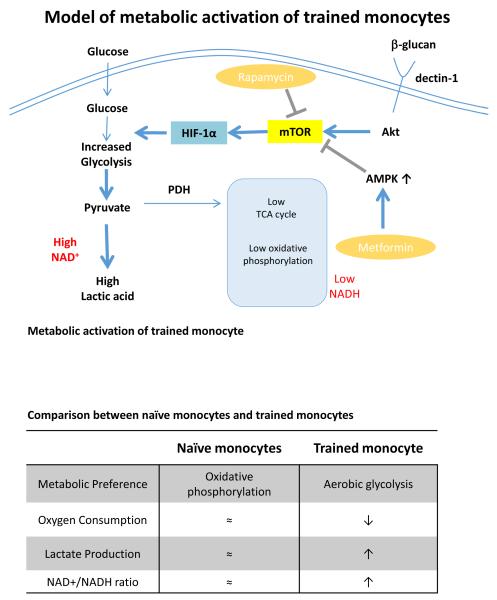
The role of histone methylation mediating short-term innate immunological memory in macrophages has been described (6) and referred to as latent enhancers for the epigenetic elements that mediate this phenomenon (28). In this study, whole-genome epigenetic profiling of histone modifications and RNA sequencing analysis has identified both immunologic and metabolic pathways stimulated during trained immunity. A novel cAMP-dependent pathway mediating trained immunity in monocytes has also been described (Saeed et al, submitted). In the present study, we identified the metabolic pathways induced in trained monocytes, demonstrating a metabolic switch towards aerobic glycolysis, which is in turn crucial for the maintenance of trained immunity (Fig. 5).
Figure 5.
Model of metabolic activation of trained monocytes, characterized by shift towards increased aerobic glycolysis and decreased oxidative phosphorylation. The potential role of rapamycin and metformin in inhibition of trained immunity is also depicted. The metabolic differences between naïve monocytes and trained monocytes is summarized in the panel.
A metabolic switch towards aerobic glycolysis has been earlier reported to be a feature of cell activation and proliferation: such an effect has been first described in neoplastic cells and termed the Warburg effect (29), while recently being reported to play a role in effector T helper lymphocytes (30) and activated macrophages (31). The elevated glycolysis metabolism in the trained monocytes might equip and prepare the cells to respond to the intruding pathogens in a robust and quicker manner in terms of proinflammatory cytokine production and possibly also through enhanced phagocytosis capacity (32).
Although our observation of the trained immunity was demonstrated in monocytes, the concept should not be exclusively restricted to the monocyte lineage. Recently, adaptive features of NK have been demonstrated by several studies (5, 33). Whether the metabolic rewiring also play a role in NK or other innate immune cells, such as dendritic cells, is warranted for further investigation. In addition, whether the training is contact-dependent or could also be induced by soluble mediators also need to be further elucidated. This aspect is of crucial interest in the field of autoinflammatory and autoimmune diseases, because these diseases are known to be worsened by the unregulated cytokine production. Our preliminary results suggest that proinflammatory cytokines such as IL-1β could also induce trained immunity in monocytes in vitro (Fig. S15). This hypothesis is further supported by previous non-specific protective effects induced by IL-1β, even when injected several days before an experimental infection is induced (34).
One important aspect to be underlined is that the molecular mechanisms investigated in the present study focused on the trained immunity in the first seven days after the initial stimulus. This is the crucial period during which trained immunity offers protection in newborn children against perinatal sepsis (35), and thus it is very relevant from a biological and clinical point of view. Longer in vivo effects in humans were also demonstrated (4), and future studies are warranted to assess whether these later effects are mediated through similar mechanisms. The later effects are however likely to be exerted also at the level of bone marrow myeloid cell progenitors, as recently demonstrated in case of TLR-induced tolerance (36).
Future studies should investigate the full consequences of this increased glycolytic process in trained immunity. Hypoxia and glycolysis enhance the proliferative response of macrophages to CSF-1 (37) and sustain the survival of activated dendritic cells (38). Soluble β–glucan from Grifola frondosa has been reported to induce macrophage proliferation (39), although we were not able to observe these effects with trained monocytes by Candida β–glucan (6). However, epigenetic profiling identified a cell cycle activation signal in β–glucan-trained cells (Saeed et al, submitted), and it is tempting to speculate that trained monocytes are not only more capable of increased cytokine production, but are also primed to respond to proliferative signals in the milieu, although this remains to be demonstrated by future studies. Finally, the identification of glycolysis as a fundamental process in trained immunity further highlights a key regulatory role for metabolism in innate host defense, and also defines a novel therapeutic target in both infectious and inflammatory diseases (8).
Materials and Methods
Isolation of primary human monocytes
Blood was collected from human healthy volunteers and two dectin-1 deficient patients after written informed consent (Ethical Committee Nijmegen-Arnhem, approval nr. NL32357.091.10). PBMCs were isolated by differential centrifugation using Ficoll-paque (GE Healthcare, Diegem, Belgium) from buffy coats obtained from Sanquin Bloodbank, Nijmegen, the Netherlands. Monocytes were purified by MACS depletion of CD3, CD19 and CD56 positive cells from the PBMCs: CD3 MicroBeads (130-050-101), CD19 MicroBeads (130-050-301) and CD56 were purchased from Miltenyi Biotec (Leiden, the Netherlands) and used according to the manufacturer’s protocol. Efficacy of depletion was controlled by flow cytometry (FC500 Beckman-Coulter, Woerden, the Netherlands) and was higher than 98%.
Genome-wide sequence analysis
For ChIP analysis or RNA sequencing, 10×106 of CD3−CD19−CD56− pure monocytes were plated on 100 mm dishes. Monocytes were pre-incubated with cell culture medium (RPMI) or β-glucan (5μg/mL) for 24 hours in a total volume of 10 mL. After 24h, cells were washed to remove the stimulus, and cells were resuspended in RPMI supplemented with 10% human pool serum. Monocytes were collected before and 6 days after the incubation for chromatin immunoprecipitation or RNA sequencing. For RNA sequencing, monocytes were collected in TRIzol Reagent (Invitrogen, Bleiswijk, the Netherlands). The purified materials were then processed to generate genomic DNA for WGBS, RNA (Trizol extraction according to manufacture instructions; Agilent BioAnalyser RIN >8) and Chromatin by fixating the cells in 1% formaldehyde.
Reagents
Candida albicans β-1,3-(D)-glucan (β-glucan) were kindly provided by Professor David Williams. Reagents used were as follows: LPS (E. coli 0B5/B5, Sigma chemical company, Diegem, Belgium), rapamycin (Sigma, R0395), metformin (R&D, AF 1730, Abingdon, UK), AICAR (Sigma, A9978), ascorbate (Sigma, A4034), wortmannin (InvivoGen, tlrl-wtm, Toulouse, France). C. albicans ATCC MYA-3573 (UC 820) were heat-inactivated for 30 min at 95°C.
Stimulation Experiments
For training, monocytes were preincubated with β-glucan (10 μg/ml) for 24 hr. After 7 days cells were re-stimulated with various microbial ligands: LPS (10 ng/ml), Pam3Cys (10 g/ml), heat-killed S. aureus or heat-killed E. coli (both at 106 microorganisms/ml). After 24 hr, supernatants were collected and stored at −20°C until cytokine measurement. All the cytokine measurement presented in the manuscript were from at least 6 donors.
To address the HIF-1a/AMPK/mTOR pathway in trained immunity, the specific inhibitors were added together with β-glucan for the first 24 hours in different doses as follows: rapamycin from 1 to 100nM, metformin from 0.3 to 30 mM, AICAR from 5 to 500 nM, ascorbate 5 and 50 μM.
Chip-Seq data analysis
H3K4me3 and H3K27ac ChIP, sequencing, and processing of the data were performed as described (6). The detailed data have been deposited in the GEO database with accession number GSE57206. Sequenced reads of 42-bp length were mapped to human genome (NCBI hg19) using bwa-alignment package mapper (40). ChiP-seq data sets were normalized as described earlier (41) and the sequenced reads were directionally extended to 300bp, corresponding to original length of sequenced DNA fragments. For each base pair in the genome, the number of overlapping sequence read was determined, averaged over 10-bp window and visualized in UCSC browser (http://genome.ucsc.edu). These normalized tracks were used to generate the genome browser screenshots. Putative H3K4me3 and H3K27ac enriched regions in the genome were identified by using MACS (42) with P<10−8. All the transcription start sites (±1kb) of genes with significant H3K4me3 signal were regarded as active promoters. H3K4me3 and H3K27ac signal at all active promoters were estimated and log2 ratio of ChiP-seq signal between treatment and control samples were calculated. Promoters that show a 2 times median absolute deviation (median ± 2×MAD) of the ratio of ChiP-seq signal (treatment/control) were regarded as regulated promoters (induced or repressed). Sequence reads counted from the normalized ChiP-seq data sets were used to generate the boxplots.
Metabolites measurements
Culture medium was collected at day 1, day 3 and day 7. The glucose and lactate concentration within the medium were determined by Glucose Colorimetric Assay Kit (Biovision, K686-100, Milpitas, CA, USA) and Lactate Colorimetric Assay Kit (Biovision, K627-100) respectively. NAD+ and NADH concentration were determined by NAD/NADH Quantification Colorimetric Kit (Biovision, K337-100) from the cell lysate according to manufacturer’s protocol. All the metabolites measurement data presented in the manuscript were from at least 6 donors.
Oxygen consumption measurement
Culture medium was collected from and 1 million cells treated with from the RPMI or β-glucan groups after which the cells were trypsinised, washed and resuspended in 60 μl of the collected culture medium. The cell suspensions cells were then used for cellular O2 consumption analysis. Oxygen consumption was measured at 37°C using polarographic oxygen sensors in a two-chamber Oxygraph (OROBOROS Instruments, Innsbruck, Austria). First, basal respiration was measured. Next, leak respiration was determined by addition of the specific Complex V inhibitor oligomycin A (OLI). Then, Maximal electron transport chain complex (ETC) capacity was quantified by applying using increasing concentrations of the mitochondrial uncoupler p-trifluoromethoxy carbonyl cyanide phenyl hydrazone (FCCP; from 1 to 14 μM final maximal concentration). Finally, Minimal respiration was assessed by adding a maximal (0.5 μM) concentration of the specific complex I inhibitor rotenone (ROT; 0.5 μM) and the complex III inhibitor antimycin A (AA; 0.5 μM).
After establishment of the baseline oxygen consumption rate, cells were treated with the ATP synthase inhibitor oligomycin to determine the rate of proton leak-dependent oxygen consumption, after which the baseline rate value was normalized to the value of the leak rate. Next, the cells were treated with the mitochondrial uncoupler FCCP to determine the maximum oxygen consumption rate. For normalization, the maximum FCCP value was ratioed to the leak value. The oxygen consumption measurement was repeated in monocytes isolated from five healthy individuals.
Western Blot
For Western blotting of AMPK, mTOR, AKT (total and phosphorylated) and actin, training was performed as described in stimulation experiments. Adherent monocytes were trained in 24 wells plate. After training and the resting period, cells were lysed in 150μl lysis buffer. Equal amounts of protein were subjected to SDS-PAGE electrophoresis using 7.5% polyacrylamide gels. Primary antibodies (1:500 and 1:50 000 (actin)) in 5% (w/v) BSA/TBST (5% bovine serum albumin/TBST) were incubated overnight at 4°C. HRP-conjugated antirabbit antibody or HRP-conjugated anti-mouse at a dilution of 1:5000 in in 5% (w/v) BSA/TBST were used for 1 hour at room temperature. Quantitative assessment of band intensity was performed by Image Lab statistical software (Bio-Rad, CA, USA). Following antibodies were used: actin antibody (Sigma, A5441), mTOR antibody (Cell Signaling, #2972, Leiden, the Netherlands), phospho-mTOR antibody (Ser2448) (Cell Signaling, #2971), AMPKα antibody (Cell Signaling, #2532), phosphor-AMPKα (Thr172) (Cell Signaling, #2531), Akt antibody (Cell Signaling, #9272), phosphor-Akt (Ser473) (Cell Signaling, #9271). At least four different individual experiments were repeated for each Western blot experiment.
Analysis of RNA-sequencing data
Sequencing reads were mapped to the mouse genome (mm10 assembly) using STAR (version 2.3.0). The aligner was provided with a file containing junctions from Ensembl GRCm38.74. In total, there are 507.5 million reads from 12 samples. Htseq-count of the Python package HTSeq (version 0.5.4p3) was used to quantify the read counts per gene based on annotation version GRCm38.74, using the default union-counting mode (The HTSeq package, http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html).
Differentially expressed genes were identified by statistics analysis using edgeR package from bioconductor. The statistically significant threshold (FDR = 0.05) was applied. For visualization, fold changes larger than 1.5 and FDR of 0.01 were used to plot expression level of protein-coding genes.
Animal experimental models
The metformin experiment was done in University of Athens with the approval of the Ethics Committee on Animal Experiments of the University of Athens (Approval No. 2550). C57BL/6J female mice (8–12 weeks) were used (Jackson Laboratories). Mice were injected with live C. albicans blastoconidia (2×104 cfu/mouse) or pyrogen-free phosphate-buffered saline (PBS) alone. Seven days later, mice were infected intravenously with a lethal dose of live C. albicans (2×106 cfu/mouse). Survival was monitored daily. To assess the involvement of AMPK-mTOR pathway in the training, metformin (250 mg/kg) or PBS were given via intravenous injection from one day before the first non-lethal dose live C. albicans challenge till 3 days post challenge on a daily basis.
Wild-type (Cre +/+, HIF flox/flox) and HIF-KO mice 8-10 weeks old were trained with 200μL i.p. of either 1 mg β-glucan particles or sterile phosphate buffered saline on days −7 and −4 prior to tail vein inoculation with 200μL of 5×106 Staphylococcus aureus strain RN4220 on day 0. Mice were monitored three times daily for survival for 14 days. Data presented are the combined survival data (Kaplan-Meier) from two independent experiments. There were 5 mice per group in the first survival experiment and 7 mice per group in the second survival experiment. A Log-Rank test was used to assess to statistical significance between the groups. For the RNA sequence analysis, both wild type and mHIF1α-KO mice were injected with PBS or β-glucan i.p. and the total RNA was extracted from spelenocytes at day 4. The total gene expression profiles were accessed by RNA sequencing.This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council. The protocol was approved by the Dartmouth IACUC (Approval No: cram.ra.2).
Metabolic activity assay
Alveolar macrophages were isolated from 6-10 week old HIF-KO and WT mice by flushing lungs ten times with 1mL PBS containing 0.5mM EDTA. Alveolar macrophages were added and allowed to adhere for 1 hr to a 96-well plate at a concentration of 8×104 in 200 μl of CO2 independent media (Leibovitz’s L-15, Life Technologies) supplemented with 10% FCS, 5 mM HEPES buffer, 1.1 mM L-glutamine, 0.5U/ml penicillin, and 50 mg/ml streptomycin. To the media, 10% Resazurin dye (Sigma) was added and the plate was incubated at 37°C for 24 hr with readings being recorded every 30 min at 600 nm. A 690nm reference wavelength was subtracted from the 600nm wavelengths and the data were normalized to wells without cells. Curdlan (100ug/mL, Sigma) was used as a stimulator of metabolic activity.
Statistical Analysis and Graph Presentation
The differences between groups were analyzed using the Wilcoxon signed rank test (unless otherwise stated). Statistical significance of survival experiment was calculated using the product limit method of Kaplan and Meier. The level of significance was defined as a p-value of < 0.05. Cytokine production as well as the band intensity ratio for Western Blot were plotted as bar chart with mean ± SEM.
Supplementary Material
Acknowledgements
S-C.C., J.Q., and M.G.N. were supported by a Vici grant of the Netherlands Organization of Scientific Research and an ERC Consolidator grant #310372 (both to M.G.N). C.W. is supported by funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013) / ERC Grant Agreement n. 2012-322698). Y.L. is supported by a Veni grant #863.13.011 of The Netherlands Organization for Scientific Research. R.A.C. and K.M.S. were supported by grants from the National Institute of Health (NIH) National Institute of General Medical Sciences 5P30GM103415-03 (Green, William PI) and 1P30GM106394-01 (Stanton, Bruce PI), and NIH/National Institute of Allergy and Infectious Diseases R01AI81838 (R.A.C. PI). R.A.C. /K.M.S. thank Dr. Brent Berwin (Dartmouth) for the S. aureus. RJX funded by DK43351, DK097485, Helmsley Trust and JDRF. The detailed data have been deposited in the GEO database with accession number GSE57206.
References
- 1.Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annual review of plant biology. 2013;64:839. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 2.Kurtz J. Specific memory within innate immune systems. Trends in immunology. 2005 Apr;26:186. doi: 10.1016/j.it.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Bowdish DM, Loffredo MS, Mukhopadhyay S, Mantovani A, Gordon S. Macrophage receptors implicated in the "adaptive" form of innate immunity. Microbes and infection / Institut Pasteur. 2007 Nov-Dec;9:1680. doi: 10.1016/j.micinf.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Kleinnijenhuis J, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proceedings of the National Academy of Sciences of the United States of America. 2012 Sep 17; doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009 Jan 29;457:557. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quintin J, et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell host & microbe. 2012 Aug 16;12:223. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benn CS, Netea MG, Selin LK, Aaby P. A small jab - a big effect: nonspecific immunomodulation by vaccines. Trends in immunology. 2013 Sep;34:431. doi: 10.1016/j.it.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Bekkering S, Joosten LA, Meer JW, Netea MG, Riksen NP. Trained innate immunity and atherosclerosis. Current opinion in lipidology. 2013 Dec;24:487. doi: 10.1097/MOL.0000000000000023. [DOI] [PubMed] [Google Scholar]
- 9.Jaskiewicz M, Conrath U, Peterhansel C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO reports. 2011 Jan;12:50. doi: 10.1038/embor.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown GD, Gordon S. Fungal beta-glucans and mammalian immunity. Immunity. 2003 Sep;19:311. doi: 10.1016/s1074-7613(03)00233-4. [DOI] [PubMed] [Google Scholar]
- 11.Oren R, Farnham AE, Saito K, Milofsky E, Karnovsky ML. Metabolic patterns in three types of phagocytizing cells. The Journal of cell biology. 1963 Jun;17:487. doi: 10.1083/jcb.17.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013 Jan 17;493:346. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 13.Tello D, et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting Complex I activity. Cell metabolism. 2011 Dec 7;14:768. doi: 10.1016/j.cmet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Jang SY, Kang HT, Hwang ES. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. The Journal of biological chemistry. 2012 Jun 1;287:19304. doi: 10.1074/jbc.M112.363747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013 Apr 11;496:238. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu TF, Vachharajani VT, Yoza BK, McCall CE. NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. The Journal of biological chemistry. 2012 Jul 27;287:25758. doi: 10.1074/jbc.M112.362343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donohoe DR, Bultman SJ. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. Journal of cellular physiology. 2012 Sep;227:3169. doi: 10.1002/jcp.24054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013 Oct 11;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nature reviews. Immunology. 2012 May;12:325. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferwerda B, et al. Human dectin-1 deficiency and mucocutaneous fungal infections. The New England journal of medicine. 2009 Oct 29;361:1760. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumder PK, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nature medicine. 2004 Jun;10:594. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 22.Hudson CC, et al. Regulation of Hypoxia-Inducible Factor 1α Expression and Function by the Mammalian Target of Rapamycin. Molecular and Cellular Biology. 2002 Oct 15;22:7004. doi: 10.1128/MCB.22.20.7004-7014.2002. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley TW, et al. Macrophage Colony-stimulating Factor Promotes Cell Survival through Akt/Protein Kinase B. Journal of Biological Chemistry. 1999 Sep 10;274:26393. doi: 10.1074/jbc.274.37.26393. 1999. [DOI] [PubMed] [Google Scholar]
- 24.Marakalala MJ, et al. Dectin-1 plays a redundant role in the immunomodulatory activities of beta-glucan-rich ligands in vivo. Microbes and infection / Institut Pasteur. 2013 Jun;15:511. doi: 10.1016/j.micinf.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gwinn DM, et al. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Molecular Cell. 30:214. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosi E. Metformin--the gold standard in type 2 diabetes: what does the evidence tell us? Diabetes, obesity & metabolism. 2009 May;11(Suppl 2):3. doi: 10.1111/j.1463-1326.2008.01031.x. [DOI] [PubMed] [Google Scholar]
- 27.Cramer T, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003 Mar 7;112:645. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013 Jan 17;152:157. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 29.Warburg O. Metabolism of tumours. Biochem. Z. 1923;142:317. [Google Scholar]
- 30.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013 Jun 6;153:1239. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Prados JC, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. Journal of immunology. 2010 Jul 1;185:605. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 32.Zinkernagel AS, Peyssonnaux C, Johnson RS, Nizet V. Pharmacologic Augmentation of Hypoxia-Inducible Factor—1α with Mimosine Boosts the Bactericidal Capacity of Phagocytes. Journal of Infectious Diseases. 2008 Jan 15;197:214. doi: 10.1086/524843. 2008. [DOI] [PubMed] [Google Scholar]
- 33.Paust S, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nature immunology. 2010 Dec;11:1127. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Meer JW, Barza M, Wolff SM, Dinarello CA. A low dose of recombinant interleukin 1 protects granulocytopenic mice from lethal gram-negative infection. Proceedings of the National Academy of Sciences of the United States of America. 1988 Mar;85:1620. doi: 10.1073/pnas.85.5.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aaby P, et al. Randomized trial of BCG vaccination at birth to low-birth-weight children: beneficial nonspecific effects in the neonatal period? The Journal of infectious diseases. 2011 Jul 15;204:245. doi: 10.1093/infdis/jir240. [DOI] [PubMed] [Google Scholar]
- 36.Yanez A, et al. Detection of a TLR2 agonist by hematopoietic stem and progenitor cells impacts the function of the macrophages they produce. European journal of immunology. 2013 Aug;43:2114. doi: 10.1002/eji.201343403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton JA, et al. Hypoxia enhances the proliferative response of macrophages to CSF-1 and their pro-survival response to TNF. PloS one. 2012:e45853. doi: 10.1371/journal.pone.0045853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Everts B, et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012 Aug 16;120:1422. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda Y, Togo T, Mizuno S, Konishi M, Nanba H. Soluble beta-glucan from Grifola frondosa induces proliferation and Dectin-1/Syk signaling in resident macrophages via the GM-CSF autocrine pathway. Journal of leukocyte biology. 2012 Apr;91:547. doi: 10.1189/jlb.0711386. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25:1754. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao NA, et al. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome research. 2011 Sep;21:1404. doi: 10.1101/gr.118042.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.