Abstract
OBJECTIVE
To assess whether the carrier status of 35 risk alleles for prostate cancer (CaP) is associated with having unfavourable pathological features in the radical prostatectomy specimen in men with clinically low risk CaP who fulfil commonly accepted criteria as candidates for active surveillance.
PATIENTS AND METHODS
We studied men of European ancestry with CaP who fulfilled the commonly accepted clinical criteria for active surveillance (T1c, prostate-specific antigen < 10 ng/mL, biopsy Gleason ≤ 6, three or fewer positive cores, ≤ 50% tumour involvement/core) but instead underwent early radical prostatectomy.
We genotyped these men for 35 CaP risk alleles. We defined ‘ unfavourable ’ pathological characteristics to be Gleason ≥ 7 and/or ≥ pT2b in their radical prostatectomy specimen.
RESULTS
In all, 263 men (median age 60 [46 – 72] years) fulfilled our selection criteria for active surveillance, and 58 of 263 (22.1%) were found to have ‘ unfavourable ’ pathological characteristics.
The frequencies of three CaP risk alleles (rs1447295 [8q24], P = 0.004; rs1571801 [9q33.2], P = 0.03; rs11228565 [11q13], P = 0.02) were significantly higher in men with ‘ unfavourable ’ pathological characteristics.
Two other risk alleles were proportionately more frequent (rs10934853 [3q21], P = 0.06; rs1859962 [17q24], P = 0.07) but did not achieve nominal statistical significance.
Carriers of any one of the significantly over-represented risk alleles had twice the likelihood of unfavourable tumour features (P = 0.03), and carriers of any two had a sevenfold increased likelihood (P = 0.001).
Receiver – operator curve analysis demonstrated an area under the curve of 0.66, suggesting that the number of single nucleotide polymorphisms carried provided discrimination between men with ‘ favourable ’ and ‘ unfavourable ’ tumour features in their prostatectomy specimen.
CONCLUSION
In potential candidates for active surveillance, certain CaP risk alleles are more prevalent in patients with ‘ unfavourable ’ pathological characteristics in their radical prostatectomy specimen.
Keywords: prostate cancer, active surveillance, single nucleotide polymorphism, risk allele
INTRODUCTION
Since implementation of widespread PSA screening, there has been a stage migration toward detecting smaller volume, lower grade prostate cancer (CaP). To address concerns of over-diagnosis and over-treatment of indolent tumours, active surveillance (AS) strategies have been implemented to manage men with low risk CaP [1]. Although favourable outcomes of men enrolled in AS programmes have been reported, no series has sufficient follow-up to permit valid conclusions to be drawn about the long-term safety and efficacy of AS [2,3], particularly in patients with a long life expectancy. The interactions between inherited germline and somatic genetic variants, environmental influences and chance factors that influence CaP aggressiveness are unknown. Therefore, it is not surprising that some apparently low risk tumours exhibit cancer progression, causing suffering and death. Despite the commonly stated perception that CaP is a slowly growing form of cancer that is often over-treated [4], most men do not choose AS when diagnosed with CaP. For example, it has been reported that ≥ 90% of men in the USA with low stage disease elect definitive treatment [5].
Genetic studies have validated approximately 35 single nucleotide polymorphisms (SNPs) that are associated with CaP susceptibility (CaP risk alleles) on a genome-wide level [6]. Each SNP has been independently associated with a modestly increased risk of developing CaP, relative to the general population (odds ratio [OR] 1.1 – 1.8) [7]. However, there also appears to be a strong cumulative effect between these SNPs [7,8], and some have been associated with clinically aggressive phenotypes [6]. As such, they may be clinically useful to stratify patients into risk profiles, such as those more likely to fail AS protocols due to the presence of higher grade or higher stage disease. To our knowledge, the clinical utility of SNPs to distinguish candidates for AS protocols has not been examined. Therefore, in men with low risk CaP who fulfil the criteria for AS, we sought to assess whether the CaP risk alleles were associated with unfavourable pathological characteristics.
PATIENTS AND METHODS
Our study group consisted of 1378 men of European ancestry diagnosed with CaP between 2003 and 2009. Of these patients, 1048 underwent radical prostatectomy (RP) by a single surgeon, and the remaining underwent surgical treatment by other urologists from the Northwestern University Specialized Program of Research Excellence (SPORE) group. The study was approved by Northwestern University's Institutional Review Board, and all participants provided written informed consent. The patients were enrolled from a prospective database. All participants filled out a questionnaire and provided a blood sample for genotype analyses.
Clinical and pathological characteristics were recorded, including preoperative PSA level, first-degree family history of CaP, clinical and pathological tumour stage, and presence of extracapsular tumour extension, seminal vesicle invasion and lymph node metastases. Organ-confined disease was defined as a tumour confined to the prostate (stage pT2). Men with extraprostatic tumour extension (≥ pT3), seminal vesicle invasion (pT3b) or lymph node metastases (N1) were categorized as having non-organ-confined disease.
For the purposes of this study, AS criteria were limited to modified Epstein criteria [9] which include biopsy Gleason score ≤ 6, PSA < 10 ng/mL, clinical stage T1c, three cores or fewer positive for cancer, and ≤ 50% tumour involvement of any one biopsy core. Unfavourable pathology was defined either by upstaging (pathological tumour stage ≥T2b) and/or upgrading (Gleason score ≥ 7 after RP).
DNA was extracted from whole blood at deCODE ® genetics Inc., in Reykjavik, Iceland. Each sample was genotyped for 35 CaP risk alleles (Table 1); the Centaurus (Nanogen) genotyping methods were employed, and their accuracy has been evaluated as previously described [10]. We genotyped for 35 SNPs, as these are published as CaP risk alleles from recent genome-wide association studies having reached genome-wide significance. Univariate logistic regression models were performed to examine dominant and recessive genetic models associated with CaP. The Akaike information criterion was used to define the carrier status of each allele [11] (Table 1).
TABLE 1.
CaP genetic risk alleles with corresponding chromosomal location, and best-fit genetic model
| SNP | Location | Risk allele | Model |
|---|---|---|---|
| rs721048 | 2q15 | A | Recessive |
| rs12621278 | 2q31.1 | G | Dominant |
| rs1465618 | 2p21 | A | Recessive |
| rs10934853 | 3q21 | A | Dominant |
| rs2660753 | 3p12.1 | T | Recessive |
| rs12500426 | 4q22.3 | A | Recessive |
| rs17021918 | 4q22.3 | T | Recessive |
| rs7679673 | 4q24 | A | Dominant |
| rs2736098 | 5p15 | A | Recessive |
| rs401681 | 5p15 | C | Dominant |
| rs9364554 | 6q25.3 | T | Recessive |
| rs10486567 | 7p15.2 | G | Recessive |
| rs6465657 | 7q21.3 | C | Recessive |
| rs16901979 | 8q24 | A | Dominant |
| rs16902094 | 8q24 | G | Dominant |
| rs445114 | 8q24 | T | Dominant |
| rs6983267 | 8q24 | G | Dominant |
| rs1447295 | 8q24 | A | Dominant |
| rs1512268 | 8p21.2 | A | Dominant |
| rs1571801 | 9q33.2 | A | Recessive |
| rs10993994 | 10q11 | T | Recessive |
| rs4962416 | 10q26.13 | C | Recessive |
| rs11228565 | 11q13 | A | Dominant |
| rs10896450 | 11q13 | G | Dominant |
| rs12418451 | 11q13.3 | A | Dominant |
| rs7127900 | 11p15.5 | A | Dominant |
| rs11649743 | 17q12 | G | Recessive |
| rs4430796 | 17q12 | A | Dominant |
| rs1859962 | 17q24 | G | Recessive |
| rs4054823 | 17p12 | T | Dominant |
| rs8102476 | 19q13 | C | Dominant |
| rs2735839 | 19q13.3 | G | Dominant |
| rs5759167 | 22q13.2 | T | Recessive |
| rs9623117 | 22q13.1 | C | Dominant |
| rs5945572 | Xp11 | A | Dominant |
We compared the clinical and pathological features of potential AS candidates with and without unfavourable RP pathology features using the chi-squared test. Per-allele ORs were estimated by logistic regression. Receiver – operating characteristic (ROC) analyses were also performed to evaluate the utility of the SNPs in discriminating between men with or without unfavourable pathology. P < 0.05 was considered significant. Additionally, multivariate analysis accounting for other significant SNPs found to be associated with unfavourable pathological characteristics was performed. All statistical analyses were performed using SAS ® 9.2.
RESULTS
Of the 1378 men, 19.1% (n = 263) had clinical and biopsy findings that would fulfil the criteria for an AS protocol. The median age of diagnosis, serum PSA concentration and frequency of first-degree relatives affected with CaP for this group was 59 (range 34 – 74) years, 4.6 ng/mL, and 60.3%, respectively. Of these men, 205 (77.9%) had favourable pathology characteristics and 58 (22.1%) had unfavourable characteristics. The baseline clinical characteristics were not significantly different between the two groups.
The genotypes of the 35 CaP risk SNPs are shown in Table 1. The frequency of each SNP was compared between those with favourable and unfavourable pathology. Of the 35 SNPs, 22 were present in an increased frequency in men with unfavourable pathological characteristics (Table 2). Three of these SNPs (rs1447295 on chromosome 8q24 [P = 0.004], rs1571801 on chromosome 9q33.2 [P = 0.03] and rs11228565 on chromosome 11q13 [P = 0.02]) were present at a significantly increased frequency in this group. After adjusting for two other significant SNPs in a multivariate analysis, all three SNPs with P < 0.05 remained significant. Two other SNPs (rs10934853 on chromosome 3q21 [P = 0.06] and rs1859962 on chromosome 17q24 [P = 0.07]) approached statistical significance. For SNP rs1447295 on chromosome 8q24, heterozygotes and homozygotes for the A allele had ORs of 2.3 (95% CI 1.21 – 4.43) and 4.6 (95% CI 0.90 – 23.90), respectively, of having unfavourable pathological characteristics compared with those homozygous for the C allele. Similarly, heterozygotes and homozygotes for the A allele of SNP rs1571801 on 9q33.2 had ORs of 1.2 (95% CI 0.62 – 2.45) and 3.9 (95% CI 1.33 – 11.26), respectively, compared with those homozygous for the C allele of having unfavourable pathological characteristics. Heterozygotes and homozygotes of the A allele of SNP rs11228565 on 11q13 had ORs of 2.0 (95% CI 1.04 – 3.65) and 2.7 (95% CI 0.90 – 7.87), respectively, of having unfavourable pathological characteristics.
TABLE 2.
CaP risk allele frequency in patients with favourable or unfavourable pathological characteristics; all would have been eligible for an AS protocol
| SNP | Chromosomal location | Favourable pathological characteristics, % (n = 205) | Unfavourable pathological characteristics, % (n = 58) | P |
|---|---|---|---|---|
| rs721048 | 2q15 | 2.5 (5) | 6.9 (4) | 0.12 |
| rs12621278 | 2q31.1 | 10.5 (21) | 14.3 (8) | 0.43 |
| rs1465618 | 2p21 | 3.0 (6) | 7.1 (7) | 0.23 |
| rs10934853 | 3q21 | 48.0 (95) | 62.1 (36) | 0.06 |
| rs2660753 | 3p12.1 | 0.5 (1) | 1.7 (1) | 0.40 |
| rs12500426 | 4q22.3 | 22.1 (44) | 24.6 (14) | 0.70 |
| rs17021918 | 4q22.3 | 5.6 (4) | 4.8 (1) | 1.0 |
| rs7679673 | 4q24 | 63.6 (124) | 61.1 (33) | 0.74 |
| rs2736098 | 5p15 | 10.5 (21) | 15.5 (9) | 0.29 |
| rs401681 | 5p15 | 85.7 (174) | 84.5 (49) | 0.82 |
| rs9364554 | 6q25.3 | 8.0 (16) | 5.2 (3) | 0.58 |
| rs10486567 | 7p15.2 | 67.2 (135) | 58.6 (34) | 0.23 |
| rs6465657 | 7q21.3 | 22.5 (45) | 20.7 (12) | 0.80 |
| rs16901979 | 8q24 | 9.4 (19) | 10.3 (6) | 0.8 |
| rs16902094 | 8q24 | 26.5 (50) | 34.6 (19) | 0.24 |
| rs445114 | 8q24 | 93.5 (186) | 89.7 (52) | 0.39 |
| rs6983267 | 8q24 | 83.7 (169) | 77.6 (45) | 0.29 |
| rs1447295 | 8q24 | 21.0 (43) | 39.7 (23) | 0.004 |
| rs1512268 | 8p21.2 | 75.0 (150) | 64.9 (37) | 0.13 |
| rs1571801 | 9q33.2 | 5.3 (9) | 16.3 (8) | 0.03 |
| rs10993994 | 10q11 | 20.7 (41) | 24.1 (14) | 0.58 |
| rs4962416 | 10q26.13 | 8.0 (16) | 5.4 (3) | 0.77 |
| rs11228565 | 11q13 | 38.4 (76) | 56.1 (32) | 0.02 |
| rs10896450 | 11q13 | 77.1 (155) | 86.2 (50) | 0.13 |
| rs12418451 | 11q13.3 | 46.7 (93) | 49.1 (28) | 0.75 |
| rs7127900 | 11p15.5 | 46.5 (94) | 51.8 (29) | 0.49 |
| rs11649743 | 17q12 | 70.8 (148) | 65.5 (38) | 0.44 |
| rs4430796 | 17q12 | 79.4 (162) | 74.1 (43) | 0.39 |
| rs1859962 | 17q24 | 33.3 (67) | 20.7 (12) | 0.07 |
| rs4054823 | 17p12 | 72.4 (147) | 79.3 (46) | 0.30 |
| rs8102476 | 19q13 | 80.8 (164) | 89.7 (52) | 0.12 |
| rs2735839 | 19q13.3 | 98.0 (198) | 98.3 (56) | 1.0 |
| rs5759167 | 22q13.2 | 14.5 (24) | 14.9 (7) | 0.94 |
| rs9623117 | 22q13.1 | 36.9 (72) | 30.9 (17) | 0.41 |
| rs5945572 | Xp11 | 37.1 (76) | 37.9 (22) | 0.91 |
We next determined whether the SNPs that were significantly over-represented in men with unfavourable pathology exhibited cumulative effects (Table 3). Carriers of any one of the three over-represented SNPs had more than twice the likelihood of unfavourable pathology (OR = 2.4, 95% CI 1.16 – 5.28; P = 0.03), and carriers of any two of them had more than a sevenfold increased likelihood of unfavourable pathology (OR = 7.2, 95% CI 2.66 – 19.41; P < 0.001) (Table 3). ROC analysis demonstrated an area under the curve of 0.66, suggesting that the number of SNPs carried provided reasonable discrimination between men with favourable and unfavourable pathology (Fig. 1). Utilizing a panel incorporating any of these three SNPs to predict unfavourable pathological features has a sensitivity of 77%, specificity of 48%, positive predictive value of 30% and negative predictive value of 88%.
TABLE 3.
Cumulative risk of having unfavourable pathological characteristics associated with carrying any one or any two to three risk alleles (rs1447295 [8q24], rs1571801 [9q33.2] and rs 11228565 [11q13]) in men who would have been eligible for an AS protocol
| Number of prostate cancer risk alleles carried | Favourable pathological characteristics, % (n) | Unfavourable pathological characteristics, % (n) | Odds ratio | 95% CI | P |
|---|---|---|---|---|---|
| None | 47.6 (79) | 22.5 (11) | Reference | Reference | - |
| Any 1 | 44.6 (74) | 51.0 (25) | 2.4 | 1.2-5.3 | 0.03 |
| Any 2-3 | 7.8 (13) | 26.5 (13) | 7.2 | 2.7-19.4 | 0.0001 |
FIG. 1.
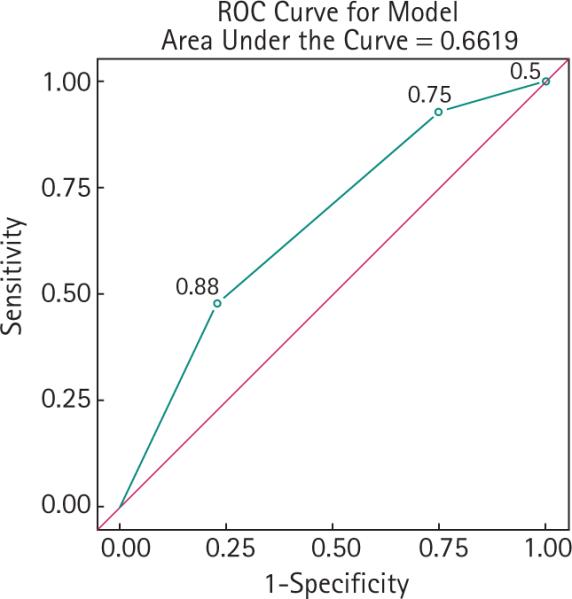
ROC analysis in potential AS candidates with favourable or unfavourable pathological characteristics.
Men who had favourable pathological characteristics had a mean and median of 26.2 and 26.0 risk alleles, respectively. In contrast, the mean (27.7; P = 0.017) and median (29; P = 0.013) were significantly higher in those with unfavourable pathology. We then used the total number of risk alleles as a continuous variable (OR = 1.09, 95% CI 1.02 – 1.18, P = 0.02) to predict the risk of adverse features. A significantly higher proportion of men with unfavourable pathological characteristics had more than 28 out of a possible 69 alleles than those with favourable pathology (50% vs 29.8%; OR = 2.36, 95% CI 1.3 – 4.28, P = 0.004) (Table 4). In contrast, carrying fewer than 20 risk alleles was associated with a protective effect, with 8.82% of men having favourable pathological features vs 1.75% of men with unfavourable features (OR 5.42, 95% CI 0.71 – 41.49, P = 0.08).
TABLE 4.
Risk allele dose out of a possible 69 alleles in men with favourable or unfavourable pathological tumour features
| Allele dose | Favourable pathological characteristics, % | Unfavourable pathological characteristics, % | Odds ratio | 95% CI | P |
|---|---|---|---|---|---|
| <28 | 70 | 50 | Reference | Reference | |
| ≥29 | 30 | 50 | 2.36 | 1.3-4.28 | 0.004 |
DISCUSSION
AS is a management strategy used for men with clinical and biopsy features characteristic of localized CaP that has a low likelihood of metastasizing, as many such men are likely to die from causes other than CaP. However, studies of AS candidates have shown that at least one-third to one-half of men managed in this way ultimately manifest evidence of disease progression within 10 years, and some ultimately suffer or die from CaP. We and others previously reported on the surgical pathology outcomes in men who met the criteria for AS protocols, showing that a substantial proportion of these men harbour histopathological evidence of aggressive disease [12 – 15]. The optimal method to identify men who will progress is yet to be defined. New strategies are needed, including biomarkers that help distinguish appropriate AS candidates.
Men with a family history of CaP are considered to have a greater genetic susceptibility of developing the disease [16]. However, the underlying mechanisms of this genetic susceptibility remain largely unknown. Reports on genes and candidate pathways that may be associated with CaP susceptibility and aggressiveness have yet to be fully elucidated [17]. A new era of genetic discovery has emerged, permitting the identification of genetic risk alleles that are associated with risk of CaP [6]. Some of these have also been associated with disease aggressiveness, e.g. SNPs on chromosome 8q24 (rs16901979, rs6983267, rs1447295, rs10993994) [18,19]. Thus, a major current focus is the discovery and validation of genetic variants that may be a surrogate for aggressive disease features and, as such, may aid in identifying men who may not be ideal candidates for AS.
Our research group has previously evaluated the associations of certain CaP risk alleles with adverse pathological features [20,21]. Specifically, we reported the differences in the carrier frequency of five SNPs on chromosome 8q24 and 17q in men who had potentially clinically ‘ insignificant ’ disease [22]. We found that men who were carriers of less than one SNP were nearly threefold more likely to have insignificant disease than carriers of more than two SNPs [22]. The results of the present study significantly expand upon these initial observations. We assessed the carrier frequency of a contemporary panel of most (n = 35) of the validated CaP risk alleles in men who elected for RP for low grade, low volume CaP on biopsy who met widely accepted eligibility criteria for AS. We then compared the carrier frequencies of the risk alleles in men whose RP specimen revealed unfavourable pathology with those who had pathologically confirmed low grade, low volume disease.
To our knowledge this is the first analysis evaluating the frequency of these 35 CaP risk alleles to predict surgical upstaging and upgrading to help identify potentially suboptimal candidates for AS. We found that three SNPs were significantly over-represented in patients with unfavourable pathology. In addition, this risk was related to the number of these SNPs carried: men who were carriers of any one SNP were twice as likely to have unfavourable pathology, and men who carried two or more had a more than sevenfold increased risk. The corollary is that men who carried none of the SNPs were more likely to be in the group that had favourable pathological characteristics (47.6% vs 22.5%), suggesting that there may be clinical utility in genotyping these men to discriminate between better or worse candidates for AS. There also appears to be a significant dose effect of the alleles. Men who were carriers of more than 29 out of a possible 69 CaP risk alleles were significantly more likely to have unfavourable pathological characteristics, and men with fewer than 29 risk alleles were more likely to have favourable pathology.
The SNP rs1227295 (region 1 on 8q24) is believed to influence the expression of the C-MYC proto-oncogene [23]. Several studies have demonstrated an association of this SNP with high-grade disease and advanced stage across ethnicities [24], although some have not [25]. While the exact mechanism remains unknown, it has been hypothesized that overexpression of C-MYC is an early oncogenic alteration in CaP. C-MYC overexpression has also been correlated with advanced characteristics and biochemical progression in RP cohorts [26]. Thus, this SNP may influence the expression of C-MYC and induce CaP susceptibility and progression.
The SNP rs1571801 on chromosome 9q33.2 lies within an intron that encodes for the gene DAB21P, a novel RAS GTPase-activating protein and putative prostate tumour suppressor gene. Variation at this SNP has been previously associated with aggressive disease features in a multi-ethnic CaP cohort [27]; here we validate this finding. Although we demonstrate that SNP rs11228565 on chromosome 11q13 is associated with unfavourable pathology in our RP cohort, this CaP susceptibility allele has not previously been found to be associated with aggressive disease features.
Our study has limitations. The sample size is underpowered for robust statistical assumptions in the usual context of genetic epidemiology. However, two of the three significant SNPs associated with unfavourable pathological features have been identified with aggressive disease features previously, supporting our findings. In addition, since all of the men were treated with RP and none were actually enrolled in an AS protocol, the study suffers from selection bias and may not be universally applicable to men undergoing AS. It is uncertain whether the same patients would have progressed beyond salvage under careful surveillance. For example, a 50-year-old man with a Gleason 6 with three biopsy cores positive for CaP would be more likely to choose RP than a 75-year-old patient with a Gleason 6 with one biopsy core positive for CaP, even though both met the modified Epstein criteria. Furthermore, we are unable to account for biopsy sampling error that may have missed higher grade disease at the time of biopsy. Since our study was conducted in men of European ancestry, these findings may not apply to other ethnic groups. Lastly, examining 35 SNPs raises a high probability of a type 1 error when the P value is set at < 0.05 for significance, and when we applied a Bonferroni correction for this none of the SNPs attained significance. This is not surprising, however, considering the sample size. Despite these limitations this study is the first to evaluate the possible clinical utility of genetic markers of CaP risk in possible candidates for AS and is hypothesis generating. Future studies aimed at determining the mechanisms of genetic interactions with tumour progression would provide critical fundamental insights into the development and progression of CaP. Therefore, ideally a large prospective trial evaluating the potential of CaP risk alleles to predict those who fail AS is required to corroborate these findings.
Our results suggest that men who are potential candidates for AS and carry three CaP risk alleles (rs1447295 [8q24], rs1571801 [9q33.2] and rs11228565 [11q13]) are more likely to have unfavourable pathological characteristics in their RP specimen.
What's known on the subject? and What does the study add?
Men fail active surveillance for a variety of reasons; however, no single reliable biomarker has been found to date which will identify these men from the outset. We know that there are about 35 prostate cancer risk alleles which have been discovered to infl uence risk of prostate cancer, from large-scale genome-wide association studies. Some of these have been associated with aggressive prostate cancer. Nobody has examined the potential for these risk alleles to predict men who might fail active surveillance.
This study adds to the growing evidence that single nucleotide polymorphisms may be able to identify men who have aggressive prostate cancers, and that this could be part of a risk algorithm used in active surveillance protocols.
ACKNOWLEDGEMENTS
Supported in part by the Urological Research Foundation, Prostate SPORE grant (P50CA90386-05S2), and the Robert H. Lurie Comprehensive Cancer Center grant (P30 CA60553). This research was presented at the 2011 American Urological Association annual meeting.
Abbreviations
- CaP
prostate cancer
- AS
active surveillance
- SNP
single nucleotide polymorphism
- OR
odds ratio
- RP
radical prostatectomy
- ROC
receiver – operating characteristic
Footnotes
CONFLICT OF INTEREST
Barry B. McGuire has received funding from GSK, Ipsen and Pfizer.
REFERENCES
- 1.Klotz LH, Choo R, Morton G, Danjoux C. Expectant management with selective delayed intervention for favorable-risk prostate cancer. Can J Urol. 2002;9(Suppl. 1):2–7. [PubMed] [Google Scholar]
- 2.Klotz L, Zhang L, Lam A, Nam R, Mamedov A, Loblaw A. Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. J Clin Oncol. 2010;28:126–31. doi: 10.1200/JCO.2009.24.2180. [DOI] [PubMed] [Google Scholar]
- 3.Soloway MS, Soloway CT, Eldefrawy A, Acosta K, Kava B, Manoharan M. Careful selection and close monitoring of low-risk prostate cancer patients on active surveillance minimizes the need for treatment. Eur Urol. 2010;58:831–5. doi: 10.1016/j.eururo.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 4.Savage CJ, Lilja H, Cronin AM, Ulmert D, Vickers AJ. Empirical estimates of the lead time distribution for prostate cancer based on two independent representative cohorts of men not subject to prostate-specific antigen screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1201–7. doi: 10.1158/1055-9965.EPI-09-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooperberg MR, Broering JM, Carroll PR. Time trends and local variation in primary treatment of localized prostate cancer. J Clin Oncol. 2010;28:1117–23. doi: 10.1200/JCO.2009.26.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindstrom S, Schumacher F, Siddiq A, et al. Characterizing associations and SNP–environment interactions for GWAS-identified prostate cancer risk markers–results from BPC3. PLoS ONE. 2011;6:e17142. doi: 10.1371/journal.pone.0017142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 8.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 9.Carter HB, Kettermann A, Warlick C, et al. Expectant management of prostate cancer with curative intent: an update of the Johns Hopkins experience. J Urol. 2007;178:2359–65. doi: 10.1016/j.juro.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 11.Helfand BT, Fought AJ, Loeb S, Meeks JJ, Kan D, Catalona WJ. Genetic prostate cancer risk assessment: common variants in. 9 genomic regions are associated with cumulative risk. J Urol. 2010;184:501–5. doi: 10.1016/j.juro.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thaxton CS, Loeb S, Roehl KA, Kan D, Catalona WJ. Treatment outcomes of radical prostatectomy in potential candidates for. 3 published active surveillance protocols. Urology. 2010;75:414–8. doi: 10.1016/j.urology.2009.07.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mufarrij P, Sankin A, Godoy G, Lepor H. Pathologic outcomes of candidates for active surveillance undergoing radical prostatectomy. Urology. 2010;76:689–92. doi: 10.1016/j.urology.2009.12.075. [DOI] [PubMed] [Google Scholar]
- 14.Griffin CR, Yu X, Loeb S, et al. Pathological features after radical prostatectomy in potential candidates for active monitoring. J Urol. 2007;178(3 Pt 1):860–3. doi: 10.1016/j.juro.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Suardi N, Briganti A, Gallina A, et al. Testing the most stringent criteria for selection of candidates for active surveillance in patients with low-risk prostate cancer. BJU Int. 2010;105:1548–52. doi: 10.1111/j.1464-410X.2009.09057.x. [DOI] [PubMed] [Google Scholar]
- 16.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 17.Ding Z, Wu CJ, Chu GC, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng I, Plummer SJ, Jorgenson E, et al. 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camp NJ, Farnham JM, Wong J, Christensen GB, Thomas A, Cannon-Albright LA. Replication of the 10q11 and Xp11 prostate cancer risk variants: results from a Utah pedigree-based study. Cancer Epidemiol Biomarkers Prev. 2009;18:1290–4. doi: 10.1158/1055-9965.EPI-08-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helfand BT, Loeb S, Cashy J, et al. Tumor characteristics of carriers and noncarriers of the deCODE 8q24 prostate cancer susceptibility alleles. J Urol. 2008;179:2197–202. doi: 10.1016/j.juro.2008.01.110. [DOI] [PubMed] [Google Scholar]
- 21.Helfand BT, Loeb S, Meeks JJ, Fought AJ, Kan D, Catalona WJ. Pathological outcomes associated with the 17q prostate cancer risk variants. J Urol. 2009;181:2502–7. doi: 10.1016/j.juro.2009.01.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helfand BT, Loeb S, Kan D, Catalona WJ. Number of prostate cancer risk alleles may identify possibly ‘insignificant’ disease. BJU Int. 2010;106:1602–6. doi: 10.1111/j.1464-410X.2010.09440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 24.Tan YC, Zeigler-Johnson C, Mittal RD, et al. Common 8q24 sequence variations are associated with Asian Indian advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:2431–5. doi: 10.1158/1055-9965.EPI-07-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pal P, Xi H, Guha S, et al. Common variants in 8q24 are associated with risk for prostate cancer and tumor aggressiveness in men of European ancestry. Prostate. 2009;69:1548–56. doi: 10.1002/pros.20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawksworth D, Ravindranath L, Chen Y, et al. Overexpression of C-MYC oncogene in prostate cancer predicts biochemical recurrence. Prostate Cancer Prostatic Dis. 2010;13:311–5. doi: 10.1038/pcan.2010.31. [DOI] [PubMed] [Google Scholar]
- 27.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–33. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]


