A remorin protein plays a role in regulating photoassimilate translocation by modulating plasmodesmata permeability in rice.
Abstract
Effective grain filling is one of the key determinants of grain setting in rice (Oryza sativa). Grain setting defect1 (GSD1), which encodes a putative remorin protein, was found to affect grain setting in rice. Investigation of the phenotype of a transfer DNA insertion mutant (gsd1-Dominant) with enhanced GSD1 expression revealed abnormalities including a reduced grain setting rate, accumulation of carbohydrates in leaves, and lower soluble sugar content in the phloem exudates. GSD1 was found to be specifically expressed in the plasma membrane and plasmodesmata (PD) of phloem companion cells. Experimental evidence suggests that the phenotype of the gsd1-Dominant mutant is caused by defects in the grain-filling process as a result of the impaired transport of carbohydrates from the photosynthetic site to the phloem. GSD1 functioned in affecting PD conductance by interacting with rice ACTIN1 in association with the PD callose binding protein1. Together, our results suggest that GSD1 may play a role in regulating photoassimilate translocation through the symplastic pathway to impact grain setting in rice.
Grain filling, a key determinant of grain yield in rice (Oryza sativa), hinges on the successful translocation of photoassimilates from the leaves to the fertilized reproductive organs through the phloem transport system. Symplastic phloem loading, which is one of the main pathways responsible for the transport of photoassimilates in rice, is mediated by plasmodesmata (PD) that connect phloem companion cells with sieve elements and surrounding parenchyma cells (Kaneko et al., 1980; Chonan et al., 1981; Eom et al., 2012). PD are transverse cell wall channels structured with the cytoplasmic sleeve and the modified endoplasmic reticulum desmotubule between neighboring cells (Maule, 2008). A number of proteins affect the structure and functional performance of the PD, which in turn impacts the cell-to-cell transport of small and large molecules through the PD during plant growth, development, and defense (Cilia and Jackson, 2004; Sagi et al., 2005; Lucas et al., 2009; Simpson et al., 2009; Stonebloom et al., 2009). For example, actin and myosin, which link the desmotubule to the plasma membrane (PM) at the neck region of PD, are believed to play a role in regulating PD permeability by controlling PD aperture (White et al., 1994; Ding et al., 1996; Reichelt et al., 1999). Callose deposition can also impact the size of the PD aperture at the neck region (Radford et al., 1998; Levy et al., 2007) and callose synthase genes such as Glucan Synthase-Like7 (GSL7, also named CalS7), GSL8, and GSL12 have been shown to play a role in regulating symplastic trafficking (Guseman et al., 2010; Barratt et al., 2011; Vatén et al., 2011; Xie et al., 2011). Other proteins that have been shown to impact the structure and function of the PD include glycosylphosphatidylinositol (GPI)-anchored proteins, PD callose binding protein1 (PDCB1), which is also associated with callose deposition (Simpson et al., 2009), and LYSIN MOTIF DOMAIN-CONTAINING GLYCOSYLPHOSPHATIDYLINOSITOL-ANCHORED PROTEIN2, which limits the molecular flux through the PD by chitin perception (Faulkner et al., 2013). Changes in PD permeability can have major consequences for the translocation of photoassimilates needed for grain filling in rice. However, the genes and molecular mechanisms underlying the symplastic transport of photoassimilates remain poorly characterized.
Remorins are a diverse family of plant-specific proteins with conserved C-terminal sequences and highly variable N-terminal sequences. Remorins can be classified into six distinct phylogenetic groups (Raffaele et al., 2007). The functions of most remorins are unknown, but some members of the family have been shown to be involved in immune response through controlling the cell-to-cell spread of microbes. StREM1.3, a remorin that is located in PM rafts and the PD, was shown to impair the cell-to-cell movement of a plant virus X by binding to Triple Gene Block protein1 (Raffaele et al., 2009). Medicago truncatula symbiotic remorin1 (MtSYMREM1), a remorin located at the PM in Medicago truncatula, was shown to facilitate infection and the release of rhizobial bacteria into the host cytoplasm (Lefebvre et al., 2010). Overexpression of LjSYMREM1, the ortholog of MtSYMREM1 in Lotus japonicus, resulted in increased root nodulation (Lefebvre et al., 2010; Tóth et al., 2012). Although a potential association between remorins and PD permeability has been proposed (Raffaele et al., 2009), the diversity observed across remorins, plus the fact that remorin mutants generated through different approaches fail to show obvious phenotypes (Reymond et al., 1996; Bariola et al., 2004), have made it challenging to characterize the function of remorins in cell-to-cell transport.
In this study, we identified a rice transfer DNA (T-DNA) insertion mutant (grain setting defect1-Dominant [gsd1-D]), with a grain setting-deficient phenotype caused by overexpression of GSD1, a remorin gene with unknown function. GSD1 is expressed specifically in phloem companion cells and is localized in the PD and PM. We provide evidence to show that overexpression of GSD1 leads to deficient grain setting in rice, likely as a consequence of reduced sugar transport resulting from decreased PD permeability in phloem companion cells.
RESULTS
Isolation and Characterization of the gsd1-D Mutant
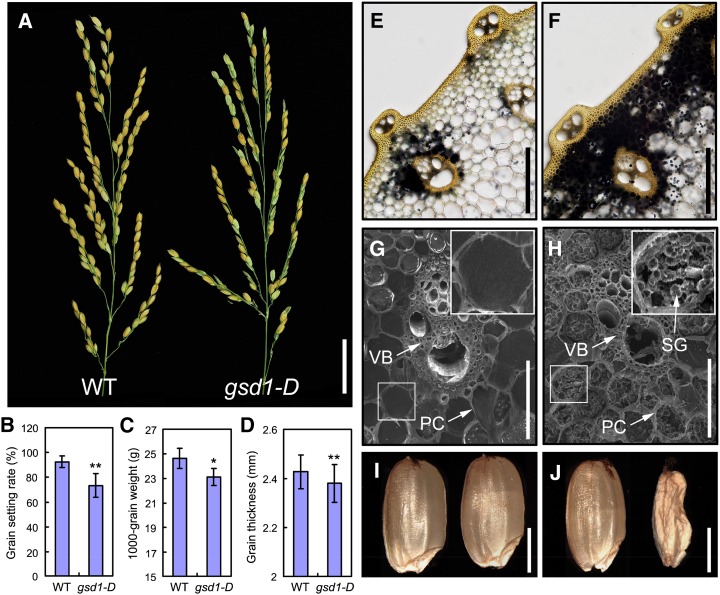
A grain setting defect mutant, named gsd1-D, was detected and isolated from our collection of T-DNA insertion rice lines that have a genetic background of O. sativa japonica cv Zhonghua 11 (ZH11). The phenotype of the mutant was examined at various stages of growth and development. During vegetative growth, the mutant gsd1-D displayed a similar phenotype to wild-type ZH11 with no apparent morphological changes. However, during the grain-filling and ripening phases, the grain setting rate in gsd1-D was reduced to 72% compared with 92% in the wild type (Fig. 1, A and B). At maturity, the 1,000-grain weight and grain thickness of gsd1-D were 6% and 3% lower, respectively, compared with the wild type (Fig. 1, C and D). Other agronomic traits such as the spikelet number, tiller number, plant height, internode length, grain length, and grain width did not differ significantly between the mutant and the wild type (Supplemental Fig. S1). The morphology of the floral organ at various stages of flower development was also examined. No distinguishable abnormalities were observed in the stamen (anther and filament) and pistil (stigma, style, and ovary) of the mutant (Supplemental Fig. S2). Observations of anther transverse sections also revealed seemly normal cellular morphology (Supplemental Fig. S3). Overall, these results suggest that GSD1 may be involved in regulating grain setting in rice but not through affecting floral development.
Figure 1.
Phenotypic comparison between the wild type and the homozygous gsd1-D mutant. A, Comparison of the wild type (left) and gsd1-D mutant panicles (right) at the grain-ripening stage. B to D, Comparison of the grain setting rate (B), 1,000-grain weight (C), and grain thickness (D) between the wild type and the gsd1-D mutant. Error bars show the means ± se (Student’s t test, n = 15 in B, n = 10 in C, and n = 25 in D). The single asterisk represents P < 0.05, and double asterisks represent P < 0.01. E and F, Iodine-potassium iodide (I2-KI) staining of starch at the second internode of the wild type (E) and gsd1-D (F) in the grain matured plants. G and H, Scanning electron microscopy observation of the second internode sections of the wild type (G) and gsd1-D (H) in the grain matured plants. I and J, Mature seeds from the wild type (I) and the gsd1-D mutant (J). PC, Parenchyma cell; SG, starch granule; VB, vascular bundle; WT, wild type. Bars = 3 cm in A; 200 μm in E and F; 50 μm in G and H; and 2 mm in I and J.
The distribution of starch granules in the parenchyma cells of the leaf sheath and culm reflect the state of photoassimilate translocation (Lian and Tanaka, 1967; Yoshida and Ahn, 1968; Perez et al., 1971). Starch accumulation in the stems of wild-type and gsd1-D plants was examined at the booting, flowering, and grain-filling stages. At the booting stage, little starch was accumulated in the stems of wild-type and gsd1-D plants (Supplemental Fig. S4, A and B). During the flowering stage, abundant starch was observed in gsd1-D and the wild type, indicating the temporary accumulation of starch in the stem for grain development (Supplemental Fig. S4, C and D). During grain filling and ripening, most of the starch that accumulated in the parenchyma cells underwent relocation for grain filling. After grain filling, little accumulated starch remained in the wild type (Fig. 1, E and G), consistent with previous studies (Cock and Yoshida, 1972). By contrast, much starch remained in the stem of gsd1-D (Fig. 1, F and H), suggesting defective translocation of photoassimilates in the mutant. Compared with the wild type (Fig. 1I), more shrunken and aborted seeds were produced by gsd1-D plants (Fig. 1J). Expression of starch synthesis-related genes, including AGPase large subunit1 (OsAGPL1), OsAGPL2, AGPase small subunit 2b,starch synthase IIa (OsSSIIa), OsSSIIIa, starch branching enzyme IIb, granule-bound starch synthaseI, and starch debranching enzyme isoamylase1 in developing grain was also affected in the gsd1-D mutant (Supplemental Fig. S5), possibly reflecting lowered starch synthesis in mutant seeds. These results indicate that the gsd1-D displayed abnormal grain development as a result of the defective translocation of photoassimilates.
Carbohydrate Export and Translocation Are Reduced in the gsd1-D Mutant
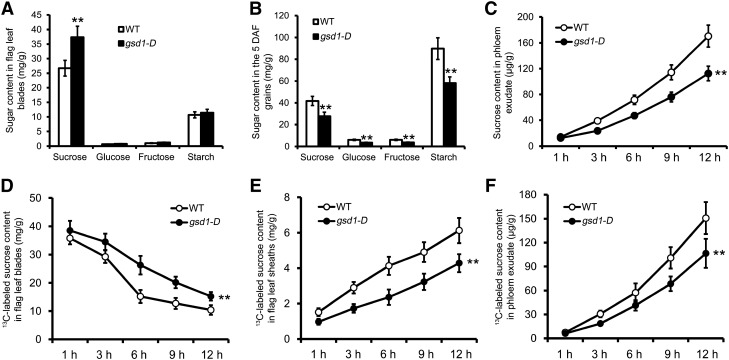
To characterize photoassimilate translocation, the content of soluble sugar (Suc, Glc, and Fru) and starch in the flag leaves and grains of the wild type and gsd1-D were measured at 5 d after flowering (DAF). In flag leaves and grains, Suc was the most abundant soluble sugar detected in both the wild type and gsd1-D. However, the Suc content was about 39% higher in gsd1-D than in the wild type (Fig. 2A). Similarly, higher levels of Glc and Fru were also detected in gsd1-D compared with the wild type, although both Glc and Fru are minor components of soluble sugars in rice (Fig. 2A). Starch content was also slightly higher in gsd1-D compared with the wild type (Fig. 2A). At 5 DAF, the content of Suc, Glc, and Fru in the grain of gsd1-D was about 34%, 42%, and 40% lower, respectively, compared with the wild type (Fig. 2B). Meanwhile, the starch content was also about 35% lower in gsd1-D compared with the wild type (Fig. 2B). To examine whether the lower soluble sugar content in the mutant was photosynthesis related, the net photosynthetic rate (PN) was measured. Results showed that both the mutant and the wild type had a similar photosynthetic rate at 5 DAF in the flag leaves (Supplemental Fig. S6, A and B). Thus, we hypothesized that the higher sugar content in the flag leaves and lower sugar content in the developing grains in the gsd1-D mutant were caused by defective sugar export from the leaf to the developing grain instead. Consistent with this, the level of soluble sugar in the phloem exudate collected from gsd1-D mutants was significantly lower than from the wild type (Fig. 2C). Collectively, these results suggest that the gsd1-D mutant is defective in the export of sugar from the photosynthetic leaf to the developing grain.
Figure 2.
Sugar content comparison between the wild type and the gsd1-D mutant at the 5 DAF stage. A, Content of Suc, Glc, Fru, and starch in the flag leaf blades of the wild type and the gsd1-D mutant at the ED. B, Content of Suc, Glc, Fru, and starch in filling grains of the wild type and the gsd1-D mutant at the ED. C, Content of sugar in phloem exudate at different collection time after the ED. D, Decline of the 13C-labeled Suc in the flag leaf blades after photosynthesis of the flag leaf blade feed with 13CO2. E, Increase of the 13C-labeled Suc in the flag leaf sheaths after photosynthesis of the flag leaf blade feed with 13CO2. F, Increase of the 13C-labeled Suc in phloem exudate after photosynthesis of the flag leaf blade feed with 13CO2. The results in A to C are means ± se from six individual plants, whereas the results in D to F are means ± se of three biological independent samples. Double asterisks indicate that the difference between the wild type and the gsd1-D mutant is statistically significant at P < 0.01 by the t test. WT, Wild type.
In addition, the loading of photoassimilates from the photosynthetic leaf to the phloem was measured using 13CO2 stable isotopic labeling combined with gas chromatography-mass spectrometry (GC-MS; Supplemental Fig. S7). The flag leaf blade of rice plants was placed in a gas-sealed glass chamber for photosynthesis and fed with 13CO2 (Supplemental Fig. S8). After 12 h of photosynthesis, the rice plants were placed in the dark for sample collection at 1, 3, 6, 9, and 12 h. Soluble sugar content was determined for the treated leaf blades, connected leaf sheath, and phloem exudates. Suc was the most abundant component in the determination. During the 12-h dark period, the 13C-labeled Suc content declined gradually in the treated flag leaf blades of both the wild type and the gsd1-D mutant (Fig. 2D), whereas the rate of decline of the 13C-labeled Suc was slower in the mutant compared with the wild type (Fig. 2D; Supplemental Fig. S7). This indicates that the rate of Suc export was reduced in the leaves of the gsd1-D mutant. Consistent with this, less 13C-labeled Suc was determined in the leaf sheath of the gsd1-D mutant (Fig. 2E; Supplemental Fig. S7). Moreover, the 13C-labeled Suc in the phloem exudate was significantly less in the gsd1-D mutant compared with the wild type (Fig. 2F). Together, the results further verified that the gsd1-D mutant is defective in exporting carbohydrates from the photosynthetic leaf.
GSD1 Encodes a Function-Unknown Remorin Protein
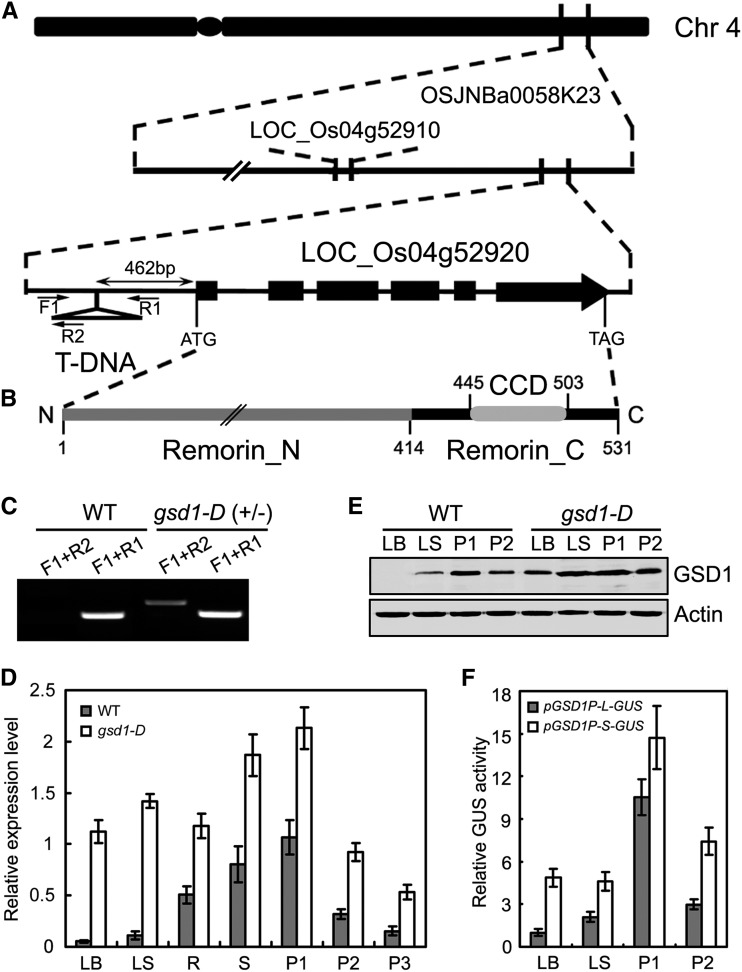
To isolate the GSD1 gene, we cloned the T-DNA left border flanking sequence of gsd1-D using thermal asymmetric interlaced PCR as previously described (Liu et al., 1995). Sequence analysis of the cloned fragment revealed that the insertion site was 462 bp upstream of the Os04g52920 gene in the bacterial artificial chromosome clone OSJNBa0058K23 of chromosome 4 (Fig. 3A). The T-DNA insertion was also confirmed by genomic DNA PCR with three PCR primers (F1, R1, and R2; Fig. 3C). Segregation analysis of the progeny of heterozygous lines showed that the T-DNA insertion was cosegregated along with mutant phenotypes with a ratio close to 1:2:1 of wild-type:heterozygous:homozygous plants (Supplemental Fig. S9). The Os04g52920 gene was designated as GSD1 and the homozygous mutant plant was used for analysis through the entire study.
Figure 3.
Molecular cloning and expression of GSD1. A, Diagram of the GSD1 (Os04g52920) gene located on chromosome 4 and the T-DNA insertion position. Exons are shown as black boxes. In the gsd1-D mutant, T-DNA is inserted at the 462 bp upstream of ATG. Primers used in the genotype analysis are indicated by black arrows. F1 and R1 are gene-specific primers, and R2 is a T-DNA-specific primer. B, Diagram of the GSD1 protein domains. Numbers indicate positions of amino acids. C, T-DNA insertion confirmation and genotype analysis. D, Quantitative reverse transcription (RT)-PCR analysis of GSD1 expression in various tissues of the wild type and the gsd1-D mutant. The rice actin1 gene was used as a reference for normalization. Results are means ± se of three individual samples. E, Comparison of GSD1 protein abundance in the leaf blade, leaf sheath, P1, and P2 between the wild type and the gsd1-D mutant. Actin is used as a loading control. The leaf blade and leaf sheath were collected from 2-month-old rice. F, Comparison of GUS activity between pGSD1-L-GUS and pGSD1-S-GUS. The results are means ± se of three biological independent samples. CCD, Coiled-coil domain; Chr, chromosome; LB, leaf blade; LS, leaf sheath; P1, booting panicle (7 d before flowering); P2, grain-filling panicle (5 DAF); P3, grain-filling panicle (10 DAF); R, root; Remorin_C, C-terminal residue; Remorin_N, N-terminal residues; S, stem.
Gene annotation showed that GSD1 encodes a remorin protein of unknown function. Remorins are unique to plants and consist of a large family of genes with six different groups, whose functions have been little characterized. The sequences of the 19 remorin members in rice are highly diverse, with a sequence identity ranging from 13% to 39% (Supplemental Fig. S10; Supplemental Table S1). GSD1, which belongs to remorin group 6, encodes a protein named remorin 6.6 following nomenclature used previously (Raffaele et al., 2007). Phylogenetically, GSD1 is close to Arabidopsis (Arabidopsis thaliana) AtREM6.2, whose function is still unknown (Supplemental Fig. S11A). To date, no remorin member in group 6 has been functionally characterized. The GSD1 gene contains six exons and five introns (Fig. 3A). The gene encodes a 531-amino acid protein with a predicted molecular mass of 58.26 kD and a pI of 8.94. Like other remorins that have been described (Raffaele et al., 2007), GSD1 protein contains a coiled-coil domain in the conserved C-terminal (Remorin_C), and nonconserved N-terminal residues (Remorin_N; Fig. 3B; Supplemental Fig. S10).
Expression of GSD1 Is Enhanced in the gsd1-D Mutant
Expression of GSD1 was examined in various tissues of the wild type and the gsd1-D mutant, including in the leaf blade, leaf sheath, root, stem, booting panicle, and grain-filling panicle. In the wild type, the expression level of GSD1 differed substantially across the various tissues. The highest expression level was observed in the booting-stage panicle, followed by the stem and root, whereas the lowest expression level was observed in the leaf blade (Fig. 3D). Expression of GSD1 in the wild-type panicle sharply decreased after the phase transition from panicle development (P1) to grain filling (P2 and P3; Fig. 3D). In the gsd1-D mutant, GSD1 expression was significantly higher in certain tissues compared with the wild type (Fig. 3D). In particular, a 31-fold higher level of GSD1 expression was observed in the leaf blade. The relative expression of GSD1 in various tissues or organs also differed between the gsd1-D mutant and the wild type. Meanwhile, expression of the GSD1 neighboring gene, Os04g52910 (Fig. 3A, annotated as a hypothetical protein), in the T-DNA mutant was not changed (data not shown).
To further analyze the protein level of GSD1 in different tissues at different developmental stages in the wild type and in the gsd1-D mutant, GSD1-specific peptides were identified and used to produce polyclonal antibodies (Supplemental Fig. S11B). Antibody specificity was verified by western blot (Supplemental Fig. S11C). The level of GSD1 protein in the leaf blade, leaf sheath, P1, and P2 was examined in both the wild type and the gsd1-D mutant. In the wild-type plant, the GSD level differed substantially between tissues and was most abundant in P1 but was undetectable in the leaf blade (Fig. 3E). Protein levels were higher in various tissues in gsd1-D plants compared with the wild type (Fig. 3E). Results from measurement of both mRNA and protein levels demonstrated that GSD1 expression was significantly increased in various tissues of the gsd1-D mutant, especially the leaf blade.
To confirm the effect of T-DNA insertion in the promoter, two lengths of the Os04g52920 promoter fragment were cloned. One fragment, designated as pGSD1-L, was 1,700 bp upstream of the ATG start codon and the other fragment, designated as pGSD1-S, was 462 bp upstream of the ATG start codon close to the T-DNA insertion site in gsd1-D. The activity of the promoter in driving GUS expression was analyzed through Agrobacterium spp.-mediated rice transformation. The pattern of relative GUS activity in the pGSD1-L-GUS transgenic lines appeared similar to the expression pattern of GSD1 across various tissues (Fig. 3, D–F). In comparison, higher activity of pGSD1-S compared with pGSD1-L was observed in the leaf blade, leaf sheath, booting panicle (P1), and filling panicle (P2; Fig. 3F). The consistency of results observed between the analysis of promoter activity and enhanced GSD1 expression in the T-DNA insertion gsd1-D mutant (Fig. 3, D and E) suggest the potential presence of cis-regulatory elements that can precisely regulate the expression level of GSD1 in various tissues at different developmental stages. The T-DNA insertion in the gsd1-D mutant may disrupt the regulation of the cis-regulatory elements in controlling GSD1 expression.
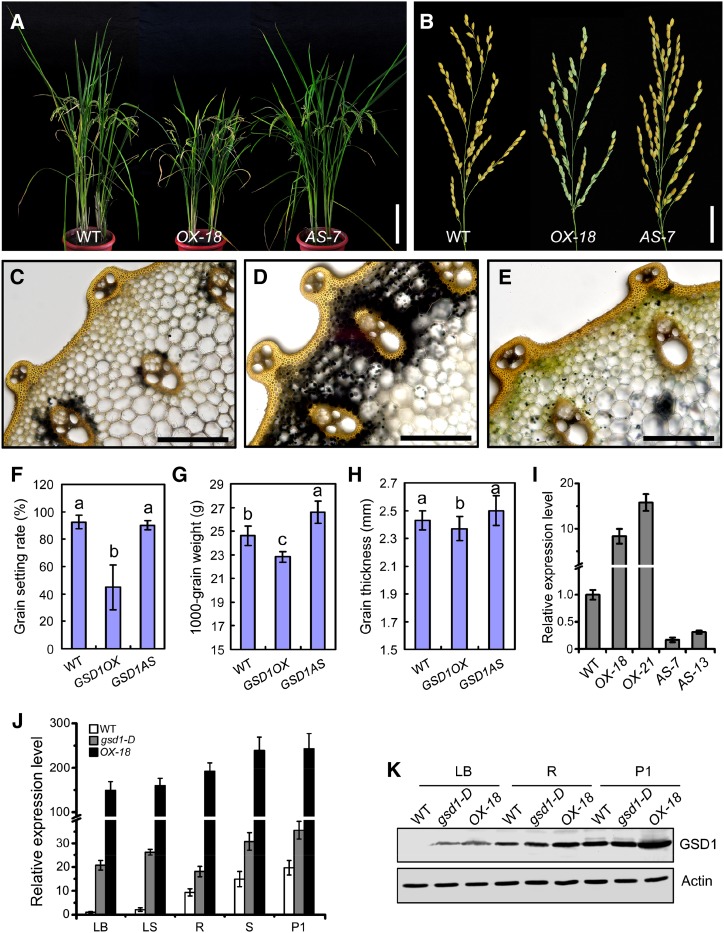
To further test whether higher expression of GSD1 leads to the reduced grain setting observed in gsd1-D, we transformed wild-type rice with a sense full coding sequence of Os04g52920 (designated GSD1OX) and an antisense coding sequence of Os04g52920 gene (designated GSD1AS) under the control of a Cauliflower mosaic virus 35S promoter. A total of 23 independent GSD1 overexpression (GSD1OX) transgenic lines and 19 independent GSD1 suppression (GSD1AS) transgenic lines were generated. Seventeen of the 23 independent GSD1OX transgenic lines displayed a significant reduction in grain setting, varying from 37% to 83% with an average of 45%. None of the GSD1AS transgenics exhibited obvious defects in grain setting compared with the wild type (Fig. 4, A, B, and F). GSD1 expression in stem tissue from overexpressed transgenic lines OX-18, OX-21, and suppressed transgenic lines AS-7 and AS-13 were measured by quantitative RT-PCR (Fig. 4I). The phenotypes of the plant and mature panicle of GSD1OX-18 and GSD1AS-7 are shown in Figure 4, A and B. A large amount of starch was found to be accumulated in the parenchyma cells around the vascular bundles in the stem of GSD1OX-18, but very little starch was detected in the wild-type or GSD1AS-7 transgenics (Fig. 4, C–E; Supplemental Fig. S12, A–C). Grain thickness, grain length, spikelet number, 1,000-grain weight, plant height, and internode length were also significantly lower in GSD1OX compared with the wild type (Fig. 4, G and H; Supplemental Fig. S12, D–G). Overexpression of GSD1 under the control of the 35S promoter resulted in a significant reduction in rice grain setting compared with the wild type. GSD1 expression in GSD1OX-18 was significantly higher compared with the wild type and gsd1-D plants in tissues including the leaf blade, leaf sheath, root, stem, and P1 (Fig. 4J). In addition, the GSD1 protein was more abundant in GSD1OX-18 compared with the wild type and gsd1-D in the leaf blade, root, and P1 (Fig. 4K). These results indicated that GSD1 overexpression resulted in a reduction in rice grain setting, providing additional evidence to suggest that the gsd1-D phenotype was caused by enhanced GSD1 expression.
Figure 4.
Comparison of wild-type and GSD1 overexpressed transgenic plants. A, Wild-type, GSD1 overexpression (GSD1OX-18), and GSD1 suppression (GSD1AS-7) plants at the grain-filling stage. B, Mature panicles of wild-type, GSD1OX-18, and GSD1AS-7 plants. C to E, I2-KI starch staining in the second internode of the wild type (C), GSD1OX-18 (D), and GSD1AS-7 (E) in the grain matured plants. F to H, Statistical analyses of the grain setting rate (F), 1,000-grain weight (G), and grain thickness (H) in the wild type, GSD1OX, and GSD1AS. Twenty panicles were analyzed for the grain setting rate. Ten independent 1,000-grain samples were analyzed for 1,000-grain weight. Sixty seeds were analyzed for seed size. Values are means ± se. Different lowercase letters indicate a significant difference at P < 0.01 by ANOVA. I, Quantitative RT-PCR analyses of the GSD1 expression in the stem of wild-type, GSD1OX, and GSD1AS transgenic plants at the tillering stage. The results are means ± se of three individual samples. J, Comparison of GSD1 expression in various tissues of the wild type, the gsd1-D mutant, and GSD1OX by quantitative RT-PCR analysis. The rice actin1 gene was used as a reference for normalization. The relative gene expression level in the wild-type leaf blade was set as 1. The results are means ± se of three individual samples. K, Comparison of GSD1 expression in protein levels in the leaf blade, root, and P1 of the wild type, the gsd1-D mutant, and GSD1OX. Actin is used as a loading control. The leaf blade, root, and P1 were collected from 2-month-old rice. LB, Leaf blade; LS, leaf sheath; P1, booting panicle (about 7 d before flowering); R, root; S, stem; WT, wild type. Bars = 20 cm in A; 3 cm in B; and 50 μm in C to E.
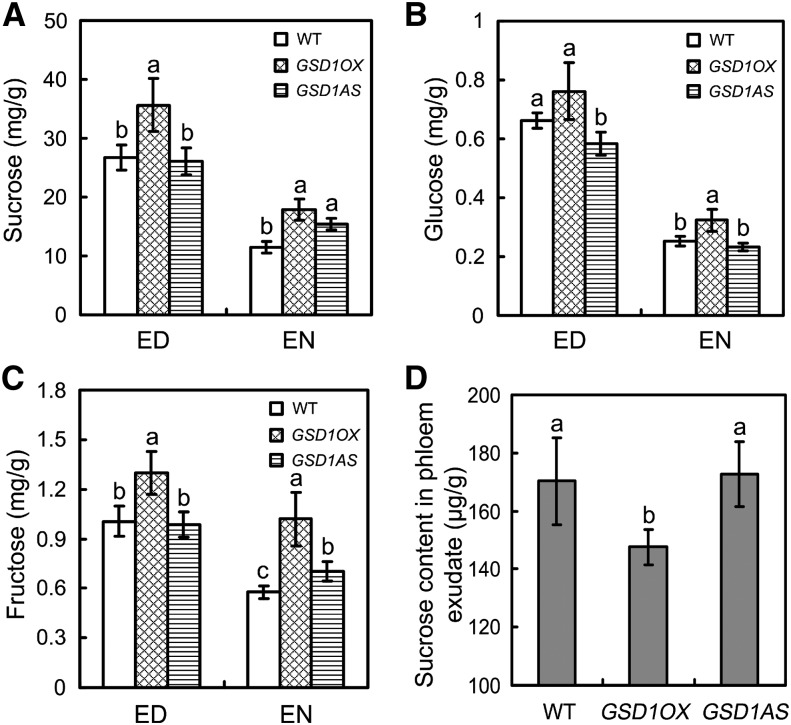
The content of soluble sugar (Suc, Glc, and Fru) was measured in the flag leaf of the GSD1AS and GSD1OX plants at 5 DAF. Compared with the wild type, Suc content was about 33% higher at the end of the day (ED) and about 55% higher at the end of the night (EN) in GSD1OX transgenics (Fig. 5A). Similarly, higher levels of Glc and Fru were also detected in GSD1OX plants compared with the wild type at the ED and the EN (Fig. 5, B and C). However, these sugar levels did not differ significantly between GSD1AS transgenic lines and the wild type. The PN in GSD1OX, GSD1AS, and the wild type was examined and no significant differences were observed between the transgenic plants and the wild type at 5 DAF in a greenhouse (Supplemental Fig. S13). The accumulation of higher soluble sugar content in GSD1OX leaves could therefore be a consequence of reduced sugar export from the leaf rather than as a result of different rates of photosynthesis. The soluble sugars in the phloem exudate of flag leaves were collected and quantified to test whether sugar export was suppressed in GSD1OX leaves. The soluble sugar content in GSD1OX plants was 14% lower compared with the wild type at 5 DAF (Fig. 5D). No significant change in sugar content was detected in GSD1AS transgenic lines (Fig. 5D). These results verify that overexpression of GSD1 reduced the efficiency of sugar export from the photosynthetic site to the phloem.
Figure 5.
Export of soluble sugars from the flag leaf at 5 DAF in transgenic rice plants. A to C, Content of Suc (A), Glc (B), and Fru (C) in wild-type, GSD1OX, and GSD1AS plants. Values are means ± se from six individual plants. D, Sugar content in phloem exudate of wild-type, GSD1OX, and GSD1AS plants. The results are means ± se of three individual samples. Different lowercase letters indicate a significant difference at P < 0.01 by ANOVA. WT, Wild type.
GSD1 Is Specifically Expressed in Phloem Companion Cells
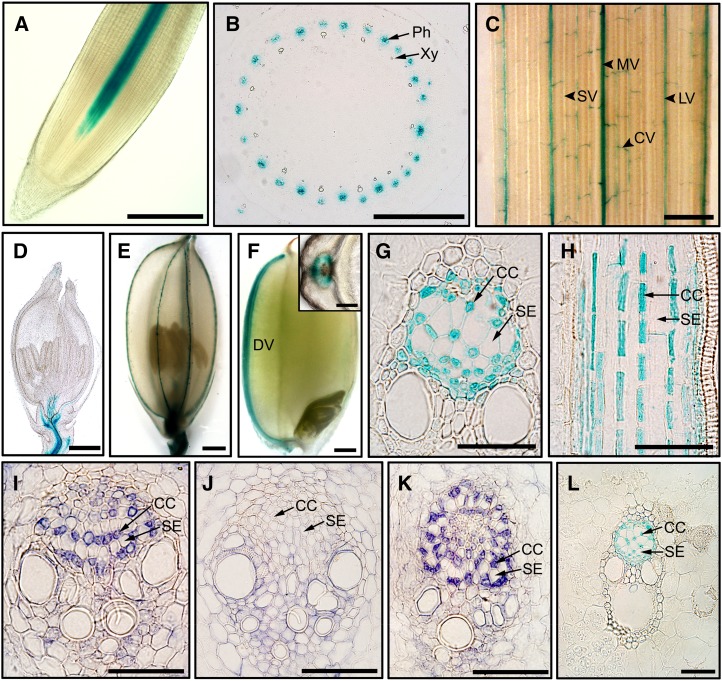
Cell type-specific expression of GSD1 was analyzed through promoter GUS activity detection and immunolocalization. The GSD1 promoter was fused to a GUS gene (pGSD1-L-GUS) and transferred into rice plants. GUS activity was examined in the root, stem, leaf, spikelet, and developing grain. GSD1 promoter activity was detected in vascular tissue, such as in root and stem vascular bundles (Fig. 6, A and B), in leaf midveins, large veins, and commissural (transverse) veins (Fig. 6C), in the vascular bundles of rachilla at the basal part of spikelet (Fig. 6D), in the nerves of lemma and palea (Fig. 6E), and in the grain dorsal vascular bundle (Fig. 6F). Further examination revealed that GSD1 promoter activity was specifically expressed in the companion cells of phloem tissue (Fig. 6, G and H). The cell type-specific expression was confirmed by immunolocalization of rice stems (6 weeks old) that were sectioned and hybridized with GSD1-specific antibodies. GSD1 was specifically localized in companion cells (Fig. 6I), consistent with the location of GSD1 promoter activity (Fig. 6G). In the gsd1-D mutant, specific localization of GSD1 in companion cells (Fig. 6K) was also observed, same as in the wild type. Furthermore, promoter pGSD1-S-GUS activity was also specifically present in companion cells (Fig. 6L), suggesting that the 462-bp promoter (pGSD1-S) is sufficient to drive GSD1 expression and that the cell-specific expression of GSD1 is not affected by the T-DNA insertion.
Figure 6.
GSD1 is expressed in companion cells demonstrated by promoter analysis and immunolocalization. A to H, Histochemical localization of GSD1 promoter-GUS activity in pGSD1-L-GUS transgenic rice. GUS expression in the root (A), cross sectioned stem (B), leaf blade (C), young spikelet (D), matured spikelet (E), and immature seed at 10 DAF with the inset for cross section (F). Cross sections of rice culm (G) and longitudinal sections of rice culm (H). I to K, Immunolocalization by GSD1-specific antibodies. GSD1 is localized in phloem companion cells in the wild type (I) and in the gsd1-D mutant (K). Detection in the wild type by preimmune IgG (J). L, GUS staining in culm of the pGSD1-S-GUS transgenic rice. CC, Companion cell; CV, commissural vein; DV, dorsal vascular bundle; LV, large vein; MV, midvein; Ph, phloem; SE, sieve element; SV, small vein; Xy, xylem. Bars = 500 μm in A to F; 100 μm in the F inset; and 50 μm in G to L.
GSD1 Is Localized to the PM and PD
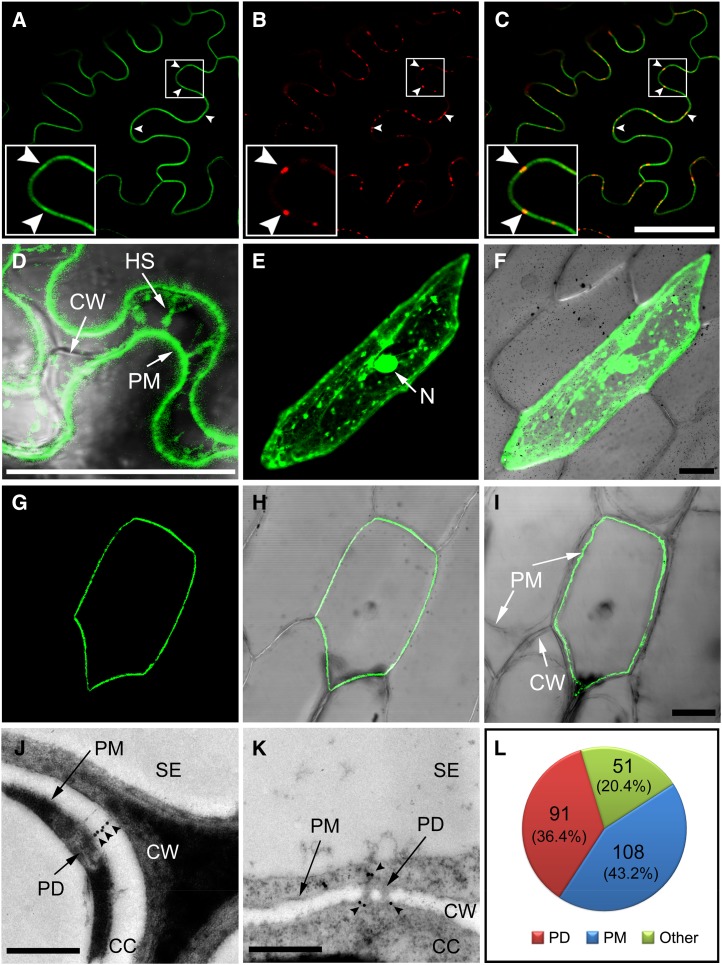
A GSD1-GFP fusion construct under the control of the 35S Cauliflower mosaic virus promoter was generated to investigate the subcellular localization of GSD1. The construct was transferred into onion (Allium cepa) epidermal cells via particle bombardment and was transferred into tobacco (Nicotiana benthamiana) leaves using Agrobacterium spp.-mediated transformation. The cells were plasmolyzed to reveal the cell wall, PM, and vacuole. In tobacco leaf abaxial epidermis cells, unevenly distributed GFP signals were detected on the PM (Fig. 7, A–C). The uneven distribution of signals colocalized with aniline blue-stained callose (Fig. 7, B and C), suggesting that GSD1 is associated with PD structure (Levy et al., 2007). The plasmolysis assay showed that the GFP signal was localized on the retracted PM and Hechtian strands (Fig. 7D). In onion epidermal cells, the GFP signal of GSD1 was specifically localized on the PM with an uneven distribution (Fig. 7, G–I), whereas the GFP signal of the control (without GSD1 fusion) was found in the cytosol and nucleus (Fig. 7, E and F).
Figure 7.
GSD1 is localized to PM and PD. A to D, GSD1 fused to GFP (GFP-GSD1) was expressed in tobacco leaf epidermal cells. GFP-GSD1 fluorescence detected on PM (A), callose staining with aniline blue indicating PD sites (B), and merged image showing that GSD1 localizes to PM and PD (C). Arrowhead indicates PD sites on the PM. D, GFP-GSD1 fluorescence detection after plasmolysis. E and F, Control GFP was expressed in onion epidermal cells. Control free GFP fluorescence signal (E). GFP control fluorescence merged with DIC image (F). G to I, GSD1 fused with GFP (GFP-GSD1) was expressed in onion epidermal cells. GFP-GSD1 fluorescence on the PM (G), GFP-GSD1 fluorescence merged with the DIC image (H), and image after plasmolysis treatment (I). J to L, Representative immunogold labeling of GSD1 and quantitative analysis of gold particles. GSD1 is localized to the channel of PD (J) and at the neck regions of PD (K) of the companion cells in 4-week-old rice shoot tissues. Quantification of gold particles is depicted in L. Arrowheads indicate immunogold labeling particles. CC, Companion cell; CW, cell wall; DIC, differential interference contrast; HS, Hechtian strand; N, nucleus; SE, sieve element. Bars = 50 μm in A to C and E to I and 500 nm in J and K.
Immunogold labeling was also used to examine the location of GSD1. Ultrathin sections of shoot tissues from 4-week-old rice were immunolabeled with the GSD1-specific antibody and observed by transmission electron microscopy. GSD1 labeling in companion cells was observed at the PD channels (Fig. 7J), PD neck regions (Fig. 7K), and PM (Supplemental Fig. S14). A quantitative analysis of the gold labels showed that among a total of 250 gold particles detected across 39 images, 91 and 108 were found at the PD and PM, respectively (Fig. 7L).
PD Permeability Is Affected by GSD1 Overexpression
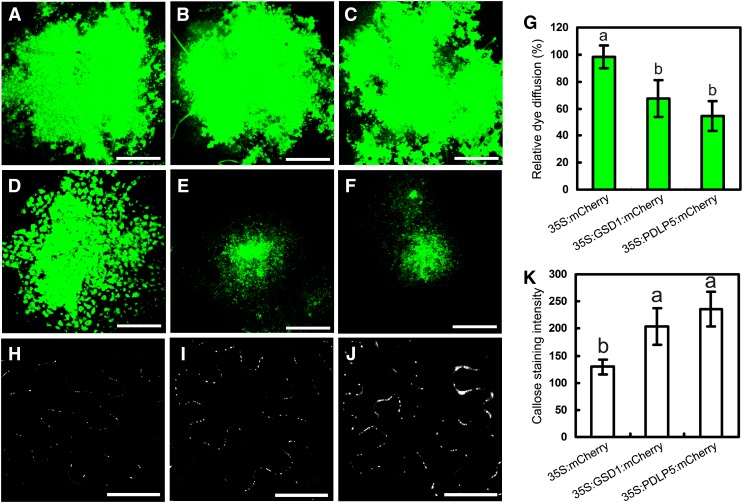
In the symplastic system, companion cells are connected to their surrounding cells by PD channels, which allow photoassimilates to load into or unload from the phloem transport system. Given the evidence presented above that suggests that sugar export is affected by overexpression of GSD1 that is localized in companion cells traversed by PD, we sought to examine whether PD permeability is affected by GSD1. The applicability of the Drop and See (DANS) dye loading assay for analyzing PD permeability was confirmed in a tobacco leaf system (Supplemental Fig. S15), according to the previously described method (Lee et al., 2011). Plasmodesmata-located protein5 (PDLP5), a PD-localized protein that acts as a negative regulator of PD permeability (Lee et al., 2011), was used as a positive control in our study. Three constructs (35S:mCherry [negative control], 35S:GSD1:mCherry, and 35S:PDLP5:mCherry) were expressed in tobacco leaves for the DANS dye loading assay. In 35S:mCherry expressed lines, abaxial epidermis was detected in a large fluorescent area, indicating that carboxyfluorescein (CF) was able to spread to many cells through PD channels (Fig. 8, A and D). By contrast, the fluorescence in 35S:GSD1:mCherry and 35S:PDLP5:mCherry lines was restricted to a small area in the abaxial epidermis, indicating inhibition of CF movement across cells (Fig. 8, B, C, E, and F). Statistical analysis showed that the CF movement was reduced by approximately 50% and 65% in 35S:GSD1:mCherry and 35S:PDLP5:mCherry lines, respectively (Fig. 8G). These results demonstrate that GSD1 overexpression inhibited PD permeability. Overexpression of PDLP5 can increase callose deposition in the PD (Lee et al., 2011). When we examined the callose distribution in the 35S:GSD1:mCherry and 35S:PDLP5:mCherry, callose deposition was increased in both PDLP5 and GSD1 overexpressed cells (Fig. 8, H–K).
Figure 8.
Overexpression of GSD1 hampers PD permeability. A to C, Confocal images showing CFDA loaded onto the adaxial surface of tobacco leaves. D to F, CF spread observed on the abaxial surface. Signals from 35S:mCherry (A and D), 35S:GSD1:mCherry (B and E), and 35S:PDLP5:mCherry (C and F) transformants. G, Quantification of the CF movement in 35S:mCherry, 35S:GSD1:mCherry, and 35S:PDLP5:mCherry transformants. More than 20 plants were used per assay, and more than three repeats were performed. The extent of dye diffusion was quantified by measuring the fluorescent diffusion area. The relative dye diffusion in 35S:mCherry transformants was set as 100%. Values are means ± se from 12 individual samples. H to J, Confocal images showing chemical staining of callose using aniline blue in 35S:mCherry (H), 35S:GSD1:mCherry (I), and 35S:PDLP5:mCherry (J) transformants. K, Quantification of the callose staining intensity in the 35S:mCherry, 35S:GSD1:mCherry, and 35S:PDLP5:mCherry transformants. Values are means ± se from 20 individual samples. Different lowercase letters in G and K indicate significant difference at P < 0.01 by ANOVA test. Bars = 500 μm in A to F and 50 μm in H to J.
GSD1 Affects PD Permeability through Interactions with OsACT1
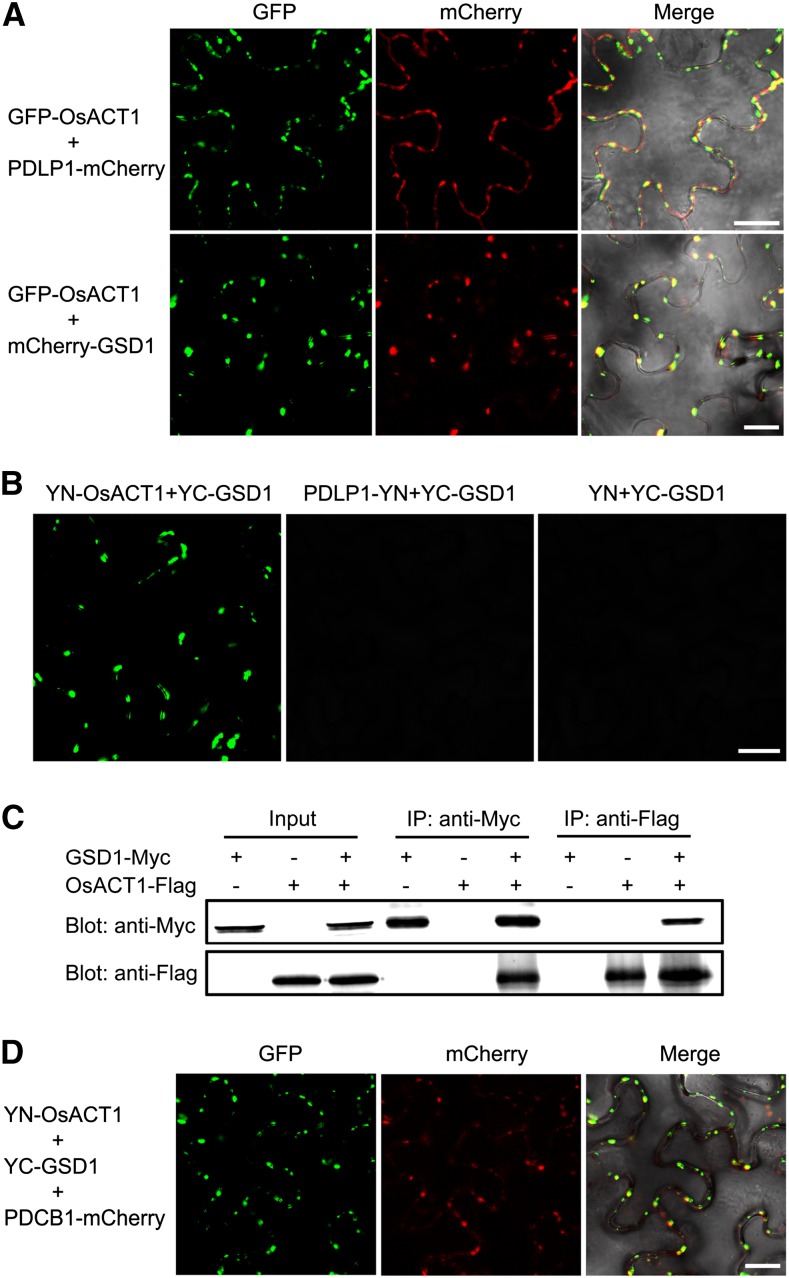
Actin has been shown to play a role in regulating PD permeability (Ding et al., 1996). We identified the location of an actin protein (designated as OsACT1, Os03g50885) that is associated with the PM when expressed in tobacco leaves (Supplemental Fig. S16). Serial z-sections of the leaf epidermal cell revealed that OsACT1 is unevenly distributed on the PM, not in the internal organelles (Supplemental Fig. S17A). Coexpression of GFP-OsACT1 with PDLP1-mCherry showed that the GFP and mCherry signals were colocalized (Fig. 9A), suggesting that OsACT1 is located to the PD. Furthermore, mCherry-GSD1 fluorescence was precisely colocalized with GFP-OsACT1 fluorescence in the PD when GFP-OsACT1 and mCherry-GSD1 were coexpressed (Fig. 9A). This was further confirmed by examining serial z-sections of an epidermal cell (Supplemental Fig. S17B). Together, the results suggest that GSD1 interacts with OsACT1 at the PD region.
Figure 9.
GSD1 interacts with OsACT1 at PD. A, mCherry-GSD1 and PDLP1-mCherry were cotransformed with GFP-OsACT1 into tobacco leaves, respectively. Confocal images show overlapping OsACT1 GFP fluorescence signals and PD marker protein PDLP1 mCherry signals. OsACT1 GFP signals and GSD1 mCherry signals also overlap. B, Interaction of GSD1 with OsACT1 was detected by BiFC. YN-OsACT1 (YN, N-terminal part of YFP), PDLP1-YN, and YN were cotransformed with YC-GSD1 (YC, C-terminal part of YFP) into tobacco leaves, respectively. YFP signals were detected in YN-OsACT1 and YC-GSD1 coexpressed tobacco leaves. No YFP fluorescence signal was detected in PDLP1-YN and YC-GSD1 or YN and YC-GSD1 coexpressed tobacco leaves. C, Interaction between GSD1-Myc and OsACT1-Flag was detected by Co-IP. GSD1-Myc and OsACT1-FLAG were expressed or coexpressed in tobacco leaves. Total protein extracts were separately immunoprecipitated with anti-Myc antibody-coupled agarose beads and anti-Flag antibody-coupled agarose beads. Proteins from the crude lysates and immunoprecipitated proteins were detected with anti-Myc antibodies and anti-Flag antibodies, respectively. D, YN-OsACT1, YC-GSD1, and PDCB1-mCherry were cotransformed into tobacco leaves. YFP and mCherry signals were detected using a confocal microscope. Overlaid YFP and mCherry fluorescence signals indicate that the GSD1/OsACT1 complex colocalized with PD callose binding protein PDCB1.
Bimolecular fluorescence complementation (BiFC) analysis was applied to examine whether GSD1 interacts with OsACT1. GSD1 was constructed with the yellow fluorescent protein (YFP) C-terminal sequence (named YC-GSD1) and OsACT1 and PDLP1 were constructed with the YFP N-terminal sequence (named YN-OsACT1 and PDLP1-YN, respectively). BiFC signals were detected in the PD (Fig. 9B) when YC-GSD1 was coexpressed with YN-OsACT1 in tobacco leaves. No signal was detected when YC-GSD1 was coexpressed with PDLP1-YN or YN (Fig. 9B). The interaction between GSD1 and OsACT1 was further examined by coimmunoprecipitation (Co-IP; Fig. 9C). The anti-Myc antibody-coupled agarose beads were able to coprecipitate the Flag-fused OsACT1 and Myc-fused GSD1. Conversely, the anti-Flag antibody-coupled agarose beads were able to coprecipitate the Myc-fused GSD1 and the Flag-fused OsACT1. These results further verified the interaction between GSD1 and OsACT1.
In addition, we coexpressed YN-OsACT1, YC-GSD1, and mCherry-PDCB1 (a callose binding protein specifically localized in PD) in tobacco leaves. Examination of the leaf epidermal cell by the median plane section and serial z-sections showed that mCherry fluorescence was precisely colocalized with YFP fluorescence (Fig. 9D; Supplemental Fig. S17C), revealing that GSD1 is also colocalized with PDCB1 in the presence of OsACT1. Thus, three proteins (GSD1, the desmotubule-associated protein OsACT1, and the callose binding protein PDCB1) may be located together in the PD region.
DISCUSSION
GSD1 Plays a Role in Regulating Grain Setting in Rice
In this study, we identified a remorin gene, GSD1, that is strictly expressed in the phloem companion cells during rice development. T-DNA insertion (462 bp upstream of the ATG start codon) causes GSD1 to be overexpressed in the gsd1-D mutant but did not change the cell type-specific expression of GSD1. The gsd1-D mutant displayed smaller panicle size, lower grain setting, and higher starch accumulation in the stem during the ripening stage. The gsd1-D mutant also exhibited reduced grain thickness but normal floral organs. Thus, grain setting is affected in the gsd1-D mutant but not by defects in floral organ development. The results from the analysis of the T-DNA mutant were corroborated in transgenic plants overexpressing GSD1, which displayed a similar or even more severe defect in grain setting than the gsd1-D mutant.
GSD1 Controls Photoassimilate Translocation through the Symplastic System in Rice by Regulating PD Permeability in Companion Cells
Previous studies showed that PD channels connect phloem companion cells, sieve elements, surrounding parenchyma cells, and mesophyll cells in the rice leaf (Kaneko et al., 1980; Chonan et al., 1981; Scofield et al., 2007). In our study, GSD1 is specifically localized in the PD channels of companion cells. Defects in carbohydrate relocation and reduced sugar export from the leaf to the phloem likely contribute to the mutant phenotype observed. Results from the DANS assay revealed that PD-mediated cell-to-cell transport is significantly attenuated by the overexpression of GSD1. Soluble sugars including Suc, Glc, and Fru were disproportionately retained in the leaves of gsd1-D mutants and GSD1OX transgenics. These results provide an array of evidence supporting the symplastic phloem loading model in rice.
The structure of the PD has an impact on the symplastic movement of photoassimilates into or out of the phloem transport system. Actin filaments associated with the desmotubule structure within the PD are believed to play a role in regulating the aperture of the cytoplasmic sleeve between the PM and desmotubule (Ding et al., 1996; Su et al., 2010). In addition, GPI-anchored protein PDCB1 is also located in the PD structure and impacts PD function (Simpson et al., 2009). Overexpression of PDCB1 resulted in increased callose accumulation and changes in PD conductivity (Simpson et al., 2009). The accumulation of a phloem-specific membrane protein, NDR1/HIN1-like26, in Arabidopsis was recently shown to have a specific effect on sugar export, resulting in slower root growth and a lower seed yield, suggesting its role in regulating PD permeability in companion cells (Vilaine et al., 2013). In this study, immunogold labeling revealed that GSD1 is localized inside the PD as well as the PM around the PD neck region. The results showed that GSD1 strongly interacted with the desmotubule-associated OsACT1 and was colocalized with PDCB1 in the presence of OsACT1. These results suggest that three proteins (GSD1, OsACT1, and PDCB1) may work together to regulate the conductivity of the PD (Fig. 10). After 48 h of transient expression in tobacco, expression of both PDCB1 and GSD1 increased callose deposition. Although the precise interaction between the colocalized GSD1 and PDCB1 is unclear, their role in regulating PD conductivity may be associated with callose deposition.
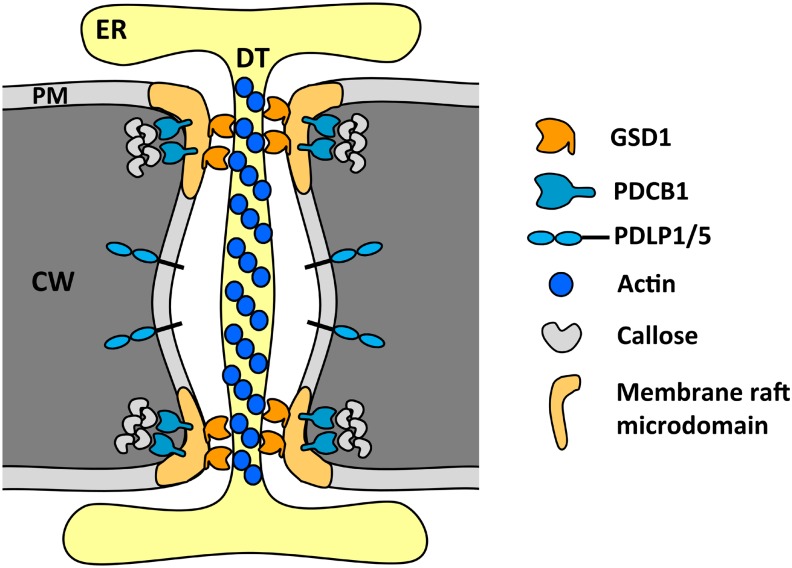
Figure 10.
A proposed model for GSD1 modulation of PD permeability. GSD1 encodes a PM protein, remorin, which is colocalized in PD with PD-specific proteins: GPI-anchored callose binding protein PDCB1, which is located at the neck region of PD (Simpson et al., 2009); and PD-located proteins PDLP1 and PDLP5 (Thomas et al., 2008; Lee et al., 2011). GSD1 interacts with OsACT1, which is associated with the desmotubule structure in PD. GSD1 may play a role in connecting the desmotubule and PM in the PD neck regions of companion cells, modulating the PD aperture through its expression levels and PM anchor. CW, Cell wall; DT, desmotubule; ER, endoplasmic reticulum.
Our data indicate that GSD1 impacts sugar translocation. Although more studies are required to verify the exact mechanism by which that occurs, overexpression of GSD1 in the gsd1-D mutant and GSD1OX transgenics may potentially affect the contractile elements that regulate PD aperture structure, leading to a slowdown in photoassimilate flux through the channel.
Suc is the predominant photoassimilate, which is loaded from the photosynthetic leaf to the phloem system and unloaded to sink cells after being transported long distances. The PD structure in the sieve element-companion cell complex could impact the loading and unloading processes in the symplastic system. Both gsd1-D mutant and GSD1OX plants with enhanced GSD1 expression show defects in carbohydrate relocation, reduced sugar export from the leaf to the phloem, and reduced grain setting rates. Abundant starch is also retained in the parenchyma cells around the vascular bundles in the gsd1-D mutant and GSD1OX transgenics after grain ripening. This evidence indicates that GSD1 likely functions in regulating symplastic phloem loading in rice. Although our evidence and discussion are focused on the loading process, the possibility that GSD1 may also impact the unloading process through the symplastic PD channels in the sieve element-companion cell complex remains a possibility open to further clarification.
Regulation of GSD1 Expression May Affect the Distribution of Photoassimilates in Rice
GSD1 (OsREM6.6) belongs to group 6 of the remorin family in rice. The function of the members in this group (OsREM6.1, OsREM6.2, OsREM6.3, OsREM6.4, OsREM6.5, and OsREM6.6) remains unknown (Supplemental Figs. S10 and S11A). Unlike other groups, group 6 remorins contain a long and highly variable N-terminal sequence ranging from 240 to 522 amino acids (Raffaele et al., 2007). The nonconserved N-terminal residues, which are predicted to be intrinsically disordered (Marín and Ott, 2012; Marín et al., 2012) with a three-dimensional structure that varies depending on physiological conditions (Dyson and Wright, 2005), are believed to play an important role in regulation, signaling, and control through high-specificity, low-affinity protein-protein interactions (Sickmeier et al., 2007). Although remorins have been postulated to affect PD conductance in previous studies, their biological functions remained ambiguous because of the lack of phenotypic consequences associated with their expression (Raffaele et al., 2009; Perraki et al., 2012). Overexpression of GSD1 from T-DNA disruption of the promoter reduced PD conductance, whereas the phenotype of the gsd1-D mutant indicates that variations in the expression level of GSD1 could regulate grain filling. This was corroborated by transgenic overexpression GSD1, which displayed a similar or even more severe grain setting defect than the gsd1-D mutant. Transgenic overexpression GSD1 also resulted in a dwarf phenotype in rice plants. This phenotype could be caused by higher expression levels of GSD1 in the phloem companion cells or GSD1 obtained expression in other cell types besides the phloem companion cells. As illustrated in Figure 10, GSD1 may function in PD channels and regulate PD conductance through the level at which it is expressed.
Expression levels of GSD1 differed across different tissues and growth stages. GSD1 expression in the panicle sharply decreased after the phase transition from panicle development to grain filling, during which a large amount of photoassimilates needs to be transported into the grain. Given that companion cells serve as the traffic control centers of the phloem in directing the transport of photoassimilates (Oparka and Turgeon, 1999), the expression level of GSD1 in companion cells could serve as a potential means by which the distribution of photoassimilates among different tissues is regulated. GSD1 expression is likely controlled by a battery of complex regulatory elements located in the GSD1 promoter and potentially in an upstream region. More in-depth studies to identify the precise transcriptional regulation of GSD1 and subsequent effect on photoassimilate distribution could have important implications for improving rice yield.
MATERIALS AND METHODS
Plant Materials
The wild-type rice (Oryza sativa) plant used in this study was Oryza sativa japonica cv Zhonghua 11. The gsd1-D mutant was isolated from a collection of T-DNA mutants with ZH11 background. The rice plant was grown in a phytotron under conditions of 60% relative humidity, 12-h photoperiod, 28°C constant temperature regime, and photon flux density at 200 to 250 µm m−2 s−1.
Soluble Sugar Analysis and Starch Measurement
The soluble sugar assay was carried out as described (Kuo et al., 1988) with some modifications. Briefly, about 1 g of samples was collected and ground into fine powder in liquid nitrogen. The ground fine powder was immediately extracted with 80% (v/v) ethanol (at a ratio of 30 mL g of meal) at 75°C for 30 min with gentle shaking. After extraction, Rib (Sigma-Aldrich) was added as the internal standard. The supernatant and debris were separated by centrifugation (12,000g for 5 min). The supernatant was transferred to a new tube and dried for the sugar assay or stored at −20°C. The debris was used to determine starch content using a starch assay kit (SA-20; Sigma-Aldrich) following the manufacturer’s instructions.
The phloem exudate assay was carried out as previously described (Eom et al., 2011) with some modifications. Briefly, the flag leaf at 5 DAF was cut at the leaf sheath region at the ED and was immediately soaked in 15 mm EDTA solution, pH 7.25. To collect phloem exudate, a fresh cut was remade at 1.5 to 2 cm above the leaf cut in the buffer. The collection was carried out in the dark at room temperature at 1, 3, 6, 9, and 12 h, respectively. The collected phloem exudates were concentrated for total soluble sugar measurement using an HPLC system (Agilent 1100 series). An amount equivalent to 5 µL of each sample was injected and separated by a 30- × 6.5-mm Sugar-PakI column (Water Associates). The column temperature was maintained at 70°C. The mobile phase was water at a constant rate of 0.6 mL min−1. The elution was monitored by a refractive index detector. Quantification of soluble sugars was based on the detected area against the standard curves, which were generated using Suc, Glc, Fru, and Rib (Sigma-Aldrich) under the same conditions.
Net Photosynthetic Analysis
PN was measured using a LI-6400 photosynthesis system (LI-COR) under conditions of 350 µmol (CO2) mol−1, photosynthetic photon flux density of 1200 µmol m−2 s−1, and air temperature of the leaf chamber at 25°C. Ten flag leaves from each line at 5 DAF were measured.
Phloem Loading Measurement
To analyze the rate of photoassimilate loading to the phloem system, we used a 13CO2 stable isotope to label the photoassimilates. The flag leaf blade was place in a gas-sealed transparent glass chamber filled with 13CO2 isotope for photosynthesis. The chamber with 20-L space was mounted with a fan, and 13CO2 was released from sodium bicarbonate-13C (Supplemental Fig. S8). The 13CO2 release was controlled by addition of 0.5 mm hydrochloric acid through a syringe connection. The concentration of CO2 was maintained approximately to 400 μL L−1. The chamber temperature was controlled at 28°C (the phytotron temperature). After 12-h photosynthesis, the treated rice plants were replaced in the dark for sample collection. The leaf blade, leaf sheath, and phloem exudate were collected at 1, 3, 6, 9, and 12 h, respectively, for soluble sugar determination as previously described (Koubaa et al., 2012). Inositol (Sigma-Aldrich) was added as the internal standard. For soluble sugar analysis, the freeze-dried sample was dissolved in 200 μL of N,N-dimethylformamide (Sigma-Aldrich) containing 0.1% pyridine (Sigma-Aldrich), and then derivatized by 50 μL of N,O-bis-(trimethylsilyl)-trifluoroacetamide (Sigma-Aldrich) at 80°C for 30 min. After derivatization, 3 μL of sample was analyzed by GC-MS. An Agilent 6890 instrument, fitted with an Agilent 5975 inert mass selective detector and an Agilent HP-5MS column (30 mm × 0.25 mm × 0.25 μm film thickness), was used for analysis. Helium was used as the carrier gas at 1.5 mL min−1, and detector temperatures were set to 310°C with electron ionization in positive mode. The gas chromatograph oven profile was as follows: holding for 5 min at an initial temperature of 120°C, gradually increasing the temperature to reach 270°C at 4°C min−1, and ramping up the temperature at 20°C min−1 to a final temperature of 310°C.
Data were acquired and processed with an Agilent ChemStation system. The soluble sugars, including Suc, Glc, and Fru, were identified through chromatographic characteristics and mass spectra. After calibration with internal standard inositol, sugar content was quantified using total ion current chromatographic peak areas against the respective authentic standard. The proportion of 13C isotope mass spectra fragments was calculated to quantify the 13C-labeled sugars.
Isolation of T-DNA Flanking Sequence and Gene Cloning
The left border flanking region of the T-DNA insertion was isolated using a thermal asymmetric interlaced PCR method described previously (Liu et al., 1995). Sequences were subcloned into pMD19T vector and sequenced using M13F and M13R primers.
Sequences were analyzed against the Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE). The T-DNA insertion was found at 462 bp upstream of the gene Os04g52920, designated as GSD1. The T-DNA insertion and genotyping were confirmed by genomic DNA PCR analysis. The gene-specific primer pairs and the T-DNA-specific primers are listed in Supplemental Table S2. The cloned GSD1 gene was sequenced and its accuracy was verified. GSD1 and its homologous sequences were retrieved from the Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu/) and analyzed using the BioEdit program (version 7.0.0, http://www.mbio.ncsu.edu/BioEdit/BioEdit.html) and the MEGA program (version 5.1, http://www.megasoftware.net).
Gene Expression Analysis
Extraction and purification of total RNA from various tissues at different growth stages were carried out as previously described (Gui et al., 2011). Complementary DNA (cDNA) was reverse transcribed from total RNA using a First-Strand cDNA Synthesis Kit (Takara). Gene expression was measured using real-time RT-PCR. Gene-specific primers (Supplemental Table S2) were designed for PCR amplification. The quantitative PCR assay was performed with a MyiQ real-time PCR detection system (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). The gene expression data were normalized using the rice actin1 gene (LOC_Os03g50885).
Antibody Production and Immunolocalization
Six GSD1-specific peptides (QQNISKGAPQ, residues 178–187; RGFIVPKISN, residues 200–209; QNMKRPSPAS, residues 218–227; ERLSFGSHQP, residues 235–244; AGVTSEYQTK, residues 258–267; and IEIRPYKDPK, residues 275–284) were used to produce polyclonal antibodies in rabbits by Abmart (http://www.ab-mart.com.cn/). Crude antisera were purified using a protein-A Sepharose CL-4B column. The antibody specificity was examined by western-blot hybridization.
For immunolocalization, samples were fixed using a microwave-accelerated, acetone-fixation, paraffin-embedding protocol developed previously (Tang et al., 2006), and immunolocalization were carried out as described (Song et al., 2010).
For immunogold labeling, samples were fixed, embedded, sectioned, and labeled as previously described (Ju et al., 2005).
Overexpression and Suppression Vector Construction and Transformation
The rice GSD1 cDNA fragment was cloned into a binary pCAMBIA1300S vector in sense or antisense orientation under the control of the 35S promoter. After the accuracy of the constructs was confirmed, they were mobilized into Agrobacterium spp. strain EHA105. Agrobacterium spp.-mediated rice transformation was carried out according to Hiei et al. (1994). Transformants (named GSD1-OX and GSD1-AS) were selected under hygromycin and verified by PCR examination.
Promoter Cloning and Analysis
To analyze the GSD1 promoter, a 1,700-bp genomic fragment upstream of the GSD1 putative translation start codon was amplified and cloned into pMD19T vector. By sequencing confirmation, the DNA fragment was subcloned into the vector pCAMBIA1301 using forward primers 5′-TTTTGAATTCACAAGACAACTAC-3′ (for the 1,700-bp promoter) and 5′-TTTTGAATTCAGACTAAAAAAGAGG-3′ (for the 462-bp promoter) with reverse primer 5′-TTTTCCATGGTTCCTCTCTCTC-3′. The constructs pGSD1-L-GUS (1,700-bp promoter) and pGSD1-S-GUS (462-bp promoter) were then transferred into Agrobacterium spp. for rice transformation. Thirty independent pGSD1-L-GUS transgenic lines and 21 independent pGSD1-S-GUS transgenic lines were generated and screened. Ten transgenic lines from each transgenic group with similar growth phenotypes were used for histochemical GUS staining according to the described protocol (Gui et al., 2011).
Quantitative GUS activity was determined as described (Blázquez, 2007). One hundred milligrams of fresh tissue was ground into a fine powder in a 1.5-mL microcentrifuge tube in liquid nitrogen and extracted with 150 μL of GUS extraction buffer. The fluorescence was measured using a microplate reader (Varioskan Flash; Thermo Scientific) with an excitation wavelength of 365 nm and an emission wavelength of 455 nm.
Microscopic Analysis
For the analysis of starch content, stems at the booting, flowering, and grain-filling stages were collected at midday. Hand-cut sections of wild-type, gsd1-D mutant, and GSD1 transgenics were stained with 1% iodine-potassium iodide, observed with a microscope, and photographed.
For scanning electron microscopy, floral organs and the second internode sections (collected at midday after grain fully ripened) from wild-type, gsd1-D mutant, and GSD1 transgenics were prepared and examined as previously described (Gui et al., 2011) with a scanning electron microscope (JSM-5610LV; JEOL) at an accelerating voltage of 10 to 20 kV in low-vacuum mode.
PD Permeability Analysis and Callose Quantification
Tobacco (Nicotiana benthamiana) leaves were infiltrated with the Agrobacterium spp. carrying 35S-mCherry, 35S-GSD1-mCherry, or 35S-PDLP5-mCherry. PD permeability was measured as described (Lee et al., 2011). Briefly, the infiltrated plant leaves were incubated for 48 to 60 h before the dye loading assay. One μL of 1 mm carboxyfluorescine diacetate (CFDA; Sigma-Aldrich) was loaded onto the adaxial leaf blade surface and incubated for 5 min before washing with water. Subsequently, the leaf was cut and mounted on a glass slide for confocal imaging of dye diffusion across the abaxial and adaxial surfaces. The extent of dye diffusion was quantified by measuring the diameters of diffusion area using the confocal microscope. The results were statistically calculated using one-way ANOVA in conjunction with Fisher’s lsd test.
Callose was stained with 0.05 mg/mL aniline blue fluorochrome (Biosupplies). Callose quantification was performed by measuring the aniline blue fluorescence spot number and spot intensity using ImageJ image processing software (version 1.40g, http://rsb.info.nih.gov/ij). Statistical differences were analyzed using one-way ANOVA in conjunction with Fisher’s lsd test.
Subcellular Localization and BiFC
To examine subcellular localization, the full-length GSD1 cDNA was amplified by PCR. The PCR product was digested and subcloned into pCAMBIA1300 vector, pA7-GFP vector (Voelker et al., 2006), and pA7-mCherry vector, respectively. Several PD marker genes or PD permeability-regulating genes were cloned and used in the study, including OsACT1, AtPDLP1, AtPDLP, and AtPDCB1. The following constructs were made. OsACT1, AtPDLP1, and AtPDLP5 were subcloned into pA7-GFP vector, respectively. AtPDLP1 was also subcloned into pA7-mCherry. 35S-GFP-GSD1, 35S-mCherry-GSD1, 35S-GFP-OsACT1, 35S-PDLP1-GFP, 35S-PDLP1-mCherry, 35S-PDLP5-mCherry, and 35S-PDCB1-mCherry constructs were mobilized into Agrobacterium spp. strain GV3101 and transformed into tobacco leaf cells, independently or in combination. After 48 h of incubation, leaf cells were examined using confocal microscopy (LSM 510 META; Zeiss). For BiFC analysis, the YFP gene was cut into two sections (YN and YC). YN and YC were subcloned into pA7-GFP vector, respectively. Then, 35S-YN-OsACT1, 35S-YC-GSD1, and 35S-PDLP1-YN were constructed to analyze their interaction.
Aniline blue fluorochrome (Biosupplies) at 0.05 mg/mL was used for callose staining. A confocal laser microscope was set under the following conditions: GFP was excited at 488 nm and the emitted light was captured at 505 to 555 nm, and aniline blue fluorochrome was excited with a wavelength of 405 nm and the emitted light was captured between 460 and 500 nm.
Co-IP
For Co-IP analysis, FLAG-tagged OsACT1 and Myc-tagged GSD1 (the C-terminal PM anchor domain deleted) were transiently expressed or coexpressed in tobacco. After 2 d of incubation, plant materials were collected and ground in liquid nitrogen to fine powder. Plant proteins were extracted with the extraction buffer (50 mm TrisHCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 1% Triton X-100, and 0.1% protease inhibitor cocktail [Promega]) for 15 min, and centrifuged at 4°C at 12,000g for 10 min. One milliliter of supernatant was incubated with 50 μL of anti-Myc antibody-coupled agarose beads (Abmart) and 50 μL of anti-FLAG antibody-coupled agarose beads (Abmart) for 2 h at 4°C, respectively. Beads were washed three times with wash buffer (50 mm TrisHCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 1 mm EDTA, and 1% Triton X-100), and bound proteins were eluted by mix washed beads with 2 × SDS loading buffer and boiled at 100°C for 5 min. Precipitated proteins were separated on SDS-PAGE and immunoblotted with anti-FLAG antibody (Abmart) and anti-Myc antibody (Abmart).
Sequence data from this article can be found in the GenBank/EMBL databases or the Rice Genome Annotation Project Database under accession numbers NM_001060440, Os04g52920 (GSD1), NM_001057621, Os03g50885 (OsACT1), At5g43980 (PDLP1), At1g70690 (PDLP5), and At5g61130 (PDCB1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypic comparison between the wild type and the gsd1-D mutant.
Supplemental Figure S2. Microscopic analyses of rice flower morphology between the wild type and the gsd1-D mutant at the heading stage.
Supplemental Figure S3. Comparison of anther development between the wild type and the gsd1-D mutant from stage 6 to stage 12 according to established anther developmental stages in rice (Zhang and Wilson, 2009).
Supplemental Figure S4. I2-KI staining of starch in the second internode of the wild type and gsd1-D at booting and flowering stages.
Supplemental Figure S5. Expression profiles of the genes involved in starch synthesis between the wild type and the gsd1-D mutant during rice grain filling.
Supplemental Figure S6. Net photosynthetic activity in the flag leaf at 5 DAF in gsd1-D mutant and wild-type plants growing in a greenhouse.
Supplemental Figure S7. GC-MS analyses of 13C-labeled soluble sugars in the leaf blade and leaf sheath of gsd1-D mutant and wild-type plants.
Supplemental Figure S8. Picture of the chamber used for flag leaf photosynthesis fed with 13CO2.
Supplemental Figure S9. Genotyping analyses of the gsd1-D T-DNA insertion mutant.
Supplemental Figure S10. Amino acid sequences of rice remorins were aligned with the remorins from dicotyledonous Arabidopsis and moss Physcomitrella patens.
Supplemental Figure S11. Phylogenic analysis and antibody developed for GSD1.
Supplemental Figure S12. Phenotypic comparison of wild-type and GSD1 transgenic plants.
Supplemental Figure S13. Net photosynthetic activity in the flag leaves at 5 DAF in wild-type and GSD1 transgenic plants growing in a greenhouse.
Supplemental Figure S14. Immunogold labeling of GSD1 in 4-week-old rice shoot tissues.
Supplemental Figure S15. CFDA-based DANS dye loading assays in tobacco leaves.
Supplemental Figure S16. Subcellular localization analysis of OsACT1 in tobacco epidermal cells.
Supplemental Figure S17. Serial images of z-sections from the top surface to the bottom of the epidermal cells.
Supplemental Table S1. Amino acid sequences similarities of rice remorin proteins.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Cheng Huang for help with vector construction, Jun Chen for HPLC analysis, Wenli Hu for GC-MS analysis, Jiqin Li for assistance with scanning electron microscopy, Xiaoyan Gao and Zhiping Zhang for assistance with transmission electron microscopy, Xiaoshu Gao for confocal laser scanning microscopy, and Genyun Chen and Zhan Shu for assistance with PN analysis.
Glossary
- PD
plasmodesmata
- PM
plasma membrane
- GPI
glycosylphosphatidylinositol
- T-DNA
transfer DNA
- ZH11
Oryza sativa japonica cv zhonghua 11
- DAF
d after flowering
- PN
net photosynthetic rate
- GC-MS
gas chromatography-mass spectrometry
- ED
end of the day
- EN
end of the night
- DANS
Drop and See
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- Co-IP
coimmunoprecipitation
- cDNA
complementary DNA
- CFDA
carboxyfluorescine diacetate
Footnotes
This work was supported by the National Key Basic Research Program of China (grant no. 2013CB127002), the Special Fund for Strategic Pilot Technology of the Chinese Academy of Sciences (grant no. XDA08020203), and the China Postdoctoral Science Foundation (grant no. 2013M530216).
The online version of this article contains Web-only data.
References
- Bariola PA, Retelska D, Stasiak A, Kammerer RA, Fleming A, Hijri M, Frank S, Farmer EE. (2004) Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol Biol 55: 579–594 [DOI] [PubMed] [Google Scholar]
- Barratt DH, Kölling K, Graf A, Pike M, Calder G, Findlay K, Zeeman SC, Smith AM. (2011) Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol 155: 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M. (2007) Quantitative GUS activity assay of plant extracts. CSH Protoc 2007: t4690. [DOI] [PubMed] [Google Scholar]
- Chonan N, Kawahara H, Matsuda T, Kaneko M. (1981) Ultrastructure of the large vascular bundles in the leaves of rice plant. Jpn J Crop Sci 50: 323–331 [Google Scholar]
- Cilia ML, Jackson D. (2004) Plasmodesmata form and function. Curr Opin Cell Biol 16: 500–506 [DOI] [PubMed] [Google Scholar]
- Cock JH, Yoshida S. (1972) Accumulation of 14C-labelled carbohydrate before flowering and its subsequent redistribution and respiration in the rice plant. Proc Crop Sci Soc Jpn 41: 226–234 [Google Scholar]
- Ding B, Kwon MO, Warnberg L. (1996) Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J 10: 157–164 [Google Scholar]
- Dyson HJ, Wright PE. (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Eom JS, Cho JI, Reinders A, Lee SW, Yoo Y, Tuan PQ, Choi SB, Bang G, Park YI, Cho MH, et al. (2011) Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol 157: 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom JS, Choi SB, Ward JM, Jeon JS. (2012) The mechanism of phloem loading in rice (Oryza sativa). Mol Cells 33: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ. (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Shen J, Li L. (2011) Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol 157: 574–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman JM, Lee JS, Bogenschutz NL, Peterson KM, Virata RE, Xie B, Kanaoka MM, Hong Z, Torii KU. (2010) Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Ju HJ, Samuels TD, Wang YS, Blancaflor E, Payton M, Mitra R, Krishnamurthy K, Nelson RS, Verchot-Lubicz J. (2005) The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol 138: 1877–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Chonan N, Matsuda T, Kawahara H. (1980) Ultrastructure of the small vascular bundles and transfer pathways for photosynthate in the leaves of rice. Jpn J Crop Sci 49: 42–50 [Google Scholar]
- Koubaa M, Mghaieth S, Thomasset B, Roscher A. (2012) Gas chromatography-mass spectrometry analysis of 13C labeling in sugars for metabolic flux analysis. Anal Biochem 425: 183–188 [DOI] [PubMed] [Google Scholar]
- Kuo TM, Vanmiddlesworth JF, Wolf WJ. (1988) Content of raffinose oligosaccharides and sucrose in various plant seeds. J Agric Food Chem 36: 32–36 [Google Scholar]
- Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, et al. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL. (2007) A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Lian S, Tanaka A. (1967) Behaviour of photosynthetic products associated with growth and grain production in the rice plant. Plant Soil 26: 333–347 [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Ham BK, Kim JY. (2009) Plasmodesmata - bridging the gap between neighboring plant cells. Trends Cell Biol 19: 495–503 [DOI] [PubMed] [Google Scholar]
- Marín M, Ott T. (2012) Phosphorylation of intrinsically disordered regions in remorin proteins. Front Plant Sci 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M, Thallmair V, Ott T. (2012) The intrinsically disordered N-terminal region of AtREM1.3 remorin protein mediates protein-protein interactions. J Biol Chem 287: 39982–39991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ. (2008) Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol 11: 680–686 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Turgeon R. (1999) Sieve elements and companion cells-traffic control centers of the phloem. Plant Cell 11: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CM, Palmiano EP, Baun LC, Juliano BO. (1971) Starch metabolism in the leaf sheaths and culm of rice. Plant Physiol 47: 404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, Cacas JL, Crowet JM, Lins L, Castroviejo M, German-Retana S, Mongrand S, Raffaele S. (2012) Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement. Plant Physiol 160: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford J, Vesk M, Overall R. (1998) Callose deposition at plasmodesmata. Protoplasma 201: 30–37 [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E, et al. (2009) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Mongrand S, Gamas P, Niebel A, Ott T. (2007) Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol 145: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt S, Knight AE, Hodge TP, Baluska F, Samaj J, Volkmann D, Kendrick-Jones J. (1999) Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J 19: 555–567 [DOI] [PubMed] [Google Scholar]
- Reymond P, Kunz B, Paul-Pletzer K, Grimm R, Eckerskorn C, Farmer EE. (1996) Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell 8: 2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi G, Katz A, Guenoune-Gelbart D, Epel BL. (2005) Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the golgi apparatus. Plant Cell 17: 1788–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Aoki N, Furbank RT. (2007) Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J Exp Bot 58: 3155–3169 [DOI] [PubMed] [Google Scholar]
- Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, et al. (2007) DisProt: the Database of Disordered Proteins. Nucleic Acids Res 35: D786–D793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. (2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Shen J, Li L. (2010) Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol 187: 777–790 [DOI] [PubMed] [Google Scholar]
- Stonebloom S, Burch-Smith T, Kim I, Meinke D, Mindrinos M, Zambryski P. (2009) Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Natl Acad Sci USA 106: 17229–17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Liu Z, Chen C, Zhang Y, Wang X, Zhu L, Miao L, Wang XC, Yuan M. (2010) Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 22: 1373–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Coughlan S, Crane E, Beatty M, Duvick J. (2006) The application of laser microdissection to in planta gene expression profiling of the maize anthracnose stalk rot fungus Colletotrichum graminicola. Mol Plant Microbe Interact 19: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. (2008) Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biology 6: 180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth K, Stratil TF, Madsen EB, Ye J, Popp C, Antolín-Llovera M, Grossmann C, Jensen ON, Schüssler A, Parniske M, et al. (2012) Functional domain analysis of the Remorin protein LjSYMREM1 in Lotus japonicus. PLoS ONE 7: e30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Vilaine F, Kerchev P, Clément G, Batailler B, Cayla T, Bill L, Gissot L, Dinant S. (2013) Increased expression of a phloem membrane protein encoded by NHL26 alters phloem export and sugar partitioning in Arabidopsis. Plant Cell 25: 1689–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker C, Schmidt D, Mueller-Roeber B, Czempinski K. (2006) Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J 48: 296–306 [DOI] [PubMed] [Google Scholar]
- White R, Badelt K, Overall R, Vesk M. (1994) Actin associated with plasmodesmata. Protoplasma 180: 169–184 [Google Scholar]
- Xie B, Wang X, Zhu M, Zhang Z, Hong Z. (2011) CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J 65: 1–14 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ahn SB. (1968) The accumulation process of carbohydrate in rice varieties in relation to their response to nitrogen in the tropics. Soil Sci Plant Nutr 14: 153–161 [Google Scholar]
- Zhang DB, Wilson ZA. (2009) Stamen specification and anther development in rice. Chinese Science Bulletin 54: 2342–2353 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.