Natural variation in the dynamic response to salt stress of the Arabidopsis root system can be divided between four distinct strategies.
Abstract
The plant root is the first organ to encounter salinity stress, but the effect of salinity on root system architecture (RSA) remains elusive. Both the reduction in main root (MR) elongation and the redistribution of the root mass between MRs and lateral roots (LRs) are likely to play crucial roles in water extraction efficiency and ion exclusion. To establish which RSA parameters are responsive to salt stress, we performed a detailed time course experiment in which Arabidopsis (Arabidopsis thaliana) seedlings were grown on agar plates under different salt stress conditions. We captured RSA dynamics with quadratic growth functions (root-fit) and summarized the salt-induced differences in RSA dynamics in three growth parameters: MR elongation, average LR elongation, and increase in number of LRs. In the ecotype Columbia-0 accession of Arabidopsis, salt stress affected MR elongation more severely than LR elongation and an increase in LRs, leading to a significantly altered RSA. By quantifying RSA dynamics of 31 different Arabidopsis accessions in control and mild salt stress conditions, different strategies for regulation of MR and LR meristems and root branching were revealed. Different RSA strategies partially correlated with natural variation in abscisic acid sensitivity and different Na+/K+ ratios in shoots of seedlings grown under mild salt stress. Applying root-fit to describe the dynamics of RSA allowed us to uncover the natural diversity in root morphology and cluster it into four response types that otherwise would have been overlooked.
Salt stress is known to affect plant growth and productivity as a result of its osmotic and ionic stress components. Osmotic stress imposed by salinity is thought to act in the early stages of the response, by reducing cell expansion in growing tissues and causing stomatal closure to minimize water loss. The build-up of ions in photosynthetic tissues leads to toxicity in the later stages of salinity stress and can be reduced by limiting sodium transport into the shoot tissue and compartmentalization of sodium ions into the root stele and vacuoles (Munns and Tester, 2008). The effect of salt stress on plant development was studied in terms of ion accumulation, plant survival, and signaling (Munns et al., 2012; Hasegawa, 2013; Pierik and Testerink, 2014). Most studies focus on traits in the aboveground tissues, because minimizing salt accumulation in leaf tissue is crucial for plant survival and its productivity. This approach has led to the discovery of many genes underlying salinity tolerance (Munns and Tester, 2008; Munns et al., 2012; Hasegawa, 2013; Maathuis, 2014). Another way to estimate salinity stress tolerance is by studying the rate of main root (MR) elongation of seedlings transferred to medium supplemented with high salt concentration. This is how Salt Overly Sensitive mutants were identified, being a classical example of genes involved in salt stress signaling and tolerance (Hasegawa, 2013; Maathuis, 2014). The success of this approach is to be explained by the important role that the root plays in salinity tolerance. Roots not only provide anchorage and ensure water and nutrient uptake, but also act as a sensory system, integrating changes in nutrient availability, water content, and salinity to adjust root morphology to exploit available resources to the maximum capacity (Galvan-Ampudia et al., 2013; Gruber et al., 2013). Understanding the significance of environmental modifications of root system architecture (RSA) for plant productivity is one of the major challenges of modern agriculture (de Dorlodot et al., 2007; Den Herder et al., 2010; Pierik and Testerink, 2014).
The RSA of dicotyledonous plants consists of an embryonically derived MR and lateral roots (LRs) that originate from xylem pole pericycle cells of the MR, or from LRs in the case of higher-order LRs. Root growth and branching is mainly guided through the antagonistic action of two plant hormones: auxin and cytokinins (Petricka et al., 2012). Under environmental stress conditions, the synthesis of abscisic acid (ABA), ethylene, and brassinosteroids is known to be induced and to modulate the growth of MRs and LRs (Achard et al., 2006; Osmont et al., 2007; Achard and Genschik, 2009; Duan et al., 2013; Geng et al., 2013). In general, lower concentrations of salt were observed to slightly induce MR and LR elongation, whereas higher concentrations resulted in decreased growth of both MRs and LRs (Wang et al., 2009; Zolla et al., 2010). The reduction of growth is a result of the inhibition of cell cycle progression and a reduction in root apical meristem size (West et al., 2004). However, conflicting results were presented for the effect of salinity on lateral root density (LRD; Wang et al., 2009; Zolla et al., 2010; Galvan-Ampudia and Testerink, 2011). Some studies suggest that mild salinity enhances LR initiation or emergence events, thereby affecting patterning, whereas other studies imply that salinity arrests LR development. The origin of those contradictory observations could be attributable to studying LR initiation and density at single time points, rather than observing the dynamics of LR development, because LR formation changes as a function of root growth rate (De Smet et al., 2012). The dynamics of LR growth and development were characterized previously for the MR region formed before the salt stress exposure, identifying the importance of ABA in early growth arrest of postemerged LRs in response to salt stress (Duan et al., 2013). The effect of salt on LR emergence and initiation was found to differ for MR regions formed prior and subsequent to salinity exposure (Duan et al., 2013), consistent with LR patterning being determined at the root tip (Moreno-Risueno et al., 2010). Yet the effect of salt stress on the reprogramming of the entire RSA on a longer timescale remains elusive.
Natural variation in Arabidopsis (Arabidopsis thaliana) is a great source for dissecting the genetic components underlying phenotypic diversity (Trontin et al., 2011; Weigel, 2012). Genes underlying phenotypic plasticity of RSA to environmental stimuli were also found to have high allelic variation leading to differences in root development between different Arabidopsis accessions (Rosas et al., 2013). Supposedly, genes responsible for phenotypic plasticity of the root morphology to different environmental conditions are under strong selection for adaptation to local environments. Various populations of Arabidopsis accessions were used to study natural variation in ion accumulation and salinity tolerance (Rus et al., 2006; Jha et al., 2010; Katori et al., 2010; Roy et al., 2013). In addition, a number of studies focusing on the natural variation in RSA have been published, identifying quantitative trait loci and allelic variation for genes involved in RSA development under control conditions (Mouchel et al., 2004; Meijón et al., 2014) and nutrient-deficient conditions (Chevalier et al., 2003; Gujas et al., 2012; Gifford et al., 2013; Kellermeier et al., 2013; Rosas et al., 2013). Exploring natural variation not only expands the knowledge of genes and molecular mechanisms underlying biological processes, but also provides insight on how plants adapt to challenging environmental conditions (Weigel, 2012) and whether the mechanisms are evolutionarily conserved. The early growth arrest of newly emerged LRs upon exposure to salt stress was observed to be conserved among the most commonly used Arabidopsis accessions Columbia-0 (Col-0), Landsberg erecta, and Wassilewskija (Ws; Duan et al., 2013). By studying salt stress responses of the entire RSA and a wider natural variation in root responses to stress, one could identify new morphological traits that are under environmental selection and possibly contribute to stress tolerance.
In this work, we not only identify the RSA components that are responsive to salt stress, but we also describe the natural variation in dynamics of salt-induced changes leading to redistribution of root mass and different root morphology. The growth dynamics of MRs and LRs under different salt stress conditions were described by fitting a set of quadratic growth functions (root-fit) to individual RSA components. Studying salt-induced changes in RSA dynamics of 31 Arabidopsis accessions revealed four major strategies conserved among the accessions. Those four strategies were due to differences in salt stress sensitivity of individual RSA components (i.e. growth rates of MRs and LRs, and increases in the number of emerged LRs). This diversity in root morphology responses caused by salt stress was observed to be partially associated with differences in ABA, but not ethylene sensitivity. In addition, we observed that a number of accessions exhibiting a relatively strong inhibition of LR elongation showed a smaller increase in the Na+/K+ ratio in shoot tissue after exposure to salt stress. Our results imply that different RSA strategies identified in this study reflect diverse adaptations to different soil conditions and thus might contribute to efficient water extraction and ion compartmentalization in their native environments.
RESULTS
Salt Stress Decreases MR and LR Length But Does Not Affect LRD
Salinity stress is known to affect root growth (West et al., 2004) and to induce the growth arrest of emerged LRs (Duan et al., 2013), yet its relative effect on individual RSA components on a larger timescale is less clear. To determine which RSA parameters are most responsive to salt stress treatment, a dose-response experiment was performed on four different natural Arabidopsis accessions (Col-0, Burren [Bur-0], C24, and Tsushima [Tsu-0]), previously described to differ in their salinity tolerance (Jha et al., 2010; Katori et al., 2010; Galpaz and Reymond, 2010). Four-day-old seedlings were transferred to media supplemented with different NaCl concentrations (0, 25, 50, 75, 100, 125, and 150 mm NaCl), scanned at 7 d after transfer (Fig. 1A), and 27 RSA traits (Table I) were quantified with EZ-Rhizo software (Armengaud et al., 2009). Next to the standard RSA parameters provided by EZ-Rhizo, lateral root length (LRL) and average lateral root length (aLRL) were calculated. In order to distinguish the effect of salt on the LR development between the MR region formed prior to and after the start of the salt stress treatment, the formation and length of LRs were studied for those two regions separately (regions A and B, respectively). Moreover, the ratios between main root length (MRL), aLRL, and total root size (TRS) were calculated for all conditions studied. For trait descriptions and an overview of salt stress-induced changes in individual trait values, see Table I. The Pearson correlation coefficients (r2) between MRL and other RSA parameters studied are listed in Table II.
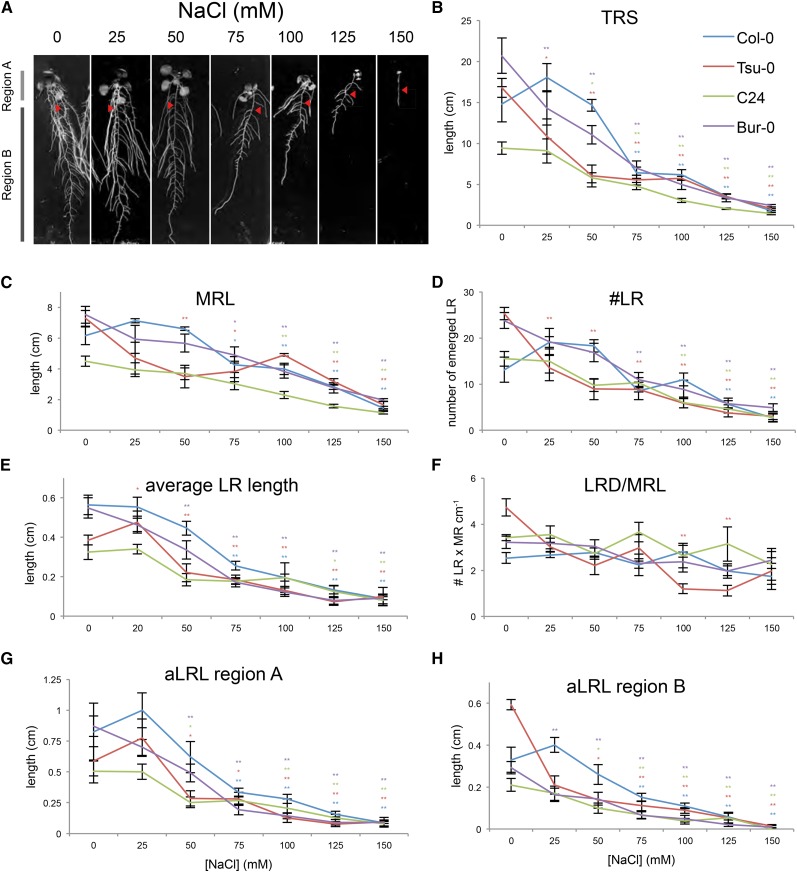
Figure 1.
Increasing salt concentrations gradually change RSA. Four-day-old seedlings of four Arabidopsis accessions (Col-0, Tsu-0, C24, and Bur-0) were transferred to media supplemented with different salt concentrations ranging from 0 to 150 mm NaCl. Eleven days after germination, the RSA of the seedlings grown at different conditions was quantified using EZ-Rhizo software. A, Representative pictures of RSA of 11-d-old Col-0 seedlings grown for 7 d on different salt concentrations. The length of MR at the transfer from standard to treatment media is indicated with red arrowheads. The effect of salt stress on individual RSA parameters was studied by comparing RSA observed at different salt stress conditions with control conditions. B to F, Increasing concentrations of salt caused a dose-dependent reduction in TRS (B), MRL (C), number of emerged LRs (#LR; D), and aLRL (E), but did not significantly affect LRD (F). G and H, The aLRL was also significantly reduced when calculated separately for both regions above (G) and below (H) the transfer point. Individual points represent the average value of eight replicates and error bars represent the ses. The effect of salt stress was defined as significant when the RSA trait value in salt stress conditions was significantly different from control conditions with Tukey’s post hoc test with significance of 0.05 (single asterisk) or 0.01 (double asterisks). Significant differences are indicated with asterisks in colors corresponding to those of the individual accessions.
Table I. Overview of RSA parameters obtained from EZ-Rhizo and their response to salinity stress.
The effect of salt stress on individual RSA parameters was studied by comparing RSA observed at different salt stress conditions with control conditions. The effect of salt stress was defined as significant when the RSA trait value in salt stress conditions was significantly different from control conditions with Scheffe’s post hoc test with significance of 0.01 (double asterisks) for at least two different accessions at three salt stress concentrations used. ns, Not significant.
| Trait Identifier | Trait Description | Unit | Salt Effect |
|---|---|---|---|
| MRL | MR length | cm | ** |
| MRVL | MR vector length | cm | ** |
| MRVA | MR vector angle | degree | ns |
| noLR | No. of LRs per MR | no. | ** |
| #LR region A | No. of LRs in region above transfer point | no. | ns |
| #LR region B | No. of LRs in region below transfer point | no. | ** |
| Basal | Length of basal zone | cm | ns |
| Branched | Length of branched zone | cm | ** |
| Apical | Length of apical zone | cm | ns |
| Basal/MRL | Length of basal zone as ratio of MRL | % | ns |
| Branched/MRL | Length of branched zone as ratio of MRL | % | ns |
| Apical/MRL | Length of apical zone as ratio of MRL | % | ns |
| Straightness | MRL divided by MR vector length | % | ns |
| TRS | Cumulative length of LR and MR | cm | ** |
| LRD/MRL | No. of LRs per cm of MR | no./cm | ns |
| LRD region A | No. of LRs per cm of region above the transfer point | no./cm | ns |
| LRD region B | No. of LRs per cm of region below the transfer point | no./cm | ns |
| LRD / BZ | No. of LRs per cm of branched zone | no./cm | ns |
| Depth | Depth | cm | ** |
| LRL | Cumulative length of LRs | cm | ** |
| LRL region A | Cumulative length of LRs in region above the transfer point | cm | ** |
| LRL region B | Cumulative length of LRs in region below the transfer point | cm | ** |
| aLRL | Average length of LR | cm | ** |
| aLRL region A | Average length of LRs in region above the transfer point | cm | ** |
| aLRL region B | Average length of LRs in region below the transfer point | cm | ** |
| MRL/TRS | MR length as a ratio of TRS | % | ** |
| aLRL/TRS | Average LR length as a ratio of TRS | % | ns |
| aLRL/MRL | Average LR length as a ratio of MR length | % | ns |
Table II. Salt stress weakens the correlation between MRL and LRL.
Pearson correlation coefficients calculated between MRL and other RSA parameters of 4-d-old seedlings transferred for 7 d to different salt concentrations. Pearson correlation coefficients were correlated on average values of eight replicates for all four accessions used with SPSS software. The significant correlations are marked with an asterisk for significance levels of 0.05 and double asterisks for 0.01. RSA trait descriptions are provided in Table I. n.a., Not applicable.
|
RSA Trait |
Concentrations of NaCl |
||||||
|---|---|---|---|---|---|---|---|
| 0 |
25 | 50 | 75 | 100 | 125 | 150 | |
| mm | |||||||
| MRVL | 0.977** | 0.993** | 0.992** | 0.991** | 0.985** | 0.962** | 0.956** |
| MRVA | 0.427* | −0.287 | 0.113 | 0.223 | 0.002 | −0.313 | −0.326 |
| #LR | 0.751** | 0.870** | 0.870** | 0.376* | 0.400* | 0.419* | n.a. |
| #LR region A | 0.333 | 0.332 | 0.367* | 0.054 | 0.267 | 0.146 | 0.432* |
| #LR region B | 0.418* | 0.803** | 0.570** | 0.209 | 0.372* | 0.435* | 0.410* |
| Basal | −0.559** | −0.092 | −0.304 | 0.244 | 0.247 | 0.129 | 0.14 |
| Branched | 0.716** | 0.821** | 0.865** | 0.656** | 0.469** | 0.691** | n.a. |
| Apical | 0.389* | 0.587** | 0.725** | 0.782** | 0.765** | 0.405* | n.a. |
| Basal/MRL | −0.595** | −0.461* | −0.621** | −0.400* | −0.237 | −0.272 | −0.078 |
| Branched/MRL | 0.175 | 0.328 | 0.369* | −0.409* | −0.239 | 0.374* | n.a. |
| Apical/MRL | 0.028 | −0.146 | 0.15 | 0.476** | 0.369* | −0.245 | 0.333 |
| Straightness | 0.06 | 0.624** | 0.403* | 0.354 | 0.113 | −0.165 | −0.32 |
| TRS | 0.920** | 0.910** | 0.906** | 0.891** | 0.893** | 0.921** | 0.821** |
| LRD/MR | −0.035 | −0.05 | 0.15 | −0.753** | −0.348 | −0.291 | 0.142 |
| LRD region A | 0.333 | 0.332 | 0.367* | 0.054 | 0.267 | 0.146 | 0.432* |
| LRD region B | −0.534** | 0.153 | −0.294 | −0.613** | −0.352* | −0.371* | 0.301 |
| LRD/BZ | −0.134 | −0.520** | −0.017 | −0.512** | 0.176 | −0.237 | 0.161 |
| Depth | 0.888** | 0.984** | 0.985** | 0.988** | 0.979** | 0.910** | 0.915** |
| LRL | 0.847** | 0.793** | 0.741** | 0.221 | 0.354* | 0.238 | 0.083 |
| LRL region A | 0.526** | 0.619** | 0.416* | −0.052 | 0.260 | 0.109 | 0.095 |
| LRL region B | 0.592** | 0.678** | 0.516** | 0.168 | 0.377* | 0.210 | 0.283 |
| aLRL | 0.418* | 0.139 | 0.350* | −0.156 | −0.245 | −0.137 | −0.017 |
| aLRL region A | 0.271 | 0.577** | 0.432* | −0.186 | −0.155 | −0.077 | 0.031 |
| aLRL region B | 0.324 | 0.628** | 0.381* | −0.169 | 0.546** | −0.054 | 0.283 |
| MRL/TRS | −0.491* | −0.177 | −0.355* | 0.605** | 0.321 | 0.171 | 0.048 |
| aLRL/TRS | −0.642** | −0.786** | −0.618** | −0.828** | −0.532** | −0.477** | −0.166 |
| aLRL/MRL | −0.504** | −0.770** | −0.529** | −0.816** | −0.546** | −0.505** | −0.24 |
Interestingly, lower salt concentrations (25 and 50 mm NaCl) slightly increased the TRS, MRL, and the number of LRs in Col-0 seedlings, whereas those traits were significantly reduced in Tsu-0 and Bur-0 seedlings and were not affected in C24 seedlings (Fig. 1, B–D). When the aLRL was studied for regions A and B separately, the differences in RSA responses between Bur-0 and Tsu-0 became more apparent. Whereas the aLRL of Bur-0 and Tsu-0 seedlings was only slightly reduced or unaffected in the region above the transfer point, the aLRL below the transfer point was significantly reduced for both accessions (Fig. 1, G and H). Those results indicate that the differences in RSA responses between the accessions at very low salt concentrations are best to be dissected into the regions formed prior to and after the start of salt stress exposure.
At higher concentrations, all accessions tested showed similar responses for all RSA components and for regions above and below the transfer point (Fig. 1, G and H; Supplemental Fig. S1). The TRS, MRL, number of LRs, and LRL were all significantly reduced in four accessions studied (Fig. 1, B–E). The aLRLs formed in regions A and B decreased in all four accessions (Fig. 1, G and H), whereas the number of LRs was observed to decrease only in region B (Supplemental Fig. S1). This decrease can be explained by a reduction in MRL, because the density of LRs in neither of the regions was altered compared with control conditions (Supplemental Fig. S1).
Correlation strength between MRL and both LRL and aLRL was observed to decrease with increasing salt stress concentrations (Table II), suggesting independent regulation of MR and LR growth under salt stress conditions. Increasing salt concentrations had no significant effect on LRD calculated either for MRL, region A, region B, or per length of branched zone (Fig. 1F; Table I; Supplemental Fig. S1). Furthermore, the number of LRs was observed to strongly correlate with the MRL among all salt concentrations tested (Table II). Although the experimental set up used here did not allow us to study the very early stages of LR development, our results suggest that the reduction in the number of visible LRs in salt stress conditions is mainly due to a decrease in MRL.
RSA Growth Dynamics Is Best Captured in Quadratic Growth Functions
Because the increasing concentrations of salt were found to affect MRL and LRL in the four accessions studied, the observed reduction in TRS could be a result of overall deceleration of root development. Decelerated MR growth by itself would have a tremendous impact on the TRS, because the number of emerged LRs depends on MRL (Table II). The dynamics of root development were examined to determine to what extent salt stress decelerates root development and whether changes in root morphology (RSA) are initiated. Four-day-old Col-0 seedlings were transferred to plates containing 0, 75, or 125 mm NaCl. RSA development was studied from 4 to 10 d after germination in control conditions and up to 12 d after germination for seedlings transferred to 75 or 125 mm NaCl by scanning the plates every other day. RSA phenotypes observed at the different developmental stages (Fig. 2A) indicated RSA reprogramming by salinity stress, rather than only retardation in root development.
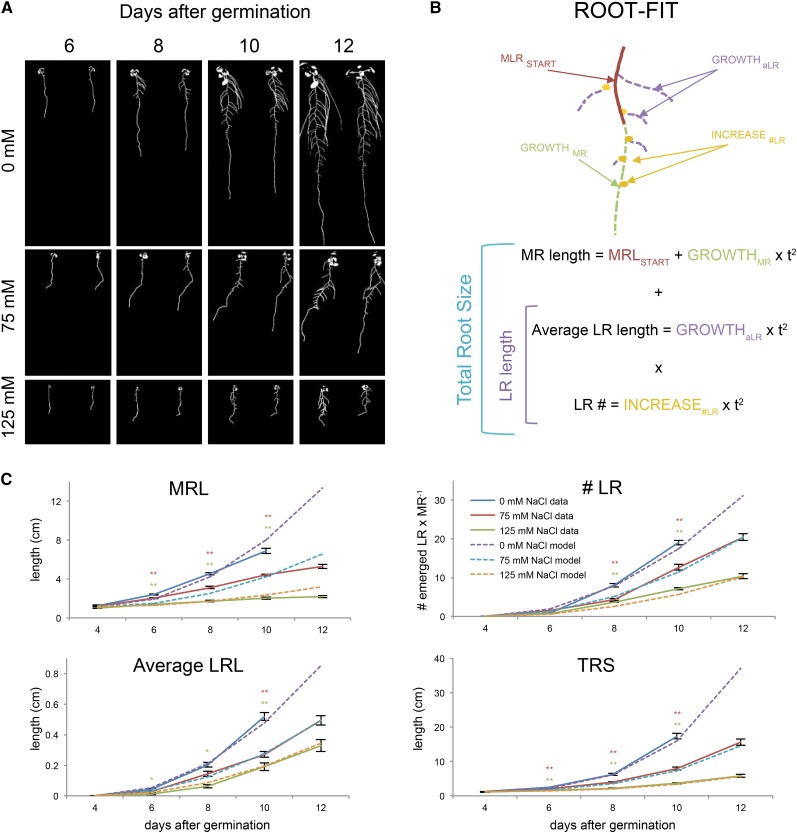
Figure 2.
Quadratic growth functions used for description of RSA growth dynamics. Four-day-old seedlings of Col-0 were transferred to media supplemented with 0, 75, or 125 mm NaCl. A, RSA of seedlings was quantified every 2 d between 4 and 12 d after germination. B, TRS is defined by the combination of MRL, aLRL, and number of emerged LRs. MRL of 4-d-old seedlings is the starting point (MRSTART) and the growth of the MR was expressed as growthMR by fitting a quadratic function on the observed MR growth. A similar approach was used to calculate the increase in number of emerged LRs and aLRL (increase#LR and growthaLR, respectively). The models describing individual RSA parameters were combined for describing cumulative LRL by multiplying number of LRs with aLRL. TRS was calculated from individual parameters by adding MRL to cumulative LRL. C, The descriptive quadratic growth models for individual parameters (dashed lines) were compared with the observed growth rates (filled lines) at growth conditions for MRL, number of emerged LRs (# LR), aLRL, and TRS. The individual data points for observed growth represent the average value of 24 replicates. Error bars represent the se. The r2 values for goodness of fit are provided in Supplemental Table S2. The effect of salt stress was defined as significant when the RSA trait value in salt stress conditions was significantly different from control conditions when tested with Tukey’s post hoc test, with significance of 0.05 (single asterisk) or 0.01 (double asterisks). The significant differences are indicated with asterisks in colors corresponding to those of the salt stress treatments.
In order to quantify the RSA development, RSA was dissected into three components: MRL, number of LRs, and aLRL (Fig. 2B). All three main components of RSA showed a reduction in response to salinity exposure (Fig. 2C, filled lines). Control and salt stress conditions were significantly different from 6 d after germination for MRL, whereas the number of LRs was significantly affected by salt stress from 8 d after germination. The difference in aLRL between 0 and 125 mm NaCl-grown seedlings was also significant from 6 d after germination, whereas the aLRL of seedlings grown at 75 mm differed significantly from control conditions only 10 d after germination (Fig. 2C). No effect on LRD calculated for the entire MRL was observed (Supplemental Fig. S2). Interestingly, when LRD was calculated for regions A and B separately, no differences in LRD were observed for region A, whereas a mild reduction in LRD was found only at 8 d after germination for region B (Supplemental Fig. S2).
Next, growth rates of MRs and LRs and increases in the number of LRs were described by fitting mathematical functions to the observed growth rates (Fig. 2B). Because the plants were 4 d old when transferred, the initial MRL was taken as a starting point. Root growth during early seedling growth is a vector of increasing meristem growth (Beemster and Baskin, 1998) and expansion, resulting in quadratic growth. By fitting quadratic functions on the individual RSA parameters, the growth rates were estimated and were subsequently used to calculate the increase in the main root length (growthMRL) the increase in the number of lateral roots (increase#LR), and the increase in the average lateral root length (growthaLRL). The functions describing individual RSA parameters were combined to describe cumulative LRL by multiplying the number of LRs with aLRL. The TRS was calculated from individual parameters by adding MRL to cumulative LRL (Fig. 2B). The values of individual parameters and the TRS as predicted by the quadratic growth models (Fig. 2C, dashed lines) were compared with the increase of root size measured (Fig. 2C, filled lines) and fits of the model with measured data were examined by calculating the coefficient of determination (r2; Supplemental Table S1). Thus, our descriptive model of fitted quadratic growth functions to RSA dynamics, which we called root-fit, was found to be in good agreement with the experimental data.
Because LR development in the region formed prior to the salt stress exposure could exhibit a different response to salinity than LRs formed after exposure to stress, an alternative model (root-fita/b) was made (Supplemental Fig. S2A) in which LRL was calculated separately for regions A and B. The average LR growth in region A was constructed as described above for growthaLR (Supplemental Fig. S2, A and B), whereas a 1-d delay was added to the model describing average LR growth and increase of LR number in region B (Supplemental Fig. S2, C and E). In this case, the increase in LR number in region A was best described using a square-root function rather than a quadratic one because the length of region A did not increase and the number of LRs was observed to reach a saturation point around six LRs per centimeter (Supplemental Fig. S2D). However, the use of different functions in the root-fita/b model hindered comparisons of the relative effect of salt stress on the individual RSA parameters, despite the good model fit to the observed increase in total LRL (Supplemental Fig. S3, F and G) as well as TRS (Supplemental Fig. S3H). Although the model based on the different regions could be valuable for other applications, we used the root-fit model describing the dynamics of the entire RSA in only three parameters (the growth rates of the MR, the average LR, and increase in LR number) for subsequent analyses of RSA remodeling and natural variation in RSA responses (Fig. 2B).
Reduction in Growth Parameters Reveals RSA Remodeling in Response to Salt
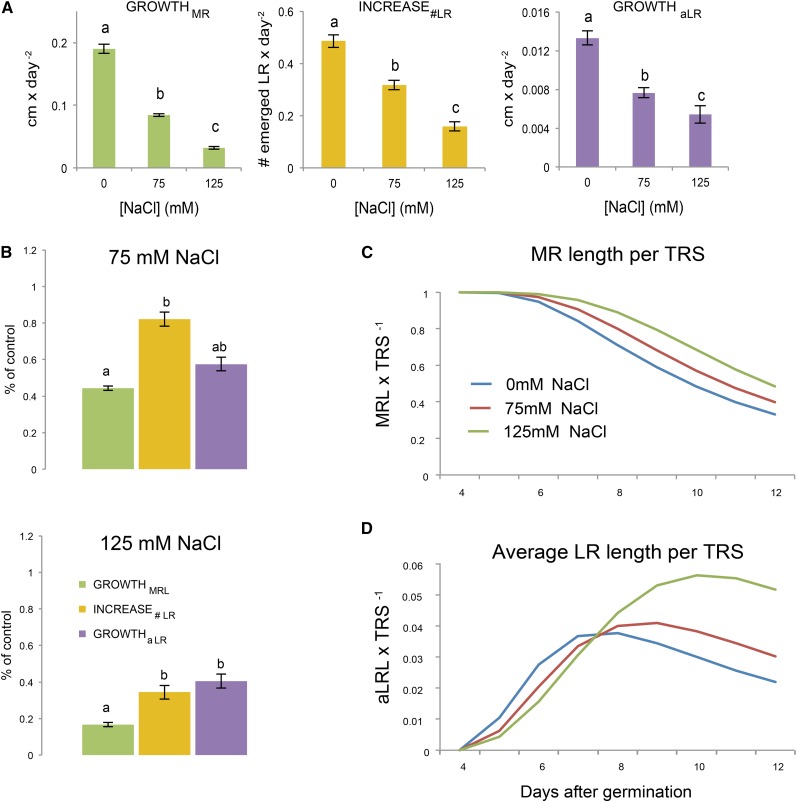
In order to study the effect of salt stress on individual components of RSA, the growth rates of the MR, the number of LRs, and the average LR were compared between the control and salt stress conditions. For all RSA parameters used, salt stress decreased the growth rate significantly (Fig. 3A). Relative MR growth was inhibited more severely than the increase in LR number or average LR growth in salt stress conditions. The effect of salt stress on reshaping the distribution of total root mass between the MR and LRs was further explored by calculating the ratios between the MR and aLRLs per TRS over the days (Fig. 3, C and D). In control conditions, the portion of TRS consisting of the MR decreased with root development as more LRs emerged and elongated from the constantly elongating MR (Fig. 3C). This progress of TRS into mainly LR-derived root mass was delayed in both salt stress conditions studied (Fig. 3C). In order to study the transition from MR-derived root mass into RSA composed mainly of LRs, the ratio between the aLRL and TRS was calculated from the growth rates of individual RSA traits (Fig. 3D). In control conditions, the aLRL as a ratio of TRS increases as the first emerged LRs start to elongate. However, this ratio drops 8 d after germination, as more newly formed LRs start to emerge, reducing the contribution of individual LRs to TRS (Fig. 3D). A similar pattern was observed for seedlings grown on media supplemented with NaCl. Interestingly, the ratio of aLRL to TRS was observed to reach a higher maximum in both NaCl conditions than in control conditions. Moreover, the maximum of aLRL/TRS was reached later and the slope after reaching the maximum was slightly more horizontal (Fig. 3D). The higher maximum value of aLRL/TRS implies that LRs of seedlings grown at mild salt stress conditions are relatively longer before the new LRs appear, when the aLRL/TRS ratio starts to decrease. The more moderate slope of the curve after the aLRL/TRS maximum in both salt stress conditions would be caused by the fewer number of new LRs emerging; therefore, the contribution of earlier formed (relatively longer) LRs remains relatively high compared with the control conditions. This shift in the aLRL/TRS ratio, as well as the more severe impact on the growth rate of the MR compared with average LR and increase in LRs (Fig. 3B), indicates that salt stress does indeed remodel the RSA of Col-0 seedlings. Salt stress thus not only retards overall root development, but also reshapes the root architecture. Our findings imply differential regulation and/or salt stress sensitivity of MR and LR growth, in agreement with the decreasing correlation between MRL and LRL in response to increasing salt stress concentrations (Table II).
Figure 3.
Salt stress decelerates and modifies root development of Col-0 seedlings. A, The growth rates of individual RSA parameters were calculated for control conditions as well as two different salt stress conditions. The bars represent the average growth rate calculated from 24 replicates. Error bars represent the se. The differences between the growth rates at different conditions were tested with one-way ANOVA with Tukey’s post hoc test of significance. Different letter codes represent distinct groups with a significance level of 0.05. B, The relative effect of salt stress on growth of individual parameters was calculated. The severity of growth reduction caused by salinity stress could be compared over the individual RSA components. Different letter codes represent distinct groups with a significance level of 0.05 as tested with Tukey’s post hoc test. To identify the changes in root mass distribution between the MR and LRs, MR and aLRL were calculated as a ratio of TRS for different growth conditions. C, The ratio between MRL and TRS changed over time as more LRs developed (blue line). This transition was delayed in plants grown at 75 and 125 mm NaCl (red and green lines, respectively). D, The ratio of aLRL to TRS was more dynamic, because initially elongating LRs increased the portion of TRS ascribed to individual LRs; however, when new LRs started to emerge, the aLRL ratio per TRS decreased (blue line). A similar pattern with a slight shift in time and maximum was observed for 75 mm NaCl (red line). This delay in maximum reach and increase in maximum height were more pronounced for the seedlings grown at 125 mm NaCl (green line). The ratios were calculated using the quadratic model for three different salt stress concentrations.
Natural Variation in RSA Strategies Reveals a Trade-Off between MR and LR Elongation
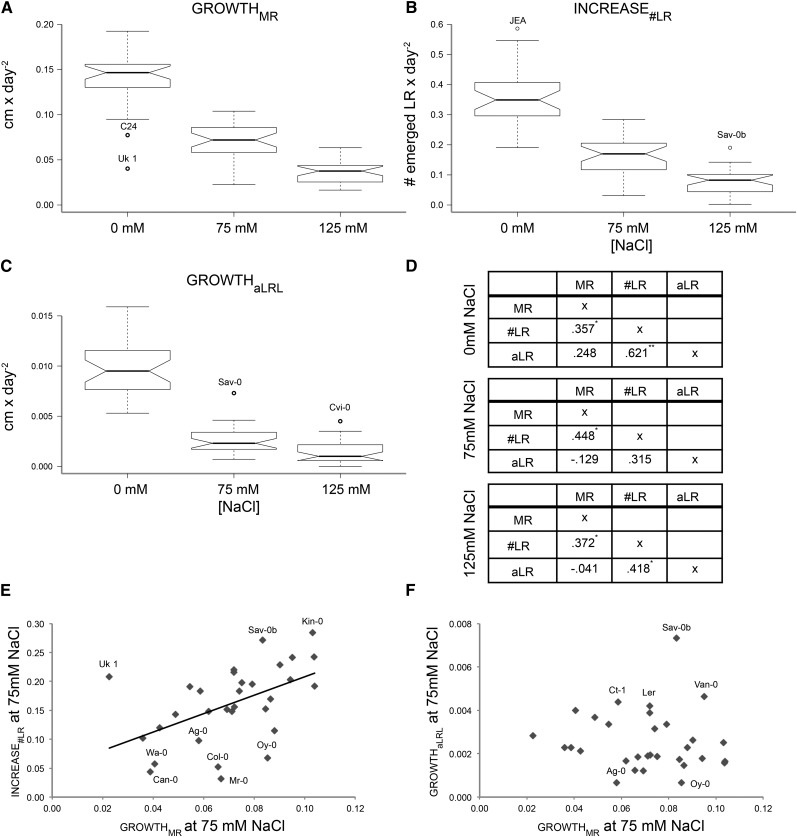
To estimate whether the strategy of RSA response to salt stress observed for Col-0 seedlings is conserved among other Arabidopsis accessions, we studied salt-induced changes in the RSA of 32 different accessions (Supplemental Table S2). These 32 accessions were chosen based on the available recombinant inbred line populations that could be used in further research identifying the genetic components underlying RSA responses to salinity stress. Phenotyping and quantification of RSA was performed as previously described for Col-0 seedlings and the dynamics in individual RSA parameters were described with root-fit for each accession individually (Supplemental Table S3). We observed significant natural variation in all parameters modeled between the accessions in control and salt stress conditions (Fig. 4, A–C; Supplemental Fig. S3). Accession Yosemite showed high variability in its average LR growth rates at 75 mm NaCl and was therefore excluded from further analysis. In general, the growth rates of the individual RSA components were observed to positively correlate between different conditions studied (Supplemental Table S4). The correlation between growthMRL and increase#LR was found to remain significant in all conditions and all accessions tested, again suggesting that the decrease in the visible LR number is mainly attributable to a reduction of the MRL rather than altered LR patterning (Fig. 4, D and E). Interestingly, under salt stress conditions, the positive r2 between the growthMRL and growthaLR changed into a weak, negative correlation for both salt stress conditions studied (Fig. 4, D and F), suggesting a trade-off between investing in either MR or average LR elongation (Fig. 4D).
Figure 4.
Natural variation reveals a trade-off between salt-induced reduction in MR and LR growth rates. We studied 32 different natural Arabidopsis accessions to examine the salt-induced changes in RSA dynamics induced by salt stress. Four-day-old seedlings were transferred to media supplemented with 0, 75, or 125 mm NaCl and the growth rates of MR, number of LRs, and average LR were described between 4 and 10 d (control conditions) or 4 and 12 d (salt) after germination using root-fit. A to C, Natural variation in growthMR (A), increase#LR (B), and growthaLR (C) was observed and several accessions were identified as outliers in MR and average LR growth rates. The boxplots represent the median growth rate as observed in 32 accessions studied. The whiskers extend to data points that are less than 1.5× from the interquartile range (IQR) away from the first and third quartiles. Notches represent 1.58 × IQR/square root (n) and give 95% confidence that two medians differ. D, To establish whether salt influenced the correlation between different RSA traits, Pearson correlation coefficients (r2) between the growth rates of individual RSA parameters were determined. Significant correlations are designed with a single asterisk (0.05) or double asterisks (0.01). E, The strong positive correlation between MR growth and increase in emerged LR number remained at 75 mm NaCl. F, The correlation between the growth of MR and average LR of accessions changed to a negative correlation at 75 mm NaCl, indicating that under salinity stress maintenance of LR growth goes to the expense of MR growth and vice versa. The growth rates per individual accession were calculated based on four replicates per condition tested. The accessions used are listed in Supplemental Table S2.
Four Strategies Guiding RSA Acclimation Responses to Salinity Stress
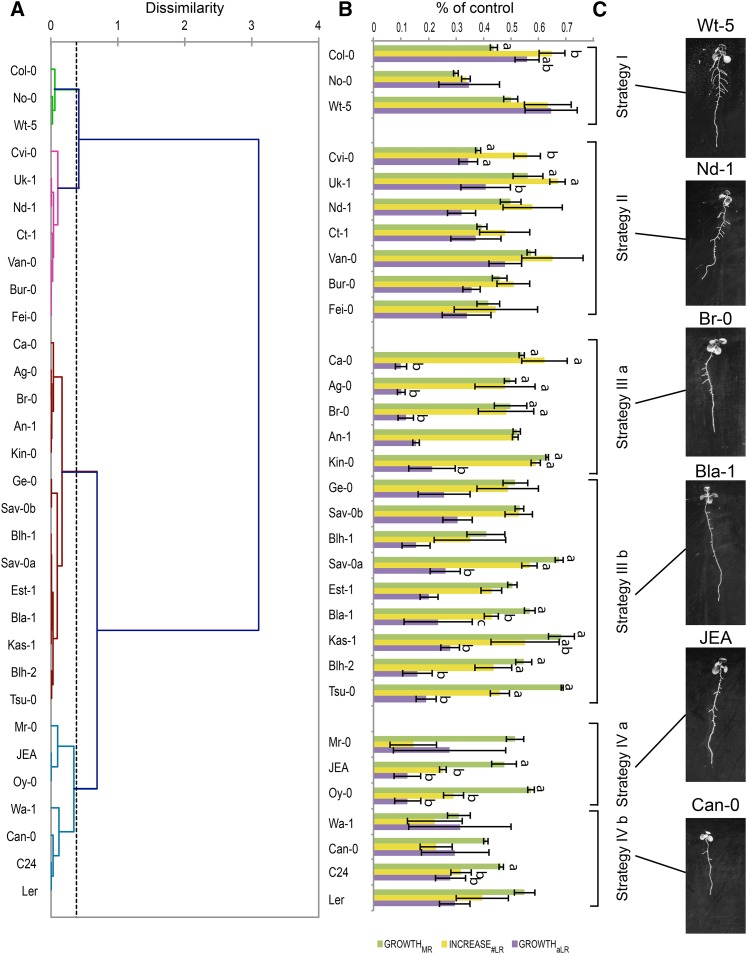
To identify common morphological strategies among the accessions, the growth rates of each accession were normalized for their growth in control conditions (Fig. 5B). Because we were interested in the redistribution of the root mass between the MR and LRs rather than the decrease in TRS per se, the relative effect of salt on growth rates of individual RSA parameters was calculated. Individual growth parameters were normalized for the general growth decrease by calculating growth rates relative to MR growth reduction. Subsequently, accessions were categorized by applying the Ward linkage hierarchical cluster method, as used by Kellermeier et al. (2013), on relative average lateral root elongation (growthaLR) and increase#LR at 75 mm NaCl scaled on MR growth rates (Fig. 5A). In this way, four major strategies were identified (Fig. 5, B and C). Accessions within strategy I showed a more severe reduction in MR elongation than in growthaLR and increase#LR. The strategy II category included accessions for which salt reduced the growth of all RSA parameters with similar impact. Accessions belonging to strategy III showed a high reduction of LR elongation compared with the reduction of main root elongation (growthMR), whereas accessions associated with strategy IV showed not only reduced growthaLR but also exhibited highly reduced increase#LR (Fig. 5, B and C). Both strategies III and IV could be divided in subcategories with different effects on growthaLR (a and b, respectively).
Figure 5.
Four distinct RSA response strategies to salt stress identified within the collection of natural accessions. Four-day-old seedlings of 32 different accessions were transferred to media supplemented with 0, 75, or 125 mm NaCl. RSA of seedlings was quantified between 4 and 12 d after germination. The changes in growth rates of MR, LR, and number of LRs were described with root-fit and the growth rates relative to control conditions were calculated, scaled for MR growth reduction, and used as an input for hierarchical clustering for discovering the most common and conserved RSA responses among tested accessions. A, Dendrogram of accessions obtained by hierarchical clustering by the Ward linkage method. B, Relative growth rates of MR, LR, and number of emerged LRs of 31 accessions tested. Growth rates were calculated by using quadratic growth models on the data collected from four seedlings per accession per treatment. The bars represent the average growth rate relative to control conditions. The error bars represent the se. Different letters are used to indicate the significant differences between the relative growth rates of MR, increase in emerged LR number, and average LR growth within the accession as tested by Tukey’s post hoc test with significance levels of 0.05, C, Pictures of RSA of representative accessions belonging to different RSA strategies grown for 8 d on medium supplemented with 75 mm NaCl. The accessions used are listed in Supplemental Table S2.
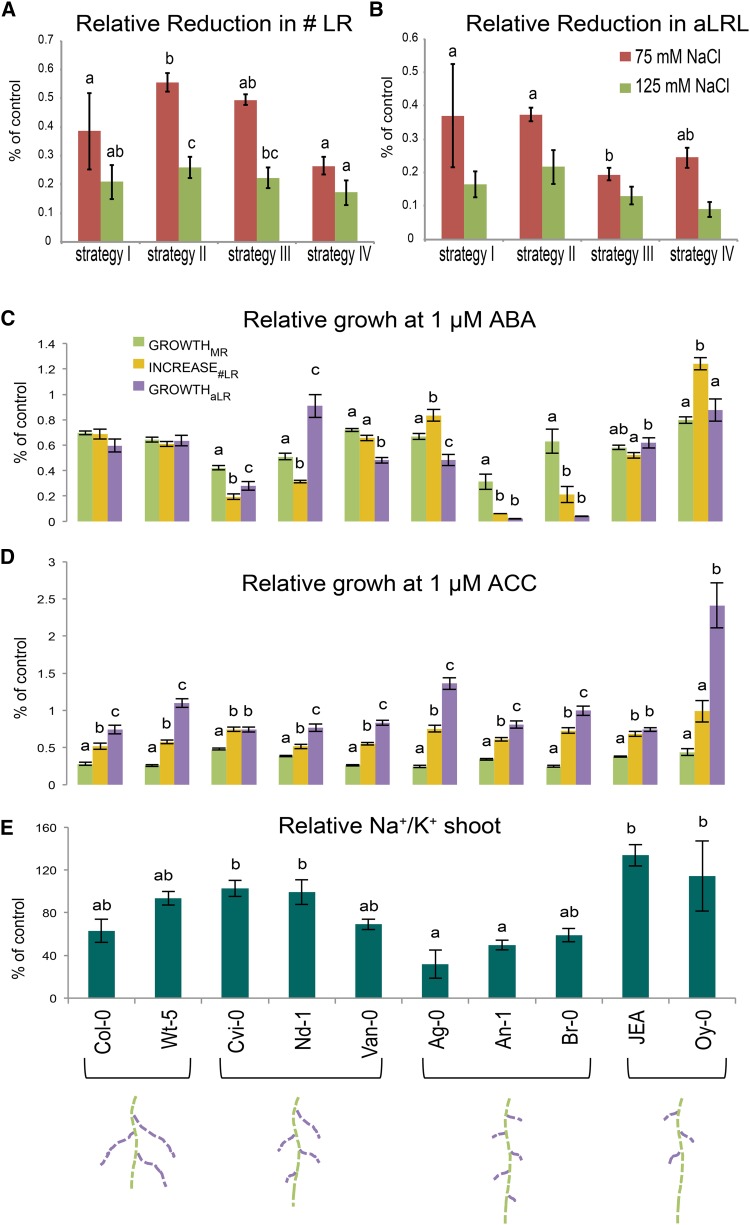
To validate the different categories, the values of RSA parameters relative to control conditions at 12 d after germination in accessions belonging to each category were pooled. The identified strategies (Fig. 5C) could be split in two groups based on the reduction in LR number (Fig. 6A). Strategies I, II, and III were only mildly affected by salt (Supplemental Fig. S4), whereas accessions belonging to strategy IV experienced a severe reduction in LR number. Moreover, concerning aLRL (Fig. 6B), accessions could be grouped into ones showing less (I and II) or more (III and IV) pronounced inhibition of LR elongation rates (Fig. 6B). The reduction in MR and TRS (Supplemental Fig. S5, A and B) as well as the measured shoot size and root/shoot ratio (Supplemental Fig. S5, C and D) were similar for all strategies defined.
Figure 6.
Different RSA strategies are partially explained by natural variation in ABA sensitivity and correlate with differences in Na+/K+ ratios in the shoot. A and B, The relative reduction in the number of LRs (# LR; A) and aLRL (B) was calculated for a set of 31 accessions representing different strategies. The reduction was calculated from 12-d-old plants based on the quadratic growth model (Fig. 2). C, and D, The differences in ABA (C) and ACC (D) sensitivity were examined by quantifying the RSA dynamics of seedlings grown between 4 d and 12 d on 1 μm ABA-supplemented media and between the 4 d and 10 d after germination on 1 μm ACC-supplemented media. The changes in growthMR, increase#LR, and growthaLR were described with a quadratic growth model and the growth rates relative to control conditions were calculated for MR growth, number of LRs, and average LR growth reduction. Different letters are used to indicate significant differences between the relative growth rates of MR, increase in emerged LR number, and average LR growth within each accession as tested by Tukey’s post hoc test with significance levels of 0.05. E, Na+ and K+ accumulation was measured in 12-d-old seedlings grown on 0 or 75 mm NaCl for 8 d. The bars represent the average change in Na+/K+ ratio compared with control conditions (Supplemental Fig. S8) as measured for four biological replicas consisting of 10 seedlings pooled together. The error bars represent the se. Different letters are used to indicate the significant differences between the accessions as tested by Tukey’s post hoc test with significance levels of 0.05. The accessions used are listed in Supplemental Table S2.
RSA Strategies Are Partially Due to Differences in ABA Sensitivity and Correlate with Different Na+/K+ Accumulation in the Shoot
To examine whether the different strategies are due to activation of similar signaling pathways, 10 Arabidopsis accessions showing different RSA strategies in response to salt stress were selected and studied for their sensitivity to two hormones involved in root growth reduction upon salt stress, ABA and ethylene (1-aminocyclopropane-1-carboxylic acid [ACC]; Achard et al., 2006; Duan et al., 2013). Four-day-old seedlings were transferred to plates supplemented with 1 μm ABA or ACC and the root growth dynamics were described using root-fit. In general, RSA of seedlings grown on 1 μm ABA were severely reduced in their LR elongation, whereas 1 μm ACC treatment mainly affected MR elongation (Fig. 6C). ABA treatment reduced the growth of MRs and average LRs as well as the increase in LR number to a similar extent for all accessions belonging to strategy I. Strategy II accessions showed diverse patterns of ABA responses. Whereas Cape Verde Islands (Cvi) and Vancouver (Van-0) showed significant reduction of growthaLR compared with growthMR, ABA treatment of Niederzenz (Nd-1) seedlings reduced growthMR more severely than growthaLR. Interestingly, two of three accessions (Antwerpen-1 [An-1] and Brunn [Br-0]) belonging to strategy III showed ABA hypersensitivity in their growthaLR and increase#LR. Strategy IV accessions exhibited no significant differences between growthMR and growthaLR in response to ABA, but showed different responses concerning the increase#LR compared with growthaLR. On the contrary, the patterns of RSA reprogramming by ethylene treatment were conserved in all accessions studied (Fig. 6D). growthMR was inhibited more severely than growthaLR in response to ACC treatment in all accessions studied. Nevertheless, some natural variation in the growth inhibition by ethylene was observed in growthaLR (Fig. 5D), with Oystese (Oy-0) and Argentat (Ag-0) being least sensitive to ACC inhibition. Yet within this natural variation, no patterns explaining different RSA strategies could be observed. These results indicate that the different RSA strategies are likely partially attributable to natural variation in ABA responsiveness.
To examine whether different RSA strategies would correspond to salt accumulation in the shoot, the Na+ and K+ contents were measured in root and shoot tissues of 10 accessions grown for 8 d on control plates or plates supplemented with 75 mm NaCl (Supplemental Fig. S6, A and B). Salt stress was observed to shift the ratio of Na+/K+ in both root and shoot tissue (Supplemental Fig. S6B), owing to increased accumulation of Na+ and a slight reduction of K+ in both tissues. The Na+/K+ ratio increased more severely in the shoot than in the root tissue. Interestingly, the relative increase in the Na+/K+ ratio measured in the shoot (Fig. 6E; Supplemental Fig. S6C) was found to be the lowest for the three accessions belonging to strategy III and the highest for strategy IV accessions. Thus, RSA strategies seem to be correlated with ion accumulation in shoot tissue, which is a measure of salinity stress tolerance.
DISCUSSION
Salt stress is known to inhibit plant growth by challenging the osmotic and ionic stress- counteracting capacities of the plant. Plant responses to salt stress involve activation of ion pumps to either restrict Na+ influx into the plant (Salt Overly Sensitive1) or to compartmentalize the excess of Na+ into tissues (High-affinity K+ Transporter1) or vacuoles (vacuolar Na+/H+ exchanger1; Munns and Tester, 2008). The initial responses to salinity stress are traditionally studied on the roots of very young seedlings. This approach was successful in the identification of genes involved in salinity tolerance in forward and reverse genetic approaches. On the other hand, the effect of salinity on more complex root traits besides MR elongation is less clear. Salt was found to induce changes in MRL and LRL, depending on the nutrient conditions (Wang et al., 2009; Zolla et al., 2010; Duan et al., 2013). Although the dynamics of initial LR growth arrest were studied in great detail by Duan et al. (2013), no studies on the complex changes in the entire RSA in response to salinity stress, especially the dynamics of the response on the longer time scale, are available. In this study, we describe key RSA parameters responsive to salt stress and reveal natural variation in growth dynamics within those traits.
Four accessions described to differ in the salinity tolerance-related traits of Na+ leaf accumulation, survival, and germination (Galpaz and Reymond, 2010; Jha et al., 2010; Katori et al., 2010) were tested for differences in their RSA under increasing salt concentrations (Fig. 1). In agreement with the previously published effect of low salinity inducing LR elongation (Zolla et al., 2010), lower concentrations of salt were found to increase the length and the number of LRs. Our results show that this increase is apparent only for Col-0 seedlings. The growth-promoting effect of low salinity could be due to an increase in the osmotic potential of the cells in the elongation zone or enhanced cell cycle activity. The Na+ ions at this range could be rapidly compartmentalized into the vacuoles without reaching the maximum capacity, thereby increasing the turgor within the cells and stimulating cell elongation. A possible effect of salt stress on meristem activity as well as on cell length in different accessions will be explored in future studies. Interestingly, accessions described to have increased salinity tolerance, Bur-0 and Tsu-0 (Rus et al., 2006; Katori et al., 2010), showed a reduction in aLRL in the root region formed after exposure to salt stress even at the lowest concentrations of salt studied. Those results suggest that efficient inhibition of root growth, even at low salt stress levels, could be linked with enhanced salt stress tolerance, similar to drought-induced inhibition of shoot growth (Claeys and Inzé, 2013).
Higher salt concentrations were found to significantly reduce MRL and TRS, in accordance with earlier observations (Wang et al., 2009; Zolla et al., 2010). However, no significant effect on LRD was observed at any concentration studied, conflicting with previous studies in which LRD in salt stress conditions was observed to be either increased, attributed to auxin signaling (Wang et al., 2009), or decreased due to ABA signaling (Zolla et al., 2010). The earlier conclusions concerning LRD could be explained by the different nutrient conditions used in different experiments. The decrease in LRD observed by Zolla et al. (2010) is consistent with a transient effect of salt stress on LRD only in the region of the MR that is formed after transfer (Supplemental Fig. S2) and the partial arrest of LR development at stages V to VI under mild salt stress conditions (McLoughlin et al., 2012). Yet by examining the density of visible LRs in different salt stress concentrations (Fig. 1C) as well as LRD dynamics in time in response to salinity stress (Supplemental Fig. S3), we could not find any effect of salt stress on emerged LRD on a longer time scale. Thus, although the resolution of our phenotyping technique does not allow distinguishing the prebranch sites or primordia and possible effects on these (Moreno-Risueno et al., 2010), we conclude that no significant changes in the LRD of emerged LRs can be observed for Col-0 seedlings in response to salinity.
Recent work by Geng et al. (2013) and Duan et al. (2013) are excellent examples highlighting the importance of studying RSA dynamics. By examining root responses during the first 24 or 72 h of salinity stress, respectively, changes in MR growth and LR emergence at high temporal and spatial resolution were identified and the importance of tissue-specific ABA signaling was revealed. Building further on this work, here we focus on dynamics of the entire RSA in response to salt stress on a lower spatial resolution but larger time scale. We were able to summarize RSA dynamics with quadratic growth functions describing the growth of MR, aLRL, and increase in LR number and combining them into one descriptive model, root-fit (Fig. 2). By comparing the growth rates of individual RSA components across different salt stress conditions as well as the relative effect of salt stress on each component individually (Fig. 3), we identified functional reprogramming of RSA in response to salt stress. Differential reduction of growthMR, increase#LR, and growthaLR in Col-0 seedlings grown on salt-containing media was observed. growthMR was affected most severely, whereas increase#LR and growthaLR showed a less severe reduction by salt stress. The quadratic growth description of growthMR and growthaLR is in good agreement with the accelerating root elongation described by Beemster and Baskin (1998), ascribed to increasing root apical meristem size. Salt stress reduces the MR growth rate by reducing cell cycle activity throughout the root apical meristem (West et al., 2004) and our data collected on a bigger spatial and temporal scale seem to be in good agreement with salt reducing meristem size. Although the observed growth rates of seedlings transferred to saline media in our conditions still exhibit accelerated root growth, although to a lower extent, the growth rates observed by West et al. (2004) followed a linear growth curve. The quadratic growth observed could be ascribed to the longer time scale used in our experimental setup, because cell cycle activity was shown to recover to near control levels at 72 h after exposure to salt stress (West et al., 2004).
Based on the root-fit model, our results provide evidence that salt stress not only retards RSA development, but also reshapes the root architecture. Interestingly, earlier observations indicated an opposite effect: at 100 mm of NaCl, LR growth was observed to be more severely reduced compared with MR growth (Duan et al., 2013). The possible explanation for this discrepancy is the basic medium, as well as the different time scales used. The severe decrease in LR elongation was observed on full-strength Murashige and Skoog (MS) medium, whereas a less severe inhibition of LR growth was observed at one-half-strength MS medium (more similar to our conditions; Duan et al., 2013). In addition, Duan et al. (2013) described LR development for 4 d after salt stress treatment, whereas the LR development in this study was examined up to 8 d after transfer into saline media. By this time, the LRs were observed to surpass the quiescent stage and root growth was largely recovered. Together, these studies highlight the importance of studying RSA responses on different basic media and different time scales to examine the full range of phenotypic plasticity in environmental stress responses (De Smet et al., 2012; Kellermeier et al., 2014).
By studying natural variation in RSA dynamics to salinity stress and the correlations between RSA traits, we found that MR growth and increase in LR number are positively correlated in all conditions, again implying little change in LR patterning due to salt stress exposure. Although correlation between growth rates of MR and average LR was not observed to be significant, the value of the correlation coefficient (r2) changed from positive in control conditions to negative in salt stress conditions, suggesting a trade-off between MR and LR elongation after exposure to salt stress. Clustering of relative changes in individual RSA parameters of all studied accessions enabled us to categorize their natural variation into four distinct RSA strategies differing mostly in their relative average LR growth and increase in LR number (Fig. 5). The majority of the accessions clustered in the strategies with heavily reduced LR elongation (strategies III and IV), few accessions did not show any substantial remodeling of RSA (strategy II) and only a couple of accessions showed an RSA response similar to Col-0, with a higher reduction in growthMR than in growthaLR or increase#LR (strategy I).
The difference in MR and LR elongation in Col-0 under salt stress conditions is mainly accomplished by differential ABA sensitivity of LR and MR (Duan et al., 2013). In addition, growth reduction by salinity has also been ascribed to increased synthesis of ethylene (Achard et al., 2006). The effect of ethylene on RSA development was observed to differ from ABA in terms of relative growth inhibition of MR and LR (Duan et al., 2013). To examine whether the differences in RSA responses to salinity stress are due to natural variation in ABA or ethylene sensitivity, the sensitivity to both phytohormones was examined on 10 accessions exhibiting different RSA responses (Fig. 6, C and D). In general, the observed patterns in ABA and ethylene treatment were in general agreement with previously published data (Duan et al., 2013), because ABA mainly inhibited LR elongation and ethylene treatment severely reduced MR growth. The observed variation in RSA responses to salt could be partially explained by their differences in ABA sensitivity, because two accessions showing a high reduction in aLRL growth on salt stress were observed to be hypersensitive to ABA with respect to aLRL growth. In addition, we identified one accession, Niederzenz, in which ABA treatment inhibited the MR elongation more severely than its aLRL growth. On the other hand, natural variation in ethylene sensitivity was conserved in terms of relative effects on MR and LR elongation, indicating that the cellular synthesis or signaling pathways regulating RSA remodeling through ethylene are more conserved in the accessions studied than those for ABA. The underlying allelic variation will be further explored by forward genetic approaches, focusing on the genetic machinery controlling the difference in ABA sensitivity between MR and LR meristems.
Although the correlation between RSA and salt accumulation or salinity tolerance is underexplored for most plant species, studies in rice (Oryza sativa; Faiyue et al., 2010a, 2010b), suggest an important role for LR formation in those traits. Sodium ions were observed to enter the transpiration stream through the sites of the MR where LRs emerged, suggesting that plants with a reduced LR number would have an advantage in terms of ion exclusion. However, in our experimental setup, the accessions showing a heavy reduction in LR elongation, rather than in LR number increase, showed the smallest increase in Na+/K+ ratio after exposure to salt stress (Fig. 6E). The discrepancy between our results and observations made in rice (Faiyue et al., 2010a, 2010b) could be due to the differential mechanisms involved in interactions between RSA and salinity tolerance in monocot and dicot plant species. Another explanation could be that our experiments were performed in nontranspiring conditions, reducing the transpiration flow and limiting the Na+ influx into the shoot tissue. Nevertheless, our results imply the importance of LR growth inhibition by salinity stress as described by Duan et al. (2013), because the accessions with the lowest increase in Na+/K+ ratio were those that showed the most severe inhibition of LR growth. Although this correlation should be examined in different experimental set ups for more accessions, this is the first study presenting a possible correlation between reduced growth of RSA and enhanced salinity stress tolerance.
In conclusion, our work illustrates how the use of a simple mathematical descriptive model, root-fit, combined with statistical methods, such as Ward linkage clustering, can uncover natural diversity in morphological changes that would normally have been overlooked. Classification into different RSA strategies rather than raw RSA traits themselves could be used for forward and reverse genetic approaches to identify genes underlying alternative RSA responses to salinity as well as other environmental stress factors. Moreover, the genetic plasticity in the RSA response to salt stress suggests that this trait could be under environmental selection, showing different acclimation strategies that are beneficial in specific natural environments.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accessions (Supplemental Table S2) were obtained from the European Arabidopsis Stock Centre (http://arabidopsis.info/). The 31 accessions were chosen based on the availability of recombinant inbred lines (Institut National de la Recherche Agronomique). Seeds used for phenotyping of RSA came from plants propagated together under long-day conditions (21°C, 70% humidity, 16-h-light/8-h-dark cycle), with 8-week-long vernalization (between 4°C and 8°C, 70% humidity, 16-h-light/8-h-dark cycle) starting at the third week after germination to ensure flowering of all accessions. Seeds used for the experiments were between 2 months and 1 year old.
Seeds were surface sterilized in a desiccator (1.6 L) using 20 mL of household bleach and 600 μL of 40% (v/v) HCl for 3 h. The seeds were stratified in 0.1% (w/v) agar at 4°C in the dark for 48 h and were sown on square petri dishes containing 50 mL of control growth medium consisting of one-half-strength MS medium, 0.5% (w/v) Suc, and 0.1% (w/v) MES. Monohydrate and 1% daishin agar, pH 5.8 (KOH), dried for 1 h in a laminar flow. Plates were placed vertically at a 70°angle. Seeds were germinated under long-day conditions (21°C, 70% humidity, 16-h-light/8-h-dark cycle). Four-day-old seedlings were transferred to square petri dishes containing control medium supplemented with NaCl as indicated per experiment. Each plate contained four seedlings of two genotypes (two seedlings per genotype). Plates were placed in the growth chamber following a random design.
Dose-Dependent Response of RSA Changes to Salt Stress
Plates were scanned 11 d after germination using an Epson Perfection V700 Scanner at a resolution of 200 dots per inch (dpi). Root phenotypes of four different accessions (Col-0, Bur-0, Tsu-0, and C24) on media supplemented with 0, 25, 50, 75, 100, 125, and 150 mm NaCl were quantified using EZ-Rhizo software (Armengaud et al., 2009). In addition to the parameters measured by EZ-Rhizo, the LRL and aLRL (LRL divided by the number of LRs) was calculated for each root and LR number, LRL, and aLRL were calculated for the regions above and below the point of transfer (regions A and B, respectively) based on the individual positions of LRs. All data were cleared from outliers. Statistical analysis was performed in SPSS software using one-way ANOVA with Tukey’s post hoc test for significance.
Descriptive Model of Salt-Induced Changes in RSA in Col-0
Plates were scanned 4, 6, 8, 10, and 12 d after germination using an Epson Perfection V700 Scanner at 200 dpi resolution. Root phenotypes on media supplemented with 0, 75, and 125 mm NaCl were quantified using EZ-Rhizo software (Armengaud et al., 2009). Root phenotypes of 12-d-old plants grown in control conditions were not quantified because the roots were too long and entangled. Data were collected from plants grown in six different experiments with four individual seedlings per salt stress concentration per experiment (n = 24 total). All data were cleared from outliers and statistical analysis was performed with Excel. The average values of MRL, aLRL, and number of LRs at 4, 6, 8, 10, and 12 d after germination were calculated for plants grown on media supplemented with different salt concentrations (n = 24).
The change in MRL was described with a quadratic function for all salt concentrations as follows: MRL = MRLSTART + growthMR × t2, where MRLSTART is the MRL 4 d after germination (same for all concentrations), t is time in days after transfer, and growthMR is a growth rate (cm × d−2), specific for the growth conditions. The dynamics in aLRL and number of LRs were described with quadratic functions for all salt concentrations as follows: noLR = increase#LR × t2 and LRL = growthaLRL × t2, where noLR is the number of LRs, t is time in days after transfer, and increase#LR and growthaLRL are growth rates for the number of LRs (number of LRs × d−2) and aLRL (cm × d−2) respectively. The growth rates for quadratic growth models were calculated on square root-transformed average values measured in a time course experiment, thereby making the growth a linear function. Linear function was calculated by using a linest function in Excel. The calculated values for linear function were subsequently squared to fit the quadratic growth of individual parameters. TRS was calculated from the three values: TRS = MRL + noLR × aLRL.
We also constructed an alternative model, which used different formulas for calculating LR increase and elongation in the MR portion above and below the point of transfer (Supplemental Fig. S2). In the alternative model, MR growth was defined as described above. The average LR elongation above the point of transfer was modeled as the dynamics in aLRL described above but fitted on the average LR growth data from region A. The average LR growth below the transfer point was modeled using the same formula, although the delay of 1 d (t − 1) was introduced as those LRs developed later. The same was done for the increase in LR number below the transfer point. Modeling of the LR increase above the transfer point was performed using a square-root function, because the length of the region A did not increase. Growth rates were calculated by squaring the raw phenotypic data, thereby making the increase in LRs in region A a linear function and fitting a linear growth function using a linest function in Excel. TRS was calculated from the five values: TRS = MRL + noLRA × aLRLA + noLRB × aLRLB.
In addition, the increase in LRD per entire MR region as well as per region above and below the transfer point (regions A and B) was calculated by using a linear growth function (Supplemental Fig. S3). The growth rates were calculated by fitting a linear growth function on the average of LRD values from individual days using a linest function in Excel.
Fit of the descriptive growth models was tested on the average of raw data collected by calculating the coefficient of determination (r2). The r2 values for individual RSA parameters are presented in Supplemental Table S1.
Describing Natural Variation in RSA Responses to Salt and Categorizing Accessions into Different Groups
The seedlings of 31 Arabidopsis accessions (Supplemental Table S2) were germinated on standard media and were transferred to plates supplemented with 0, 75, or 125 mm NaCl 4 d after germination. The plates were scanned 4, 6, 8, 10, and 12 d after germination using an Epson Perfection V700 Scanner at 200 dpi resolution. Root phenotypes on media supplemented with 0, 75, and 125 mm NaCl were quantified using EZ-Rhizo software (Armengaud et al., 2009). The descriptive models for each accession were calculated as described above for Col-0 on data collected from four individual seedlings per accession per condition. The fit of the descriptive models with the data was tested on average values per RSA trait per condition by calculating the coefficient of determination (r2). The growth rates calculated were scaled for MR growth, by dividing the relative growth rates of average LR and the number of LRs by MR growth rate, and were used for hierarchical cluster analysis performed using the Xlstat add-in for Excel by using Ward linkage clustering, as described in Kellermeier et al. (2013).
Examining ABA and Ethylene Sensitivity of Selected Accessions with Different Salt Stress-Induced RSA Response Types
The seedlings of 10 Arabidopsis accessions exhibiting different RSA responses to salt stress were germinated on standard media and were transferred to standard plates supplemented with 1 μm ABA or 1 μm ACC or to control plates at 4 d after germination. Root phenotypes on media supplemented with 1 μm ACC and control plates were quantified between 4 d and 10 d after germination, whereas RSA of plants grown on 1 μm ABA was quantified between 4 d and 12 d after germination using EZ-Rhizo software (Armengaud et al., 2009). The descriptive models for each accession were calculated as described above for Col-0 on data collected from 16 individual seedlings per accession per condition.
Salt Accumulation in Root and Shoot Tissue of Accessions with Different Salt Stress-Induced RSA Response Types
The seedlings of 10 Arabidopsis accessions exhibiting different RSA responses to salt stress were germinated on standard media and were transferred to standard plates supplemented with 0 or 75 mm NaCl at 4 d after germination. After 8 d, 10 plants of the same genotype were pooled together. The seedlings were divided into shoot and root, dried, and analyzed for Na+ and K+ concentrations using an atomic absorption spectrometer (Aanalyst 200; PerkinElmer) as described by Wei et al. (2014). The average of four biological replicas was used for the ion concentration calculations per gram of dry weight. All data were cleared from outliers and statistical analysis was performed in Excel. SPSS was used for one-way ANOVA with Tukey’s post hoc test for significance.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Salt treatment has a different effect on emerged LR number above and below the transfer point but does not affect LR density, straightness, or MR zonation of 11-d-old seedlings of Col-0, Tsu-0, C24, and Bur-0 Arabidopsis accessions.
Supplemental Figure S2. Dynamics of emerged LRD development calculated per entire MR as well as for the region above and below the transfer point.
Supplemental Figure S3. Describing RSA dynamics of LR development above and below the transfer point yields similar predictions of TRS as the simplified (root-fit) model.
Supplemental Figure S4. Reduction in MR, LR number, and average LR length by salt stress for the individual Arabidopsis accessions clustered into four distinct responses.
Supplemental Figure S5. Decrease in overall root and shoot mass does not vary between four different strategies identified.
Supplemental Figure S6. Accumulation of Na+ and K+ in root and shoot tissue of 10 individual Arabidopsis accessions exhibiting different RSA strategies grown in standard and mild salt stress conditions.
Supplemental Table S1. Values of RSA growth parameters and r2 values as used in the descriptive growth model.
Supplemental Table S2. List of Arabidopsis accessions used for screening salt-induced changes in RSA.
Supplemental Table S3. Growth parameters for individual RSA traits calculated per accessions.
Supplemental Table S4. Correlations between root-fit components estimated for 31 Arabidopsis accessions studied between different salt stress concentrations.
Supplementary Material
Acknowledgments
We thank Joost Keurentjes (Wageningen University) for providing seeds of Arabidopsis accessions, and Bondien Dekker and Marthe Noordhoorn Boelen (University of Amsterdam) for practical assistance during BSc student internships.
Glossary
- RSA
root system architecture
- MR
main root
- LR
lateral root
- ABA
abscisic acid
- LRD
lateral root density
- Col-0
ecotype Columbia of Arabidopsis
- LRL
lateral root length
- aLRL
average lateral root length
- MRL
main root length
- TRS
total root size
- growthMRL
increase in main root length
- increase#LR
increase in the number of lateral roots
- growthaLRL
the increase in the average lateral root length
- growthaLR
average lateral root elongation
- growthMR
main root elongation
- ACC
1-aminocyclopropane-1-carboxylic acid
- MS
Murashige and Skoog
- dpi
dots per inch
Footnotes
This work was supported by the Netherlands Organisation for Scientific Research (STW Learning from Nature project no. 10987).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Genschik P. (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. (2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 [DOI] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI. (1998) Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Pata M, Nacry P, Doumas P, Rossignol M. (2003) Effects of phosphate availability on the root system architecture: largescale analysis of the natural variation between Arabidopsis accessions. Plant Cell Environ 26: 1839–1850 [Google Scholar]
- Claeys H, Inzé D. (2013) The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol 162: 1768–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X. (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12: 474–481 [DOI] [PubMed] [Google Scholar]
- De Smet I, White PJ, Bengough AG, Dupuy L, Parizot B, Casimiro I, Heidstra R, Laskowski M, Lepetit M, Hochholdinger F, et al. (2012) Analyzing lateral root development: how to move forward. Plant Cell 24: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G, Van Isterdael G, Beeckman T, De Smet I. (2010) The roots of a new green revolution. Trends Plant Sci 15: 600–607 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR. (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiyue B, Al-Azzawi MJ, Flowers TJ. (2010a) The role of lateral roots in bypass flow in rice (Oryza sativa L.). Plant Cell Environ 33: 702–716 [DOI] [PubMed] [Google Scholar]
- Faiyue B, Vijayalakshmi C, Nawaz S, Nagato Y, Taketa S, Ichii M, Al-Azzawi MJ, Flowers TJ. (2010b) Studies on sodium bypass flow in lateral rootless mutants lrt1 and lrt2, and crown rootless mutant crl1 of rice (Oryza sativa L.). Plant Cell Environ 33: 687–701 [DOI] [PubMed] [Google Scholar]
- Galpaz N, Reymond M. (2010) Natural variation in Arabidopsis thaliana revealed a genetic network controlling germination under salt stress. PLoS ONE 5: e15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Julkowska MM, Darwish E, Gandullo J, Korver RA, Brunoud G, Haring MA, Munnik T, Vernoux T, Testerink C. (2013) Halotropism is a response of plant roots to avoid a saline environment. Curr Biol 23: 2044–2050 [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Testerink C. (2011) Salt stress signals shape the plant root. Curr Opin Plant Biol 14: 296–302 [DOI] [PubMed] [Google Scholar]
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PMY, Tham C, Duan L, Dinneny JR. (2013) A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25: 2132–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Banta JA, Katari MS, Hulsmans J, Chen L, Ristova D, Tranchina D, Purugganan MD, Coruzzi GM, Birnbaum KD. (2013) Plasticity regulators modulate specific root traits in discrete nitrogen environments. PLoS Genet 9: e1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RFH, Friedel S, von Wirén N. (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujas B, Alonso-Blanco C, Hardtke CS. (2012) Natural Arabidopsis brx loss-of-function alleles confer root adaptation to acidic soil. Curr Biol 22: 1962–1968 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM. (2013) Sodium (Na+) homeostasis and salt tolerance of plants. Environ Exp Bot 92: 19–31 [Google Scholar]
- Jha D, Shirley N, Tester M, Roy SJ. (2010) Variation in salinity tolerance and shoot sodium accumulation in Arabidopsis ecotypes linked to differences in the natural expression levels of transporters involved in sodium transport. Plant Cell Environ 33: 793–804 [DOI] [PubMed] [Google Scholar]
- Katori T, Ikeda A, Iuchi S, Kobayashi M, Shinozaki K, Maehashi K, Sakata Y, Tanaka S, Taji T. (2010) Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J Exp Bot 61: 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A. (2014) Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Chardon F, Amtmann A. (2013) Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol 161: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM. (2014) Sodium in plants: perception, signalling, and regulation of sodium fluxes. J Exp Bot 65: 849–858 [DOI] [PubMed] [Google Scholar]
- McLoughlin F, Galvan-Ampudia CS, Julkowska MM, Caarls L, van der Does D, Laurière C, Munnik T, Haring MA, Testerink C. (2012) The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J 72: 436–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijón M, Satbhai SB, Tsuchimatsu T, Busch W. (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat Genet 46: 77–81 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, et al. (2012) Wheat grain yield on saline soils is improved by an ancestral Na⁺ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Testerink C. (2014) The art of being flexible: how to escape from shade, salt and drought. Plant Physiol 166: 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U, Cibrian-Jaramillo A, Ristova D, Banta JA, Gifford ML, Fan AH, Zhou RW, Kim GJ, Krouk G, Birnbaum KD, et al. (2013) Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc Natl Acad Sci USA 110: 15133–15138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SJ, Huang W, Wang XJ, Evrard A, Schmöckel SM, Zafar ZU, Tester M. (2013) A novel protein kinase involved in Na(+) exclusion revealed from positional cloning. Plant Cell Environ 36: 553–568 [DOI] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trontin C, Tisné S, Bach L, Loudet O. (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol 14: 225–231 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li K, Li X. (2009) Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. J Plant Physiol 166: 1637–1645 [DOI] [PubMed] [Google Scholar]
- Wei Z, Julkowska MM, Laloë JO, Hartman Y, de Boer GJ, Michelmore RW, van Tienderen PH, Testerink C, Schranz ME. (2014) A mixed-model QTL analysis for salt tolerance in seedlings of crop-wild hybrids of lettuce. Mol Breed 34: 1389–1400 [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G, Inzé D, Beemster GTS. (2004) Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol 135: 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla G, Heimer YM, Barak S. (2010) Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J Exp Bot 61: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.