A drought stress-inducible receptor-like kinase is important for plant growth and grain yield in rice.
Abstract
Rice (Oryza sativa) is the primary food source for more than one-half of the world’s population. Because rice cultivation is dependent on water availability, drought during flowering severely affects grain yield. Here, we show that the function of a drought-inducible receptor-like cytoplasmic kinase, named GROWTH UNDER DROUGHT KINASE (GUDK), is required for grain yield under drought and well-watered conditions. Loss-of-function gudk mutant lines exhibit sensitivity to salinity, osmotic stress, and abscisic acid treatment at the seedling stage, and a reduction in photosynthesis and plant biomass under controlled drought stress at the vegetative stage. The gudk mutants interestingly showed a significant reduction in grain yield, both under normal well-watered conditions and under drought stress at the reproductive stage. Phosphoproteome profiling of the mutant followed by in vitro assays identified the transcription factor APETALA2/ETHYLENE RESPONSE FACTOR OsAP37 as a phosphorylation target of GUDK. The involvement of OsAP37 in regulating grain yield under drought through activation of several stress genes was previously shown. Our transactivation assays confirmed that GUDK is required for activation of stress genes by OsAP37. We propose that GUDK mediates drought stress signaling through phosphorylation and activation of OsAP37, resulting in transcriptional activation of stress-regulated genes, which impart tolerance and improve yield under drought. Our study reveals insights around drought stress signaling mediated by receptor-like cytoplasmic kinases, and also identifies a primary regulator of grain yield in rice that offers the opportunity to improve and stabilize rice grain yield under normal and drought stress conditions.
The contemporary climate and increasing demand for limited fresh water threatens agriculture in the future. Rice (Oryza sativa), in particular, is very sensitive to even milder water stress both at vegetative and reproductive stages (Centritto et al., 2009). In Asia alone, water stress affected nearly 23 million hectares of rice growing area (Serraj et al., 2008). As an important food crop that feeds more than one-half of the world’s population, unstable rice production due to recurring drought can have potential global socioeconomic impact. In the face of these challenges, enhanced rice yield under normal as well as stress conditions is an ideal trait that will have a huge impact on rice productivity.
Plants have evolved several morphophysiological and biochemical strategies to cope with environmental stress (Umezawa et al., 2006). These adaptive strategies are regulated by an intricate signaling network and orchestrated stress-responsive gene expression (Shinozaki and Yamaguchi-Shinozaki, 2007). The components of stress signaling and gene regulation have been widely studied with an aim to improve crop productivity under stressful environments (Baena-González et al., 2007). There has recently been an emphasis on drought transcriptome analysis in rice to identify candidate genes that can contribute to drought tolerance (Lenka et al., 2011; Cal et al., 2013; Minh-Thu et al., 2013). Reverse genetics approaches, such as transfer DNA (T-DNA) or transposon tagging resulting in the complete loss of gene function, have allowed major progress to be made in functional analysis of genes and determine traits contributing to drought tolerance (Zhang et al., 2013; Zhu and Xiong, 2013).
In plants, the activation of protein phosphorylation cascades is considered a key factor in early stress response. Under drought, several protein kinases have been described as signal transducers (Marshall et al., 2012). In particular, plasma membrane-anchored receptor-like kinases (RLKs) are known for early recognition of external signals and translation into cellular responses (Xiong et al., 2002). Structurally, RLKs are characterized by having an extracellular domain, a transmembrane domain, and an intracellular kinase domain (Torii, 2000). Mechanistically, environmental stimuli are sensed by the extracellular domain of RLKs, leading to homodimerization or heterodimerization followed by autophosphorylation of the cytoplasmic kinase domain and subsequent activation of downstream signaling components by transphosphorylation (Morris and Walker, 2003). However, there are plant-specific RLKs without an extracellular domain and these RLKs contain only the transmembrane domain with an intracellular kinase domain or only an intracellular kinase domain. These RLKs are referred to as receptor-like cytoplasmic kinases (RLCKs; Shiu and Bleecker, 2001, 2003; Jurca et al., 2008; Vij et al., 2008). Some RLCKs were recently shown to interact with RLKs to form a receptor complex and mediate signal transduction (Murase et al., 2004; Veronese et al., 2006; Tanaka et al., 2012). Approximately 200 genes encoding RLCKs have been reported in the Arabidopsis (Arabidopsis thaliana) genome (Shiu and Bleecker, 2003; Jurca et al., 2008). In the rice genome, 379 RLCKs have been identified, 82 of which are differentially expressed under abiotic stresses (Vij et al., 2008). However, only a few RLCKs have been functionally characterized to be involved in abiotic stress response. In Arabidopsis, abscisic acid (ABA)- and osmotic stress-inducible receptor-like cytoplasmic kinase1 was shown to negatively regulate abiotic stress signaling. In addition, ABA- and osmotic stress-inducible receptor-like cytoplasmic kinase1 interacts with cysteine-rich receptor-like kinase36 forming a receptor complex leading to abiotic stress signal transduction (Tanaka et al., 2012). A calcium-binding receptor-like cytoplasmic kinase identified from Glycine soja improved tolerance of Arabidopsis plants to high salinity and ABA with increased expression of stress-responsive genes (Yang et al., 2010a). Arabidopsis calmodulin-binding receptor-like cytoplasmic kinase1 was suggested to have a role in stress signaling (Yang et al., 2010b). The phosphorus-starvation tolerance1 gene from traditional rice (aus-type variety Kasalath) improved root growth and grain yield of rice in phosphate-deficient soil with constitutive induction of drought stress-responsive genes (Gamuyao et al., 2012). Another RLCK from rice (OsRLCK253) was suggested to interact with stress-associated proteins (OsSAP1/11) and activate stress response. Overexpression of OsRLCK253 in Arabidopsis improved tolerance to drought and salt stress (Giri et al., 2011).
In this study, we identified a RLCK gene GROWTH UNDER DROUGHT KINASE (GUDK) from rice and elucidated the signaling pathway controlled by it under drought. GUDK is drought inducible and its loss of function resulted in the reduction of plant growth under vegetative drought and grain yield under reproductive drought. GUDK transphosphorylates the transcription factor OsAP37, which has been shown by overexpression studies to be important for yield under drought (Kim and Kim, 2009; Oh et al., 2009). Here we show that activation of OsAP37-inducible stress genes requires GUDK function.
RESULTS
GUDK Is a Drought-Inducible Kinase
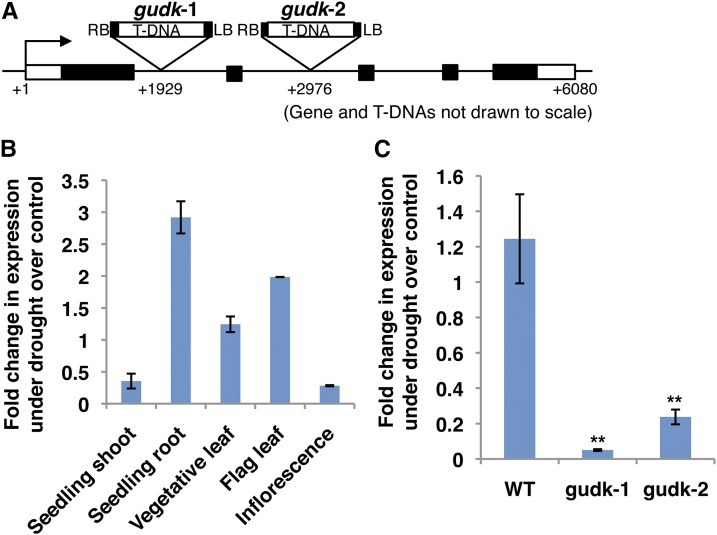
In our previous study, drought transcriptome analysis of rice (O. sativa ssp. japonica ‘Nipponbare’) at the seedling, vegetative, and reproductive stages was used to mine a Rice Environmental Coexpression Network (Ambavaram et al., 2011) and derive subnetworks of drought transcriptional clusters enriched for drought stress-responsive genes (https://plantstress-pereira.uark.edu/RECoN2/). From these drought transcriptional clusters, genes with annotated regulatory functions were selected for further functional characterization through reverse genetics analysis using publicly available rice knockout mutant resources under several abiotic stresses. A kinase gene, designated as GUDK (LOC_Os03g08170), that exhibited the sensitive phenotype under several abiotic stresses was selected to characterize its role under drought stress. GUDK, with a gene size of 6,080 nucleotides and deduced protein length of 425 amino acids, showed 64% identity with an Arabidopsis uncharacterized protein kinase superfamily protein (At1g61590). The structural predictions using the deduced amino acid sequence showed RLCK features with only an intracellular kinase domain and no transmembrane or extracellular domains. Two independent T-DNA insertion mutant lines (gudk-1 and gudk-2, termed as mutant lines) identified for the gene were found to have both insertions in the intronic region (Fig. 1A). To examine the expression of GUDK at seedling, vegetative, and reproductive stages, drought stress was imposed on 7-d-old, 35-d-old, and reproductive R3-stage plants, respectively, until wilting, and a set of control plants was maintained at flooded conditions for all stages. Quantitative PCR (qPCR) analysis at different growth stages of wild-type rice plants revealed drought induction of GUDK expression in the seedling root, vegetative leaf, and flag leaf with 3-fold, 1.5-fold, and 2-fold increases in transcript levels compared with respective controls maintained at flooded conditions (Fig. 1B). Although variation in the expression pattern of rice RLCKs has been observed under several abiotic stresses (Vij et al., 2008), the regulatory nature and phosphorylation function of GUDK suggested that a small increase in transcript levels could be sufficient to mediate a drought response function. Expression of GUDK was examined in the mutant lines and wild-type plants in leaf tissue under drought stress, showing no detectable GUDK transcripts and verifying a loss of function of GUDK in both the mutant lines (Fig. 1C).
Figure 1.
Expression of GUDK in wild-type and gudk mutant lines. A, Schematic diagram of the GUDK gene (LOC_Os03g08170) showing two independent T-DNA insertions (gudk-1 and gudk-2 insertion mutant lines of GUDK). B, qPCR analysis of GUDK at different growth stages of wild-type plants under drought stress. Drought stress was imposed on wild-type seedlings as well as vegetative-stage and reproductive-stage plants at 7 d, 35 d, and the R3 stage, respectively, and samples were collected upon wilting. A set of plants was also maintained at flooded conditions at all stages as controls. C, qPCR analysis of GUDK in wild-type and mutant line leaf tissue under drought stress. Drought stress was imposed on 35-d-old wild-type and gudk mutant lines (gudk-1 and gudk-2, two independent T-DNA insertion mutant lines of the GUDK gene) and samples were collected upon wilting. A set of plants was also maintained at flooded conditions as controls. Gene expression was normalized with reference to Ubiquitin and was expressed as the fold change in expression under drought over the control samples. The values are the mean ± se of three biological replicates. Double asterisks indicate significance at P ≤ 0.01 compared with the wild type as analyzed by the Student’s t test. WT, Wild type. [See online article for color version of this figure.]
Loss of Function of GUDK Increases Sensitivity of Rice Seedlings to Abiotic Stress
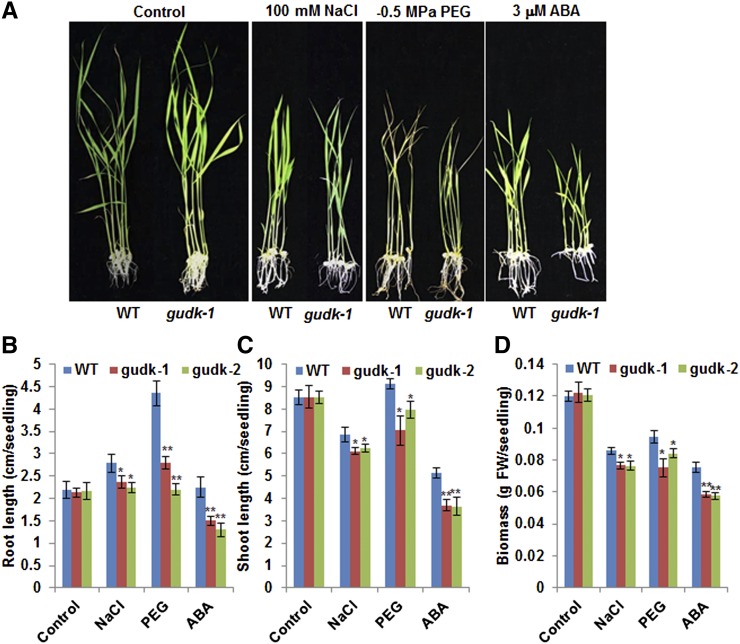
To test the growth performance of the mutant lines to salinity, osmotic stress, and ABA treatments, seedlings were transferred to growth media containing 100 mm NaCl, −0.5 MPa polyethylene glycol (PEG) 6,000, and 3 μm ABA, respectively. After 7 d of stress, both mutant lines showed poorer growth phenotypes (Fig. 2A), with a significant reduction in root length, shoot length, and biomass under salinity, PEG, and ABA (Fig. 2, B–D). The highest reduction in the root length of mutant lines was seen under PEG (Fig. 2B) compared with wild-type plants in which the root length was higher than that observed under control conditions. Under control conditions, there was no significant difference in seedling morphology of both mutant lines compared with the wild type, suggesting that the observed stress response was attributable to loss of function of GUDK. Taken together, the results indicate that loss of function of GUDK resulted in increased sensitivity of seedlings to salinity, osmotic stress, and ABA treatment.
Figure 2.
Abiotic stress sensitivity of gudk mutant lines at the seedling stage. Dehusked seeds derived from wild-type and gudk (gudk-1 and gudk-2) mutant plants were pregerminated on Whatman filter paper wetted with water in petri dishes. Two days after pregermination, equal-sized seedlings were transferred onto growth medium supplemented with 100 mm NaCl, −0.5 MPa PEG 6,000, and 3 μm ABA. A, Phenotype of the wild-type and gudk-1 mutant line grown for 7 d under salinity, osmotic stress, and ABA treatments. B to D, Root length (B), shoot length (C), and biomass (fresh weight; D) of wild-type and mutant lines grown for 7 d under salinity, osmotic stress, and ABA treatments. Values are the mean ± se (n = 10). Single and double asterisks indicate significance at P ≤ 0.05 and P ≤ 0.01, respectively, compared with the wild type as analyzed by the Student’s t test. FW, Fresh weight; WT, wild type. [See online article for color version of this figure.]
Drought Tolerance at the Vegetative Stage Requires GUDK Function
To test the involvement of GUDK in drought tolerance, both mutant lines and wild-type plants were tested for their drought stress response at the vegetative stage. After 12 d of withholding water, most of the leaves of mutant plants were brittle and dry with only a few rolled young leaves, whereas most of the leaves in the wild-type plants were rolled with a few brittle and dry leaves (Fig. 3A). A week after rewatering, the survival rate was measured by counting the number of plants with at least one fully expanded leaf. Approximately 57% of the wild-type plants recovered but only 26% to 33% recovered in the mutant lines (Fig. 3).
Figure 3.
Drought stress sensitivity of gudk mutant lines at the seedling stage. Wild-type and gudk-1 and gudk-2 mutant seedlings were grown for 20 d under well-watered conditions by maintaining 15 equal-sized seedlings per pot. Irrigation was withheld on the 20th day for 12 d, followed by rewatering for a week. A, Phenotype of drought-stressed plants followed by recovery. Photographs were taken before stress (0 d), at the end of the stress period (12 d), and a week after recovery (7 d recovery). B, Seedling survival. A week after recovery, the number of seedlings with at least one fully expanded leaf was counted and the percentage of survival was calculated. Values are the mean ± se of three biological replicates (n = 15). Double asterisks indicate significance at P ≤ 0.01 compared with the wild type as analyzed by the Student’s t test. WT, Wild type. [See online article for color version of this figure.]
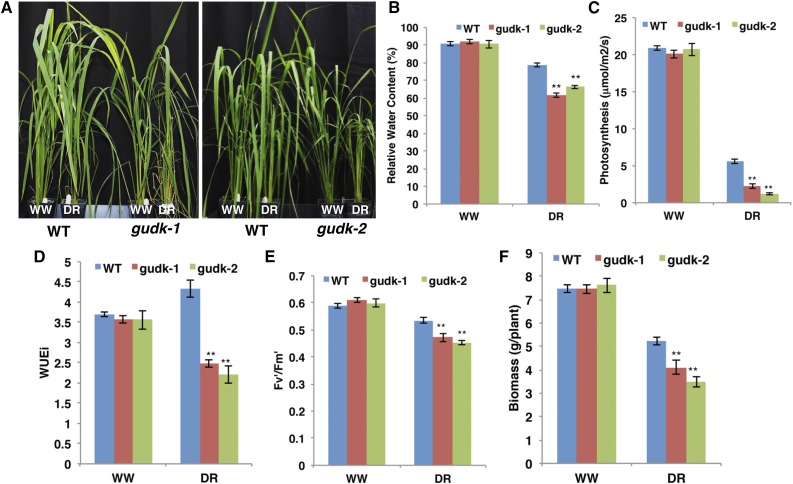
Detailed analysis of drought response was done by controlled drought stress treatment, maintaining plants at 40% field capacity (FC), which revealed the high sensitivity of the gudk mutant lines (Fig. 4A). At 10 d of controlled drought, mutant lines showed a reduction in relative water content (RWC) of 12% to 17% compared with drought-stressed wild-type plants (Fig. 4B). Similarly, mutant lines showed significant reductions in the photosynthesis rate (Fig. 4C), instantaneous water use efficiency (WUEi; Fig. 4D), and efficiency of PSII measured in light-adapted leaves (Fig. 4E). The reduction in photosynthesis parameters translated to a significant reduction in biomass (Fig. 4F). There was a 1.1- to 1.8-g reduction in the biomass of mutant lines under drought stress conditions compared with wild-type plants, supporting the role of GUDK in drought tolerance for growth of rice plants at the vegetative stage.
Figure 4.
Sensitivity of gudk mutant lines to vegetative drought. Drought stress was applied to 45-d-old wild-type and gudk mutant plants by maintaining a set of plants at 40% FC, whereas the well-watered plants were maintained at 100% FC for 10 d. During the stress period, pots were weighed daily at a fixed time of the day, and water lost was replenished to maintain the required FC. At the end of the stress period, growth and physiological performance were measured. A, Phenotype of mutant lines maintained at well-watered and drought stress conditions for 10 d. B, RWC of leaves at the end of 10 d of drought stress. C to E, Assimilation rate (C), WUEi (D), and efficiency of PSII (E) in light-adapted leaves, measured at 10 d of drought stress. F, Biomass (dry weight) measured at 10 d of drought stress. Values are the means ± se (n = 10). Double asterisks indicate significance at P ≤ 0.01 compared with the wild type as analyzed by the Student’s t test. DR, Drought stress; DW, dry weight; Fv′/Fm′ , maximum photochemical efficiency of PSII during steady-state illumination; WT, wild type; WW, well watered. [See online article for color version of this figure.]
Rice Grain Yield Production Requires GUDK Function
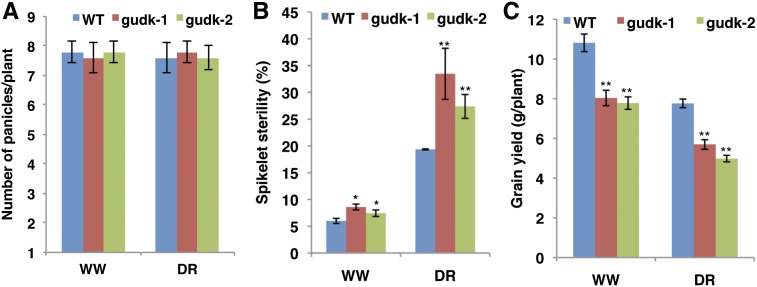
To test the role of GUDK under drought at the reproductive stage, water was withheld at the preanthesis stage until leaf rolling occurred, followed by maintaining the plants under well-watered conditions until physiological maturity. There was no significant difference in the panicle number between mutant lines and wild-type plants under both the well-watered and drought stress conditions (Fig. 5A). The spikelet number per panicle was also not significantly different between wild-type and mutant plants under both well-watered and drought stress conditions. However, a marginal reduction in the spikelet number per panicle was observed under drought stress both in wild-type and mutant plants (Supplemental Fig. S1). Under drought stress, the mutant lines showed a significant 8% to 14% increase in spikelet sterility over wild-type plants (Fig. 5B). A significant reduction in spikelet sterility was also observed under well-watered treatments of mutant lines compared with wild-type plants (Fig. 5B). Most importantly, grain yield of mutant lines was reduced under well-watered as well as drought stress conditions (Fig. 5C). Under well-watered conditions, there was 2.8 to 3.1 g (25%–28%) less yield in mutant lines compared with the wild type. Under drought, the yield reduction in mutant lines was 2.1 to 2.8 g compared with wild-type plants. These results clearly indicate that GUDK is important for grain yield production not only under drought but also under well-watered conditions.
Figure 5.
Yield reduction in mutant lines under drought and well-watered conditions. Drought stress was applied to both wild-type and gudk insertion mutant lines at the R3 stage by withholding irrigation until all of the leaves rolled and wilted, followed by rewatering and maintaining the plants at well-watered conditions until physiological maturity. A set of plants was maintained under flooded conditions as well-watered plants. At physiological maturity, panicles were harvested individually and the yield components were evaluated. A to C, Panicle number (A), spikelet sterility (expressed in percentages; B), and grain yield measured at physiologically maturity (C). Values are the means ± se (n = 6). Single and double asterisks indicate significance at P ≤ 0.05 and P ≤ 0.01, respectively, compared with the wild type as analyzed by the Student’s t test. DR, Drought stress; WT, wild type; WW, well watered. [See online article for color version of this figure.]
GUDK Is a Dual-Specificity, Autophosphorylating Protein Kinase
To determine the biochemical activity and substrate specificity of GUDK, kinase assays were performed with the GUDK-glutathione S-transferase (GST) fusion protein, as well as with synthetic kinase substrates and inhibitors. Both substrates, Arg-Arg-Sarcoma (RR-SRC; Arg-Arg-Leu-Ile-Glu-Asp-Ala-Glu-Tyr-Ala-Ala-Arg-Gly, a Tyr-specific kinase substrate) and casein (a Ser/Thr-specific protein kinase substrate), were found to be substrates of GUDK (Supplemental Fig. S2A). The highest activity was found at an RR-SRC concentration of 0.3 μm and a casein concentration of 0.4 μm. GUDK had no effect on histone and myelin basic protein, the other two Ser/Thr-specific protein kinase substrates used in the assay (Supplemental Fig. S2A). Furthermore, genistein and AG1478 (triphostin; Tyr kinase inhibitors) and staurosporine (a protein kinase C inhibitor) inhibited GUDK activity by 30% to 67% (Supplemental Fig. S2B), whereas 3-(4-chlorophenyl)-1-(1,1-dimethylethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine (PP2 a Src-family kinase inhibitor) had no effect on GUDK activity (Supplemental Fig. S2B). Kinase assays using RR-SRC as the substrate and different divalent cations suggested the absolute requirement of Mg2+ for GUDK activity with an optimum concentration of 8 mm (Supplemental Fig. S2C). GUDK showed a marginally higher activity at 8 mm Ca2+, but the kinase activity was completely independent of Mn2+. Moreover, the assay with GUDK alone showed phosphorylation activity, suggesting that GUDK has the ability to autophosphorylate (Supplemental Fig. S3A), with the highest autophosphorylation activity found after 30 min. Immunoblot analysis using a phospho-Tyr antibody confirmed the autophosphorylation of GUDK (Supplemental Fig. S3B). The autophosphorylation activity was Mg2+ dependent, with Ca2+ also having a marginal effect on GUDK autophosphorylation activity although Mn2+ had no effect (Supplemental Fig. S3, C and D). Taken together, these results suggest that GUDK is a Mg2+-dependent, dual-specificity, autophosphorylating kinase.
Phosphoproteome Analysis and in Vitro Assays Identify OsAP37 as a Transphosphorylation Target of GUDK
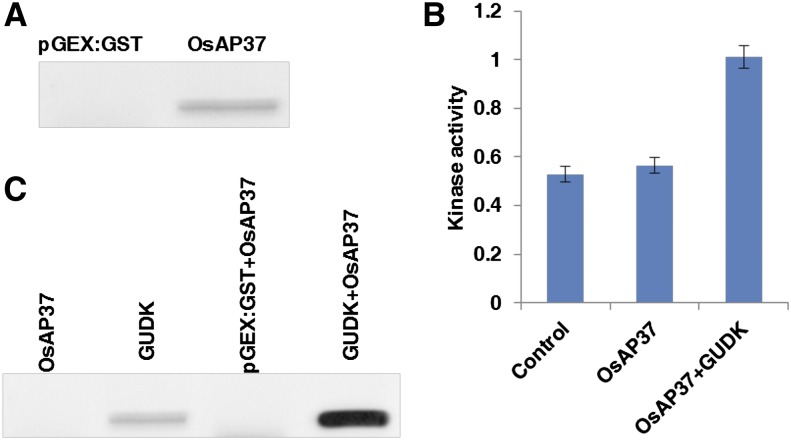
To gain insight into how GUDK modulates plant response to drought, a phosphoproteomic approach was used to identify phosphorylation targets (Supplemental Fig. S4). The total proteins isolated from the wild-type and the gudk-1 mutant lines under well-watered and drought stress conditions, constituting two biological replicates per sample, were extracted and enriched for phosphoproteins by passing through Pierce Graphite Spin columns and were visualized by the ProQ Diamond Phosphoprotein Gel Stain on SDS-PAGE. The enriched phosphoproteins were tryptic digested (in solution) and separated by liquid chromatography (LC) coupled to tandem mass spectrometry (MS/MS) for peptide identification. The data files containing MS/MS spectra were searched against the rice genome database using the Mascot search engine (http://www.matrixscience.com/) to identify the peptides. From the acquired data, only peptides meeting a 5% false discovery rate cutoff (Mascot score > 40; P < 0.05) and observed in both of the replicates of each sample were included in our analysis. Furthermore, proteins that were present in the wild-type samples under well-watered or drought stress conditions and were absent in mutant samples were selected for the in vitro phosphorylation assay (Table I). To study the interaction of GUDK with all 14 putative target proteins, GST pull-down assays were conducted with GUDK purified as GUDK-GST fusion protein, and target proteins, which were purified as target-6xHis fusion proteins. The GST pull-down assays showed that GUDK interacts only with transcription factor OsAP37 (LOC_ Os01g58420) in vitro as identified by immunoblot analysis using 6xHis antibody (Fig. 6A). GUDK did not show any interaction with other proteins in the GST pull-down assay, and hence only OsAP37 was used for further analysis. To study the phosphorylation property of GUDK, an in-solution kinase assay was performed using OsAP37 as the substrate. The in-solution kinase assay showed that kinase activity doubled in the presence of OsAP37 and GUDK together (Fig. 6B). Furthermore, in-gel assays confirmed the transphosphorylation of OsAP37 by GUDK, which was detected by immunoblot analysis using an phospho-Tyr antibody (Fig. 6C). In addition, the assays also showed autophosphorylation of GUDK (Fig. 6C).
Table I. Target proteins selected for in vitro phosphorylation assays.
The Mascot score, relative intensity, and number of peptides with percentage of sequence coverage are taken directly from the Mascot report after a rice genome database search using Mascot. The Mascot score represents those proteins that were significantly represented in the ion spectrum after fragmentation with a score > 40 (P < 0.05). Relative intensity is the measure of total concentration of the protein in the sample that is directly related to the number of peptides eluting from the HPLC column and plotted against time. —, Nondetection of protein in the sample.
| Proteins Identified by LC-MS/MS | Locus ID | Predicted Molecular Mass | Mascot Score |
Relative Intensity |
Peptides
(Sequence Coverage) |
|||
|---|---|---|---|---|---|---|---|---|
| Well Watered | Drought Stress | Well Watered | Drought Stress | Well Watered | Drought Stress | |||
| kDa | n (%) | |||||||
| Chlorophyll a/b-binding preprotein | Os01g52240 | 28.04 | — | 95.45 | — | 58,260 | — | 8 (23.65) |
| Granule-bound starch synthaseI | Os06g04200 | 66.99 | 46.78 | — | 50,471 | — | 7 (17.62) | — |
| Putative chloroplast phosphoglycerate kinase | Os01g58610 | 32.48 | 178.81 | 234.54 | 206,862 | 221,079 | 14 (32.08) | 16 (38.78) |
| AP2-1 protein (OsAP37) | Os01g58420 | 48.09 | 52.36 | 64.98 | 87,345 | 108,765 | 8 (28.78) | 12 (36.87) |
| Wall-associated kinase 4-like | Os01g49529 | 77.7 | 114.54 | 125.98 | 69,321 | 88,076 | 9 (18.54) | 12 (24.67) |
| Stress-induced receptor-like kinase1 | Os07g08860 | 57.7 | — | 99.77 | — | 82,349 | — | 9 (28.07) |
| αNAC-like protein | Os01g50360 | 93.5 | 79.02 | — | 89,095 | — | 9 (16.3) | — |
| GLABRA2-type homeodomain protein | Os04g48070 | 57.1 | 55.12 | — | 54,389 | — | 7 (19.67) | — |
| GA insensitive (GAI), repressor of GAI and scarecrow family transcription factor | Os11g31100 | 87.6 | 58.86 | — | 61,274 | — | 7 (19.85) | — |
| Myeloblastosis (MYB)-LIKE DNA-binding domain | Os03g51220 | 81.5 | 48.82 | — | — | 59,315 | — | 7 (21.20) |
| Putative F-box protein | Os02g10700 | 61.16 | 71.15 | — | — | 75,715 | — | 8 (18.03) |
| Basic helix-loop-helix protein-like | Os02g48060 | 30.82 | 42.45 | — | — | 215,417 | — | 13 (19.46) |
| Zinc finger CCCH domain-containing protein | Os02g35150 | 78.63 | 69.27 | — | 54,583 | — | 6 (19.81) | — |
| GA- and ABA-regulated MYB transcription factor | Os01g59660 | 60.79 | 43.21 | — | — | 53,567 | — | 8 (24.98) |
Figure 6.
In vitro phosphorylation of OsAP37 by GUDK. A, GST pull-down assay showing the interaction of GUDK with OsAP37. The OsAP37-6xHis fusion protein was incubated with GUDK-GST fusion protein on Glutathione Sepharose 4B resin and the protein-protein interaction was detected by immunoblot analysis using His-antibody (1:1,000 dilution). The purified pGEX:GST (vector) induced with 1 mm isopropylthio-β-galactoside was used as the control. B, In-solution kinase assay showing the phosphorylation of OsAP37 by GUDK. The assay was performed with GUDK-GST fusion protein and OsAP37-6xHis fusion protein using the Kinase-Glo Luminescent Kinase Assay Platform and the kinase activity was expressed as the reciprocal of the luminescence signal. A control assay was run without OsAP37 and GUDK. Each bar represents the means ± se of three independent experiments. C, In vitro phosphorylation assay conducted using GUDK-GST fusion protein and OsAP37-6xHis fusion protein. The interaction was detected by immunoblot analysis using an phospho-Tyr antibody. [See online article for color version of this figure.]
OsAP37-Dependent Induction of Stress Genes Requires GUDK
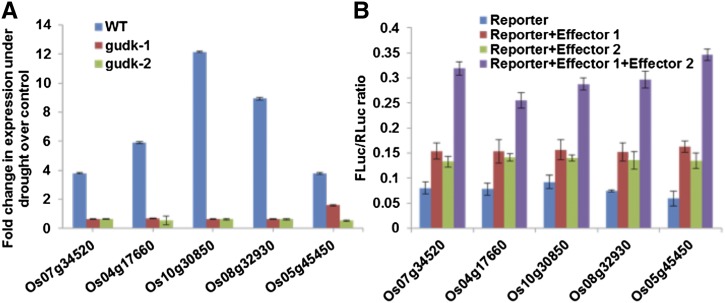
A previous study by Oh et al. (2009) showed that OsAP37 regulates the expression of several stress-responsive genes under drought, salinity, and low temperature conditions. From these OsAP37-regulated genes, we selected six genes that contain the GCC core recognition sequence. qPCR analysis of isocitrate lyase (ICL; Os07g34520), rhodanese-like domain-containing protein (RLD; Os04g17660), RING domain-containing E3 ubiquitin ligase (E3UL; Os10g30850), RcbX (Os08g32930), and DUF584 domain-containing protein (Dfu584; Os05g45450) in the mutant lines under drought stress showed a significant reduction in the transcript levels (Fig. 7A), whereas the genes were highly induced under drought in the wild-type plants (Fig. 7A). To investigate whether activation of these stress genes requires GUDK function, dual firefly-renilla luciferase-based in planta transactivation assays were performed using rice protoplasts with renilla luciferase as the internal control. The promoters (1 kb upstream of the transcriptional start site) of OsAP37 target genes linked to the firefly luciferase gene were used as reporter constructs. The coding regions of OsAP37 and GUDK cloned under the Cauliflower mosaic virus (CaMV) 35S promoter, in two separate constructs, were used as effector-1 and effector-2, respectively. Rice protoplasts were transfected with reporter constructs alone as controls to subtract background luminescence. To test whether OsAP37 or GUDK alone can activate stress genes, rice protoplasts were cotransfected with reporter and effector-1 or effector-2 constructs, respectively. Furthermore, to test the requirement of GUDK for activation of genes downstream to OsAP37, rice protoplasts were cotransfected with reporter, effector-1, effector-2, and renilla luciferase constructs. The results were expressed as the ratio of firefly luminescence normalized to luminescence obtained from renilla. The results showed that luminescence was low in the presence of reporters and OsAP37 or GUDK alone. Luminescence increased by nearly 2-fold in the presence of OsAP37 along with GUDK, suggesting that GUDK function is essential for OsAP37 activation as well as regulation of the downstream targets (Fig. 7B).
Figure 7.
GUDK regulates the expression of OsAP37 target genes. A, qPCR analysis of OsAP37 downstream target genes in the wild type and gudk-1 and gudk-2 insertion mutant lines under well-watered as well as drought stress conditions, using target gene primers (Supplemental Table S1D). Gene expression was normalized with reference to Ubiquitin and is expressed as the fold change in drought stress over well-watered samples. B, Dual firefly-renilla luciferase transactivation assays with reporter (target promoters fused to firefly luciferase), effector-1 (CaMV 35S:OsAP37), and effector-2 (CaMV 35S:GUDK) plasmids transfected into rice protoplasts along with renilla luciferase plasmid as the internal control. Results are expressed as the ratio of luminescence obtained from firefly luciferase to the luminescence obtained from renilla luciferase (FLuc/RLuc). The data points are the means ± se of three independent experiments with significance tested at P ≤ 0.01 compared with the relevant controls. WT, Wild type. [See online article for color version of this figure.]
DISCUSSION
High grain yield under drought stress as well as optimal conditions is a stability trait of interest for crop improvement. To understand the basis of yield stability under drought, we used a reverse genetics strategy in rice to screen knockout mutants of drought-responsive genes for phenotypes related to growth under drought and grain yield under drought component traits. Here we show that the GUDK gene of rice is required for grain yield under normal well-watered and drought stress conditions. We analyzed the GUDK downstream phosphorylation targets and studied the particular interaction with APETALA2/ETHYLENE RESPONSE FACTOR OsAP37, which was previously shown by overexpression studies to increase grain yield under drought stress (Oh et al., 2009).
Rice, a major global food crop and a model crop for cereals, is an excellent system to develop a better understanding of drought stress signaling pathways and to improve growth and yield under limited water conditions. The analysis of stress-responsive transcription factors identified many genes that can provide drought stress tolerance in multiple plant systems. However, there is less information available on the signaling pathways responsible for drought tolerance and grain yield that are upstream of transcription factors. Protein kinases such as cell surface RLKs and RLCKs are suggested to play a crucial role in drought stress signaling (Osakabe et al., 2010; Marshall et al., 2012). In this study, we identified a drought stress-induced RLCK, GUDK, from rice drought subnetworks and unraveled its role in drought stress response. Two independent T-DNA insertion lines with no obvious phenotypic difference at the vegetative stage under normal growth conditions, compared with wild-type plants, showed sensitivity to both vegetative-stage and reproductive-stage drought stress treatments.
GUDK Is Necessary for Drought Tolerance in Rice
GUDK is induced by drought in the seedling root, vegetative leaf, and flag leaf, indicating function in drought stress signaling in various tissues at the vegetative and reproductive stages. Roots play a crucial role in sensing water availability and transmitting appropriate signals from root to shoot (Schachtman and Goodger, 2008). Root-specific protein kinases, such as SNF1-related protein kinase 2C, were previously suggested to be the early sensors of water (Umezawa et al., 2004). GUDK was highly induced in roots under drought stress (Fig. 1B), with a concomitant reduction in the root growth of mutant seedlings under salinity, osmotic stress, and ABA treatment (Fig. 2), indicating an important role of GUDK in early stress sensing. The induction of GUDK under drought in leaves at the vegetative stage (Fig. 1B) suggests a role in this tissue, and corresponds to reduced photosynthesis, WUEi, and biomass in the gudk loss-of-function mutant lines under drought stress (Fig. 4), suggesting that GUDK functions in drought stress response and is required for drought tolerance at the vegetative stage.
GUDK Regulates Rice Yield by Potentially Transphosphorylating Transcription Factor OsAP37
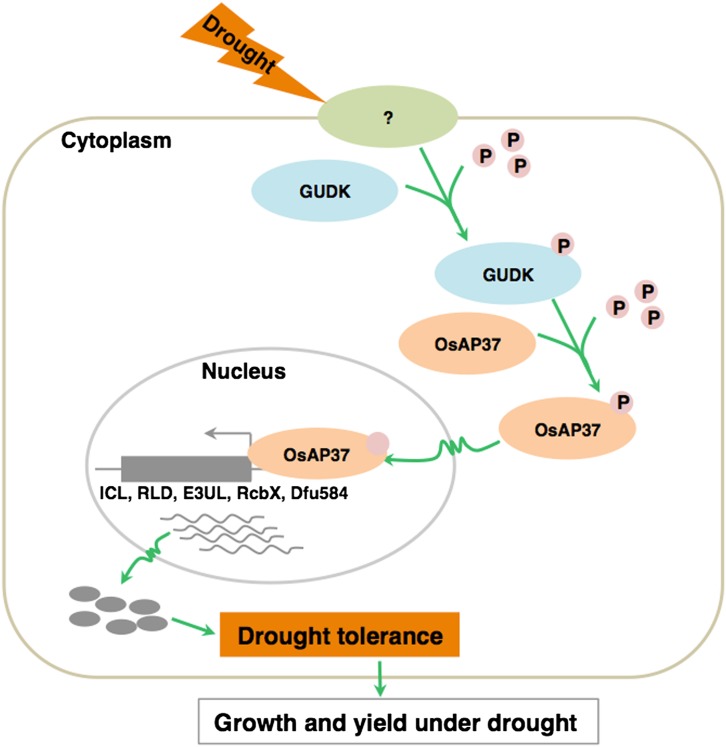
Overall drought tolerance of plants cannot be inferred from performance of plants under vegetative drought alone because drought stress during the reproductive stage can lead to a significant reduction in rice yield (Venuprasad et al., 2007; Centritto et al., 2009; He et al., 2010). Therefore, sustained growth and yield under drought will be the major criterion for developing drought-tolerant rice plants. In addition, it is also important to develop rice plants that yield better under milder stress as well as normal growth conditions (Guan et al., 2010; Marshall et al., 2012). In gudk mutant lines, a reduction in grain yield was found not only under drought stress but also under well-watered normal conditions (Fig. 5). The reduced grain yield of mutant plants under well-watered conditions was mainly attributable to the reduction in the number of filled spikelets because the number of panicles per plant and the spikelet number per panicle did not significantly change between mutant and wild-type plants (Fig. 5; Supplemental Fig. S1). In rice, grain yield reduction is mainly caused by the reduced number of filled spikelets per panicle without a substantial change in the number of spikelets per panicle (Ekanayake et al., 1989; Wei et al., 2014). Therefore, the reduced grain yield of gudk mutant plants was mainly attributable to higher spikelet sterility. Phosphoproteome analysis of wild-type and mutant plants under drought and well-watered conditions and in vitro kinase assays identified OsAP37 as the potential target of GUDK (Fig. 6; Table I). Overexpression of OsAP37 was previously shown to increase the tolerance of rice plants to drought stress at the vegetative stage and significantly enhanced grain yield (16%–57%) in drought under field conditions compared with wild-type plants (Kim and Kim, 2009; Oh et al., 2009). This improved drought tolerance was associated with the induction of several drought stress-responsive genes in the OsAP37 overexpression plants. In gudk mutant lines, there was no induction of these stress genes under drought stress (Fig. 7A). Furthermore, transactivation assays confirmed the requirement of GUDK for OsAP37-mediated activation of stress genes (Fig. 7B). OsAP37-activated genes such as ICL, RLD, E3UL, RcbX, and Dfu584 are known to play a crucial role in carbon metabolism and photosynthesis under drought stress (Kwak et al., 2007; Lee et al., 2009; Kolesiński et al., 2011; Luhua et al., 2013; Wang et al., 2014). The high induction under drought of GUDK in the flag leaf and reduced photosynthesis in the mutant lines suggest that GUDK regulates photosynthesis and carbon metabolism genes by phosphorylating OsAP37, resulting in sustained yield. Figure 8 shows a working model of GUDK under early events of drought stress signaling and transcriptional activation of drought-regulated genes.
Figure 8.
Model showing the proposed drought stress signaling pathway mediated by GUDK. In response to drought, signals are transmitted to GUDK and via phosphorylation (P) to transcription factor OsAP37, which is activated and in turn transcriptionally activates genes involved in photosynthetic carbon metabolism and drought tolerance (e.g. ICL, RLD, E3UL, RcbX, and Dfu584). [See online article for color version of this figure.]
In conclusion, this study highlights GUDK as a drought stress-inducible, dual-specificity autophosphorylating protein kinase that is required for transphosphorylation of transcription factor OsAP37, which controls growth and yield under drought. The significantly reduced growth and yield under drought in gudk mutant lines indicates that GUDK is a promising candidate gene for improving yield under drought in rice.
MATERIALS AND METHODS
Characterization of gudk Mutant Lines
Two independent mutant lines with T-DNA insertion in intronic regions, named as gudk-1 (ARAC10) and gudk-2 (ARPF01), were identified in the Oryza Tag Line Database (http://orygenesdb.cirad.fr/tools.html) and the mutant lines were procured for analysis. T-DNA insertions were confirmed by PCR using a combination of gene-specific primers and a T-DNA border primer (Supplemental Table S1). Homozygous mutant plants were identified by PCR using a pair of gene-specific primers and a combination of gene-specific and T-DNA border primers (Supplemental Table S1A). Null mutation of the gene was confirmed by conducting qPCR with RNA from drought-stressed mutant lines using gene-specific primers (Supplemental Table S1B).
Gene Expression Analysis
Total RNA was isolated from specific tissue of both wild-type and mutant lines using TRIzol reagent (Invitrogen) and was treated with RNase-free DNase I (Promega) per the manufacturers’ instructions. First-strand complementary DNA (cDNA) was synthesized from 1 μg of total RNA for each sample using the GoScript Reverse Transcription System (Promega) according to the manufacturer’s instructions. cDNA was diluted to 1:10 (v/v) and used for qPCR analysis along with Ubiquitin as the reference gene. qPCR reactions were performed using GoTaq qPCR Master Mix in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) according to the manufacturer’s instructions. The cycle threshold values of each sample were normalized to the Ubiquitin reference and the relative fold change in gene expression was calculated following the 2-ΔΔCt method (Livak and Schmittgen, 2001). Three independent biological replicates for each sample were maintained.
Abiotic Stress Treatment and Stress Response Measurements at Seedling Stage
Seeds of wild-type and mutant lines were dehusked and pregerminated on wet Whatman filter paper kept in petri plates. Two days after pregermination, 10 seedlings of an equal-sized radicle and plumule were transferred onto one-half-strength Murashige and Skoog medium in Magenta boxes supplemented with 100 mm NaCl, −0.5 MPa PEG 6,000, or 3 μm ABA. PEG medium was prepared as described (Verslues et al., 2006). Boxes were kept in a growth chamber for 7 d at day/night temperature of 26°C/22°C ± 1, light intensity of 600 μmol m−2 s−1, and 60% relative humidity. At the end of the stress period, seedlings were uprooted from the media and blotted on paper towels to remove any media, and the root length, shoot length, and fresh weight were measured immediately.
Drought Stress Treatment and Stress Response Measurements at the Vegetative Stage
For expression analysis, seeds were germinated on pots filled with Redi-earth potting mix (Sun Gro Horticulture Distribution). Twenty-five days after germination, drought stress was applied by withholding water until leaf rolling and leaf samples were collected for total RNA isolation.
To determine the seedling survival under drought stress, 25 seeds were germinated and grown in 1-gallon plastic pots filled with an equal amount of Redi-earth potting mix. A week after germination, seedlings were thinned to 15 seedlings per pot by retaining only equal-sized seedlings. Twenty days after germination, water was withheld for 12 d, followed by rewatering. Photographs were taken before stress (0 d), at the end of stress period (12 d), and a week after recovery (7 d recovery) using a Canon EOS REBEL T4i digital camera. Three biological replicates were maintained for both the wild-type and mutant lines.
To determine the growth response of mutant lines under controlled drought stress conditions, 1-week-old equal-sized individual seedlings were transplanted into 4-square-inch plastic pots filled with Redi-earth potting mix of known weight and water-holding capacity. Thirty-five days after transplanting, controlled drought stress was imposed on 10 pots by following a gravimetric approach. Another 10 pots were maintained at well-watered conditions (100% FC) as controls. For drought stress, the soil water content was brought down to 40% FC over a period of 3 to 4 d and plants were maintained at that level for 10 d by weighing the pots daily at a fixed time of the day and replenishing the water lost through evapotranspiration. At the end of the stress period, measurements of photosynthesis and maximum photochemical efficiency of PSII during steady-state illumination were taken on the second fully expanded leaf from the top, using a portable photosynthesis meter (LI-6400XT; LI-COR) at a CO2 concentration of 370 μmol mol−1, light intensity of 1,000 μmol m−2 s−1, and 55% to 60% relative humidity. WUEi was calculated using photosynthesis measurements and the transpiration rate as follows: WUEi = (photosynthesis/transpiration rate). Leaf RWC was measured as described (Barr and Weatherley, 1962) in the leaves used for photosynthesis measurements. The leaf fragments of the same length were excised and fresh weight was measured immediately. Leaf fragments were hydrated to full turgidity by floating them on deionized water for 6 h. The fragments were then blotted on paper towels, and the fully turgid weight was taken. Leaf samples were then oven dried at 80°C for 72 h and weighed to determine the dry weight. The percentage of RWC was calculated as follows: RWC (%) = (fresh weight − dry weight)/(turgid weight − dry weight) × 100. To determine biomass, shoots were harvested, oven dried at 80°C for 72 h, and weighed.
Unless otherwise stated, all plants were grown in greenhouse conditions with a day/night temperature of 26°C/22°C ± 1, and light intensity of 600 μmol m−2 s−1 with light/dark cycles of 10/14 h. All pots were placed in water-filled trays to simulate flooded conditions and were fertilized every week with 24-8-16 Miracle-Gro (Scotts Miracle-Gro Products) until drought stress.
Drought Stress Treatment at the Reproductive Stage and Analysis of Yield Components
Five individual plants in 4-square-inch plastic pots were grown under well-watered conditions. Drought stress was applied by withholding water at the preanthesis stage for 4 to 8 d until the leaves rolled, followed by rewatering. Panicles exposed to drought stress during the 4-d to 8-d window were marked and used for yield component analysis. A set of well-watered plants was also maintained as the control. Plants were further grown at well-watered conditions until physiological maturity. Drought-exposed panicles were harvested, and the numbers of filled and unfilled spikelets were counted to determine spikelet sterility (in percentages). The filled spikelets were dried at 37°C for 5 d and weighed to determine yield per plant.
Phosphoprotein Enrichment and Identification of Phosphopeptides Using LC-MS/MS
Total protein was extracted from wild-type and gudk-1 well-watered plants as well as drought-stressed plants (Roy et al., 2007). Approximately 10 g of leaf tissue was frozen in liquid nitrogen and ground in 5 mL of ice-cold protein extraction buffer (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 400 mm Suc, 10% [v/v] glycerol, 2.5 mm EDTA, pH 8.0, 1 mm phenylmethylsulfonyl fluoride, 1 mm 2-mercaptoethanol, and 5 μg mL−1 leupeptin). Protein extract was transferred to a sterile microfuge tube and centrifuged at 2,000g for 5 min at 4°C to pellet the debris. The supernatant was transferred to a fresh tube and the protein concentration in each sample was determined by the Bradford assay (Bradford, 1976) using bovine serum albumin as the standard (Fraction V; Sigma). Total protein samples were loaded onto Pierce Graphite Spin columns and eluted according to the manufacturer’s instructions (Thermo Scientific). SDS-PAGE was performed and the ProQ Diamond Phosphoprotein Gel Stain was used for staining specific phosphorylated proteins (Invitrogen). In-solution trypsin digestion and protein identifications were performed as described (Alvarez et al., 2006) using Mascot by searching the rice database.
Expression and Purification of Proteins in Escherichia coli and in Vitro Phosphorylation Assays
GUDK full-length cDNA amplified from rice (Oryza sativa) was cloned into the pGEX4T3 (Invitrogen) bacterial expression vector at BamHI/EcoRI sites (Supplemental Table S1C) and was transformed into E. coli BL21(DE3)pLysS (Novagen) competent cells. Cells were grown at 37°C (optical density 0.6) and expression was induced by further incubation with 1 mm isopropylthio-β-galactoside at 37°C for 5 h. Cells were harvested and lysed in 1× phosphate-buffered saline containing lysozyme to the final concentration of 1 mg mL−1. The lysate was incubated on ice for 2 h followed by sonication on ice for 10 s, repeated six times at 20-s intervals. After centrifugation, the soluble fraction was purified by using Glutathione Sepharose 4B resin following the manufacturer’s instructions (GE Healthcare). Immunoblotting was performed as previously described (Richard et al., 1991) with modifications using an phospho-Tyr antibody (1:1,000 dilution) as a primary antibody and goat anti-rabbit IgG conjugated with alkaline phosphatase (1:1,000 dilution) as a secondary antibody. For biochemical characterization of the kinase, the kinase assays were performed as described (Shen et al., 2004) with some modifications. Kinase activity was assayed using the Kinase-Glo Luminescent Kinase Assay Platform (Promega) and was expressed as the reciprocal of the luminescence signal. For the autophosphorylation assay, 1 µg of purified GUDK protein was preincubated in a buffer (50 mm HEPES, pH 7.4, 10 mm MgCl2, 1 mm dithiothreitol, and 2 mm ATP) for 0 to 60 min at 37°C (Yoshida et al., 2005). Autophosphorylation or in vitro kinase activity was detected by immunoblotting with an phospho-Tyr antibody or by an in-solution kinase assay using the Kinase-Glo Luminescent Kinase Assay Platform.
GST Pull-Down Assay
The full-length cDNAs of putative targets for GUDK were cloned into the pET28a (Novagen) expression vector, which has a 6xHis-tag at the N terminus. The 6xHis-tag fusion proteins were incubated with GUDK-GST fusion protein on Glutathione Sepharose 4B resin and the complex was purified following the manufacturer’s instructions (GE Healthcare). The protein-protein interactions were determined by immunoblot analysis using 6xHis antibodies (1:1,000 dilution; Rockland Immunochemicals) as previously described (Pan et al., 1999).
Transactivation Assay
The firefly and renilla luciferase genes derived from pGL3-Basic and pGL4 vectors (Promega), respectively, were cloned into pUC19 at the BamHI and KpnI sites. Firefly luciferase was cloned under promoters (1 kb) of OsAP37 target genes (Supplemental Table S1E) to serve as the reporter constructs, whereas renilla luciferase was cloned under the CaMV 35S promoter to serve as the internal control. The coding region of OsAP37 was cloned into pUC19 under the CaMV 35S promoter to serve as the effector-1 construct. The coding region of GUDK was also cloned into pUC19 under the CaMV 35S promoter to serve as the effector-2 construct. Rice protoplasts were isolated from leaf tissue as described (Hellens et al., 2005; Zhang et al., 2011) and were transfected with the reporter alone as the control, cotransfected with reporter and effector-1 constructs or cotransfected with reporter, effector-1, and effector-2 constructs. In all of the assays, the renilla luciferase construct was cotransfected to serve as the internal control. Protoplast transfections were performed by electroporation (Saunders et al., 1995; Bates, 1999). After 20 h of incubation, the protoplasts were lysed and used for the dual luciferase assay (Chen et al., 2008; Shi et al., 2011). Luminescence was measured by the GloMax 20/20 Luminometer (Promega) by mixing 80 µL of sample extract with 80 µL of Luciferase Assay Reagent (Promega). The data were collected as ratios and relative luciferase activity was calculated for each transfection by dividing firefly luminescence by renilla luminescence. The data are presented as the means of three biological replicates with significance tested at P ≤ 0.01 (Student’s t test).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Number of spikelets in wild-type and mutant plants under well-watered and drought stress conditions.
Supplemental Figure S2. Kinase assays to identify the substrate specificity and cofactor requirements of GUDK.
Supplemental Figure S3. In vitro autophosphorylation assays for GUDK.
Supplemental Figure S4. Schematic representation of the workflow for the identification of GUDK targets.
Supplemental Table S1. Primers used in the study.
Supplementary Material
Acknowledgments
We thank John Guerber for multiplying mutant seeds at the U.S. Department of Agriculture quarantine facility and Dr. Rohana Liyanage of the University of Arkansas Statewide Mass Spectrometry Facility for help during LC-MS/MS phosphoproteome analysis.
Glossary
- T-DNA
transfer DNA
- RLK
receptor-like kinase
- RLCK
receptor-like cytoplasmic kinase
- ABA
abscisic acid
- PEG
polyethylene glycol
- FC
field capacity
- RWC
relative water content
- WUEi
instantaneous water use efficiency
- GST
glutathione S-transferase
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- cDNA
complementary DNA
Footnotes
This work was supported by the National Science Foundation (award no. DBI–0922747).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alvarez S, Goodger JQD, Marsh EL, Chen S, Asirvatham VS, Schachtman DP. (2006) Characterization of the maize xylem sap proteome. J Proteome Res 5: 963–972 [DOI] [PubMed] [Google Scholar]
- Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A. (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 155: 916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Barr HD, Weatherley PE. (1962) A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci 15: 413–428 [Google Scholar]
- Bates G. (1999) Plant transformation via protoplast electroporation. In Hall R, ed, Plant Cell Culture Protocols, Vol 111 Humana Press, Totowa, NJ, pp 359–366 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cal AJ, Liu D, Mauleon R, Hsing YI, Serraj R. (2013) Transcriptome profiling of leaf elongation zone under drought in contrasting rice cultivars. PLoS ONE 8: e54537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centritto M, Lauteri M, Monteverdi MC, Serraj R. (2009) Leaf gas exchange, carbon isotope discrimination, and grain yield in contrasting rice genotypes subjected to water deficits during the reproductive stage. J Exp Bot 60: 2325–2339 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayake IJ, De Datta SKD, Steponkus PL. (1989) Spikelet sterility and flowering response of rice to water stress at anthesis. Ann Bot (Lond) 63: 257–264 [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539 [DOI] [PubMed] [Google Scholar]
- Giri J, Vij S, Dansana PK, Tyagi AK. (2011) Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol 191: 721–732 [DOI] [PubMed] [Google Scholar]
- Guan YS, Serraj R, Liu SH, Xu JL, Ali J, Wang WS, Venus E, Zhu LH, Li ZK. (2010) Simultaneously improving yield under drought stress and non-stress conditions: a case study of rice (Oryza sativa L.). J Exp Bot 61: 4145–4156 [DOI] [PubMed] [Google Scholar]
- He YX, Zheng TQ, Hao XB, Wang LF, Gao YM, Hua ZT, Zhai HQ, Xu JL, Xu ZJ, Zhu LH, et al. (2010) Yield performances of japonica introgression lines selected for drought tolerance in a BC breeding programme. Plant Breed 129: 167–175 [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurca ME, Bottka S, Fehér A. (2008) Characterization of a family of Arabidopsis receptor-like cytoplasmic kinases (RLCK class VI). Plant Cell Rep 27: 739–748 [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim JK. (2009) Rice transcription factor AP37 involved in grain yield increase under drought stress. Plant Signal Behav 4: 735–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesiński P, Piechota J, Szczepaniak A. (2011) Initial characteristics of RbcX proteins from Arabidopsis thaliana. Plant Mol Biol 77: 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak KJ, Kim JY, Kim YO, Kang H. (2007) Characterization of transgenic Arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress. Plant Cell Physiol 48: 221–231 [DOI] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT. (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenka SK, Katiyar A, Chinnusamy V, Bansal KC. (2011) Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luhua S, Hegie A, Suzuki N, Shulaev E, Luo X, Cenariu D, Ma V, Kao S, Lim J, Gunay MB, et al. (2013) Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol Plant 148: 322–333 [DOI] [PubMed] [Google Scholar]
- Marshall A, Aalen RB, Audenaert D, Beeckman T, Broadley MR, Butenko MA, Caño-Delgado AI, de Vries S, Dresselhaus T, Felix G, et al. (2012) Tackling drought stress: Receptor-like kinases present new approaches. Plant Cell 24: 2262–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh-Thu PT, Hwang DJ, Jeon JS, Nahm BH, Kim Y-K. (2013) Transcriptome analysis of leaf and root of rice seedling to acute dehydration. Rice (N Y) 6: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris ER, Walker JC. (2003) Receptor-like protein kinases: the keys to response. Curr Opin Plant Biol 6: 339–342 [DOI] [PubMed] [Google Scholar]
- Murase K, Shiba H, Iwano M, Che FS, Watanabe M, Isogai A, Takayama S. (2004) A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK. (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150: 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Mizuno S, Tanaka H, Maruyama K, Osakabe K, Todaka D, Fujita Y, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J Biol Chem 285: 9190–9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Sehnke PC, Ferl RJ, Gurley WB. (1999) Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell 11: 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard MC, Litvak S, Castroviejo M. (1991) DNA polymerase B from wheat embryos: a plant δ-like DNA polymerase. Arch Biochem Biophys 287: 141–150 [DOI] [PubMed] [Google Scholar]
- Roy S, Sarkar SN, Singh SK, Sengupta DN. (2007) A dideoxynucleotide-sensitive DNA polymerase activity characterized from endoreduplicating cells of mungbean (Vigna radiata L.) during ontogeny of cotyledons. FEBS J 274: 2005–2023 [DOI] [PubMed] [Google Scholar]
- Saunders JA, Lin CH, Hou BH, Cheng J, Tsengwa N, Lin JJ, Smith CR, McIntosh MS, Van Wert S. (1995) Rapid optimization of electroporation conditions for plant cells, protoplasts, and pollen. Mol Biotechnol 3: 181–190 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQD. (2008) Chemical root to shoot signaling under drought. Trends Plant Sci 13: 281–287 [DOI] [PubMed] [Google Scholar]
- Serraj R, Bennett J, Hardy B, eds (2008) Drought Frontiers in Rice: Crop Improvement for Increased Rainfed Production. World Scientific Publishing, Singapore, and International Rice Research Institute, Los Baños, Philippines [Google Scholar]
- Shen YY, Duan CQ, Liang XE, Zhang DP. (2004) Membrane-associated protein kinase activities in the developing mesocarp of grape berry. J Plant Physiol 161: 15–23 [DOI] [PubMed] [Google Scholar]
- Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A. (2011) SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet 7: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2012) Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J 70: 599–613 [DOI] [PubMed] [Google Scholar]
- Torii KU. (2000) Receptor kinase activation and signal transduction in plants: an emerging picture. Curr Opin Plant Biol 3: 361–367 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol 17: 113–122 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. (2004) SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 17306–17311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad R, Lafitte HR, Atlin GN. (2007) Response to direct selection for grain yield under drought stress in rice. Crop Sci 47: 285–293 [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK. (2008) The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant 1: 732–750 [DOI] [PubMed] [Google Scholar]
- Wang WH, Chen J, Liu TW, Chen J, Han AD, Simon M, Dong XJ, He JX, Zheng HL. (2014) Regulation of the calcium-sensing receptor in both stomatal movement and photosynthetic electron transport is crucial for water use efficiency and drought tolerance in Arabidopsis. J Exp Bot 65: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Hu W, Deng X, Zhang Y, Liu X, Zhao X, Luo Q, Jin Z, Li Y, Zhou S, et al. (2014) A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol 14: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl): S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ji W, Zhu Y, Gao P, Li Y, Cai H, Bai X, Guo D. (2010a) GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J Exp Bot 61: 2519–2533 [DOI] [PubMed] [Google Scholar]
- Yang T, Chaudhuri S, Yang L, Du L, Poovaiah BW. (2010b) A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem 285: 7119–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. (2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Q, Wu J, Zheng X, Zheng S, Sun X, Qiu Q, Lu T. (2013) Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 8: e57472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Xiong L. (2013) Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc Natl Acad Sci USA 110: 17790–17795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.