A zinc deficiency transcription factor moderates oxidative stress resistance.
Abstract
Identification of viable strategies to increase stress resistance of crops will become increasingly important for the goal of global food security as our population increases and our climate changes. Considering that resistance to oxidative stress is oftentimes an indicator of health and longevity in animal systems, characterizing conserved pathways known to increase oxidative stress resistance could prove fruitful for crop improvement strategies. This report argues for the usefulness and practicality of the model organism Brachypodium distachyon for identifying and validating stress resistance factors. Specifically, we focus on a zinc deficiency B. distachyon basic leucine zipper transcription factor, BdbZIP10, and its role in oxidative stress in the model organism B. distachyon. When overexpressed, BdbZIP10 protects plants and callus tissue from oxidative stress insults, most likely through distinct and direct activation of protective oxidative stress genes. Increased oxidative stress resistance and cell viability through the overexpression of BdbZIP10 highlight the utility of investigating conserved stress responses between plant and animal systems.
Global food security is dependent on both the availability of arable land as well as its agricultural production potential (Long et al., 2006; Fedoroff et al., 2010; Conforti, 2011; Wheeler and von Braun, 2013). Though roughly one-third of all arable land is currently in use, by 2050, estimated increases in agricultural production and yield of over 70% are predicted to be necessary to meet the growing demands for food (Conforti, 2011). This corresponds to an annual global increase of an additional one billion tons of cereal, including roughly 300 million tons of wheat (Conforti, 2011). To address food security, multiple research strategies are being implemented to identify or create stress-resistant, high-yielding crop varieties, including traditional breeding through germplasm mining of cultivated crops and wild relatives (Witcombe et al., 2008; Redden, 2013), as well as biotechnological approaches including genetic modification (Serageldin, 1999; Long, 2014). To improve upon these strategies, basic research into the mechanisms of abiotic stress resistance will be essential.
In the last decade, the genetically tractable model organism Brachypodium distachyon has been introduced as a viable biotechnological model for temperate cereal crops (Draper et al., 2001; Vogel and Bragg, 2009). When compared with Arabidopsis (Arabidopsis thaliana) as a model system, B. distachyon is similar in that its genome is also relatively small (300 Mbp), and its life cycle entails a short generation time (8–12 weeks) that requires simple growing conditions that can be easily scaled up in the greenhouse (1,000 plants m–2; Draper et al., 2001; Vogel and Bragg, 2009). Further, B. distachyon is more closely related to wheat (Triticum aestivum) and barley (Hordeum vulgare) than rice (Oryza sativa). However, unlike wheat, with a complicated genome structure and genome size of greater than 16,000 Mbp, many B. distachyon lines are diploid and more easily transformable (Vogel and Bragg, 2009). Recently, the genome, transcriptome, and other interactive bioinformatics tools for B. distachyon have been made publically available, thus further adding to the usefulness of B. distachyon as a model system (International Brachypodium Initiative, 2010; Brkljacic et al., 2011; Huo et al., 2011; Li et al., 2012). Combined, these tools will allow for systematic analysis of gene function followed by applied research toward cold cereal crop improvement for increased abiotic stress resistance.
Plant abiotic stress exposure from increased soil salinity, temperatures, and drought results in tissue damage that dramatically alters cellular metabolism (Miller et al., 2008; Cramer et al., 2011). This cellular damage releases and produces reactive oxygen species (ROS), which in turn activate the oxidative stress response for either cellular recovery or programmed cell death (PCD; Gill and Tuteja, 2010). Consistent with this functional link between abiotic stress and oxidative stress, overexpression of many conserved oxidative stress protective factors, including glutathione S-transferases (GSTs), superoxide dismutases (SODs), aldehyde dehydrogenases, and thioredoxins, increase abiotic stress tolerance (Gill and Tuteja, 2010), though the extent to which overexpression of oxidative stress transcription factors is protective is not clear. In plants, exploration of specific basic leucine zipper (bZIP) oxidative stress transcription factors has mostly focused on the TGA bZIP family, which bears some resemblance to the yeast (Saccharomyces cerevisiae) phase II detoxification factor YEAST AP-1-LIKE (Johnson et al., 2001; Després et al., 2003). In the animal kingdom, many stress transcription factors are utilized during oxidative stress to activate protective responses (bZIP transcription factors X-BOX BINDING PROTEIN1, ACTIVATING TRANSCRIPTION FACTOR4 [ATF4], NF-E2-RELATED FACTOR1-3, and ATF6; An and Blackwell, 2003; Harding et al., 2003; Glover-Cutter et al., 2013), with many of these factors playing intersecting roles in related cellular stresses, including protein homeostasis and aging (Henis-Korenblit et al., 2010; Kenyon, 2010; Hetz, 2012).
The intersection of oxidative stress resistance and longevity is a well-researched field in the animal kingdom (Tullet et al., 2008; Ristow and Schmeisser, 2011; Calabrese et al., 2012; Gems and Partridge, 2013), with many stress and longevity pathways conserved in the plant kingdom. For example, micronutritional depravation of essential nutrients is associated with aging and cognitive decline in animal systems (Chen et al., 2013) and growth and mass decline in plant systems. Specifically, zinc deficiency has been linked to Alzheimer’s and Parkinson’s disease in humans and in model systems for these diseases, including mouse, Caenorhabditis elegans, and Drosophila spp. (Chen et al., 2013; Szewczyk, 2013). For plants, zinc deficiency results in poorer yields due to plant death and necrosis (Assunçáo et al., 2010a; Lin and Aarts, 2012). Considering that many soils used for agriculture in developing countries are zinc deficient and that the majority of animal zinc uptake is through plant consumption and diet (Assunçáo et al., 2010a), a better understanding of the zinc deficiency pathway in cereal crops is imperative.
Zinc is essential for many enzymatic activities across the plant and animal kingdoms (Sinclair and Krämer, 2012; Miao et al., 2013). Many abiotic stress factors in plants rely on zinc for function (Miller et al., 2008), and disruption of the zinc deficiency response in mice, yeast, and Drosophila spp. results in decreased resistance to oxidative stress (Higgins et al., 2002; Swindell, 2011). Reciprocally, dietary supplementation of zinc to oxidative stressed cells reverses oxidative stress sensitivity (Ha et al., 2006; Günther et al., 2012). Zinc deficiency increases the level of cellular ROS, resulting in the activation of zinc transporters for increased influx of intracellular zinc (Günther et al., 2012). Though this response also activates glutathione synthesis in animals and plants for detoxification of ROS (Cakmak, 2000; Günther et al., 2012), the precise mechanism for how cells respond to zinc deficiency-mediated oxidative stress is not fully understood.
In yeast, Drosophila spp., and mammals, the zinc deficiency response is controlled by zinc finger transcription factors ZINC-RESPONSIVE ACTIVATOR PROTEIN1 (ZAP; Eide, 2009), Drosophila homolog of METAL-RESPONSIVE TRANSCRIPTION FACTOR1 (dMTF-1; Zhang et al., 2001), and METAL-REGULATORY TRANSCRIPTION FACTOR1 (MTF-1; Günther et al., 2012), respectively. MTF-1 is autoregulated and activated during multiple stresses, including hypoxia and oxidative stress, in addition to zinc deficiency. MTF-1 can both positively and negatively regulate gene expression (Zhang et al., 2001), and most likely requires cofactors such as SPECIFICITY PROTEIN1 or MEDIATOR15. Both ZAP1 and MTF-1 have defined DNA-binding motifs (Eide, 2009; Gunther et al., 2012), though direct binding and genomewide identification of targets during multiple stresses have not been fully determined.
In plants, using Arabidopsis as a model, two functionally redundant and highly homologous transcription factors have been identified that regulate the zinc deficiency response, AtbZIP19 and AtbZIP23 (Assunçáo et al., 2010b). Unlike the zinc finger transcription factors ZAP1 and MTF-1, Arabidopsis bZIP19/bZIP23 use a bZIP domain to bind to DNA and potentially dimerize with other bZIP transcription factors. AtbZIP19 and AtbZIP23 and their primary zinc response targets are most likely conserved in cereals based on sequence homology (Assunçáo et al., 2010b). However, the connection between zinc homeostasis and oxidative stress resistance and the potential role of these transcription factors in mediating oxidative stress have yet to be investigated in plants.
Highlighting the utility of stress studies in animal systems, we have identified a new oxidative stress B. distachyon bZIP transcription factor (BdbZIP10) that is highly homologous to AtZIP19/AtZIP23. Through characterization of oxidative stress resistance reporter genes in B. distachyon, we determined that overexpression of BdbZIP10 activates a protective transcriptional response that results in enhanced oxidative stress resistance and increased viability. Thus, germplasm screening for endogenous hyperactivation of this zinc deficiency pathway or genetic engineering of this pathway may be a viable direction for increased stress resistance in commercial cereal crops.
RESULTS AND DISCUSSION
Identification of Oxidative Stress Transcriptional Reporters in B. distachyon
To investigate the oxidative stress resistance potential in B. distachyon, conserved stress factors were first selected to assess the oxidative stress response. Transcriptional reporters were selected from publications of oxidative stress-induced gene expression datasets from multiple species followed by homolog identification using bioinformatics to compare predicted B. distachyon protein sequences to Arabidopsis, C. elegans, and Homo sapiens protein sequences (using BLASTP, CLUSTALW, and ALIGN; Desikan et al., 2001; Harding et al., 2003; Abercrombie et al., 2008; Oliveira et al., 2009). For this study, six conserved detoxification factors (GST24, COPPER/ZINC SUPEROXIDE DISMUTASE2 [CSD2], GLUTATHIONE PEROXIDASE6 [GPX6], GLUTATHIONE S-CONJUGATE TRANSPORTING ATPase1 [GSC1], SUPEROXIDE DISMUTASE1 [SOD1], and MANGANESE SUPEROXIDE DISMUTASE1 [MSD1]), two conserved stress recovery factors (ASPARAGINE SYNTHETASE1 [ASN1] and SIRTUIN1 [SIRT1]), and one plant specific antinecrosis factor (LESION STIMULATING DISEASE1 [LSD1]) were selected as transcriptional reporters for assessment of stress resistance in B. distachyon (Supplemental Table S1).
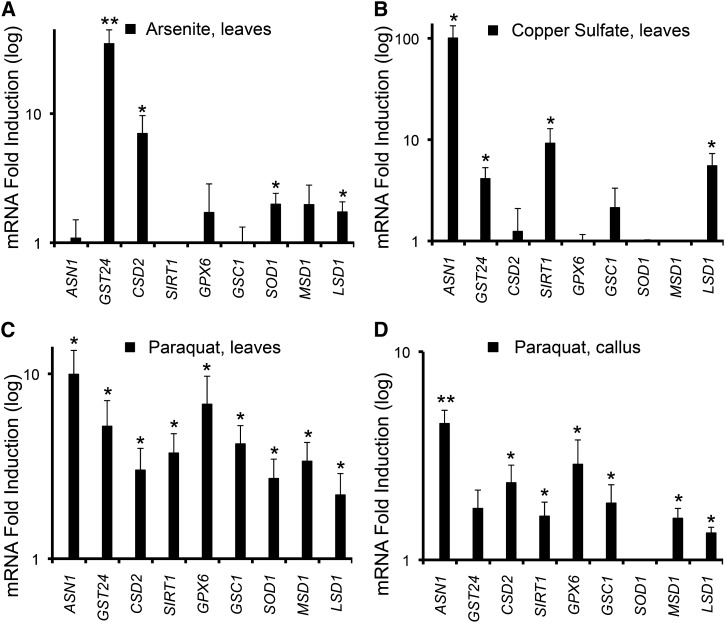
To validate the transcriptional response of our candidate reporter genes, we performed quantitative reverse transcriptase (QRT)-PCR after treatment with three distinct oxidative stress stimuli, arsenite (Hei et al., 1998; Abercrombie et al., 2008), copper sulfate (Gaetke and Chow, 2003), and paraquat (Mittler, 2002). The highly conserved detoxification factors BdGST24 (Board and Menon, 2013), BdCSD2 (Kliebenstein et al., 1998; Alscher et al., 2002; Chu et al., 2005), BdGSC1 (Arabidopsis thaliana MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN1 [MRP1] homolog; Lu et al., 1997), BdGPX6 (Brigelius-Flohé, Maiorino, 2013), BdSOD1 (Kliebenstein et al., 1998; Alscher et al., 2002), and BdMSD1 (Kliebenstein et al., 1998; Alscher et al., 2002) were transcriptionally up-regulated to various levels in B. distachyon leaf or callus tissue under conditions of oxidative stress through sodium arsenite treatment (Fig. 1A, soil application), heavy-metal copper sulfate treatment (Fig. 1B, leaf float), or paraquat treatment (Fig. 1, C and D, leaf float and callus submersion). Consistent with observations of tailored transcriptional responses for specific stress stimuli (Reichert and Menzel, 2005; Oliveira et al., 2009; Sahu et al., 2013), each oxidative insult resulted in a specific pattern of induction among the detoxification genes (Fig. 1, A–D).
Figure 1.
Identification of oxidative stress-induced reporter genes in B. distachyon. A, QRT-PCR analysis of leaf tissue after treatment with 5 mm arsenite for 1 h. B, QRT-PCR analysis of leaf tissue after treatment with 250 μm copper sulfate for 48 h. C, QRT-PCR analysis of leaf tissue after treatment with 160 μm paraquat for 24 h. D, QRT-PCR analysis of callus tissue after 160 μm paraquat treatment for 8 h. Values are plotted as increases in mRNA fold induction in log scale after normalization to control (nontreated) samples with n ≥ 3. Error bars represent the se of the mean (***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student’s t test).
In addition to these canonical oxidative stress reporters, we identified conserved transcriptional reporters representative of cellular health and recovery efforts, such as BdASN1, BdSIRT1, and BdLSD1. ASN1 initiates cellular recovery through ASN synthesis, with expression up-regulated during conditions of reduced translation, such as endoplasmic reticulum stress (Balasubramanian et al., 2013), amino acid starvation (Richards and Kilberg, 2006), and oxidative stress in various organisms (Abercrombie et al., 2008). As expected, transcription of BdASN1 was mildly induced when exposed to the oxidative stressor arsenite under our conditions (Fig. 1A) and strongly induced with copper sulfate (Fig. 1B) and paraquat (Fig. 1, C and D).
The deacetyltransferase SIRT1 has been implicated in proactive roles for oxidative stress resistance (Michan and Sinclair, 2007) and lifespan (Houtkooper et al., 2012) in animals, though these observations have yet to be validated in a plant system. Similar to published observations in mouse cardiac tissue (Alcendor et al., 2007), BdSIRT1 expression was up-regulated after treatment with paraquat in both leaf (Fig. 1C) and callus tissue (Fig. 1D). Additionally, copper sulfate treatment resulted in a roughly 10-fold increase in BdSIRT1 transcript levels (Fig. 1B). However, increased expression of BdSIRT1 after arsenite exposure was not observed under our conditions (Fig. 1A).
Prolonged oxidative stress will eventually result in PCD and plant necrosis. To monitor this pathway under our oxidative stress conditions, we chose to focus on the expression of the anti-PCD factor, BdLSD1. LSD1 is a constitutively and ubiquitously expressed ROS-regulated factor that is transcriptionally up-regulated during oxidative stress (Van Breusegem and Dat, 2006). As expected, transcript levels of BdLSD1 were increased with sodium arsenite, copper sulfate, and paraquat treatments (Fig. 1, A–D). In sum, the identified detoxification response and cellular protective factors in Figure 1 were sufficiently induced with oxidative stress and can thus serve as endogenous transcriptional reporters for oxidative stress resistance in the model organism B. distachyon.
Conserved B. distachyon Zinc Deficiency Transcription Factor Is Induced with Oxidative Stress
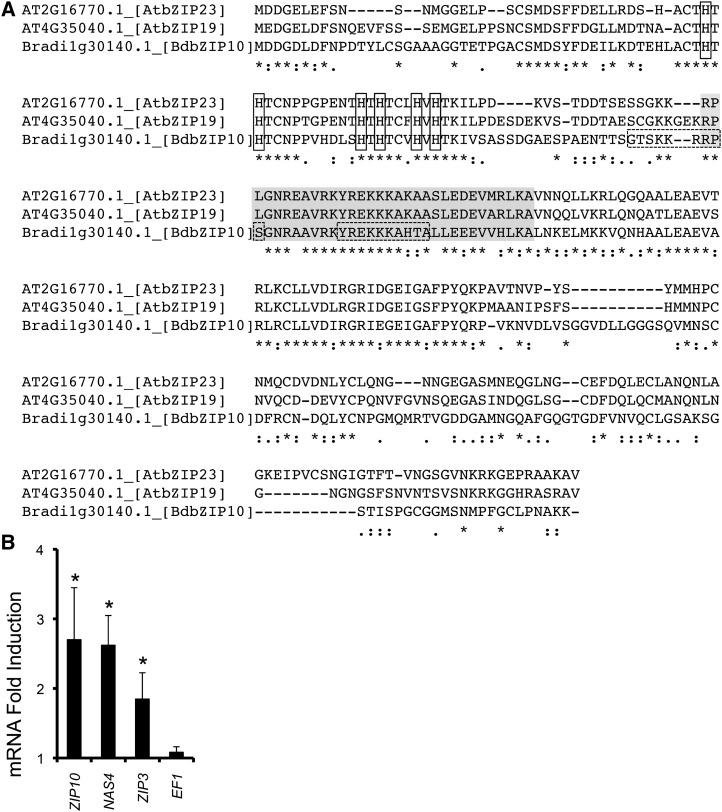
In Arabidopsis, two transcription factors regulate the response to zinc deficiency, AtZIP23 and AtZIP19 (Assunçáo et al., 2010b). To determine the B. distachyon homolog(s), bioinformatic analysis followed by analysis of transcription during zinc stress was performed. First, forward and reverse BLASTP analysis identified the top B. distachyon candidate Bradi1g30140.1 (Gramene BdbZIP10) as a possible zinc deficiency transcription factor homolog. To supplement this BLASTP analysis, we performed CLUSTALW analysis (Fig. 2A), which revealed a high degree of protein sequence conservation, including the two sequential conserved N-terminal His-rich motifs specific for zinc deficiency bZIP transcription factors across plant species (Assunçáo et al., 2010b). Similar to our CLUSTALW result, ALIGN analysis revealed 47.7% and 43.9% sequence identity to AtZIP23 and AtZIP19, respectively. Further, consistent with activity during zinc stress, zinc deficiency activated transcriptional expression of BdbZIP10 and predicted BdbZIP10 targets BdZIP3 (ZIP metal transporter) and BdNAS4 (metal homeostasis gene, nicotianamine synthase), similar to Arabidopsis expression analysis of AtZIP19 and AtZIP23 (Assunçáo et al., 2010b). As expected, zinc deficiency did not activate the housekeeping gene TRANSLATION ELONGATION FACTOR1 (BdEF1; Fig. 2B). Taken together, these results suggest that BdbZIP10 is homologous to zinc deficiency transcription factors AtZIP23 and AtZIP19.
Figure 2.
Identification of the zinc deficiency transcription factor in B. distachyon. A, CLUSTALW-aligned sequences of Arabidopsis zinc deficiency transcription factors AtbZIP19/23 with BLASTP identified B. distachyon protein Bradi1g30140.1 (BdbZIP10, using Gramene numbering system). Note the conserved His characteristic of zinc deficiency bZIP transcription factors in the N terminus (boxed), in addition to the bZIP domain (gray highlight), and the predicted nuclear localization signals (dotted box). Asterisk indicates single fully conserved residue, colon indicates conservation of strong groups, period indicates conservation of weak groups, and dash indicates no consensus (SDSC San Diego Supercomputer Center Biology Workbench [http://workbench.sdsc.edu]). B, QRT-PCR analysis of transcript levels after 15 d without zinc. Values are plotted as increases in mRNA fold induction after normalization to control (nontreated) samples with n ≥ 3. Error bars represent the se of the mean (***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student’s t test).
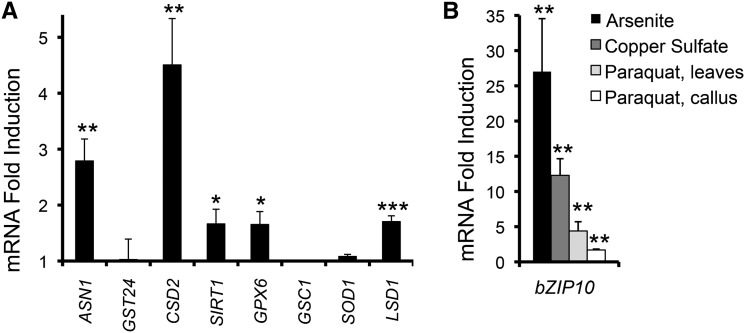
Before testing whether transgenic activation of the zinc deficiency response elicits oxidative stress resistance, we first investigated the possibility that zinc deficiency activates oxidative stress in B. distachyon, similar to observations in animal systems. Consistent with a tailored response to oxidative stress, ASN1, CSD2, SIRT1, GPX6, and LSD1 transcripts were significantly increased with zinc deficiency, whereas oxidative stress reporters GST24, SOD1, and GSC1 were not induced (Fig. 3A). Activation of these oxidative stress reporters during zinc deficiency suggests activation of a transcription factor(s) capable of orchestrating the oxidative stress response during both zinc and oxidative stress.
Figure 3.
Zinc deficiency activates the oxidative stress pathway in B. distachyon. A, QRT-PCR analysis of oxidative stress reporter genes after 15 d of zinc deficiency. B, QRT-PCR analysis of BdbZIP10 transcript levels after treatment with copper sulfate (250 μm, 48 h), arsenite (5 mm, 1 h), and paraquat (160 μm, 24 h) normalized to control samples (nontreated). Values are plotted as increases in mRNA fold induction after normalization to control (nontreated) samples with n ≥ 3. Error bars represent the se of the mean (***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student’s t test).
Many stress transcription factors are transcriptionally induced with their respective stressors (Estruch, 2000). To test whether BdbZIP10 transcript levels increase with oxidative stress similar to induction during zinc deficiency, we performed QRT-PCR after treatment with sodium arsenite, copper sulfate, and paraquat. Each of these oxidative stress conditions significantly increased transcript levels of BdbZIP10 (Fig. 3B), suggesting that this transcription factor may be involved with the oxidative stress response in B. distachyon.
BdbZIP10 Overexpression Enhances Transcription of Protective Response Factors
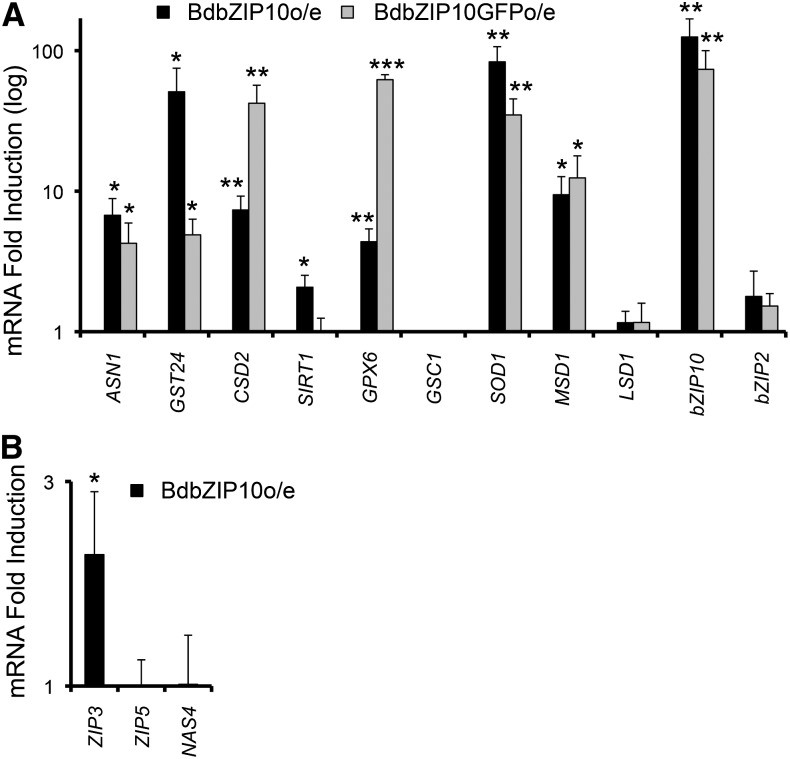
To determine whether constitutive overexpression of BdbZIP10 activates transcription of oxidative stress resistance genes, two vectors containing full-length BdbZIP10 coding sequences driven by the constitutive maize (Zea mays) ubiquitin (Ubi-1) promoter were constructed, with one containing a C-terminal GFP tag. Agrobacterium tumefaciens-mediated transformation was used to introduce these genes into B. distachyon calli, creating multiple plant lines and callus lines selected for overexpressed BdbZIP10 (BdbZIP10o/e) or BdbZIP10::GFP (BdbZIP10::GFPo/e; Fig. 4). Gene expression changes were analyzed using RNA extracted from leaf tissue of these transgenic lines using QRT-PCR (n > 3). Both BdbZIP10o/e and BdbZIP10::GFPo/e plants exhibited similar transcriptional profiles from our selected reporter genes (Fig. 4A). As expected, oxidative stress reporter genes were not activated in callus transformed with GFP alone (Supplemental Fig. S1A). For BdbZIP10-overexpressing plant lines, of the six detoxification reporter genes, BdCSD2, BdGPX6, BdSOD1, and BdMSD1 were up-regulated when BdbZIP10 is overexpressed, with BdCSD2 and BdGPX6 showing greater expression in the BdbZIP10::GFP lines. The protective factor BdASN1 was also up-regulated in both overexpressing lines. However, the detoxification factor BdGSC1 and the plant antinecrosis factor BdLSD1 were not induced with BdbZIP10 overexpression in BdbZIP10o/e and BdbZIP10::GFPo/e plants. On the other hand, BdSIRT1 transcript levels were increased to a lesser extent than other oxidative stress resistance reporters in BdbZIP10o/e, though this increase was not detectable in our BdbZIP10::GFPo/e plants. Although differences in the degree of expression of these reporter genes exist between the two BdbZIP10-overexpressing transgenic lines, the general observation of enhanced oxidative stress reporter gene transcription in the absence of zinc or oxidative stress strongly implicates BdbZIP10 as a cross-functional stress transcription factor.
Figure 4.
Overexpression of BdbZIP10 activates expression of protective oxidative stress genes. A, QRT-PCR analysis of oxidative stress-protective reporter genes in plants overexpressing BdbZIP10 (BdbZIP10o/e; leaf tissue), graphed in log scale after normalization to control (wild-type) sample transcript levels. B, QRT-PCR analysis of zinc deficiency response genes in BdbZIP10o/e leaf tissue, normalized to wild-type transcript levels. Error bars represent the se of the mean (*** P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student’s t test, with n ≥ 3).
Of note, BdbZIP10 transgenic plants did not exhibit stressed phenotypes and developed similar to wild-type plants (Supplemental Fig. S1B), suggesting that overexpression of BdbZIP10 did not simply elicit oxidative stress. Additionally, though overexpression of BdbZIP10 activated expression of its predicted zinc deficiency target reporter BdZIP3 (Assunçáo et al., 2010b; Fig. 4B), overexpression did not activate other zinc deficiency pathway components such as BdZIP5 and BdNAS4 (Fig. 4B). These observations, together with similar oxidative stress gene induction patterns of plants transformed with two independent BdbZIP10 constructs (BdbZIP10o/e and BdbZIP10::GFPo/e), strongly suggest that BdbZIP10 regulates expression of oxidative stress resistance genes in B. distachyon.
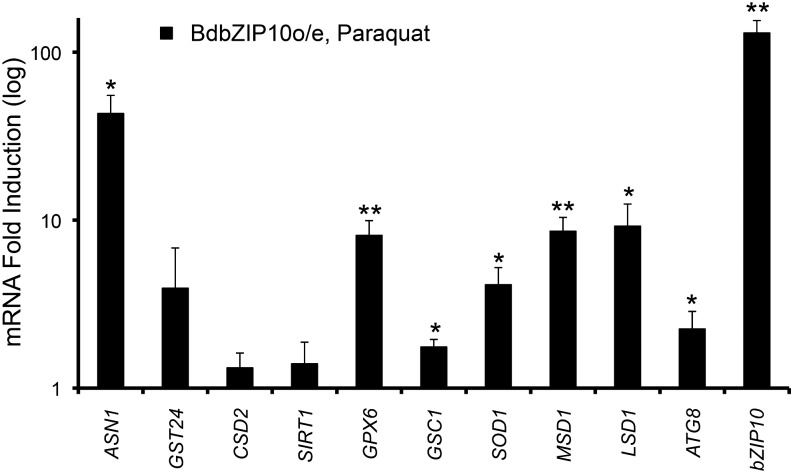
To further characterize the extent of the transcriptional stress response of overexpressing BdbZIP10, we examined the expression of our oxidative stress resistance reporter genes after treatment with an oxidative stressor. Of the three oxidative stressors tested, paraquat treatment initiated the most robust and consistent induction for each of our reporters (Fig. 1, C and D). Wild-type and BdbZIP10o/e plants were treated with paraquat, and QRT-PCR was performed. To focus on the effects of BdbZIP10 overexpression, results were then normalized to wild-type paraquat-treated expression levels (Fig. 5). Of the six detoxification reporters, transcript levels of four factors were significantly increased with BdbZIP10 overexpression (BdGPX6, BdGSC1, BdSOD1, and BdMSD1). Of the three cellular protective reporter genes, both BdASN1 and plant-specific BdLSD1 transcript levels increased with paraquat treatment of BdbZIP10o/e plants compared with paraquat treated wild-type plants. These results differ slightly from untreated conditions, where GSC1 and LSD1 transcriptional induction was not observed with BdbZIP10 overexpression (Fig. 4A). Possible explanations for these differences include additional stress-based signals for transcriptional activation of LSD1 and GSC1, or perhaps this indicates that BdbZIP10 indirectly activates their respective transcription through an unknown stress factor specific to paraquat stress.
Figure 5.
Oxidative stress genes are primed for enhanced induction in BdbZIP10-overexpressing plants (BdbZIP10o/e). QRT-PCR analysis of oxidative stress reporter genes in BdbZIP10o/e leaf tissue normalized to wild-type levels after parallel paraquat treatment (160 μm, 24 h), graphed in log scale. Error bars represent the se of the mean (***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student’s t test, with n ≥ 3).
To further explore potential transcriptional activation of cellular protective pathways in BdbZIP10o/e plants, we also focused on transcriptional activation of the autophagy pathway, which has been associated with enhanced longevity in animals (Madeo et al., 2010) and oxidative stress in plants (Pérez-Pérez et al., 2012). B. distachyon AUTOPHAGY-RELATED PROTEIN8 (BdATG8), the plant homolog of mammalian microtubule-associated LIGHT CHAIN3, is up-regulated in transgenic BdbZIP10o/e plants (Fig. 5). Consistent with the role of ATG8 during abiotic stress (Xiong et al., 2007) as well as its transcriptional induction under heat stress (Zhou et al., 2013), paraquat treatment in BdbZIP10o/e plants significantly increased BdATG8 levels above that of wild-type treated plants (Fig. 5). Taken together, transcriptional activation of oxidative stress resistance and cellular protective reporters by overexpression of BdbZIP10 both in the absence and presence of oxidative stress strongly suggest that the zinc deficiency transcription factor homolog in B. distachyon, BdbZIP10, influences oxidative stress resistance, potentially directly as a transcription factor.
BdbZIP10 Localizes to the Nucleus with Oxidative Stress
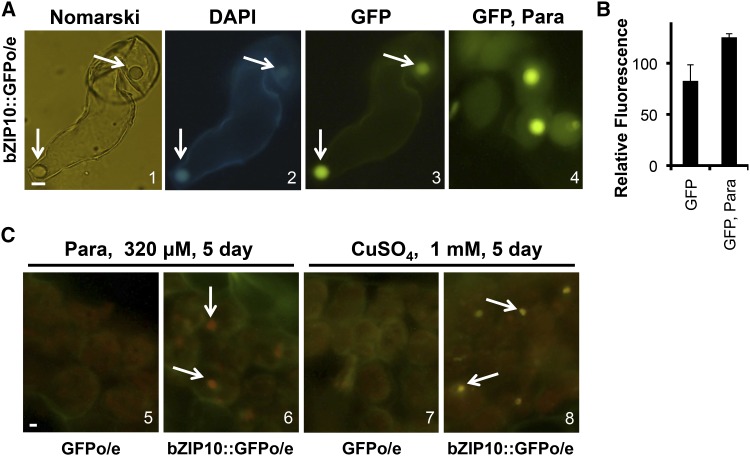
If BdbZIP10 acts as a transcription factor, detection of this protein in the nucleus should be possible given its nuclear localization signal (Fig. 2A). We next investigated whether BdbZIP10 localized to the nucleus using fluorescence microscopy. BdbZIP10::GFPo/e callus tissue was homogenized and examined under Nomarski light, and the nucleus was detected with 4′,6-diamidino-2-phenylindole (DAPI) staining and UV light (Fig. 6A, sections 1 and 2; Zink et al., 2003; Hunt et al., 2013). Consistent with its potential role as a transcription factor, BdbZIP10::GFP localized to the nucleus at detectable levels (Fig. 6A, section 3). Further, GFP intensity visually increased with oxidative stress by paraquat treatment (Fig. 6, A and B, section 4 and quantification by ImageJ).
Figure 6.
BdbZIP10 is localized to the nucleus. A, Fluorescence micrographs of BdbZIP10::GFPo/e callus tissue, Nomarski differential interference contrast image (1), DAPI staining for nuclei (2), and GFP fluorescence under normal (nontreated, 3) and oxidative stress conditions with paraquat (Para; 4, 320 μm paraquat for 24 h). B, ImageJ quantification of fluorescence in 3 and 4. Error bars represent the se of the mean. C, Fluorescence micrographs of leaf tissue from plants transformed with GFP alone or BdbZIP10::GFP after oxidative stress treatment with paraquat (5 and 6) and copper sulfate (7 and 8). Arrows indicate predicted nuclei structures. Bar = 6 μm.
To further verify nuclear localization of BdbZIP10::GFP, cryocut leaf sections of mature leaf tissue that was treated with either paraquat (Fig. 6C, sections 5 and 6) or copper sulfate (Fig. 6C, sections 7 and 8) were analyzed by microscopy. Nuclear localization of BdbZIP10::GFP was consistently observed in leaf sections from both oxidative stress treatments. Importantly, this localization was not simply a function of the GFP tag, as transgenic plants expressing GFP alone did not show a significant amount of nuclear localization. These results strongly suggest that the BdbZIP10 protein may function as a transcription factor similar to bZIP domain family members across species (Jakoby et al., 2002). Also, similar to many bZIP transcription factors in animals, BdbZIP10 is localized in the nucleus with stress and thus primed for transcriptional activity during oxidative stress (Ameri and Harris, 2008; Ma, 2013).
BdbZIP10 Localizes to the Site of Transcription
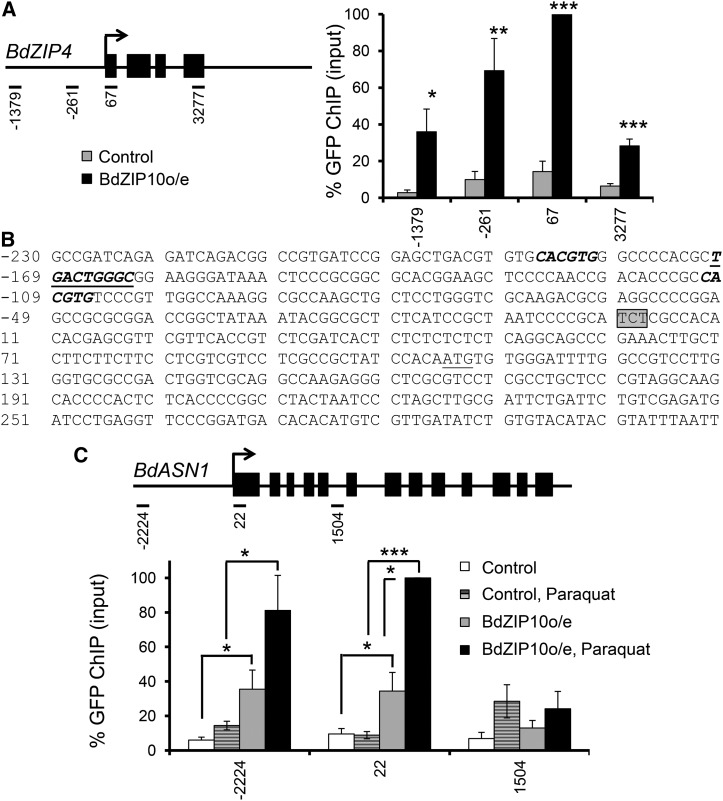
Our observation of nuclear-localized BdbZIP10::GFP strongly suggests that this protein acts as a transcription factor. As many transcription factors can be captured at the site of transcription by chromatin immunoprecipitation (ChIP), we first tested whether BdbZIP10 could be found at the site of transcription of BdZIP4, a known transcriptional target of BdbZIP10 Arabidopsis homologs AtZIP19/AtZIP23, by quantitative PCR. As predicted, BdbZIP10::GFP localized along the BdZIP4 gene locus at levels well above the wild-type control background (Fig. 7A).
Figure 7.
BdbZIP10::GFP localizes to distinct gene loci. A, ChIP analysis by quantitative PCR of the predicted BdbZIP10 reporter BdZIP4. Gray bars represent amplification signal from control (wild-type) extract, and black bars represent amplification signal from BdbZIP10::GFPo/e extract. Amplicons are indicated on the gene map with exons denoted as black boxes. B, Sequence of BdASN1 with predicted bZIP-binding domains marked in bold italics and underlined bold italics. The beginning of the 5′ untranslated region is boxed in gray, and the ATG start site is underlined. C, ChIP analysis by quantitative PCR of the BdASN1 gene locus before and after induction of oxidative stress with paraquat treatment. Control samples are marked white (nontreated) or striped (paraquat treated), and BdbZIP10::GFPo/e samples are represented with gray (nontreated) or black (paraquat treated) columns. Note the enhanced recruitment of GFP signal with paraquat treatment. Error bars represent the se of the mean (*** P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student’s t test, with n ≥ 3).
Given the degree and variety of oxidative stress reporter gene induction with BdbZIP10 overexpression (Figs. 4 and 5), we next investigated whether BdbZIP10 could be found at the site of transcription of a common oxidative stress reporter gene. Specifically, we tested whether BdbZIP10 could be found at the gene locus of BdASN1. Arabidopsis ASN1 is up-regulated with both zinc deficiency (van de Mortel et al., 2006) and oxidative stress (Abercrombie et al., 2008), and its mammalian homolog (ASNS) is directly regulated by the bZIP nutrient/endoplasmic reticulum/oxidative stress transcription factor ATF4 (Ameri and Harris, 2008; Han et al., 2013). Though BdASN1 does not contain the zinc Deficiency Response Element associated with AtZIP23/AtZIP19 binding, like the reporter BdZIP4 (Assunçáo et al., 2010b), the promoter region of BdASN1 does contain both Box A and Box G elements typical for plant bZIP DNA-binding motifs (Izawa et al., 1993; Fig. 7B).
ChIP analysis by quantitative PCR along the BdASN1 gene revealed significant BdbZIP10::GFP recruitment around the promoter region compared with downstream regions (+1,504, representative of background signal) and wild-type extracts (Fig. 7C, light gray versus white bars). Upon treatment with the oxidative stressor paraquat, recruitment density of BdbZIP10::GFP increased, suggesting selective recruitment at the site of transcription (Fig. 7C, black bars). This observation of increased BdbZIP10::GFP signal near the BdASN1 promoter by ChIP is consistent with our fluorescence microscopy observation of enhanced BdbZIP10::GFP nuclear localization with stress (Fig. 6). Combined with our BdASN1 transcriptional induction data from BdbZIP10o/e plants (Figs. 4 and 5), these results implicate BdbZIP10 as a transcription factor with the potential to directly regulate oxidative stress response genes.
BdbZIP10 Enhances Oxidative Stress Resistance and Survival
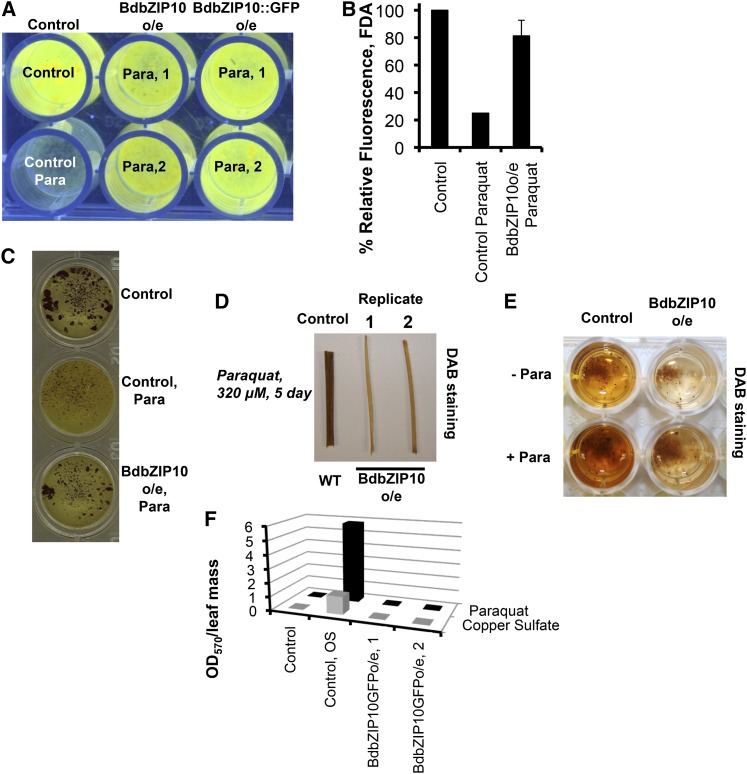
Considering the transcription factor potential of BdbZIP10 (Fig. 7) and the increased transcription levels of protective genes with BdbZIP10 overexpression (Figs. 4 and 5), we next determined whether transgenic overexpression of BdbZIP10 has a biological effect on oxidative stress resistance of B. distachyon. While there is no phenotypic difference observed between paraquat-treated and nontreated wild-type and BdbZIP10-overexpressing plants (Supplemental Fig. S1C), treatment of cut leaf tissue from BdbZIP10-overexpressing plants showed a marked reduction in paraquat-induced leaf bleaching compared with wild-type plants (Supplemental Fig. S1D).
To further determine whether differences in cell viability exist between wild-type and BdbZIP10o/e plants, fluorescein diacetate staining (FDA) was used to stain living B. distachyon callus. Cell-permeable FDA staining identifies viability by utilizing the nonspecific esterase activity of living cells, which hydrolyzes the nonfluorescent FDA to form green fluorescent fluorescein (Schwab and Hulskamp, 2008). Both BdbZIP10o/e and BdbZIP10::GFPo/e callus lines exhibited considerably higher levels of FDA staining with paraquat treatment (Fig. 8A) that were quantified using ImageJ software (Fig. 8B). These results are consistent with a protective effect of increased stress resistance and survival with BdbZIP10 overexpression.
Figure 8.
Overexpression of BdbZIP10 increases oxidative stress resistance. A, Cell viability analysis by FDA after treatment of homogenized callus tissue with paraquat (320 μm, 24 h). Fluorescence of live tissue is visualized under UV lighting. Note the high levels of fluorescence during oxidative stress in BdbZIP10o/e and BdbZIP10::GFPo/e cells compared with the control cells treated with paraquat (Para). B, ImageJ quantification of fluorescence observed in A, with error bars represented as the se of the mean. C, Representations of cell viability by MTT staining of live callus tissue after treatment with paraquat (320 μm, 24 h). D and E, DAB staining of hydrogen peroxide after paraquat treatment (320 μm) comparing wild-type (WT) control leaf (D) or callus (E) tissue to BdbZIP10o/e leaf tissue. Note the reduction in staining in BdbZIP10o/e leaves and calli. F, Quantification of ROS levels using NBT staining and thiazol measurement by spectrometer A570. Leaf tissue exposed to either paraquat (black bars) or copper sulfate (gray bars) normalized to leaf mass for either control (wild type, nontreated), control oxidative stress (Control, OS; wild type, treated), or oxidative stress-treated BdbZIP10o/e leaf replicates are plotted. OD570, Optical density at 570 nm.
Though FDA has been used to reliably identify viable tissue, autohydrolysis in the absence of living cells has been reported (Clarke et al., 2001; Boyd et al., 2008). To address this possibility and to further verify increased cell viability of BdbZIP10o/e tissue during oxidative stress, we treated callus tissue with paraquat and visualized cell viability by dimethylthiazol (MTT) staining (Fig. 8C). Callus tissues undergoing paraquat-induced oxidative stress treatment showed a marked reduction in live tissue staining (reddish brown) in wild-type cells (Fig. 8C, top two wells). By contrast, paraquat treatment had very little effect on cell viability measured by MTT staining in BdbZIP10o/e callus (Fig. 8C, bottom well). Both FDA and MTT cell viability results strongly implicate BdbZIP10 as a stress resistance factor during oxidative stress.
Having determined that BdbZIP10 regulates transcription of detoxification and protective factors with known roles in oxidative stress resistance (Figs. 4 and 5), we next investigated whether cell viability of BdbZIP10 overexpression correlated with decreased levels of cellular ROS. To measure changes in the levels of ROS, we first focused on visualizing hydrogen peroxide accumulation in leaf tissue by 3,3′-diaminobenzidine (DAB) staining, which polymerizes as a red/brown polymer with peroxidase activity (Thordal-Christensen et al., 1997; Fryer et al., 2002). Whereas paraquat treatment of wild-type control leaf cuts resulted in characteristic brown DAB staining, this staining was greatly reduced in leaves overexpressing BdbZIP10 (Fig. 8D). Similar observations were seen with callus tissue, with overexpressing BdbZIP10 homogenized callus showing less DAB staining after paraquat treatment compared with wild-type control callus tissue (Fig. 8E). These observations suggest that overexpression of BdbZIP10 results in reduced accumulation of hydrogen peroxide during oxidative stress.
To further confirm reduction in the levels of ROS in BdbZIP10o/e tissue during oxidative stress, we measured levels of superoxide by the reduction of nitro-blue tetrazolium (NBT). Leaves were treated with either paraquat or copper sulfate (Fig. 8F) and incubated with NBT, followed by extraction, solubilization, and detection of formazan at 570 nm (Piddington et al., 2001). When normalized to leaf mass, extracts of BdbZIP10o/e leaf samples showed a marked reduction in signal compared with wild-type controls, consistent with decreased production of ROS during oxidative stress. Taken together, these results suggest that BdbZIP10 acts as a stress resistance transcription factor, making this factor a viable candidate for either transgenic manipulation for increased stress resistance or as a transcription node for screening germplasm for commercial cereal crops.
CONCLUSION
BdbZIP10 Is the B. distachyon Zinc Deficiency Transcription Factor
Though the structure and mode of activation for zinc deficiency transcription factors varies between the plant and animal kingdoms, the nutritional requirement for zinc as a micronutrient is universal (Frassinetti et al., 2006). We have demonstrated that the B. distachyon zinc deficiency factor is BdbZIP10 by sequence homology (Fig. 2A), by its induction under zinc-deficient conditions (Fig. 2B), and by functional analysis through activation of specific zinc-responsive reporter genes (Figs. 2B, 4B, and 7A). Like other stress transcription factors, BdbZIP10 localizes to the nucleus with stress (Figs. 6 and 7), possibly due to posttranslational modifications enhancing stability or directing subcellular localization (An and Blackwell, 2003; Hetz, 2012). Further supporting evidence that BdbZIP10 acts as a stress transcription factor includes the activation of stress-responsive genes (Figs. 4 and 5) and the observation that BdbZIP10 can be found at gene loci by ChIP (Fig. 7).
Zinc deficiency and oxidative stress are tightly correlated in both plant and animal systems (Cakmak, 2000; Higgins et al., 2002; Swindell, 2011). Reduced levels of zinc or loss of zinc deficiency factors results in increased ROS, decreased oxidative stress resistance, and reduced viability. However, little is known about the protective effect of enhanced modulation of the zinc deficiency pathway. In Drosophila spp., the viability of parkin mutants (model for Parkinson’s disease) is extended and ROS levels reduced with exogenous overexpression of dMTF-1. Overexpression of dMTF-1 also extends Drosophila spp. lifespan (Saini et al., 2011). Similarly, we now demonstrate in the B. distachyon plant model that overexpression of BdbZIP10 activates protective genes involved in oxidative stress resistance (Figs. 4 and 5). Most importantly, overexpression of BdbZIP10 enhances oxidative stress resistance and improves plant cell viability during oxidative stress (Fig. 8, A–C), most likely through the reduction of ROS (Fig. 8, D–F).
Though overexpression of B. distachyon and Drosophila spp. zinc transcription factors is beneficial for these organisms, an overabundance of the heavy metal zinc is itself lethal (Lin and Aarts, 2012). Given the toxicity and growth effects of too much zinc, we do not believe that the mechanism of action of BdbZIP10 overexpression is solely the result of enhanced activation of the zinc deficiency pathway increasing zinc levels in the plant, as these plants develop normally and show no signs of toxic insults (Supplemental Fig. S1B). Additionally, transcriptional analysis reveals that only a select subset of zinc-responsive genes is activated with BdbZIP10 overexpression (Fig. 4B). Future studies identifying genomewide direct gene targets of BdbZIP10 will be important in teasing out the mechanism of this enhanced resistance to oxidative stress.
Considering that abiotic stress results in oxidative stress and that enhanced oxidative resistance increases viability from yeast to mammalian systems (Gems and Partridge, 2013), we propose a systematic analysis of conserved oxidative stress-resistant pathways using the monocot model organism B. distachyon to select viable strategies for stress-resistant crop identification through germplasm screening and/or genetic modification. To this end, we have identified protective oxidative stress reporter genes for assessment of the oxidative stress response (Fig. 1). In this report, we focused on the zinc deficiency pathway as a potential mediator of oxidative stress. We report BdbZIP10 as a viable candidate for further investigation for abiotic and biotic stress resistance in forage grasses and temperate cereals.
MATERIALS AND METHODS
Brachypodium distachyon Growing Conditions
Mature seeds of B. distachyon (Bd21-3 or seeds from transformed plants) were planted in Sunshine Growing Mix (Sun Gro Horticulture). Plants were fertilized once with Osmocote Plus 15-9-12 slow-release fertilizer. Pots were placed in a cold chamber (4°C with 8 h of fluorescent light for 5 d) for seed stratification. Plants were then grown either under greenhouse conditions with supplemental light (high-pressure sodium lamps, 400 or 430 W; Philips Lighting) to provide 16 h light with 20°C day and 17°C night (these are the settings, but they were much warmer during the summer) or in a growth chamber with 20-h-light/4-h-dark cycles at 24°C day and 18°C night in a Conviron PGV36 walk-in growth chamber. Plants were grown for at least 4 weeks postgermination before leaf tissue was extracted for RNA, ChIP, and ROS analysis.
Stress Assays
Approximately 6- to 8-week-old (Lehmann et al., 2012; Martin et al., 2012) potted B. distachyon plants measuring, on average, 15 20 cm in height, arranged four plants per 500-mL pot, were treated with sodium arsenite, copper sulfate, or paraquat. For arsenite treatments, 100 mL of 5 mm arsenite (Sigma) was added directly to the pot at the roots using modified protocols (Dombrowski et al., 2008; Lehmann et al., 2012), and leaf tissue was extracted after 1 h of treatment. Paraquat and copper sulfate treatments used a modified version of existing leaf toxicity protocols (Zhang et al., 2006; Wan et al., 2009). For copper sulfate treatment, 7.5- to 10.2-cm leaves were harvested and submerged in a dilution of either 250 μm (gene expression stress assays) or 1 mm (nuclear localization and cell assays) under continuous light for either 48 h (gene expression assays) or 5 d (cell assays and nuclear localization). For paraquat leaf treatment, 3- to 4-inch leaves were harvested and submerged in fresh dilutions of either 160 μm (gene expression stress assays) or 320 μm (nuclear localization and cell viability assays) under continuous light for up to 24 h (gene expression assays), 72 h (cell assays), or 5 d (nuclear localization assays), unless otherwise indicated. For paraquat treatment of callus tissue, callus was briefly homogenized (Ultra-Turrax T25, setting 4) and nutated in 160 μm (gene expression assays, 8 h) or 320 μm paraquat (cell assays, 24 h) diluted in phosphate-buffered saline (PBS), unless otherwise indicated.
For zinc deficiency studies, plants were grown under greenhouse conditions as above in moistened hydroton clay balls, and nutrients were supplied daily as a modified Hoagland’s medium (Yordem et al., 2011; full-strength Yordem’s medium: 1 mm KH2PO4, 3.75 mm KOAc, 5 mm Ca(NO3)2, 1.25 mm KNO3, 2 mm MgSO4, 3.75 mm NH4OAc, 46 μm H3BO3, 9.1 μm MnCl2, 0.77 μm ZnSO4, 0.32 μm CuSO4, 0.83 μm H2Mo4, 100 μm FeSO4, and 100 μm EDTA). Plants were watered with one-half-strength Yordem’s medium for the first week and then full-strength Yordem’s medium with or without zinc for control or zinc-deficient plants, respectively. Leaf tissue samples for RNA extraction were collected after applying full-strength Yordem’s medium with/without zinc for 15 d.
Vector Construction
RNA was extracted from leaf tissue stressed with arsenite (5 mm, 1 h), paraquat (160 μm, 24 h), and copper sulfate (250 μm, 48 h) and DNase treated with TurboDNase (Ambion), and complementary DNA (cDNA) was synthesized by reverse transcriptase (RT)-PCR (5 μg of RNA, Superscript III; please see RNA extraction method for more detail). cDNAs were collected and diluted 5-fold for PCR cloning. BdbZIP10 nomenclature (protein name, Gene ID [Bradi1g30140], and nucleotide sequence) was obtained through the transcription factor database collected in http://www.grassius.org. Primers were designed using the Clontech Web design for In-Fusion cloning into a modified pART (Gleave, 1992; without and with C-terminal GFP tag) driven by the ZmUbi1 promoter (Supplemental Fig. S1E; Wang et al., 1997, 1998; Murray et al., 2004). Forward primer 5′-CGACTCTAGAGGATCCATGGACGACGGGGACC and reverse primer 5′-ATGAATTCGAGCTCGGTACCCTACTTCTTTGCATTCGGCAAACA were used for insert amplification of the coding region of BdbZIP10 from stress B. distachyon cDNA. Clontech In-Fusion vector preparation was performed according to manufacturer’s instructions, and the vector was transformed into competent DH5α cells. Plasmid was purified using Qiagen midiprep columns, sequenced to verify the construct, and then transformed into Agrobacterium tumefaciens AGL1 (Lazo et al., 1991).
B. distachyon Transformations
Bd21-3 transformations for GFP, BdbZIP10, and BdbZIP10::GFP were performed as previously described (Draper et al., 2001; Vogel et al., 2006; Vogel and Hill, 2008). The complete protocol for embryogenic tissue development from immature seeds and transformation of the embryogenic callus is available at http://Brachypodium.pw.usda.gov/files/BrachyTransformProtocol_Feb2009.pdf (Vogel and Hill, 2008). Briefly, after seed dissection, embryogenic callus tissue was subcultured for approximately 6 weeks before transformation with A. tumefaciens. For transformation, callus tissue was suspended in A. tumefaciens AGL1 (Lazo et al., 1991) containing the plasmids of interest for up to 15 min, the liquid was removed, and the calli were placed on dry Whatman filter paper in the dark for 3 d. Transformed callus was selected on callus induction medium containing 40 mg L–1 hygromycin and 150 mg L–1 Timentin following the protocol described in the above linked PDF. Transformed callus was moved to regeneration media after 3 to 5 weeks and grown in light chamber conditions of 16-h light/8-h dark cycles at 28°C. Shoots were transferred to magenta boxes and placed in the light chamber. Plantlets were transferred to soil and placed under a plastic wrap for about a week to help acclimate the plants, which were then grown in the greenhouse or in growth chambers as described above. Experiments were performed with at least three T0 or three T1 BdbZIP10-overexpressing lines.
RNA Extraction and Quantification
RNA extraction was performed as previously described (Martin et al., 2013). Briefly, 1- to 3-inch leaf pieces were homogenized in Trizol (Ultra-Turrax T25, setting 4) for 30 s followed by quick freezing at –80°C. Samples were then thawed, debris removed by centrifugation, and RNA extraction completed using the manufacturer’s recommended conditions (Trizol, Invitrogen). Extracted RNA was then treated with DNase per manufacturer’s recommended conditions (Turbo DNase, Ambion), and at least 1 ug of cDNA was synthesized using Superscript III Reverse Transcriptase kit (Invitrogen, manufacturer’s conditions).
ChIP
Extracts from control (wild type) or BdbZIP10::GFPo/e plants were prepared using a modified protocol from Ricardi et al. (2010). Diced leaf tissue (1–5 g) was immersed in 37 mL of 1% (v/v) formaldehyde in extraction buffer 1 (0.44 m Suc, 10 mm Tris, pH 8.0, 5 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride) and vacuum infiltrated for 15 min (100 in Hg), followed by 5-min vacuum infiltration after addition of 2.5 mL of 2 m Gly. Leaf tissue was then washed twice with ice water, quick frozen in liquid nitrogen, ground to a fine frozen powder with a mortar and pestle, and resuspended in extraction buffer 1. The solution was then filtered through four layers of miracloth and centrifuged at 2,880g for 20 min. The pellet was resuspended in extraction buffer 2 (0.25 m Suc, 10 mm Tris, pH 8.0, 10 mm MgCl2, 1% [v/v] TritonX 100, 0.1 mm phenylmethylsulfonyl fluoride, and 5 mm β-mercaptoethanol) and incubated on ice for 10 min. The suspension was then centrifuged at 2,100g for 20 min, followed by resuspension of the pellet in extraction buffer 2 minus Triton-X, and then centrifuged at 2,100g for 20 min. The pellet was suspended in radioimmunoprecipitation assay buffer, sonicated at 5 × 15-s intervals at 20% amplitude, treated with Micrococcal Nuclease (Roche) at 37°C for 10 min, and then quick frozen at –80°C. Samples were then prepared for immunoprecipitation with GFP antibody (Abcam ab290) as previously described (Glover-Cutter et al., 2008).
Quantitative PCR
Quantification of both cDNA and ChIP samples was performed using BioRad Real-Time PCR detection system using primers designed with Roche LightCycler Probe Design Software 2.0, as described previously (Glover-Cutter et al., 2008; Martin et al., 2012, 2013). For RT analysis, B. Distachyon UBIQUITIN C18 and/or BdEF1 were used as normalization housekeeping control genes (Hong et al., 2008). Where indicated, RT values were graphed in log(10) scale and denoted as mRNA Fold Induction (log). For ChIP analysis, amplicons from the upstream intergenic region, promoter, 5′, and middle of the gene were chosen based on probability score and Δ G values for optimized quantitative PCR, with the middle of the amplicon given as the name for each primer pair. Immunoprecipitation values were normalized to input values for each condition and presented as percentage ChIP of the maximal signal value as described previously (Glover-Cutter et al., 2008).
Light Microscopy
Nuclear localization analysis of BdbZIP10 was performed using light microscopy, annotated visually from n ≥ 3 independent experiments, similar to previously described transcription factors (An and Blackwell, 2003; Sun et al., 2013). Either homogenized callus tissue or leaf cuts were used for light microscopy analysis. For callus tissue, approximately 5 μL of homogenized liquid was mounted and examined with an Olympus BH-2 microscope with Nomarski differential interference contrast lighting for cellular definition and details. Leaf tissues samples were macerated on a glass slide in a drop of water and covered with a cover glass. DAPI-stained calli were examined with an Olympus BH-2 Microscope with UV light and Chroma 11000v3 UV filter cube (Ex 350 nm). Samples for GFP localization were examined with a BP490 filter (Ex 490 nm). Images were captured at 125× or 250× magnification using a Moticam 2500 digital camera (Motic Instruments) connected to an Hewlett Packard laptop running Motic Images Plus software version 2.0. The software ImageJ was used to roughly estimate GFP intensity.
Biological Analysis for Viability and ROS Accumulation
Cell viability assays for oxidative stress resistance included staining of live tissue with FDA (Widholm, 1972) or MTT (Mosmann, 1983). After paraquat treatment, callus tissue was incubated with 0.1 μg mL–1 of FDA diluted in PBS (Cold Spring Harbor Protocols) and photographed on a UV light box. For MTT treatment, homogenized callus was incubated with 0.5 mg mL–1 MTT diluted in PBS for 4 h in 24-well plates and scanned for visual observation of red dye accumulation. ROS was either visualized through hydrogen peroxide accumulation by DAB staining after vacuum infiltration of DAB (in water, pH 3.8; Sunkar, 2010) or by NBT metabolism after NBT vacuum infiltration (0.06% [w/v] NBT, 10 mm NaN3, and 50 mm KH2PO4; Sunkar, 2010). Briefly, leaf tissues from equivalent developmental stages and leaf positions were vacuum infiltrated at 100 inches of mercury for five 1-min intervals and cut into approximately 1-inch leaf segments. DAB samples were then incubated in the dark for 6 h, briefly air dried, observed for brown staining characteristic of DAB accumulation, and photographed. NBT samples were incubated in the light for 1 h, followed by ethanol extraction and quantification of formazan by spectrophotometric analysis at 570 nm (Piddington et al., 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Overexpression of BdbZIP10 and GFP-only control reveals best phenotype with leaf float.
Supplemental Table S1. Primers used for quantitative PCR and QRT-PCR.
Supplementary Material
Acknowledgments
We thank Thomas Lockwood (U.S. Department of Agriculture-Agricultural Research Service) for assistance in B. distachyon transformation and general lab and ordering assistance, Dr. John Vogel (U.S. Department of Agriculture-Agricultural Research Service) for the generous gift of B. distachyon seeds, Drs. Ming-Bo Wang and Richard Brettell (Commonwealth Scientific and Industrial Research Organization) for providing the pVec8 plasmid, and Dr. Steve Strauss (Oregon State University) for providing the pArt vector. Experimental methods performed in this research complied with current laws and regulations of the United States of America. The use of trade, firm, or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable.
Glossary
- ROS
reactive oxygen species
- PCD
programmed cell death
- QRT
quantitative reverse transcriptase
- FDA
fluorescein diacetate staining
- DAPI
4′,6-diamidino-2-phenylindole
- ChIP
chromatin immunoprecipitation
- MTT
dimethylthiazol
- DAB
3,3′-diaminobenzidine
- NBT
nitro-blue tetrazolium
- PBS
phosphate-buffered saline
- cDNA
complementary DNA
- RT
reverse transcriptase
Footnotes
This work was supported by the U.S. Department of Agriculture-Agricultural Research Service Administrator’s Postdoctoral Research Associate Program (to R.C.M. to fund K.M.G.-C.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abercrombie JM, Halfhill MD, Ranjan P, Rao MR, Saxton AM, Yuan JS, Stewart CN., Jr (2008) Transcriptional responses of Arabidopsis thaliana plants to As (V) stress. BMC Plant Biol 8: 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. (2007) Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res 100: 1512–1521 [DOI] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS. (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53: 1331–1341 [PubMed] [Google Scholar]
- Ameri K, Harris AL. (2008) Activating transcription factor 4. Int J Biochem Cell Biol 40: 14–21 [DOI] [PubMed] [Google Scholar]
- An JH, Blackwell TK. (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17: 1882–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assunção AG, Schat H, Aarts MG. (2010a) Regulation of the adaptation to zinc deficiency in plants. Plant Signal Behav 5: 1553–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assunção AGL, Herrero E, Lin YF, Huettel B, Talukdar S, Smaczniak C, Immink RGH, van Eldik M, Fiers M, Schat H, et al. (2010b) Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci USA 107: 10296–10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MN, Butterworth EA, Kilberg MS. (2013) Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am J Physiol Endocrinol Metab 304: E789–E799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board PG, Menon D. (2013) Glutathione transferases, regulators of cellular metabolism and physiology. Biochim Biophys Acta 1830: 3267–3288 [DOI] [PubMed] [Google Scholar]
- Boyd V, Cholewa OM, Papas KK. (2008) Limitations in the use of fluorescein diacetate/propidium iodide (FDA/PI) and cell permeable nucleic acid stains for viability measurements of isolated islets of Langerhans. Curr Trends Biotechnol Pharm 2: 66–84 [PMC free article] [PubMed] [Google Scholar]
- Brigelius-Flohé R, Maiorino M. (2013) Glutathione peroxidases. Biochim Biophys Acta 1830: 3289–3303 [DOI] [PubMed] [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, Brutnell T, Sibout R, Bevan M, Budak H, et al. (2011) Brachypodium as a model for the grasses: today and the future. Plant Physiol 157: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I. (2000) Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol 146: 185–205 [DOI] [PubMed] [Google Scholar]
- Calabrese V, Cornelius C, Dinkova-Kostova AT, Iavicoli I, Di Paola R, Koverech A, Cuzzocrea S, Rizzarelli E, Calabrese EJ. (2012) Cellular stress responses, hormetic phytochemicals and vitagenes in aging and longevity. Biochim Biophys Acta 1822: 753–783 [DOI] [PubMed] [Google Scholar]
- Chen P, Martinez-Finley EJ, Bornhorst J, Chakraborty S, Aschner M. (2013) Metal-induced neurodegeneration in C. elegans. Front Aging Neurosci 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, Jinn TL. (2005) A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol 139: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JM, Gillings MR, Altavilla N, Beattie AJ. (2001) Potential problems with fluorescein diacetate assays of cell viability when testing natural products for antimicrobial activity. J Microbiol Methods 46: 261–267 [DOI] [PubMed] [Google Scholar]
- Conforti P (2011) Looking Ahead in World Food and Agriculture: Perspectives to 2050. Food and Agriculture Organization of the United Nations, Rome [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, A-H-Mackerness S, Hancock JT, Neill SJ. (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski JE, Baldwin JC, Martin RC. (2008) Cloning and characterization of a salt stress-inducible small GTPase gene from the model grass species Lolium temulentum. J Plant Physiol 165: 651–661 [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM. (2001) Brachypodium distachyon: A new model system for functional genomics in grasses. Plant Physiol 127: 1539–1555 [PMC free article] [PubMed] [Google Scholar]
- Eide DJ. (2009) Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem 284: 18565–18569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F. (2000) Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24: 469–486 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV, Battisti DS, Beachy RN, Cooper PJ, Fischhoff DA, Hodges CN, Knauf VC, Lobell D, Mazur BJ, Molden D, et al. (2010) Radically rethinking agriculture for the 21st century. Science 327: 833–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frassinetti S, Bronzetti G, Caltavuturo L, Cini M, Croce CD. (2006) The role of zinc in life: a review. J Environ Pathol Toxicol Oncol 25: 597–610 [DOI] [PubMed] [Google Scholar]
- Fryer MJ, Oxborough K, Mullineaux PM, Baker NR. (2002) Imaging of photo-oxidative stress responses in leaves. J Exp Bot 53: 1249–1254 [PubMed] [Google Scholar]
- Gaetke LM, Chow CK. (2003) Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 189: 147–163 [DOI] [PubMed] [Google Scholar]
- Gems D, Partridge L. (2013) Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol 75: 621–644 [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–930 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. (2008) RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter KM, Lin S, Blackwell TK. (2013) Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet 9: e1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther V, Lindert U, Schaffner W. (2012) The taste of heavy metals: gene regulation by MTF-1. Biochim Biophys Acta 1823: 1416–1425 [DOI] [PubMed] [Google Scholar]
- Ha KN, Chen Y, Cai J, Sternberg P., Jr (2006) Increased glutathione synthesis through an ARE-Nrf2-dependent pathway by zinc in the RPE: implication for protection against oxidative stress. Invest Ophthalmol Vis Sci 47: 2709–2715 [DOI] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 15: 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Hei TK, Liu SX, Waldren C. (1998) Mutagenicity of arsenic in mammalian cells: role of reactive oxygen species. Proc Natl Acad Sci USA 95: 8103–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M, Kenyon C. (2010) Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci USA 107: 9730–9735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13: 89–102 [DOI] [PubMed] [Google Scholar]
- Higgins VJ, Alic N, Thorpe GW, Breitenbach M, Larsson V, Dawes IW. (2002) Phenotypic analysis of gene deletant strains for sensitivity to oxidative stress. Yeast 19: 203–214 [DOI] [PubMed] [Google Scholar]
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM. (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. (2012) Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D, Chambers JP, Behpouri A, Kelly SP, Whelan L, Pietrzykowska M, Downey F, McCabe PF, Ng CKY. (2013) Brachypodium distachyon cell suspension cultures: establishment and utilisation. Cereal Res Commun 42: 58–69 [Google Scholar]
- Huo N, Garvin DF, You FM, McMahon S, Luo MC, Gu YQ, Lazo GR, Vogel JP. (2011) Comparison of a high-density genetic linkage map to genome features in the model grass Brachypodium distachyon. Theor Appl Genet 123: 455–464 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH. (1993) Plant bZIP protein DNA binding specificity. J Mol Biol 230: 1131–1144 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Johnson C, Boden E, Desai M, Pascuzzi P, Arias J. (2001) In vivo target promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J 28: 237–243 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. (2010) The genetics of ageing. Nature 464: 504–512 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL. (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Lehmann M, Laxa M, Sweetlove LJ, Fernie AR, Obata T. (2012) Metabolic recovery of Arabidopsis thaliana roots following cessation of oxidative stress. Metabolomics 8: 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Rudi H, Stockinger EJ, Cheng H, Cao M, Fox SE, Mockler TC, Westereng B, Fjellheim S, Rognli OA, et al. (2012) Comparative analyses reveal potential uses of Brachypodium distachyon as a model for cold stress responses in temperate grasses. BMC Plant Biol 12: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Aarts MG. (2012) The molecular mechanism of zinc and cadmium stress response in plants. Cell Mol Life Sci 69: 3187–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP. (2014) We need winners in the race to increase photosynthesis in rice, whether from conventional breeding, biotechnology or both. Plant Cell Environ 37: 19–21 [DOI] [PubMed] [Google Scholar]
- Long SP, Ainsworth EA, Leakey AD, Nösberger J, Ort DR. (2006) Food for thought: lower-than-expected crop yield stimulation with rising CO2 concentrations. Science 312: 1918–1921 [DOI] [PubMed] [Google Scholar]
- Lu YP, Li ZS, Rea PA. (1997) AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc Natl Acad Sci USA 94: 8243–8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. (2013) Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 53: 401–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Tavernarakis N, Kroemer G. (2010) Can autophagy promote longevity? Nat Cell Biol 12: 842–846 [DOI] [PubMed] [Google Scholar]
- Martin RC, Glover-Cutter K, Baldwin JC, Dombrowski JE. (2012) Identification and characterization of a salt stress-inducible zinc finger protein from Festuca arundinacea. BMC Res Notes 5: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Glover-Cutter K, Martin RR, Dombrowski JE. (2013) Virus induced gene silencing in Lolium temulentum. Plant Cell Tissue Organ Cult 113: 163–171 [Google Scholar]
- Miao X, Sun W, Fu Y, Miao L, Cai L. (2013) Zinc homeostasis in the metabolic syndrome and diabetes. Fr Medecine 7: 31–52 [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. (2007) Sirtuins in mammals: insights into their biological function. Biochem J 404: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133: 481–489 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65: 55–63 [DOI] [PubMed] [Google Scholar]
- Murray F, Brettell R, Matthews P, Bishop D, Jacobsen J. (2004) Comparison of Agrobacterium-mediated transformation of four barley cultivars using the GFP and GUS reporter genes. Plant Cell Rep 22: 397–402 [DOI] [PubMed] [Google Scholar]
- Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. (2009) Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8: 524–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Lemaire SD, Crespo JL. (2012) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddington DL, Fang FC, Laessig T, Cooper AM, Orme IM, Buchmeier NA. (2001) Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect Immun 69: 4980–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redden R. (2013) New approaches for crop genetic adaptation to the abiotic stresses predicted with climate change. Agronomy 3: 419–432 [Google Scholar]
- Reichert K, Menzel R. (2005) Expression profiling of five different xenobiotics using a Caenorhabditis elegans whole genome microarray. Chemosphere 61: 229–237 [DOI] [PubMed] [Google Scholar]
- Ricardi MM, González RM, Iusem ND. (2010) Protocol: fine-tuning of a chromatin immunoprecipitation (ChIP) protocol in tomato. Plant Methods 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards NG, Kilberg MS. (2006) Asparagine synthetase chemotherapy. Annu Rev Biochem 75: 629–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S. (2011) Extending life span by increasing oxidative stress. Free Radic Biol Med 51: 327–336 [DOI] [PubMed] [Google Scholar]
- Sahu SN, Lewis J, Patel I, Bozdag S, Lee JH, Sprando R, Cinar HN. (2013) Genomic analysis of stress response against arsenic in Caenorhabditis elegans. PLoS ONE 8: e66431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N, Georgiev O, Schaffner W. (2011) The parkin mutant phenotype in the fly is largely rescued by metal-responsive transcription factor (MTF-1). Mol Cell Biol 31: 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab B, Hülskamp M. (2008) Vital stain for plant cytoplasm. CSH Protoc 2008: t4936. [DOI] [PubMed] [Google Scholar]
- Serageldin I. (1999) Biotechnology and food security in the 21st century. Science 285: 387–389 [DOI] [PubMed] [Google Scholar]
- Sinclair SA, Krämer U. (2012) The zinc homeostasis network of land plants. Biochim Biophys Acta 1823: 1553–1567 [DOI] [PubMed] [Google Scholar]
- Sun L, Yang ZT, Song ZT, Wang MJ, Sun L, Lu SJ, Liu JX. (2013) The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J 76: 274–286 [DOI] [PubMed] [Google Scholar]
- Sunkar R (2010) Plant Stress Tolerance: Methods and Protocols. Humana Press, New York [Google Scholar]
- Swindell WR. (2011) Metallothionein and the biology of aging. Ageing Res Rev 10: 132–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk B. (2013) Zinc homeostasis and neurodegenerative disorders. Front Aging Neurosci 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. (2006) Reactive oxygen species in plant cell death. Plant Physiol 141: 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel JE, Almar Villanueva L, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, Ver Loren van Themaat E, Koornneef M, Aarts MG. (2006) Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol 142: 1127–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Bragg J (2009) Brachypodium distachyon, a new model for the Triticeae. In C Feuillet, GJ Muehlbauer, eds, Genetics and Genomics of the Triticeae. Springer, New York, pp 427–449 [Google Scholar]
- Vogel J, Hill T. (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27: 471–478 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Leong OM, Hayden DM. (2006) Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult 84: 100179–100191 [Google Scholar]
- Wan X, Tan J, Lu S, Lin C, Hu Y, Guo Z. (2009) Increased tolerance to oxidative stress in transgenic tobacco expressing a wheat oxalate oxidase gene via induction of antioxidant enzymes is mediated by H2O2. Physiol Plant 136: 30–44 [DOI] [PubMed] [Google Scholar]
- Wang MB, Matthews PR, Upadhyaya MN, Waterhouse PM. (1998) Improved vectors for Agrobacterium tumefaciens-mediated plant transformation. Acta Hortic 461: 401–407 [Google Scholar]
- Wang MB, Upadhyaya MN, Brettell RIS, Waterhouse PM. (1997) Intron mediated improvement of a selectable marker gene for plant transformation using Agrobacterium tumefaciens. J Genet Breed 51: 325–334 [Google Scholar]
- Wheeler T, von Braun J. (2013) Climate change impacts on global food security. Science 341: 508–513 [DOI] [PubMed] [Google Scholar]
- Widholm JM. (1972) The use of fluorescein diacetate and phenosafranine for determining viability of cultured plant cells. Stain Technol 47: 189–194 [DOI] [PubMed] [Google Scholar]
- Witcombe JR, Hollington PA, Howarth CJ, Reader S, Steele KA. (2008) Breeding for abiotic stresses for sustainable agriculture. Philos Trans R Soc Lond B Biol Sci 363: 703–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordem BK, Conte SS, Ma JF, Yokosho K, Vasques KA, Gopalsamy SN, Walker EL. (2011) Brachypodium distachyon as a new model system for understanding iron homeostasis in grasses: phylogenetic and expression analysis of Yellow Stripe-Like (YSL) transporters. Ann Bot (Lond) 108: 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Jiang M, Zhang J, Tan M, Hu X. (2006) Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol 141: 475–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Egli D, Georgiev O, Schaffner W. (2001) The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol Cell Biol 21: 4505–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z. (2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Sadoni N, Stelzer E. (2003) Visualizing chromatin and chromosomes in living cells. Methods 29: 42–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.