Mapping-by-sequencing identifies a naturally occurring variant of a kinesin that causes root hairs to branch.
Abstract
In Arabidopsis (Arabidopsis thaliana), branched root hairs are an indicator of defects in root hair tip growth. Among 62 accessions, one accession (Heiligkreuztal2 [HKT2.4]) displayed branched root hairs, suggesting that this accession carries a mutation in a gene of importance for tip growth. We determined 200- to 300-kb mapping intervals using a mapping-by-sequencing approach of F2 pools from crossings of HKT2.4 with three different accessions. The intersection of these mapping intervals was 80 kb in size featuring not more than 36 HKT2.4-specific single nucleotide polymorphisms, only two of which changed the coding potential of genes. Among them, we identified the causative single nucleotide polymorphism changing a splicing site in ARMADILLO REPEAT-CONTAINING KINESIN1. The applied strategies have the potential to complement statistical methods in high-throughput phenotyping studies using different natural accessions to identify causative genes for distinct phenotypes represented by only one or a few accessions.
Root hairs are tubular tip outgrowths of single root epidermal cells (trichoblasts). They are an excellent genetic system and serve as a model to study the molecular components regulating tip growth (Carol and Dolan, 2002; Samaj et al., 2004; Lee and Yang, 2008). One of the main regulators of tip growth in root hairs is the small G protein RHO OF PLANTS2 (ROP2; Jones et al., 2002; Payne and Grierson, 2009). ROP2 determines the position of root hairs in incipient epidermal root hair cells and remains localized in the emerging tip during root hair tip growth (Molendijk et al., 2001; Jones et al., 2002). In addition, other factors have been identified to be important for growth and its directionality including phosphoinositides, cytoplasmic [Ca2+] gradients and their oscillation, reactive oxygen species, the RAB GTPase homolog A4B, and the cytoskeleton (Foreman et al., 2003; Preuss et al., 2004, 2006; Carol et al., 2005; Thole et al., 2008; Heilmann, 2009).
Defects in essential processes for the establishment and maintenance of tip growth lead to deviations in root hair morphology such as branching and waviness (Samaj et al., 2004; Lee and Yang, 2008). Both the microtubules (MTs) and actin are important regulators of tip growth with MTs maintaining one growth point (Bibikova et al., 1999; Miller et al., 1999; Baluska et al., 2000). The latter is evident from the finding that artificially induced [Ca2+] gradients can induce additional growth tips when MTs are destroyed by drug treatments (Bibikova et al., 1999).
Although the genetic and molecular analysis revealed a well-understood working model for root hair growth, little is known about the natural variation of the underlying processes. Which are the adaptive processes of relevance to specific environmental cues and which have already been selected for in natural accessions? One way to address this question is to link genotype and phenotype by association mapping using various Arabidopsis (Arabidopsis thaliana) accessions. This is greatly facilitated by the 1001 Genomes Project (http://1001genomes.org), providing an increasing number of sequenced accessions. In some cases, phenotypes are only found in one or a few accessions. When the minor allele frequency (AF) is low, the identification of such rare causative alleles with genome-wide association mapping studies is challenging because they cannot be discriminated from false positives (e.g. sequencing errors or synthetic associations; Korte and Farlow, 2013), they are not detectable because of chosen thresholds, or they do not support a statistically significant value (Cantor et al., 2010). Other time-consuming approaches with low mapping resolution, such as quantitative trait loci mapping, need to be followed to identify the causative gene. For this, mapping-by-sequencing, which was originally developed for the identification of mutagen-induced changes in model species (Schneeberger et al., 2009b), can help to rapidly identify causal polymorphisms including nonmodel and nonreference strains (Nordström et al., 2013; Takagi et al., 2013). The resolution of mapping-by-sequencing experiments in Arabidopsis mapping populations is typically between multiple hundreds of kilobase pairs up to a few megabase pairs (James et al., 2013). Although intervals of this size allow the identification of causal mutations in forward genetic screens, they are problematic for the analysis of diverse Arabidopsis accessions because the single nucleotide polymorphism (SNP) density is very high; consequently, hundreds of polymorphisms have to be considered.
Here we report on a modification of the mapping-by-sequencing strategy providing a shortcut from distinct, monogenic accession-specific phenotypes to the causative SNP. When studying root hair morphology in 62 accessions for which the genome sequences were released by the 1001 Genomes Project (Cao et al., 2011), we found one accession (Heiligkreuztal2 [HKT2.4]) in which almost all root hairs were branched. To identify the causative gene, we used an approach based on mapping-by-sequencing. Instead of one outcross, we used outcrosses with three different accessions. We selected F2 seedlings exhibiting the distinct, monogenic, recessive root hair branching phenotype for sequencing. Combining the intersection of the three resulting mapping intervals with a selection for accession-specific SNPs revealed two primary candidate genes responsible for the root phenotype. We demonstrate that the causative SNP renders a splicing site in ARMADILLO REPEAT-CONTAINING KINESIN1 (ARK1) inactive and therefore leads to a defective ARK1/MORPHOGENESIS OF ROOT HAIR2 (MRH2) protein that is thought to coordinate actin microfilaments and MTs during tip growth of root hairs (Yang et al., 2007; Yoo and Blancaflor, 2013).
RESULTS AND DISCUSSION
HKT2.4 Has a Root Hair Branching Phenotype
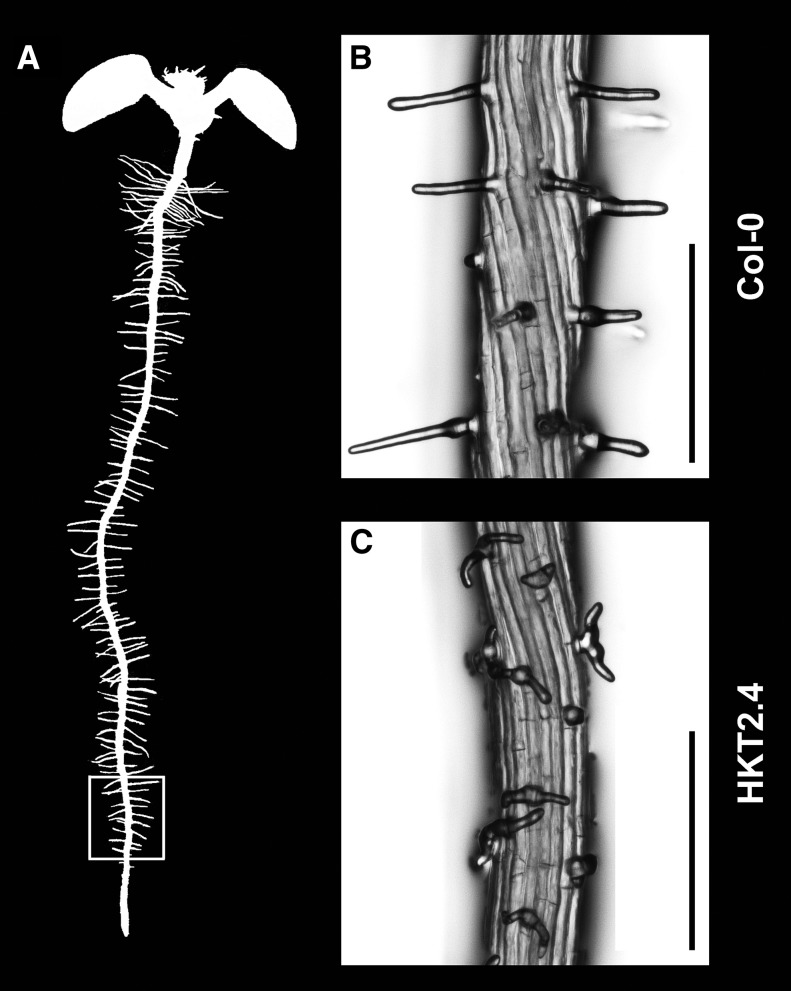
In this study, we analyzed the root hair morphology of 62 Arabidopsis accessions in the root hair patterning zone. Among the analyzed accessions, only HKT2.4 that originated from the area around Tuebingen, Germany, showed branched root hairs (Fig. 1; Supplemental Table S1).
Figure 1.
Root hair morphology in Col-0 and HKT2.4 plants. A, Scheme of Arabidopsis roots. The analyzed region is marked with a white square. This region is named the lower region in the text. B, Root hairs of Col-0 plants are tubular. C, Root hairs of HKT2.4 are wavy and branched. Scale bars = 250 µm.
To determine whether root hair branching is caused by one or multiple loci, we performed segregation analyses. We chose the Arabidopsis ecotypes Columbia-0 (Col-0), Landsberg erecta (Ler), and Wassilewskija-2 (Ws-2) as crossing partners because various root hair mutants have been described in these backgrounds, thereby enabling a direct phenotypic comparison in subsequent experiments. First, we determined the percentage of branched root hairs in all four accessions in the lower zone of the root (Fig. 1A). Whereas almost all root hairs were branched in HKT2.4 (96.8% ± 4.1%), less than 5% were branched in Col-0 and Ws-2 and 10.9% ± 4.1% were branched in Ler (Supplemental Table S2). In all F1 seedlings from all three crosses, we observed an intermediate root hair branching phenotype in the upper region of the root (Supplemental Fig. S1). In this region, F2 seedlings segregated in a 1:2:1 ratio (n > 100; Table I) for Col-0/Ler/Ws-2-like, intermediate, and HKT2.4-like phenotypes, indicating semidominance for this phenotype (Col-0, 0.8:2.3:0.9; Ler, 0.9:2.1:1; and Ws-2, 0.9:2.1:1). Because all seedlings with an intermediate root hair branching phenotype in the upper region showed no branching in the lower region, a 3:1 segregation ratio for nonbranching versus branching could be deduced for the lower region.
Table I. Segregation of the root hair branching phenotype in F2 seedlings.
For the segregation analysis, root hairs along the whole root were analyzed. The intermediate branching phenotype was only observed in the upper zone (below the embryonic root hairs; Supplemental Fig. S1).
Taken together, the genetic data indicate that the distinct HKT2.4 root hair branching phenotype in the lower region is monogenetic and therefore accessible to a mapping approach using Col-0, Ler, and Ws-2.
Study Design Aiming at a High Mapping Resolution
Because quantitative trait locus mapping is a powerful but time-consuming method with low mapping resolution, we searched for mapping alternatives to unravel the underlying mutation of the HKT2.4-specific root hair branching phenotype. Mapping-by-sequencing combining bulk segregant analyses with next-generation sequencing is one recently established method for rapid ethyl methanesulfonate mutant identification in Arabidopsis, Arabis alpina, and other species (Schneeberger et al., 2009b; Hartwig et al., 2012; Nordström et al., 2013).
Simulations of mapping-by-sequencing experiments using mapping populations generated by crossing Arabidopsis Ler and Col-0 revealed relevant criteria for the experimental design (James et al., 2013). On the basis of these simulations, four main aspects were considered.
First, we decided to use paired-end sequencing because this is expected to result in an increased fraction of informative read alignments (James et al., 2013). Second, because 1% to 2% of false-scored plants can lead to 82% to 145% more candidate SNPs in the simulation (James et al., 2013) and given that roots exhibit high phenotypical plasticity (Müller and Schmidt, 2004), we performed pre-experiments and segregation analysis to ensure precise phenotyping. Third, because the coverage and the number of recombinants are correlated and both have an impact on the size of the mapping interval (James et al., 2013), we aimed for a 25-fold coverage. In addition, we phenotypically selected about 200 seedlings from the F2 mapping population. This strategy is expected to define a 400-kb mapping interval (James et al., 2013). Fourth, we used three populations to take advantage of different recombination hotspots in different accessions (Drouaud et al., 2006, 2007; Kim et al., 2007; Salomé et al., 2012) and to increase the number of recombinants.
Identification of an 80-kb Mapping Interval
In light of the above-mentioned four aspects, we followed the procedure summarized in Supplemental Figure S2, whereby HKT2.4 was crossed to each of Col-0, Ler, and Ws-2, respectively. Because the success of a particular cross can depend on which accession was used as a mother (Autran et al., 2011; Fitz Gerald et al., 2014), reciprocal crosses were performed. F2 seedlings with HKT2.4-specific phenotypes were selected from all three segregating populations and pooled for both directions of the crossings. These seedlings were expected to carry the causative SNP and to vary in other SNPs due to recombination events. This resulted in F2 pools with at least 199 seedlings each (Table II). The parental and bulked segregants’ genomes were whole-genome sequenced with a minimum of 25-fold mean coverage, resulting in at least 110.39 million positions with sufficient and unique sequencing information. More than 95% of the reads were mapped (Table II; Supplemental Fig. S3).
Table II. Summary of genome resequencing.
N.A., Not available.
| Sample and Resequencing Information | HKT2.4 | Col-0 | Ler | Ws-2 | HKT2.4 × Col-0 | HKT2.4 × Ler | HKT2.4 × Ws-2 |
|---|---|---|---|---|---|---|---|
| No. of pooled F2 seedlings | 200 | 200 | 200 | 200 | 205 | 214 | 199 |
| Coverage (mean) | 39× | 25× | 33× | 36× | 28× | 42× | 33x |
| No. of reads | 5.68 × 107 | 3.65 × 107 | 4.89 × 107 | 5.31 × 107 | 4.20 × 107 | 6.28 × 107 | 5.00 × 107 |
| No. of unique positions | 1.11 × 108 | 1.18 × 108 | 1.10 × 108 | 1.12 × 108 | 1.18 × 108 | 1.14 × 108 | 1.15 × 108 |
| No. of markers | N.A. | N.A. | N.A. | N.A. | 178,042 | 43,204 | 21,164 |
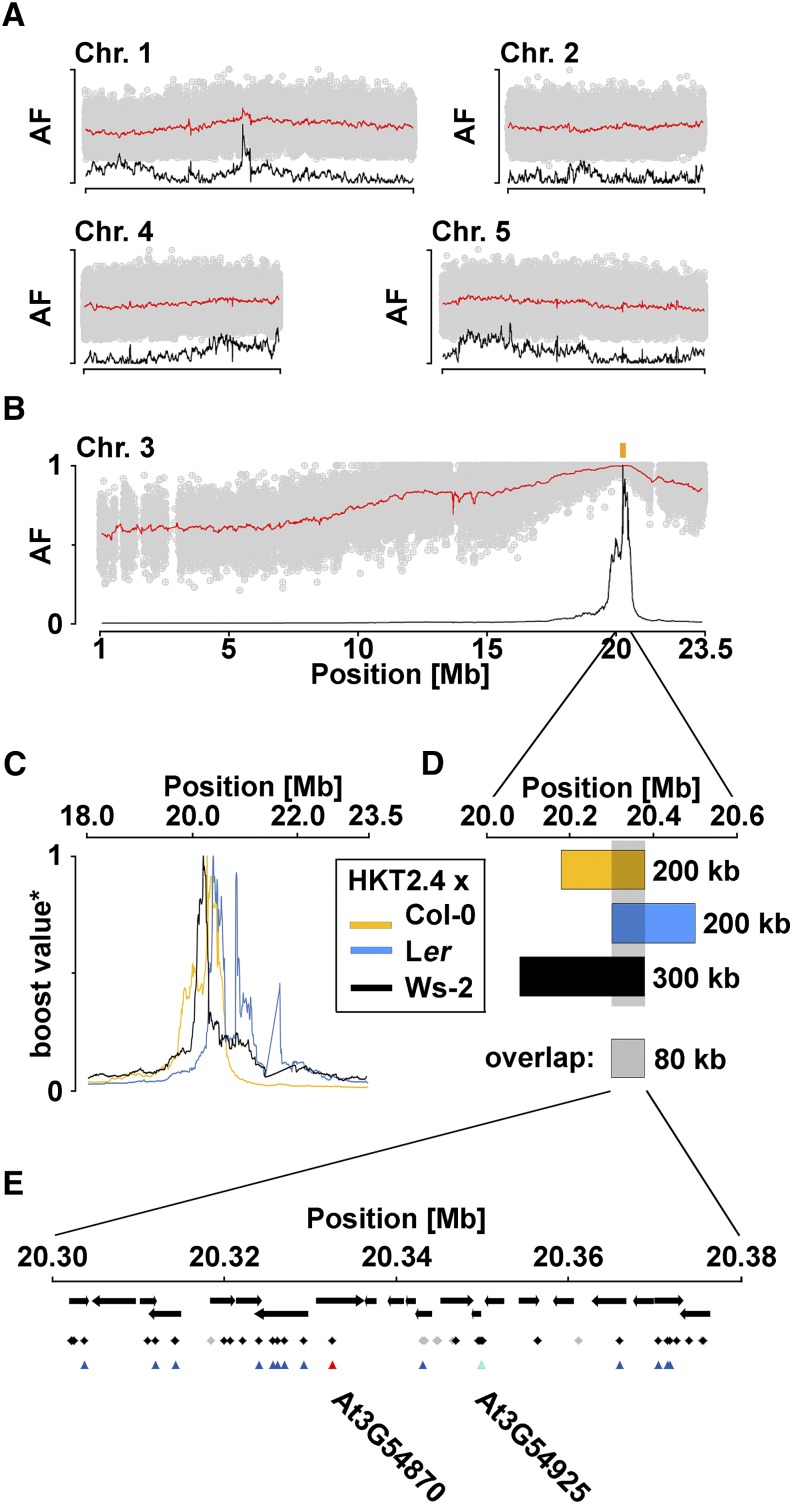
For all three crossings, we used the resequencing information of the parental lines to generate genome-wide lists of markers used for the estimation of AFs with SHOREmap software. Average AFs, as assessed in sliding windows, and SHOREmap-derived boost values revealed linkage between a region on chromosome 3 and the phenotype, thereby providing evidence that chromosome 3 harbors the causative locus (Fig. 2, A–C; Supplemental Figs. S4 and S5). The corresponding mapping intervals had sizes of 200 kb (crossings with Col-0 and Ler) and 300 kb (crossing with Ws-2) but their intersection revealed a final mapping interval of not more than 80 kb between 20.30 and 20.38 megabase pairs on chromosome 3 (Fig. 2D).
Figure 2.
Identification of candidate SNPs through a combined mapping-by-sequencing and HKT2.4-specific SNP-selection approach. A, Visualization of AFs at the noncausal chromosomes from the phenotypically selected F2 mapping population of HKT2.4 crossed to Col-0 (scheme in Supplemental Fig. S2). The points in gray are individual AFs at the markers, which were created by using SHOREmap according to the resequencing information of parental lines, namely HKT2.4 and Col-0. The red line shows the average AFs at the markers within 200-kb sliding windows (10-kb step). The black line is the sliding window-based boost value, a slightly modified version of the r value as introduced with SHOREmap. B, The same visualization reveals that chromosome 3 carries the causal mutation for the selected phenotype. The peak of boost values is expected to be linked to the causal mutations. The yellow rectangle marks the SHOREmap-derived mapping interval. C, Enlargement for the boost values derived from HKT2.4 crossed to Col-0, Ler, and Ws-2 on chromosome 3. D, Intersection of SHOREmap-derived mapping intervals from the three crossings leads to a reduced region of 80 kb (20,300,000–20,379,999), which harbors the causal mutation. E, The 80-kb overlapping mapping interval with all genes annotated by arrows according to TAIR10 (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_gff3/). Black and gray diamonds are HKT2.4-specific intragenic SNPs identified from the sequencing approach. Gray indicates SNPs with a coverage below 10. Triangles indicate SNPs that are also HKT2.4 specific for the set of 62 initially analyzed accessions. Only two of these change the coding DNA sequence as annotated by TAIR10. Cyan indicates nonsynonymous SNPs. Red indicates change of splicing site. On the basis of this selection, the two most likely causal genes are given. See Supplemental Figures S4 and S5 for the crossings with Ler and Ws-2. The HKT2.4-specific SNP identification from the sequencing and from the accession-based approach is documented in Supplemental Table S3. Chr, Chromosome; Mb, megabase pair.
To summarize, mapping-by-sequencing can be used to map Arabidopsis accession-specific phenotypes. In this study, our strategy outperformed the prediction and reduced the mapping interval to 80 kb. This suggests that, depending on the focal accession, different crossing partners (namely those with the highest SNP density and different recombination landscapes) can further increase the mapping resolution.
Identification of Candidate Genes through HKT2.4-Specific SNP Selection
The identified 80-kb mapping interval contained 21 genes (The Arabidopsis Information Resource release 10 [TAIR10], http://www.arabidopsis.org; Fig. 2E). To further narrow down the number of candidate genes, we made use of the sequencing data and extracted all SNPs that were HKT2.4 specific and therefore did not occur in Col-0, Ws-2, or Ler (Supplemental Table S3). This step resulted in 166 candidate SNPs. Using the Col-0 TAIR10 annotation data, we identified 72 intragenic SNPs, 13 of which change the amino acid sequence of the corresponding proteins, reducing the number of candidate genes to 5 (Supplemental Table S3).
Because none of the 62 accessions showed the HKT2.4-specific phenotype, it is conceivable that they do not have the causative polymorphism. We therefore extracted the sequences of the mapping interval for the analyzed 62 accessions from the 1001 Genomes Project (http://signal.salk.edu/atg1001/3.0/gebrowser.php), called the SNPs, and selected for HKT2.4-specific SNPs (Supplemental Table S3). When selecting only those HKT2.4-specific SNPs that were identified by both approaches (sequencing and an accession-based approach), 36 SNPs were considered, 15 of which are intragenic and two of the intragenic SNPs change the amino acid sequence of two proteins (Fig. 2E; Supplemental Table S3).
For the latter two SNPs, we extracted the nucleotide at the corresponding position for all available accessions from the 1001 Genomes Project. For the first SNP at position 20,332,575 on chromosome 3, none of the accessions shared the SNP with HKT2.4 (Supplemental Table S4). For the second SNP, only one accession (Kastel-1) shared the same SNP allele with HKT2.4 at position 20,349,799 on chromosome 3. Moreover, 11 additional accessions featured a third allele at the same position (Supplemental Table S5). Neither Kastel-1 nor one of the other accessions (Taynuilt) showed root hair branching (Supplemental Fig. S6). The SNP at position 20,332,575 on chromosome 3 changes a splicing site in At3g54870 according to the TAIR10 annotation. The gene encodes ARK1/MRH2. Root hair branching has been reported for mutants of ARK1 (Jones et al., 2006).
Without additional information, we could identify the causative SNP exclusively by the analysis of wild accessions and their genomes.
Interestingly, no other accession carries the causative SNP in the worldwide collection of more than 800 sequenced accessions, although several accessions around Tuebingen are among these. There might have been either selection against this SNP or the SNP has recently arisen.
A Modification in an ARK1 Splicing Site Causes the HKT2.4-Specific Root Hair Branching Phenotype
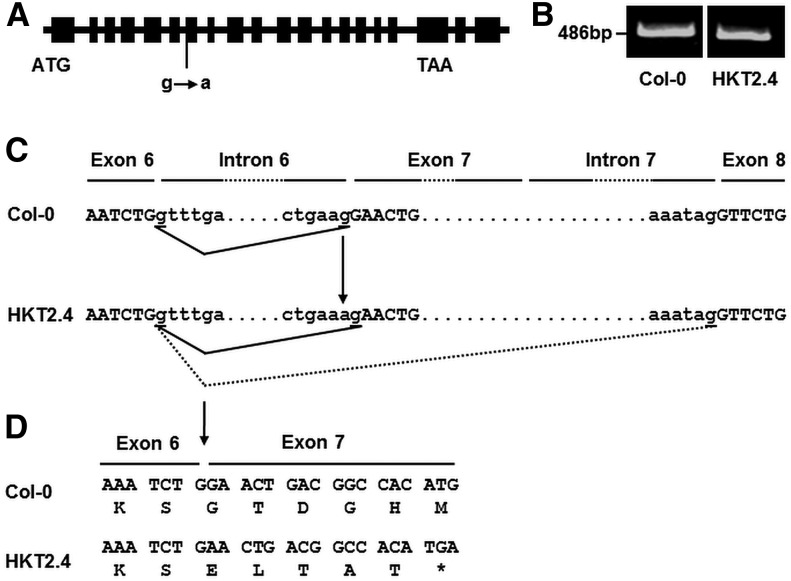
Compared with the Col-0 TAIR10 annotation, the splicing acceptor site in intron 6 of ARK1 differs in HKT2.4 (Fig. 3A). We generated complementary DNA from the relevant root tips including the lower zone analyzed in this study. Using primers located in exon 6 and exon 7, we obtained a PCR product for Col-0 and HKT2.4 of similar size (Fig. 3B). A control PCR using primers for the translation elongation factor EF1αA4 (Kirik et al., 2007) verified the absence of genomic DNA. According to splicing site prediction by NetGene2 (Brunak et al., 1991; Hebsgaard et al., 1996), no PCR product was expected for HKT2.4 because the splicing acceptor site of intron 7 was predicted to be used instead of the splicing acceptor site in intron 6 (Fig. 3C). This prompted us to sequence the HKT2.4 complementary DNA. As a result, the splicing site in intron 6 of HKT2.4 was found to be shifted by one nucleotide leading to a frame shift and premature stop codon in exon 7 (Fig. 3D).
Figure 3.
ARK1 gene structure and splicing site in Col-0 and HKT2.4. A, A scheme of the Col-0 ARK1 gene structure (annotated according to TAIR10, http://www.arabidopsis.org). Boxes represent exons. HKT2.4 has a G-to-A nucleotide exchange in the splicing acceptor site of intron 6. B, Real-time PCR for the region surrounding the mutated splicing site on ARK1 showing that Col-0 and HKT2.4 both have a transcript of similar size. C, A scheme showing the actual splicing sites based on sequencing (black lines) and prediction based on NetGene2 (dashed lines). Note that the splicing site in HKT2.4 is shifted only by one nucleotide compared with Col-0. D, Sequencing revealed that the differing splicing sites in ARK1 of HKT2.4 causes a frame shift resulting in a premature stop codon in exon 7 (asterisk).
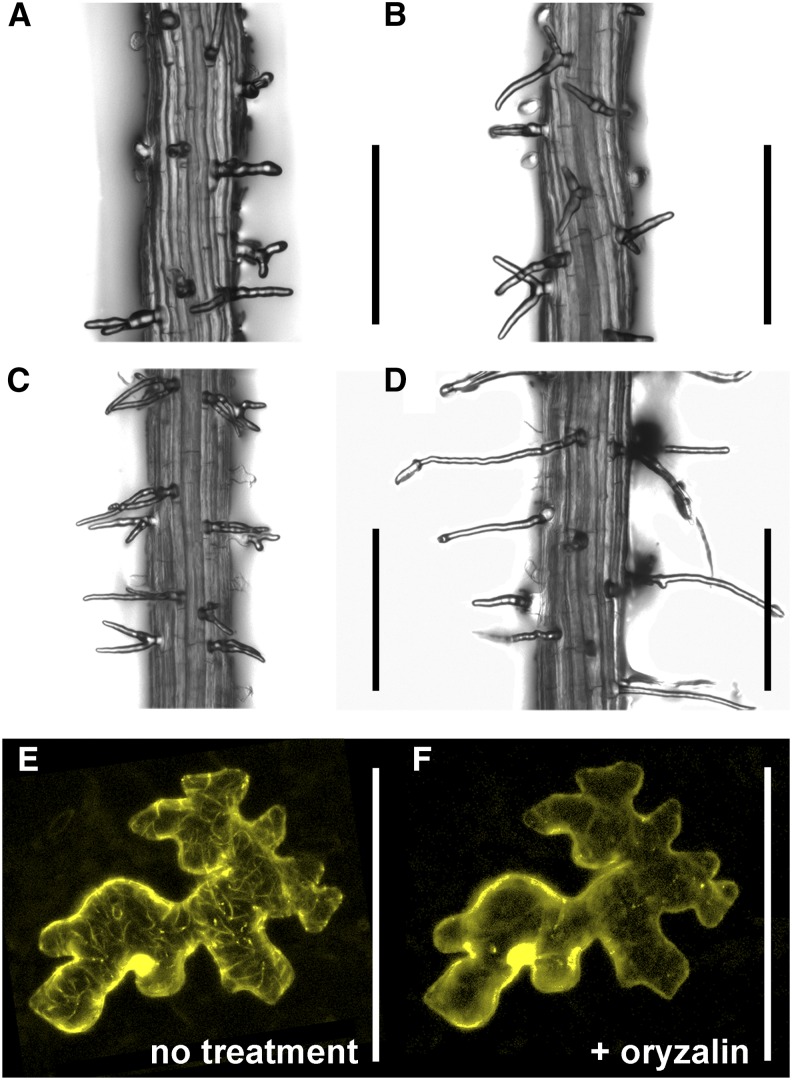
The ark1-1 line is a transfer DNA mutant in the Col-0 background for which a branching phenotype has been reported (Jones et al., 2006). We determined the percentage of branched root hairs in ark1-1 in the lower root region and found no significant difference from HKT2.4 (Supplemental Table S2). Then the causative nature of the ARK1 allele from HKT2.4 was verified by crossing ark1-1 with HKT2.4. F1 seedlings showed branched root hairs similar to HKT2.4 and ark1-1 confirming the allelism (Fig. 4; Supplemental Table S2).
Figure 4.
ARK1Col-0 complements root hair branching in HKT2.4 and localizes to MTs. A and B, The ark1-1 mutant (A) and HKT2.4 (B) have branched root hairs in contrast with Col-0 (Fig. 1 shows Col-0). C, F1 plants of HKT2.4 crossed to ark1-1 have branched root hairs. D, Overexpression of the genomic region of ARK1 from Col-0 rescues the HKT2.4 branched root hair phenotype in T1 plants. The plant with the strongest rescue is shown . See Supplemental Table S6 for the analysis of additional seedlings. For all bright-field pictures, z-stacks of the upper zone (Supplemental Fig. S1) of 7-d-old seedlings were used. E and F, Localization of YFP-ARK1Col-0 before (E) and after (F) oryzalin treatment. Transient expression using the YFP-ARK1Col-0 rescue construct in Arabidopsis leaf epidermal cells. CLSM z-stacks (step size, 2 µm) are shown. Scale bars = 250 µm in A to D and 100 µm in E and F. [See online article for color version of this figure.]
To test whether ARK1 from Col-0 (ARK1Col-0) can rescue the HKT2.4 root hair branching phenotype, yellow fluorescent protein (YFP)-ARK1Col-0 and ARK1Col-0 were overexpressed under the Cauliflower mosaic virus 35S promoter (Pro35S) in HKT2.4 background. The transformation efficiency of HKT2.4 was low. We obtained four Pro35S:YFP-ARK1Col-0 (HKT2.4) lines and eight Pro35S:ARK1Col-0 (HKT2.4) lines, respectively, which showed partial rescue (Fig. 4; Supplemental Table S6). The Pro35S:YFP-ARK1Col-0 (HKT2.4) rescue lines did not show a detectable YFP signal; therefore, we tested the construct in a transient expression assay in Arabidopsis (Col-0) rosette leaf epidermal cells. As previously shown in infiltrated tobacco (Nicotiana tabacum) leaves (Yoo et al., 2008) and supported by oryzalin treatment, YFP-ARK1Col-0 can decorate MTs in Arabidopsis (Fig. 4, E and F). Whereas oryzalin abolished YFP-ARK1Col-0 localization to cytoskeletal structures, this was not seen upon latrunculin B treatment (Supplemental Fig. S7). ARK1 function has been well characterized in root hairs of the Arabidopsis Col-0 ecotype (Sakai et al., 2008; Yoo et al., 2008). The ARK1 protein was recently shown to be a MT plus-end directed catastrophe factor (Eng and Wasteneys, 2014). ARK1 is additionally proposed to specify root hair polarity through interactions with actin and proper targeting of ROP2 and RAB GTPase homolog A4B to the root hair tip (Yang et al., 2007; Yoo and Blancaflor, 2013).
Together, these data show that the candidate SNP in ARK1, as identified by the combined mapping-by-sequencing and HKT2.4-specific SNP-selection approach, leads to a splicing site shift causing the root hair branching phenotype of HKT2.4.
The presented pipeline can also be applied for causative SNPs that change regulatory regions. In this case, the SNP-selection process starting with Supplemental Table S3 would be conducted as described for SNPs in the coding DNA sequence but focus on SNPs in the regulatory regions. Subsequent verification can be done through expression analysis.
This study describes a pipeline for the identification of causal SNPs for phenotypes occurring only in one or a few accessions. We present strategies that narrowed down the final mapping interval for the unique root hair branching phenotype of HKT2.4. Application of an unbiased accession-specific SNP-selection approach led to the identification of the causative SNP in ARK1.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The following Arabidopsis (Arabidopsis thaliana) lines were used in this study: Col-0, Ws-2, Ler, HKT2.4 (CS76404), Kastel-1 (CS22807), Taynuilt (CS1572), ark1-1 (Salk_035063C; Alonso et al., 2003; Jones et al., 2006), Pro35S:ARK1Col-0(HKT2.4), and Pro35S:YFP-ARK1Col-0(HKT2.4). Plant transformation (Clough and Bent, 1998) and growth conditions were previously described (Rishmawi et al., 2014).
Plasmid Construction
For ARK1Col-0, the genomic region (from the start codon to stop codon with the introns) was amplified from Col-0 (primers in Supplemental Table S7), cloned into pDONR20,1 and subsequently transferred to pFAST-R02 (Shimada et al., 2010) and pENSG-YFP using Gateway technology (Invitrogen). pENSG-YFP originates from N. Medina-Escobar (unpublished data). A version for C-terminal fusions is published (Feys et al., 2005).
Material for Sequencing
Reciprocal crosses between Arabidopsis ecotypes were done to obtain the F2 mapping populations. About 200 F2 seedlings of the reciprocal crosses showing HKT2.4-like branched root hairs were pooled. DNA was extracted using an DNeasy Plant Mini Kit (Qiagen). Samples of at least 2 µg of total genomic DNA with high purity (260:280 ratio ≥ 1.8) were sequenced by the Cologne Center for Genomics using one lane for paired-end Illumina HiSeq sequencing.
Resequencing and SHOREmap Analysis
Resequencing was performed with the SHORE short-read analysis pipeline (Ossowski et al., 2008). Raw reads of all seven samples were aligned against the Col-0 reference genome (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_chromosome_files/) using GenomeMapper (Schneeberger et al., 2009a). Short-read alignments were corrected for read-pair information. Consensus calling was performed by using SHORE consensus. SHOREmap v2.1 (http://shoremap.org) was used to predict mapping intervals for each of the three bulked mapping populations (Schneeberger et al., 2009b). SNP variants identified in the resequencing analyses of the parental strains were filtered by SHOREmap create and those passing the filtering were used as markers. SHOREmap outcross calculates the AF estimates at each marker position (i.e. the ratio of the reads of mutant alleles divided by all reads at that marker locus) and determines mapping intervals according to the average AFs within sliding windows and the corresponding coefficients of variation. In addition to AFs, SHOREmap visualizes a different metric for the AFs (boost value [Bv]). If mobs is the observed mean of AFs within a sliding window and t is the AF at which the causal mutation segregates, then Bv = 1/|1 − max(t, 1 − t)/max(mobs,1 − mobs)|. Polymorphisms in the 80-kb intersection of the three mapping intervals (i.e. chromosome 3:20,300,000–20,379,999) were functionally annotated based on TAIR10 gene annotations (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_gff3/).
HKT2.4-Specific SNP Selection
Sequences of the accession (Supplemental Table S1) were extracted from the Arabidopsis 1001 Genomes GEBrowser (http://signal.salk.edu/atg1001/3.0/gebrowser.php) for the region of the mapping interval on chromosome 3-covering positions 20,300,000 to 20,379,999 (Cao et al., 2011) and were aligned with R software (http://www.r-project.org/) using the adegenet and seqinr package (Charif and Lobry, 2007; Jombart, 2008; R Core Team, 2013). Single-candidate SNPs were similarly extracted from the GEBbrowser for all available accessions. Sequences were available from the U.S. Department of Energy Joint Genome Institute, the Gregor Mendel Institute of Molecular Plant Biology, Institute for Genome Research and Systems Biology of the University of Bielefeld Center for Biotechnology, Max Planck Institute for Developmental Biology, Monsanto Company, the Salk Institute Genomic Analysis Laboratory, or published sources, respectively (Cao et al., 2011; Gan et al., 2011; Long et al., 2013; Schmitz et al., 2013).
Splicing Site Determination
RNA was extracted from the root using an RNeasy Plant Mini Kit. The fragment flanking the splicing site was amplified using Pf-splicing MRH2 and Pr-splicing MRH2 (Supplemental Table S7). NetGene2 was used for prediction of splicing sites (Brunak et al., 1991; Hebsgaard et al., 1996).
Microscopy, Root Hair Branching Analysis, and Transient Assay
Root hairs were analyzed in the elongation zone (lower zone) by bright-field microscopy with a Leica DM5000B microscope equipped with a Leica DFC360FX camera and Leica Application Software AF (Leica Microsystems). Pictures were acquired as previously described (Failmezger et al., 2013). For transient expression, 3-week-old Arabidopsis (Col-0) rosette leaf epidermal cells were biolistically transformed with ARK1Col-0-pENSG-YFP or the F-actin marker Lifeact-eGFP (Era et al., 2009) as previously described (Steffens et al., 2014) and analyzed with confocal laser-scanning microscopy (CLSM) after 19 to 21 h. CLSM was performed using a Leica TCS-SPE confocal microscope (DM5500Q) equipped with Leica Lite 2.05 software (LCS; Leica Microsystems). Z-stacks (step size: 2 µm) were projected with the software’s max projection. Drug treatments with oryzalin (40 µm, 2 h) and latrunculin B (100 µm, 1.5–2 h) were previously described (Steffens et al., 2014).
Statistical Analysis and Software Used
Statistical analysis, except for the resequencing and mapping, was conducted using Origin (OriginLab) applying the Anderson-Darling test (normal distribution) and the Mann-Whitney U test (significance). Images were processed using ImageJ software (http://imagej.nih.gov/ij/, 1997–2012; U.S. National Institutes of Health) and Photoshop Elements software (version 7.0.1; Adobe Systems) using brightness and contrast adjustments for z-stacked pictures only. Z-stacking was done using CombineZM (Alan Hadley, http://web.archive.org/web/20090122072134/http://hadleyweb.pwp.blueyonder.co.uk/CZM/News.htm).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number ARK1 (AT3G54870; Gene ID 824652).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Selection of seedlings for F2 pools used for mapping-by-sequencing.
Supplemental Figure S2. Scheme of mapping-by-sequencing using accessions to generate a mapping interval.
Supplemental Figure S3. Alignment statistics of short reads of parental lines and the mapping populations.
Supplemental Figure S4. AF plots from the phenotypically selected F2 mapping population of HKT2.4 × Ler.
Supplemental Figure S5. AF plots from the phenotypically selected F2 mapping population of HKT2.4 × Ws-2.
Supplemental Figure S6. Root hair phenotypes of accessions with different candidate SNP alleles in AT3G54925.
Supplemental Figure S7. Latrunculin B treatment cannot abolish YFP-ARK1Col-0 cytoskeletal localization.
Supplemental Table S1. List of accessions analyzed for root hair branching phenotype in the lower zone.
Supplemental Table S2. Root hair branching percentages.
Supplemental Table S3. HKT2.4-specific SNPs and their sequence quality and annotation.
Supplemental Table S4. Candidate SNP in AT3G54870 extracted for 855 Arabidopsis accessions.
Supplemental Table S5. Candidate SNP in AT3G54925 extracted for 855 Arabidopsis accessions.
Supplemental Table S6. Root hair branching in rescued T1 seedlings.
Supplemental Table S7. List of primers.
Supplementary Material
Acknowledgments
We thank Janine Altmüller and the Cologne Center for Genomics for sequencing; Tamara Blyszcz, Caroline Bley, and Heike Wolff for help with plant work, Benjamin Jaegle and Jonas R. Klasen for support in using R software; Philipp Thomas for providing the Lifeact-eGFP plasmid; and Bastian Welter for technical assistance.
Glossary
- MT
microtubule
- AF
allele frequency
- SNP
single nucleotide polymorphism
- Col-0
ecotype Columbia-0 of Arabidopsis
- Ler
ecotype Landsberg erecta of Arabidopsis
- Ws-2
ecotype Wassilewskija-2 of Arabidopsis
- TAIR10
The Arabidopsis Information Resource release 10
- CLSM
confocal laser-scanning microscopy
Footnotes
This work was supported by SFB680 (to M.H.), the Cluster of Excellence on Plant Sciences (grant no. EXC 1028 to M.H. and a fellowship to L.R.), and the International Max Planck Research Schools (fellowship to L.R.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. (2011) Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 [DOI] [PubMed] [Google Scholar]
- Baluska F, Salaj J, Mathur J, Braun M, Jasper F, Samaj J, Chua NH, Barlow PW, Volkmann D. (2000) Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev Biol 227: 618–632 [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. (1999) Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J 17: 657–665 [DOI] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S. (1991) Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol 220: 49–65 [DOI] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. (2010) Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am J Hum Genet 86: 6–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Dolan L. (2002) Building a hair: tip growth in Arabidopsis thaliana root hairs. Philos Trans R Soc Lond B Biol Sci 357: 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L. (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry J. (2007) SeqinR 1.0-2: a contributed package to the R Project for Statistical Computing devoted to biological sequences retrieval and analysis. In Bastolla U, Porto M, Roman HE, Vendruscolo M, eds, Structural Approaches to Sequence Evolution. Springer, Berlin, Heidelberg, pp 207–232 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Drouaud J, Camilleri C, Bourguignon PY, Canaguier A, Bérard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, et al. (2006) Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res 16: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M, Martin O, Zanni V, Brunel D, Mézard C. (2007) Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet 3: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng RC, Wasteneys GO. (2014) The microtubule plus-end tracking protein ARMADILLO-REPEAT KINESIN1 promotes microtubule catastrophe in Arabidopsis. Plant Cell 26: 3372–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Era A, Tominaga M, Ebine K, Awai C, Saito C, Ishizaki K, Yamato KT, Kohchi T, Nakano A, Ueda T. (2009) Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failmezger H, Jaegle B, Schrader A, Hülskamp M, Tresch A. (2013) Semi-automated 3D leaf reconstruction and analysis of trichome patterning from light microscopic images. PLoS Comput Biol 9: e1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz Gerald JN, Carlson AL, Smith E, Maloof JN, Weigel D, Chory J, Borevitz JO, Swanson RJ. (2014) New Arabidopsis advanced intercross recombinant inbred lines reveal female control of nonrandom mating. Plant Physiol 165: 175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig B, James GV, Konrad K, Schneeberger K, Turck F. (2012) Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol 160: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouzé P, Brunak S. (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24: 3439–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I. (2009) Using genetic tools to understand plant phosphoinositide signalling. Trends Plant Sci 14: 171–179 [DOI] [PubMed] [Google Scholar]
- James GV, Patel V, Nordström KJ, Klasen JR, Salomé PA, Weigel D, Schneeberger K. (2013) User guide for mapping-by-sequencing in Arabidopsis. Genome Biol 14: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. (2008) adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405 [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS. (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Kirik V, Schrader A, Uhrig JF, Hulskamp M. (2007) MIDGET unravels functions of the Arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell 19: 3100–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Farlow A. (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Yang Z. (2008) Tip growth: signaling in the apical dome. Curr Opin Plant Biol 11: 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjálmsson BJ, Korte A, Nizhynska V, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DD, De Ruijter NCA, Bisseling T, Emons AmC. (1999) The role of actin in root hair morphogenesis: studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J 17: 141–154 [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K. (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Schmidt W. (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJ, Albani MC, James GV, Gutjahr C, Hartwig B, Turck F, Paszkowski U, Coupland G, Schneeberger K. (2013) Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat Biotechnol 31: 325–330 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RJ, Grierson CS. (2009) A theoretical model for ROP localisation by auxin in Arabidopsis root hair cells. PLoS ONE 4: e8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Kovar DR, Lee YR, Staiger CJ, Delmer DP, Liu B. (2004) A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiol 136: 3945–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Schmitz AJ, Thole JM, Bonner HK, Otegui MS, Nielsen E. (2006) A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol 172: 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rishmawi L, Pesch M, Juengst C, Schauss AC, Schrader A, Hülskamp M. (2014) Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol 165: 186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Honing Hv, Nishioka M, Uehara Y, Takahashi M, Fujisawa N, Saji K, Seki M, Shinozaki K, Jones MA, et al. (2008) Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J 53: 157–171 [DOI] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Fitz J, Laitinen RA, Warthmann N, Yant L, Weigel D. (2012) The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb) 108: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J, Baluska F, Voigt B, Schlicht M, Volkmann D, Menzel D. (2004) Endocytosis, actin cytoskeleton, and signaling. Plant Physiol 135: 1150–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. (2013) Patterns of population epigenomic diversity. Nature 495: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Hagmann J, Ossowski S, Warthmann N, Gesing S, Kohlbacher O, Weigel D. (2009a) Simultaneous alignment of short reads against multiple genomes. Genome Biol 10: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU. (2009b) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I. (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Steffens A, Jaegle B, Tresch A, Hülskamp M, Jakoby M. (2014) Processing-body movement in Arabidopsis depends on an interaction between myosins and DECAPPING PROTEIN1. Plant Physiol 164: 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, et al. (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74: 174–183 [DOI] [PubMed] [Google Scholar]
- Thole JM, Vermeer JE, Zhang Y, Gadella TW, Jr, Nielsen E. (2008) Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gao P, Zhang H, Huang S, Zheng ZL. (2007) A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS ONE 2: e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CM, Blancaflor EB. (2013) Overlapping and divergent signaling pathways for ARK1 and AGD1 in the control of root hair polarity in Arabidopsis thaliana. Front Plant Sci 4: 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CM, Wen J, Motes CM, Sparks JA, Blancaflor EB. (2008) A class I ADP-ribosylation factor GTPase-activating protein is critical for maintaining directional root hair growth in Arabidopsis. Plant Physiol 147: 1659–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.