Two Arabidopsis genes with a role in thermotolerance are oriented in a head-to-head manner in the genome and the intergenic region between these genes functions as a heat-inducible bidirectional promoter.
Abstract
In Arabidopsis (Arabidopsis thaliana), the At1g74310 locus encodes for caseinolytic protease B-cytoplasmic (ClpB-C)/heat shock protein100 protein (AtClpB-C), which is critical for the acquisition of thermotolerance, and At1g74320 encodes for choline kinase (AtCK2) that catalyzes the first reaction in the Kennedy pathway for phosphatidylcholine biosynthesis. Previous work has established that the knockout mutants of these genes display heat-sensitive phenotypes. While analyzing the AtClpB-C promoter and upstream genomic regions in this study, we noted that AtClpB-C and AtCK2 genes are head-to-head oriented on chromosome 1 of the Arabidopsis genome. Expression analysis showed that transcripts of these genes are rapidly induced in response to heat stress treatment. In stably transformed Arabidopsis plants harboring this intergenic sequence between head-to-head oriented green fluorescent protein and β-glucuronidase reporter genes, both transcripts and proteins of the two reporters were up-regulated upon heat stress. Four heat shock elements were noted in the intergenic region by in silico analysis. In the homozygous transfer DNA insertion mutant Salk_014505, 4,393-bp transfer DNA is inserted at position −517 upstream of ATG of the AtClpB-C gene. As a result, AtCk2 loses proximity to three of the four heat shock elements in the mutant line. Heat-inducible expression of the AtCK2 transcript was completely lost, whereas the expression of AtClpB-C was not affected in the mutant plants. Our results suggest that the 1,329-bp intergenic fragment functions as a heat-inducible bidirectional promoter and the region governing the heat inducibility is possibly shared between the two genes. We propose a model in which AtClpB-C shares its regulatory region with heat-induced choline kinase, which has a possible role in heat signaling.
High temperature stress constitutes a major environmental stress, particularly for sessile plant systems. On experiencing heat stress (HS), plants respond by vigorously transcribing heat shock protein (Hsp) genes, which function largely as molecular chaperones in protecting cells from damage caused by misfolding of proteins (Kotak et al., 2007a; Richter et al., 2010; Sarkar et al., 2014). Plants harbor several classes of Hsps, such as Hsp100, Hsp90, Hsp70, Hsp40, and small Hsps (Rajan and D’Silva, 2009; Sarkar et al., 2009, 2013a, 2013b; Singh et al., 2012). Hsp100 class proteins, referred to as caseinolytic protease (Clp) family proteins, are exclusively involved in dissolution of toxic protein aggregates (Agarwal et al., 2001; Katiyar-Agarwal et al., 2001; Singh and Grover, 2010). This family comprises ClpA, ClpB, ClpC, ClpD, ClpX, ClpM, ClpN, and ClpY subfamilies (Singh et al., 2010). Unlike others, ClpB/Hsp100 proteins, which are primarily heat inducible, function solely in renaturation of aggregated proteins. Plants comprise three ClpB homologs that are localized to the cytoplasm/nucleus (ClpB-C), mitochondria (ClpB-M), and chloroplast (ClpB-P). ClpB-C protein is considered critical for survival under conditions of high temperature stress. Maize (Zea mays) mutant null for ClpB-C/Hsp100 protein is highly sensitive to extreme temperatures compared with wild-type plants (Nieto-Sotelo et al., 2002). The rice (Oryza sativa) heat stress-associated 32-kD (Hsa32) protein knockout mutant, in which decay of heat-induced ClpB-C is faster, has defective long-term acquired thermotolerance (Lin et al., 2014). In addition to heat-induced regulation, ClpB/Hsp100 genes are developmentally regulated. Uninduced levels of rice ClpB-C/Hsp100 are particularly high in mature grains and decline during the seed germination phase (Singla et al., 1998). Seeds/grains of several other plant genera, such as wheat (Triticum durum), maize, and mustard (Brassica juncea), also contain high uninduced levels of ClpB-C/Hsp100 proteins. In maize, ClpB-C is expressed at a high level in the premeiosis stage of tassel development in the ears, silks, endosperm, and embryo, but is present at only a low level in foliar leaves and roots (Young et al., 2001).
Past studies show that at the transcriptional level, the ability of heat shock promoters to sense heat is largely attributed to heat shock elements (HSEs) located in the promoter region (Sugio et al., 2009; Akerfelt et al., 2010). The consensus sequence that demarcates HSEs in eukaryotic heat-inducible promoters is defined by altering units of 5′-nGAAn-3′, upstream of the TATA box (Mittal et al., 2011; Scharf et al., 2012). The organization of this consensus sequence separates HSEs into three groups: perfect, gapped, and stepped (Yamamoto et al., 2005). HSEs provide the binding targets for the transactive heat shock factors (Hsfs), which mediate the expression of Hsp genes (Zhu et al., 2006; Mittal et al., 2009). The maize ClpB-C promoter contains five HSEs present within the proximal 289-bp promoter region (Nieto-Sotelo et al., 1999). Heat-inducible expression of rice ClpB-C is mediated specifically by OsHsfA2C, which binds to the only HSE (with configuration of nnnnnnGAAnnTTC) present in the ClpB-C promoter (Singh et al., 2012). Yeast (Saccharomyces cerevisiae) ClpB/Hsp100 (Hsp104) is induced after exposure to heavy metals such as arsenite (Sanchez et al., 1992). Metal inducibility of Hsp genes has been attributed to stress-responsive elements (STREs) and animal proto-oncogene1 (AP-1) cis elements noted in the Hsp promoters (Ruis and Schüller, 1995; Grably et al., 2002). At the post-transcriptional level, Hsp mRNAs are preferentially translated, whereas normal cellular mRNAs are translationally suppressed during HS conditions. The 5′ untranslated regions (UTRs) of Hsp mRNAs play a key regulatory role in this process by allowing sustained translation of Hsp transcripts during HS by virtue of the internal ribosome entry site (IRES) element located in this region (Rubtsova et al., 2003; Hernández et al., 2004; Spriggs et al., 2008). It is indicated that Hsp mRNAs are less dependent on cap-dependent mechanisms and have alternative, cap-independent mechanisms for the translation process, which involve the IRES in their 5′ UTRs. Drosophila melanogaster Hsp70, human Hsp70, and Hsp90 (Vivinus et al., 2001; Rubtsova et al., 2003) as well as maize alcohol dehydrogenase (Adh; Mardanova et al., 2008) genes are shown to contain IRES sequences in their 5′ UTRs. Furthermore, studies show that the IRES is preferably located toward the 3′ end of the 5′ UTR, just upstream of the AUG codon (Pelletier et al., 1988; Komar and Hatzoglou, 2005).
Plant ClpB-C transcripts are strongly induced by HS in vegetative tissues in Arabidopsis (Arabidopsis thaliana), maize, tobacco (Nicotiana tabacum), wheat, rice, and soybean (Glycine max; Lee et al., 1994; Schirmer et al., 1994; Wells et al., 1998; Campbell et al., 2001; Young et al., 2001; Agarwal et al., 2002; Nieto-Sotelo et al., 2002; Singh et al., 2010). By expressing the Gus reporter downstream of the rice ClpB-C promoter, Singh et al. (2012) showed heat and metal stress-induced expression in vegetative tissues and constitutive expression in the anther, style, ovary, and embryonal one-half of seeds. The presence of the IRES in the 5′ UTR of maize ClpB-C mRNAs has been shown (Dinkova et al., 2005). Most genetic work on plant ClpB-C protein has been carried out on the Arabidopsis plant model. Mutant Arabidopsis with nonfunctional ClpB-C (hot1-1) or null for ClpB-C (hot1-3) is defective in acquisition of tolerance to high temperature stress (Hong and Vierling, 2000, 2001). Young et al. (2005) analyzed a 390-bp AtClpB-C promoter fragment, incorporated in a mutator-like element, for its expression pattern. Barring the latter study, the AtClpB-C promoter has not been deeply analyzed. In this work, we aimed to elucidate further details on the genetic regulation of the 1kb region upstream of ATG of the AtClpB-C gene. We show that the 1kbAtClpB-C promoter harbors elements for HS and metal stress-inducible transcript expression and for constitutive expression of the transcripts in young developing pollen and mature seeds. The 1kbAtClpB-C promoter sequence contains 222-bp 5′ UTRs with an intervening intron of 106 bp. There was no adverse effect noted on the expression of the downstream reporter gene with respect to both HS inducibility and developmental cues when 77 bp (including 22 bp from the UTR and 55 bp from the intron) were deleted from the 3′ end of the 1kb promoter sequence.
In some studies, the expression of adjacent genes is suggested to be highly correlated. In prokaryotes, it is invariably the case that a single regulatory unit controls two or more functionally related genes (Kruglyak and Tang, 2000). Cho et al. (1998) showed that genes expressed in the same phase of the cell cycle are adjacently located. Several functionally related genes are adjacently located and coexpressed in yeast (Zhang and Smith, 1998; Kruglyak and Tang, 2000). The similarity in expression of clustered genes has also been documented in Caenorhabditis elegans (Lercher et al., 2002), D. melanogaster (Herr and Harris, 2004), and humans (Lercher et al., 2003). Williams and Bowles (2004), while analyzing the effect of genome organization on expression of genes in Arabidopsis, reported that neighboring genes are coexpressed. Bidirectional gene organization is a common architectural feature in the human genome, in which approximately 10% of genes are organized head-to-head (Trinklein et al., 2004). In Arabidopsis, calmodulin methyl transferase (At4g35987) and the senescence-associated gene (At4g35985) are located in a head-to-head orientation with their 1,258-bp intergenic sequence serving as a tissue-specific and stress-inducible bidirectional promoter (Banerjee et al., 2013).
While analyzing the AtClpB-C promoter in this study, we noted that a choline kinase (CK) gene (At1g74320) is present upstream of the AtClpB-C gene in head-to-head orientation on chromosome 1 of the Arabidopsis genome. The CK enzyme catalyzes the first reaction in the Kennedy pathway for phosphatidylcholine (PC) biosynthesis (Gibellini and Smith, 2010). PC is the major phospholipid in the eukaryotic cell membranes and has a considerable structural and functional role (Gibellini and Smith, 2010). It is a precursor for the synthesis of glycerolipids such as monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and sulfoquinovosyldiacylglycerol, which constitute the plastid membranes of plants (Ohlrogge and Browse, 1995). PC also serves as a pool for lipid second messengers, such as lysophosphatidylcholine, phosphatidic acid (PA), diacylglycerol, and lysophosphatidic acid (Mishkind et al., 2009). In higher plants, PC participates in the abiotic stress adaptive response (Zheng et al., 2011). As a response to HS, a gradual but significant increase in CK activity was noted in barley (Hordeum vulgare) aleurone layers (Johnston et al., 2007). The first step of choline phosphorylation in the PC biosynthesis pathway, which is catalyzed by CK, is considered as a rate-controlling step (Tasseva et al., 2004). The Arabidopsis genome contains five CK genes (At1g34100, At1g71697, At1g74320, At2g26830, and At4g09760). At1g71697 (AtCK1) is induced in response to wounding, whereas At1g74320 and At4g09760 are induced by high salt and mannitol (Tasseva et al., 2004). We report that AtClpB-C and At1g74320 (hereafter referred to as AtCk2) genes, both of which are critical in governing the phenotype of heat tolerance (Larkindale and Vierling, 2008), possibly share the requisite genetic regulation machinery under cellular conditions.
RESULTS
Expression Characteristics of the 1kbAtClpB-C Promoter Region
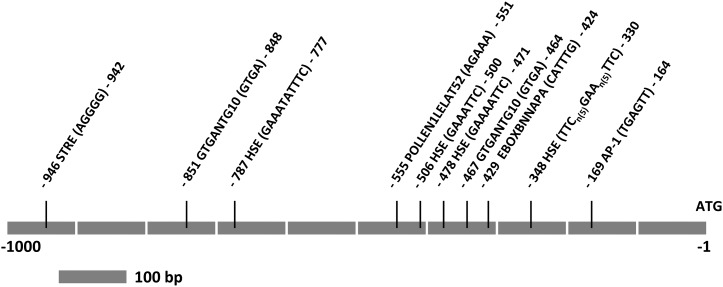
The nucleotide sequence of the 1kbAtClpB-Cpro region was obtained from The Arabidopsis Information Resource (www.arabidopsis.org) database. A search using in silico tools indicated the presence of several stress- and development-related elements in this sequence. Four HSE-like elements were noted with the following configurations: TTCCAGATGAATCTCCTTC (position −330 to −348; appears as the stepped type with TTCnnnnnGAAnnnnnTTC configuration), GAAAATTC (position −471 to −478; variant of the perfect type with configuration of nnnnnnGAAnnTTC), GAAATTC (position −500 to −506; GAAnTTC configuration), and GAAATATTTC (position −777 to −787; GAAnnnnTTC configuration; Fig. 1). A single STRE (position −942 to −946) and one POLLEN1LELAT52 (position −555 to −560) sequence were noted. In addition, two GTGA motifs (positions −464 to −467 and −848 to −851), an E-BOX element (position −424 to −429), and an AP-1 binding element (position −164 to −169) were marked. A canonical TATA box was found at the −235 to −240 position, which is 3′ to the predicted transcription start site (TSS), indicating that transcription initiates without TATA mediation and the promoter is TATA less. All of the above elements are marked taking A of the ATG translation initiation codon as +1 (Fig. 1).
Figure 1.
Linear representation of the 1kb upstream promoter region of the AtClpB-C gene. Sequences for ATG, the HSE (−330 to −348, −471 to −478, −500 to −506, and −777 to −787), the STRE (−942 to −946), the AP-1 element (−164 to −169), the E-BOX element (−424 to −429), the GTGA element (−464 to −467, and −848 to −851), and POLLEN1LELAT52 (−551 to −555) are indicated. All elements are marked taking A of ATG as +1.
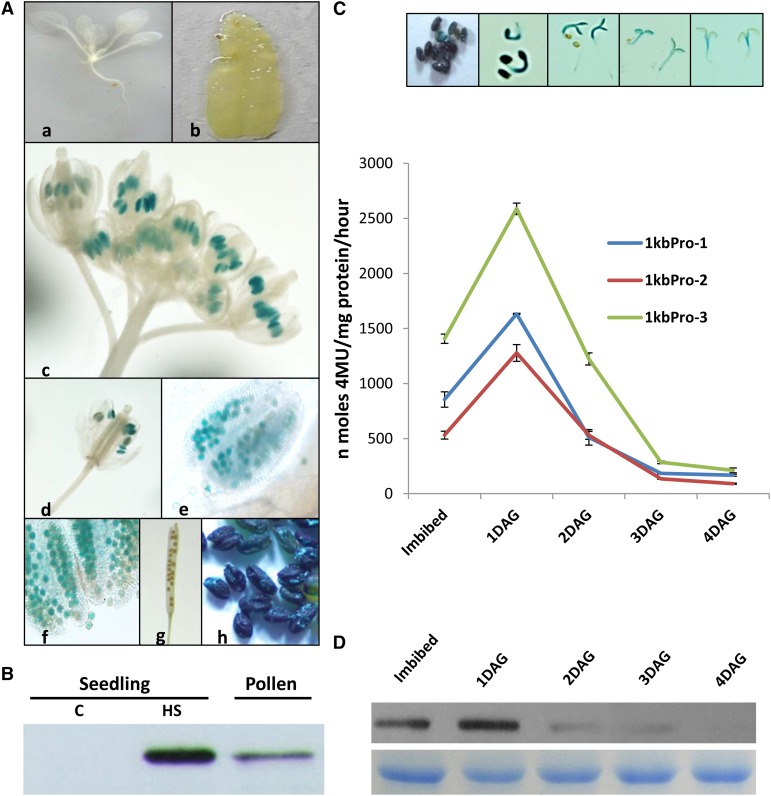
PCR-amplified 1kbAtClpB-Cpro (Supplemental Fig. S1) was cloned upstream of the Gus gene to make a 1kbAtClpB-Cpro::Gus construct (Supplemental Fig. S2A). Ecotype Columbia-0 of Arabidopsis (Col-0) plants stably transformed with this binary construct yielded 1kbpro plants. T0 plants were analyzed for integration of transfer DNA (T-DNA) by PCR using primers specific for Gus and neomycin phosphotransferaseII genes. Three PCR-positive T2 homozygous transgenic lines (1kbpro-1, 1kbpro-2, and 1kbpro-3) were obtained. Gus expression analysis was carried out on homozygous T3 generation plants. Ten-day-old seedlings of the 1kbpro lines did not show Gus staining under unstressed control conditions, whereas seedlings exposed to 38°C for 2 h showed heat-inducible Gus staining in histochemical assays (Supplemental Fig. S2B). Similar observations were made for the leaves of 1-month-old plants. In quantitative estimation of Gus protein, heat-stressed 1kbpro-1, 1kbpro-2, and 1kbpro-3 seedlings showed a remarkable increase in Gus activity in contrast with control seedlings (Supplemental Fig. S2C). Gus expression was induced under arsenic and cadmium stress in 1kbpro plants (Supplemental Fig. S2D). In unstressed (control) 1kbpro plants, Gus expression was prominent in microspores (pollen grains) within the undehisced anthers (Fig. 2Ae). Young developing siliques did not show Gus staining, whereas imbibed mature seeds showed intense Gus staining (Fig. 2Ah). Western blotting using anti-AtClpB-C antibodies showed a remarkable level of AtClpB-C protein in young pollen of unstressed wild-type Arabidopsis plants (Fig. 2B). Mature seeds of 1kbpro plants showed high levels of Gus protein (Fig. 2C). 4-Methylumbelliferyl-beta-d-glucuronide assays showed a remarkable increase in Gus activity during the first 24 h of seed germination. In the subsequent period, Gus activity showed a gradual decline with negligible Gus activity at 5 d after germination stage (Fig. 2C). In western-blot analysis, the level of AtClpB-C protein in the 24-h germination sample was higher compared with the imbibed seeds (Fig. 2D). With progressing day-after-germination stages, the levels of AtClpB-C protein declined.
Figure 2.
Histochemical staining for Gus expression analysis at different developmental stages of 1kbpro plants under unstressed (control) conditions. A, Ten-day-old seedlings (a), mature leaf from 1-month-old plant (b), inflorescence (c), single flower (d), anther (e), pollen grains (f), young silique (g), and mature imbibed seeds (h). B, Western blot showing the levels of AtClpB-C proteins in pollen of Arabidopsis. Control and heat-stressed (38°C/2 h) protein samples from seedlings were loaded as controls. Two micrograms of total protein from seedlings and 10 µg of total protein from pollen were loaded. C, Gus activity at different stages of seed germination. The graph shows the quantitative estimation through the 4-methylumbelliferyl-beta-d-glucuronide assay, whereas the photograph shows the histochemical Gus staining. D, Western-blot analysis showing the levels of AtClpB-C protein during seed germination. Five micrograms of protein was analyzed, and the bottom row represents the Coomassie Blue-stained protein bands showing equal loading. C, Control; DAG, days after germination; HS, heat stressed. [See online article for color version of this figure.]
Expression Characteristics of the ∆AtClpB-C Promoter
The 1kbAtClpB-C promoter sequence contains a 222-bp 5′ UTR with an intervening intron of 106 bp (Supplemental Fig. S3A). A promoter fragment with deletion of 77 bp from the 3′ end of the 1kbAtClpB-C promoter was PCR amplified. The deleted 77-bp region included 55 bp of the intron and 22 bp of the 3′ end of the 5′ UTR sequences (Supplemental Fig. S3A). Construct ∆AtClpB-Cpro:Gus contained a 923-bp AtClpB-C promoter fragment (672-bp sequence of promoter, 200 bp of 5′ UTR with 51 bp of intron sequence) upstream of the Gus reporter gene. Stable homozygous transgenic Arabidopsis lines (∆pro-1, ∆pro-2, and ∆pro-3 lines) were confirmed for T-DNA insertion by PCR. Gus expression in vegetative tissues (10-d-old seedlings and leaves from 1-month-old plants) of T3 ∆pro plants was strictly heat inducible (Supplemental Fig. S4, A and B). The levels of Gus activity were comparable in ∆pro and 1kbpro seedlings. Gus transcript analysis by semiquantitative RT-PCR (sqRT-PCR) showed a similar pattern of induction in ∆pro and 1kbpro seedlings (Supplemental Fig. S3C). Arsenic- and cadmium-induced Gus expression also appeared comparable in ∆pro and 1kbpro seedlings (Supplemental Fig. S4C). Unstressed reproductive tissues of ∆pro plants showed constitutive expression of Gus in pollen and mature seeds like in 1kbpro plants (Supplemental Fig. S4D). From the above work, we observed that deletion of 77 bp from the 3′ end of 1kbAtClpB-C promoter (which includes 22 bp of the 5′ UTR and 55 bp of the intervening intron) has no effect in terms of qualitative and quantitative Gus expression profiling.
Genomic Sequence Further 5′ Upstream of the AtClpB-C Gene
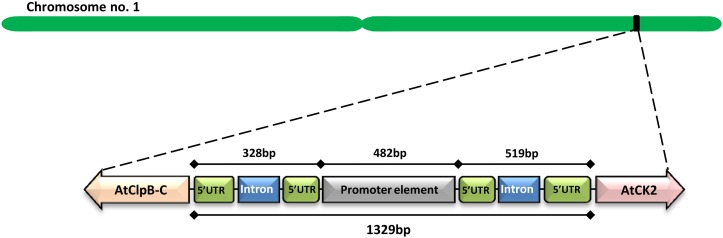
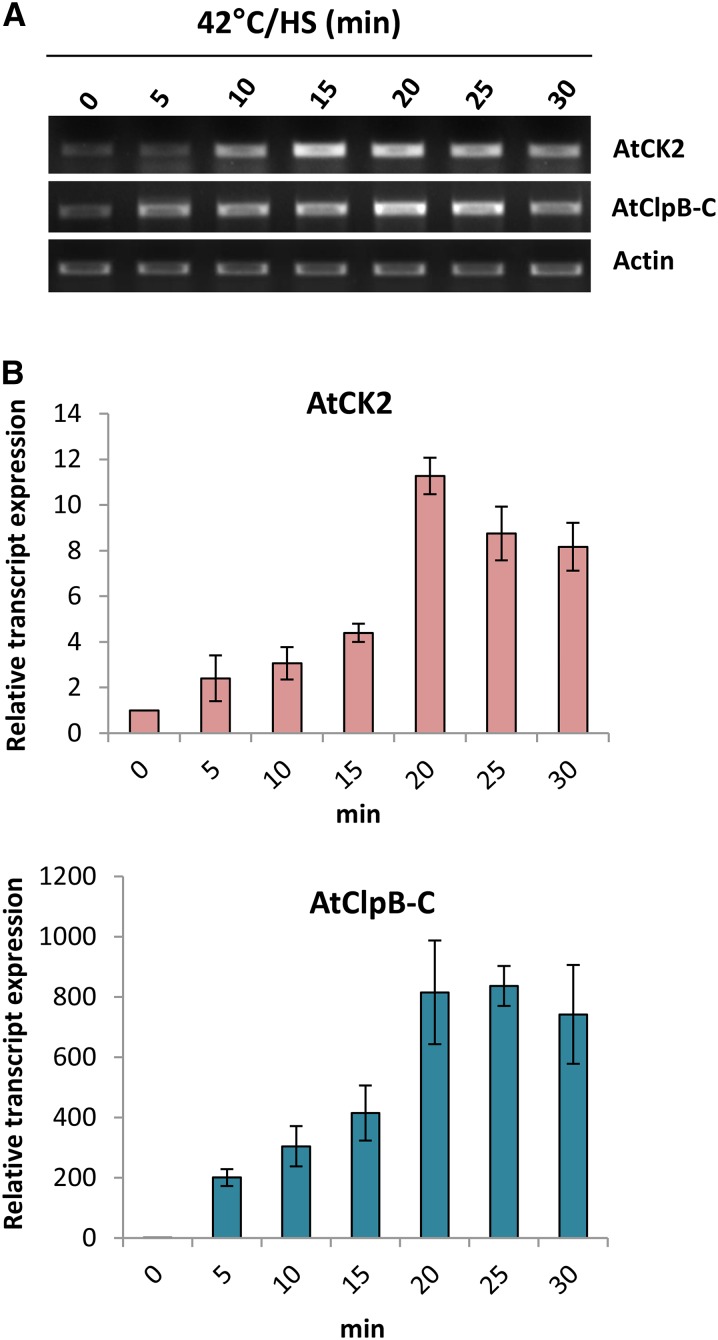
Analysis of genomic organization of the AtClpB-C (At1g74310) gene in The Arabidopsis Information Resource (www.arabidopsis.org) database showed that a putative CK gene (At1g74320; AtCK2) lies immediately upstream of it. Both genes are head-to-head oriented on chromosome 1 of the Arabidopsis genome (Fig. 3). In silico analysis revealed that the translation initiation sites of the two genes are distanced 1,329 bp apart. Although AtClpB-C has a 222-bp 5′ UTR with a 106-bp intervening intron sequence, the AtCK2 gene contains a 5′ UTR of 231 bp with a 228-bp intervening intron. The core promoter region between the two genes leaving the 5′ UTRs is 482 bp. All four HSEs located in the 1kb promoter of the AtClpB-C gene, as mentioned earlier, lie in this core 482-bp fragment (Fig. 3; Supplemental Fig. S5). Transcripts of AtClpB-C and AtCK2 genes were highly HS inducible in semiquantitative and quantitative PCR (Fig. 4). Induction was relatively rapid and predominant for AtClpB-C compared with the AtCK2 gene.
Figure 3.
Schematic representation showing genomic organization of AtClpB-C (At1g74310) and AtCK2 (At1g74320) genes on chromosome 1 of Arabidopsis. The 1,329 bp of the intergenic region with the core 482-bp promoter are marked. The 519 bp upstream of the AtCK2 gene contains 231 bp of the 5′ UTR with an intervening 228-bp intron, whereas the 328 bp upstream of the AtClpB-C gene represents 222 bp of the 5′ UTR with an intervening 106-bp intron. [See online article for color version of this figure.]
Figure 4.
A, sqRT-PCR data showing the transcript levels of AtClpB-C and AtCK2 genes under 0, 5, 10, 15, 20, 25, and 30 min of HS treatment (42°C). The bottom row shows the standardization with actin primers. B, Real-time PCR data showing the relative transcript abundance of both the genes under similar stress regimes. [See online article for color version of this figure.]
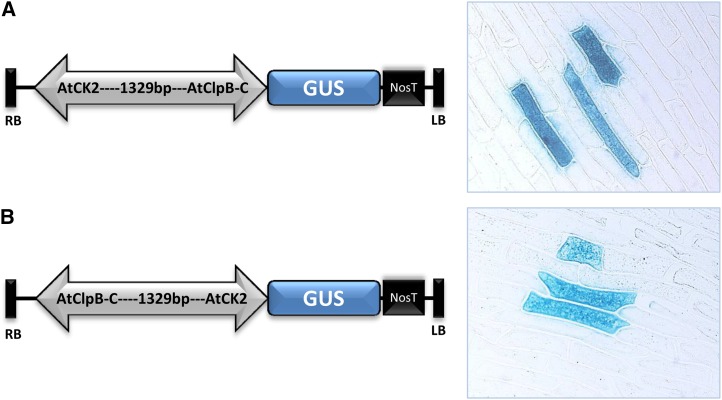
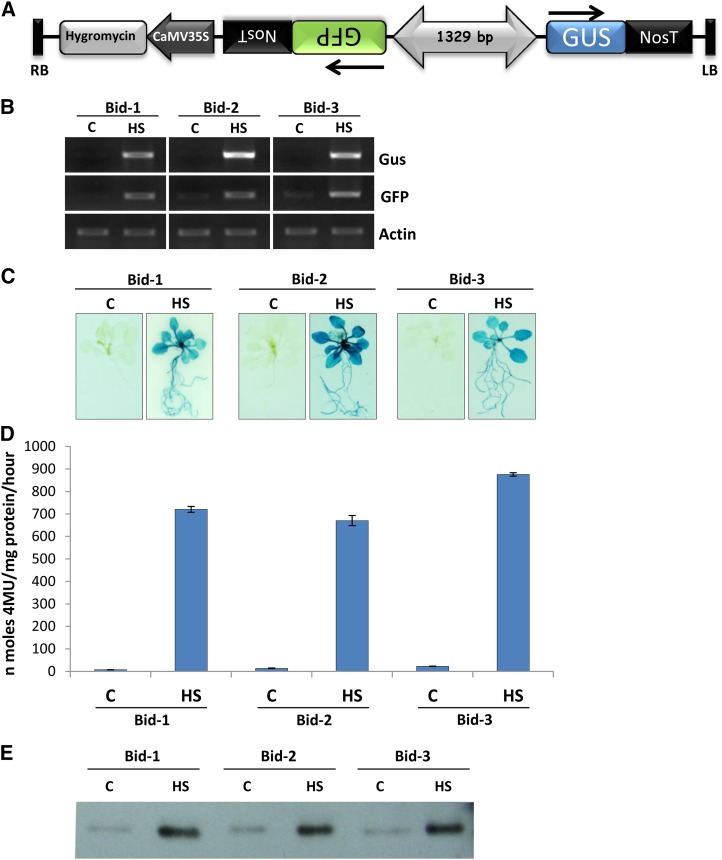
To determine whether the 1,329-bp sequence can drive expression from both sides, this sequence was cloned upstream of the Gus reporter in either orientation and the resulting plasmids were bombarded in onion (Allium cepa) epidermal peels. Several cells expressing Gus were seen in peels bombarded with the two clones (Fig. 5). Next, the construct containing a 1,329-bp intergenic sequence between head-to-head oriented GFP and Gus reporter genes (Fig. 6A) was stably transformed in Arabidopsis (Col-0) plants. Ten-day-old Bid-1, Bid-2, and Bid-3 seedlings stressed at 42°C for 15 min showed HS-inducible transcript expression of both Gus and GFP reporters (Fig. 6B). The levels of transcripts in unstressed control seedlings were negligible. Bid seedlings showed heat-induced expression of Gus protein (Fig. 6, C and D). Total soluble proteins were isolated from control and stressed Bid seedlings (38°C for 2 h) and were analyzed for GFP protein using anti-GFP antibodies. Heat-inducible increases in GFP levels were clearly noted in the three Bid lines (Fig. 6E).
Figure 5.
Transient expression of the Gus reporter in onion epidermal cells for confirmation of promoter activity of the 1,329-bp fragment in either direction. A, Construct in which the Gus reporter is driven by the AtClpB-C gene promoter. B, Construct in which the promoter is oppositely oriented and the Gus reporter is driven by the AtCK2 gene promoter. Respective onion peels expressing Gus are shown on the right. LB, Left border; NosT, nopaline synthase terminator; RB, right border. [See online article for color version of this figure.]
Figure 6.
Analysis of Gus and GFP protein expression in Bid-1, Bid-2, and Bid-3 lines under control and high-temperature stress conditions. A, Diagrammatic representation of the bidirectional GFP::AtCK2/1,329/AtClpB-C::Gus construct. B, sqRT-PCR showing heat-inducible expression of GFP and Gus reporter transcripts in three Bid lines. C, Qualitative analysis of Gus protein. The photograph shows the Gus-stained seedlings under both control and heat-stressed (38°C/2 h) conditions. D, Graph showing the quantification of Gus expression level through a 4-methylumbelliferyl-beta-d-glucuronide assay. E, Western blot showing the levels of GFP protein under control and heat-stressed conditions. Five micrograms of protein was analyzed. C, Control. [See online article for color version of this figure.]
Analysis of the Effect of T-DNA Insertion in the Intergenic Region
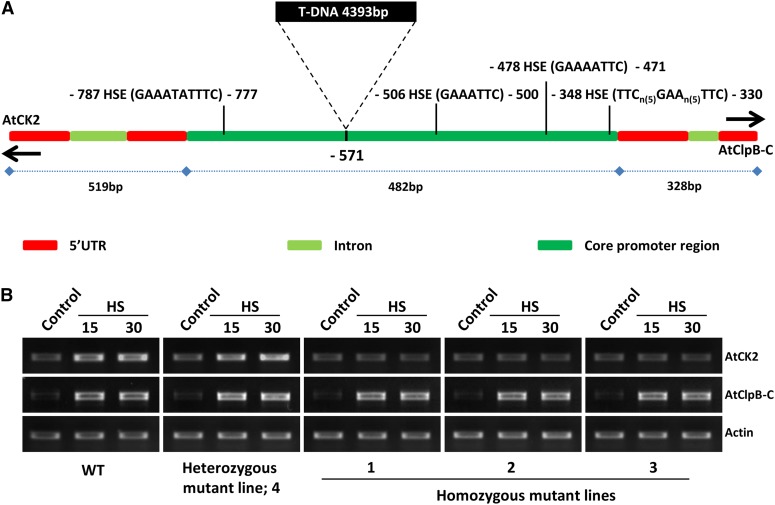
We searched the Arabidopsis Biological Resource Center database for polymorphism in the 1,329-bp intergenic region. In single T-DNA insertion mutant Salk_014505, a 4,393-bp T-DNA (from the pROK2 vector) was present in the core promoter region at position −517 upstream of ATG of the AtClpB-C gene (Fig. 7A). Insertion of T-DNA divided the core promoter sequences of 482 bp into two regions, one of 239 bp toward the AtCK2 side (containing one of the four putative HSEs) and another of 243 bp toward the AtClpB-C side (containing three of the four putative HSEs). Zygosity of the mutant T3 plants was checked (Supplemental Fig. S6) and three homozygous and one heterozygous mutant plants were analyzed. Heat-inducible expression of the AtCK2 transcript was completely lost, whereas the expression of AtClpB-C was not affected in the homozygous mutant lines. In the heterozygous and wild-type plants, expression of both AtCK2 and AtClpB-C genes was distinctly heat inducible (Fig. 7B).
Figure 7.
A, Schematic representation of the 1,329-bp intergenic region between AtClpB-C and AtCK2 genes with point of T-DNA insertion (in Salk_014505 mutant) and HSE distribution. The 4,393-bp T-DNA is inserted within the core promoter region. Of the four HSEs, the distal one (considering upstream of the AtClpB-C gene) is separated from the other three by T-DNA insertion. The configuration of the one HSE present within the 1kb promoter region of the AtCK2 gene and the three HSEs within the 1kb promoter region of AtClpB-C gene is shown. B, sqRT-PCR analysis of the mutant plants for expression of AtCK2 and AtClpB-C genes under HS of 38°C for 15 and 30 min. Three homozygous lines (named as 1, 2, and 3), one heterozygous line (4), and wild-type plants were analyzed. WT, Wild type. [See online article for color version of this figure.]
DISCUSSION
Previous studies have shown that the transcript expression of Hsp genes by heat and heavy metal stress in vegetative tissues as well as their constitutive expression in pollen and seeds is governed by Hsp promoters (Haralampidis et al., 2002; Singh et al., 2010, 2012). The 1kb rice ClpB-C promoter contains a single functional HSE, whereas five HSEs are noted within the proximal 289 bp of the ClpB-C promoter region in maize (Nieto-Sotelo et al., 1999). Through in silico searching, we noted four putative HSEs of variable types in the 1kbAtClpB-C promoter. Gus transcript and protein levels were rapidly induced in 1kbPro plants in response to HS conditions. The cooperative binding of the Hsfs to four HSEs may be a reason for the rapid transcript induction noted in this gene. Congruently, Young et al. (2005) noted that the mutator-like element transposon fragment with 390 bp of the AtClpB-C promoter containing only one of the four HSEs retained a relatively lower HS induction. Heat and metal stresses are shown to have an overlap in their signal reception by involving STRE and AP-1 binding elements besides HSEs (Haralampidis et al., 2002). In line with this, we noted that the AtClpB-C promoter contains an STRE and an AP-1 binding element and showed arsenic- and cadmium-inducible Gus expression. In the case of the ClpB-C promoter-driven Gus gene in rice plants, Gus expression was noted under unstressed control conditions in reproductive tissues with a significantly higher expression in the embryonal one-half of seeds (Singh et al., 2012). Likewise, Gus expression was noted in seeds of 1kbPro plants. In addition, constitutive expression of Gus in young developing microspores within the undehisced anthers was also noted in this study. In parallel, detectable levels of AtClpB-C protein were noted in young Arabidopsis pollen. Developmental expression of AtClpB-C thus far has been extensively discussed with reference to its accumulation during seed maturation; however, to our knowledge, no reports specifically comment on the expression of this protein in the pollen grains (Boston et al., 1996; Hong and Vierling, 2000, 2001; Queitsch et al., 2000; Hong et al., 2003; Kotak et al., 2007b). In this study, we report that Arabidopsis pollen contains AtClpB-C protein from the early stages. Flowering and microsporogenesis are the most susceptible stages to high temperatures (Satake and Yoshida, 1978; Farrell et al., 2006; Jagadish et al., 2007, 2008, 2010). The presence of ClpB-C protein in pollen may have a role in pollen HS response biology. Pollen-related expression of AtClpB-C can possibly be attributed to the presence of GTGANTG10 (GTGA) and POLLEN1LELAT52 elements in the 1kbAtClpB-C promoter. These elements have been implicated in pollen-related gene expression in earlier studies (Rogers et al., 2001; Khurana et al., 2013). In an earlier study, AtClpB-C was detected specifically 4 d after pollination in developing siliques, after which it disappeared and then began to reaccumulate during later stages of seed maturation (Hong and Vierling, 2001). In addition, a gradual decline in levels of this protein were reported during germination. By contrast, an increase in Gus activity in 1kbpro plants during the first 24 h of seed germination was noted in this study. There was a parallel increase in AtClpB-C protein levels in wild-type plants during the first 24 h of seed germination. We thus report that the expression of ClpB-C/Hsp100 increases during the early stages of seed germination, indicating a possible role of ClpB-C in regulation of the initial seed germination process in Arabidopsis. The detailed relationship of various cis-acting sequences in governing heat, metal, and developmental stage-dependent inducibility of HS promoters needs to be fully unveiled.
Using a genome-wide approach for understanding the components of heat response in Arabidopsis, Larkindale and Vierling (2008) reported that the ClpB-C (Hsp101, At1g74310) and AtCK2 (At1g74320) genes are critical for governing high temperature tolerance in Arabidopsis. The latter study showed that the AtClpB-C mutant plant is extremely heat sensitive and the AtCK2 mutant plant is heat sensitive to moderate levels. While analyzing the AtClpB-C genomic region for its promoter analysis, we strikingly noted that AtClpB-C and AtCK2 genes are head-to-head oriented on chromosome 1 of the Arabidopsis genome. The translation start sites of these genes are 1,329 bp apart. We noted that the transcripts of the AtClpB-C and AtCK2 genes are coordinately regulated during high temperature stress (Fig. 4). The 1,329-bp intergenic region of the two genes could drive Gus expression in either orientation in transient assays with onion peel epidermal cells (Fig. 5). Furthermore, this region cloned between the Gus and GFP reporters showed heat-inducible expression of both of the reporter genes in stably transformed Arabidopsis plants (Fig. 6), indicating that this region acts as a bidirectional promoter. Genome-wide analysis in humans indicates that the distance between the two TSSs in a bidirectional promoter is largely within 1,000 bp, whereas this distance is generally longer than 1,000 bp in plants (Trinklein et al., 2004; Mitra et al., 2009). The distance between the TSSs of two rice chymotrypsin protease inhibitor genes (OCPI1 and OCPI2), sharing a bidirectional promoter, is 1,126 bp (Singh et al., 2009). The distance between the TSSs of AtClpB-C and AtCK2 is only 482 bp. Interestingly, the four putative HSEs present in the intergenic region between these two genes are located within this 482-bp fragment. In the Salk_014505 mutant, T-DNA insertion is located at position −517 upstream of ATG of the AtClpB-C gene, almost at the center of the 482-bp core promoter region. Heat-inducible expression of the AtCK2 gene was lost, whereas the AtClpB-C gene retained its usual level of expression in this mutant line (Fig. 7). As a result of insertion, AtCK2 loses proximity to three of the four putative HSEs. This may be the reason for the loss of heat inducibility of the AtCK2 promoter in the mutant plants. This study suggests that the regulatory elements that govern the HS inducibility of the 1,329-bp bidirectional promoter may lie within the region upstream of ATG of AtClpB-C and −517 bp. Combining this finding with the work of Young et al. (2005), it can be concluded that the three HSEs are crucial and sufficient for sensing heat stimuli for induction of AtClpB-C and AtCK2 genes. It was interesting to note that the divergent organization of AtClpB-C and AtCK2 genes is conserved among the homologs of these genes in the Brassicaceae family of dicots (Supplemental Fig. S7; Supplemental Table S1). Furthermore, we noted that among the five AtCK genes, only AtCK2 is HS induced (Gene Expression Omnibus microarray data accession no. GSE51879; Merret et al., 2013). In light of this context, the physical linking of only the AtCK2 gene with AtClpB-C may bear significance.
Many functionally related genes are adjacently located and coexpressed in yeast (Zhang and Smith, 1998; Kruglyak and Tang, 2000). Singh et al. (2009) showed that the rice protease inhibitor genes OCPI1 and OCPI2 are coordinately regulated and share a bidirectional promoter for their regulation. In rice, two small Hsp genes, OsHsp17.3 and OsHsp18.0, share a 356-bp putative bidirectional promoter (Guan et al., 2004). Xu et al. (2012) performed a comparative study on gene expression and biological functions between mouse and human bidirectional genes. Their analysis indicated that the potential selective constraints for conservation of bidirectional architecture are gene function, not the coregulation of paired gene expression. We note that a bidirectional promoter is shared between two functionally dissimilar proteins that are involved in a common phenotype of heat tolerance. AtClpB-C is a chaperone protein, whereas AtCK2 encodes for the CK enzyme that has a role in plasma membrane synthesis.
Overall, there are 5,763 divergent gene pairs in Arabidopsis (Krom and Ramakrishna, 2008). Merret et al. (2013) recently noted that 680 Arabidopsis genes are up-regulated by more than 2-fold under HS. Analyzing these two data sets, we identified that there are 16 divergent gene pairs (with distance between TSSs < 1.5 kb) in which both genes show heat up-regulation (Supplemental Fig. S8; Supplemental Table S2). This study focused on one such case of AtClpB-C and AtCK2 among the 16 gene pairs. The above account further reveals that the heat-induced fold up-regulation for the paired genes differs significantly for members of the divergent gene pairs, suggesting that the bidirectional promoter may generate transcripts in differential amounts. The difference in the heat-induced fold up-regulation of AtClpB-C and AtCK2 transcripts as noted in this study is corroborated by the above observation.
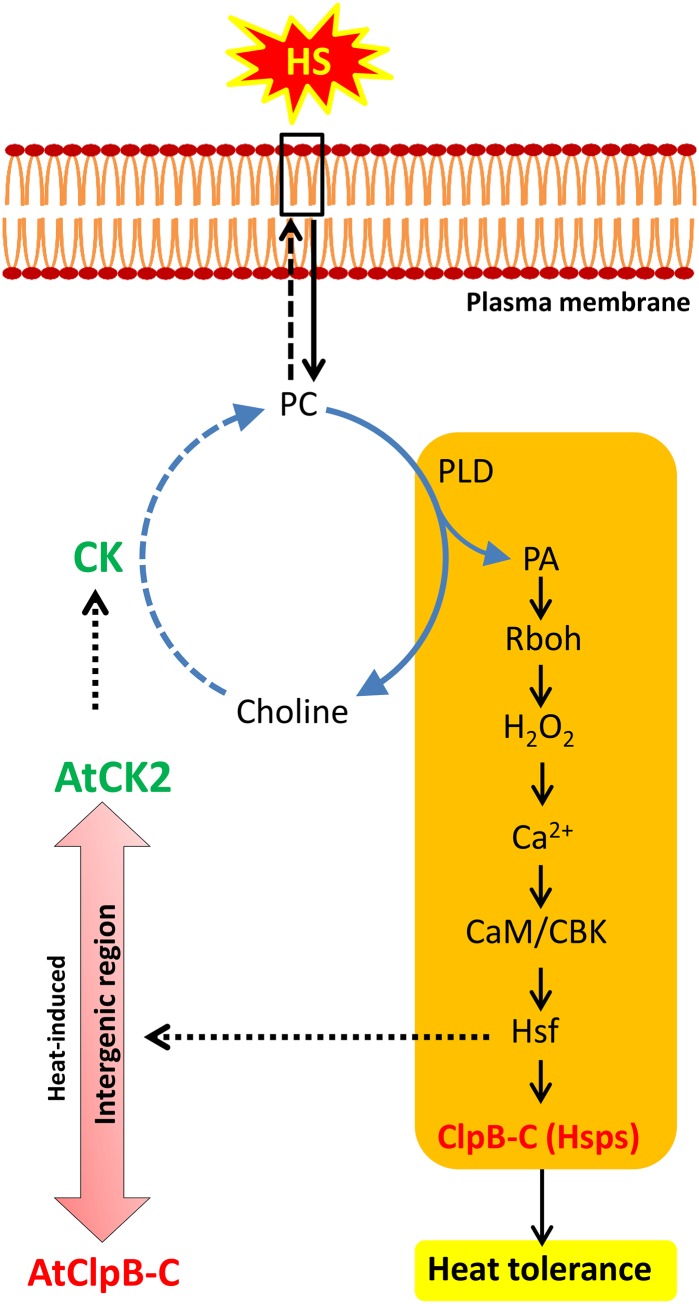
Recent studies show involvement of the membrane in the sensing of HS. It is suggested that after the membrane senses the HS, a series of changes are elicited resulting in induction of Hsps (Hofmann, 2009; Horváth et al., 2012). Saidi et al. (2009) showed that HS-induced elevation of membrane fluidity activates the expression of Hsp genes in a Ca2+-dependent manner. It has also been noted that HS activates the phospholipid-based signaling pathways (Mishkind et al., 2009). High temperature-triggered phospholipase D activity results in the generation of PA that acts as a second messenger, increasing hydrogen peroxide levels and inducing Ca2+ influx. The Ca2+ influx in turn regulates the activation of calmodulin binding kinase, in a calmodulin-dependent fashion. Active calmodulin binding kinase aids the phosphorylation of Hsfs, which successively modulate the expression of Hsps (Horváth et al., 2012). PA is majorly formed by the breakdown of PC and is accumulated in plant cell membranes after the onset of HS. However, the increase in PA is not associated with a corresponding decrease in PC levels (Mishkind et al., 2009). The heat-inducible CK appears to be involved in the generation of PC for maintenance of membrane integrity during HS and generation of PA molecules (model in Fig. 8). Clearly, there is a need to understand the relevance of CK in HS biology in greater depth. Our results suggest that a common regulatory module may be involved in AtClpB-C and AtCK2 heat induction. In rice, the ClpB-C promoter is specifically driven by OsHsfA2C, which is shown to interact with its only HSE (Singh et al., 2012). The Arabidopsis genome has 21 Hsf genes (Fragkostefanakis et al., 2014); however, which Hsf interacts with the AtClpB-C promoter is not yet identified.
Figure 8.
Model highlighting the possible function and position of heat-induced CK among the key players of the PA-mediated heat-signaling pathway, elicited by the plasma membrane during HS. It is previously shown that heat-induced PA accumulation causes a series of reactions ultimately resulting in induction of Hsps (see text for details). The step-wise components involved in this cascade are shown in the rectangular box. Heat-induced CK might compensate for the loss of PC by actively phosphorylating choline generated by PC breakdown, thereby increasing PC biosynthesis. In the AtClpB-C:AtCK2 divergent pair, CK induction during HS by Hsf may help in generation of the required PC for uninterrupted signaling. The steps in this model indicated by filled arrows are based on published evidence. The dotted arrows represent the hypothesis forwarded in this study. The cycle shown with circular arrows represents the Kennedy pathway for PC biosynthesis. CaM, Calmodulin; CBK, calmodulin binding kinase; PLD, phospholipase D; Rboh, respiratory burst oxidase homolog. [See online article for color version of this figure.]
The interesting fact emerging from this study is that nature has packed together two genes involved in heat shock response. Where AtCK2 is placed at the top of HS signaling, AtClpB-C is placed toward the downstream of the cascade. The possible relevance of this compactness might be the fine-tuning of these two genes. We propose that a common regulatory module in heat inducibility of AtCK2 and AtClpB-C genes may provide a basis for interdependence of these two processes and may prove to be a novel gene regulatory mechanism employed by plants under physiological stress. Finally, the heat-inducible bidirectional promoter reported in this study can be used to construct a binary vector for gene stacking in which two genes can be expressed in a heat-inducible manner in a transgenic system.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Transgenic Arabidopsis (Arabidopsis thaliana) lines were raised in the Col-0 background. Seeds were placed in pots containing Soilrite. The pots were kept in a cold room (4°C) for 48 h for stratification and were subsequently placed in a culture room maintained at 22°C ± 1°C with 16-h-light/8-h-dark regimes with light intensity of 100 to 125 µmol m−2 s−1 for raising the full-grown plants. For raising seedlings through tissue culture, Arabidopsis seeds were first surface sterilized with 70% (v/v) ethanol for 30 s followed by a treatment of 2% (v/v) sodium hypochlorite with 1 drop of Tween 20 for 5 min and then washed with sterile water three or four times and finally suspended in 0.1% (w/v) agar. The seeds were plated onto Murashige and Skoog medium containing 1% (w/v) Suc and 0.8% (w/v) agar, pH 5.8. The plates were kept for stratification (48–72 h) and finally shifted to the culture room. For the selection of transformants, surface-sterilized seeds were plated on one-half-strength Murashige and Skoog medium supplemented with the appropriate antibiotic (50 mg L−1 kanamycin or 15 mg L−1 hygromycin) and Augmentin (150 mg L−1). Positive seedlings were transferred to pots in the culture room for further growth.
Mutant Analysis
T-DNA insertion mutant (Salk_014505; Alonso et al., 2003) seeds were ordered from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Individual seeds were grown in pots and plants were analyzed for their zygosity status by PCR (Supplemental Fig. S6). Information on the primers required for the PCR (Supplemental Table S3) was obtained from the T-DNA Express Web site. Three homozygous and one heterozygous mutant lines were selected for analysis.
Construct Designing
For genetic regulation studies, two different promoter fragments, 1,012 bp (1kbAtClpB-Cpro) upstream of ATG codon and 935 bp (∆AtClpB-Cpro; excluding a 77-bp region from the 3′ end of the 1,012-bp fragment), were PCR amplified from Arabidopsis genomic DNA, sequenced (Supplemental Fig. S1), and cloned upstream of Gus in pBT2Gus vector. The promoter-Gus fragments were cleaved from the resulting plasmids and introduced in the pCAMBIA2300 binary vector. The resulting constructs (1kbAtClpB-Cpro::Gus and ∆AtClpB-Cpro::Gus, respectively) were transformed in Arabidopsis to raise stable homozygous transgenic plants (referred as 1kbpro plants and ∆pro plants, respectively). To analyze the bidirectionality, the 1,329-bp region upstream of ATG (AtCK2/1,329/AtClpB-C) of AtClpB-C was cloned in a bidirectional vector (Singh et al., 2009) in between head-to-head oriented Gus and GFP in the background of pCAMBIA1381Z binary vector. The resulting binary plasmid (GFP::AtCK2/1,329/AtClpB-C::Gus construct; Fig. 6A) in each case was introduced in the AGL1 strain of Agrobacterium tumefaciens and was stably transformed in Arabidopsis (GFP::AtCK2/1,329/AtClpB-C::Gus plants; referred to as Bid plants). Plant transformation was carried out employing the floral dip method (Clough and Bent, 1998). Genomic DNA was isolated following the protocol described by Kobayashi et al. (1998). For confirmation of T-DNA integration, PCR for the Gus gene was performed using specific primers. Primers utilized for PCR amplifications are listed in Supplemental Table S3.
Protein Isolation from Seeds, Seedlings, and Pollen Tissues
Soluble proteins from Arabidopsis seedlings were isolated in HEPES buffer solution (25 mm HEPES, pH 7.5, 0.5% [v/v] Triton X-100, 200 mm NaCl, 0.5 mm EDTA, 10 mm MgCl2, and 1× protease inhibitor cocktail [G-Biosciences]). Seedlings were ground in liquid N2 with a pinch of polyvinylpyrrolidone. HEPES extraction buffer was added to the powdered tissue. The homogenate was centrifuged, and the supernatant was used as protein extract. Total seed proteins were isolated in Tris buffer (30 mm Tris-Cl, pH 8.5, 1 mm EDTA, 1%[w/v] SDS, and 1× protease inhibitor cocktail). Dry, imbibed, or germinating Arabidopsis seeds were homogenized with a glass pestle in a microcentrifuge tube (MCT) with the required volume of buffer. Homogenates were centrifuged, and the supernatant was used as seed protein extract. Proteins from Arabidopsis pollen were isolated using HEPES extraction buffer. Young flowers from Arabidopsis inflorescences were taken in the MCT containing the required quantity of HEPES extraction buffer. MCT was gently tapped, allowing the pollen from the anther to mix in the buffer. Pollen was ground with a glass pestle and the homogenate was centrifuged. Clear lysate was used as the protein sample.
Gus Analysis
A protocol described by Jefferson et al. (1987) was employed for histochemical Gus staining. Qualitative estimation of Gus activity was performed using 5-bromo-4-chloro-3-indolyl-β-glucuronic acid as substrate. Control tissues (10-d-old seedlings, leaves from 1-month-old mature plants, inflorescence, siliques, and seeds) were directly stained overnight at 37°C. For HS treatment, seedlings were first exposed to 38°C for 2 h and then stained overnight at 37°C. For each construct, an assay was performed with three independent replicates. For metal stress, seedlings were floated in metal solutions of required concentration for 6 h and stained as above. After Gus staining, chlorophyll was removed from the tissues with ethanol:acetic acid (3:1). For fluorometric analysis, three transgenic lines positive for histochemical Gus staining were analyzed as described by Jefferson et al. (1987). Total protein from seed and seedling tissues was extracted in Gus extraction buffer. Reaction was set in replicates of three for each biological sample analyzed. Fluorescence was recorded by a DyNA Quant TM 200 fluorimeter (Hoefer Pharmacia Biotech).
Western-Blot Analysis
Protein samples were separated on 10% (w/v) SDS gel. For immunoblots, gels were processed by electroblotting on nitrocellulose membrane (Hybond C-super membrane; Amersham-Pharmacia) as described by Katiyar-Agarwal et al. (2003). For analyzing the levels of AtClpB-C, anti-AtClpB-C primary antibodies (raised against N-terminal region; Agrisera) and anti-rabbit secondary antibodies (Sigma-Aldrich) with 1:5,000 and 1:10,000 dilutions, respectively, were used. For GFP protein analysis, anti-GFP primary antibodies (Sigma-Aldrich) and antimouse secondary antibodies (Sigma-Aldrich) with 1:10,000 and 1:10,000 dilutions, respectively, were used.
RNA Isolation, Complementary DNA Synthesis, and Transcript Analysis
Isolation of total RNA from the control and stressed Arabidopsis seedlings was carried out using guanidinium thiocyanate extraction buffer (Chomczynski and Sacchi, 1987). For the synthesis of first-strand complementary DNA (cDNA), 2 µg of total RNA was taken and M-MLV reverse transcriptase (MBI Fermentas) was used. Actin was amplified as an internal control for the normalization of mRNA levels in different cDNA preparations. For quantitative PCR analysis, primers were designed using PRIMER EXPRESS (version 2.0; PE Applied Biosystems) with default parameters (list of primers in Supplemental Table S3). First-strand cDNA was synthesized by reverse transcription using 2 µg of total RNA of 10-d-old Arabidopsis seedlings in 50 µL of reaction volume using a high-capacity cDNA archive kit (Applied Biosystems). The reaction was put in 96-well optical reaction plates (Applied Biosystems), using an ABI Prism 7000 sequence detection system and software (PE Applied Biosystems). For normalization, actin was used as an internal control. Relative expression values were calculated after normalizing against the maximum expression value. For final analysis, two biological replicates and three technical replicates were used.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide sequence of 1kb promoter upstream to ATG of AtClpB-C gene.
Supplemental Figure S2. Analysis of Gus expression in 1kbpro plants under stress conditions.
Supplemental Figure S3. Schematic representation of 1kbAtClpB-Cpro and ΔAtClpB-Cpro fragments and heat-induced Gus transcript expression analysis in 1kbPro and Δpro lines.
Supplemental Figure S4. Gus expression analysis in Δpro plants.
Supplemental Figure S5. Nucleotide sequence of 1,329-bp intergenic region between AtClpB-C and AtCK2 genes.
Supplemental Figure S6. PCR confirmation of the status of 4 individual mutant plants raised from Salk_014505 T3 seeds.
Supplemental Figure S7. Analysis of the organization of AtClpB-C and AtCK2 gene homologs in several plant genera covering different families across the angiosperm.
Supplemental Figure S8. Genomic organization of the 16 heat up-regulated divergent gene pairs in Arabidopsis.
Supplemental Table S1. Information on the AtClpB-C and AtCK gene homologs in selected plant genera covering different families across the angiosperm.
Supplemental Table S2. List of heat-up-regulated divergent gene pairs in Arabidopsis.
Supplemental Table S3. Primers used in the study.
Supplementary Material
Acknowledgments
We thank Dr. Ramakrishna Wusirika (Michigan Technological University) for providing the list of divergent gene pairs in Arabidopsis.
Glossary
- HSE
heat shock element
- STRE
stress-responsive element
- UTR
untranslated region
- IRES
internal ribosome entry site
- CK
choline kinase
- PC
phosphatidylcholine
- PA
phosphatidic acid
- TSS
transcription start site
- Col-0
Ecotype Columbia-0 of Arabidopsis
- T-DNA
transfer DNA
- cDNA
complementary DNA
Footnotes
This work was supported by the Government of India Department of Biotechnology, Center for Advanced Research and Innovation on Plant Stress and Developmental Biology (grant), the Government of India Department of Science and Technology (J.C. Bose fellowship award to A.G.), and the Government of India Council of Scientific and Industrial Research (fellowship award to R.C.M.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Agarwal M, Katiyar-Agarwal S, Grover A. (2002) Plant Hsp100 proteins: structure, function and regulation. Plant Sci 163: 397–405 [Google Scholar]
- Agarwal M, Katiyar-Agarwal S, Sahi C, Gallie DR, Grover A. (2001) Arabidopsis thaliana Hsp100 proteins: kith and kin. Cell Stress Chaperones 6: 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Banerjee J, Sahoo DK, Dey N, Houtz RL, Maiti IB. (2013) An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic Arabidopsis and tobacco plants. PLoS ONE 8: e79622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston RS, Viitanen PV, Vierling E. (1996) Molecular chaperones and protein folding in plants. Plant Mol Biol 32: 191–222 [DOI] [PubMed] [Google Scholar]
- Campbell JL, Klueva NY, Zheng HG, Nieto-Sotelo J, Ho TD, Nguyen HT. (2001) Cloning of new members of heat shock protein HSP101 gene family in wheat (Triticum aestivum (L.) Moench) inducible by heat, dehydration, and ABA(1). Biochim Biophys Acta 1517: 270–277 [DOI] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell 2: 65–73 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dinkova TD, Zepeda H, Martínez-Salas E, Martínez LM, Nieto-Sotelo J, de Jiménez ES. (2005) Cap-independent translation of maize Hsp101. Plant J 41: 722–731 [DOI] [PubMed] [Google Scholar]
- Farrell TC, Fox KM, Williams RL, Fukai S. (2006) Genotypic variation for cold tolerance during reproductive development in rice: screening with cold air and cold water. Field Crops Res 98: 178–194 [Google Scholar]
- Fragkostefanakis S, Röth S, Schleiff E, Scharf KD. (July 3, 2014) Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ http//dx..org/10.1111/pce.12396 [DOI] [PubMed] [Google Scholar]
- Gibellini F, Smith TK. (2010) The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62: 414–428 [DOI] [PubMed] [Google Scholar]
- Grably MR, Stanhill A, Tell O, Engelberg D. (2002) HSF and Msn2/4p can exclusively or cooperatively activate the yeast HSP104 gene. Mol Microbiol 44: 21–35 [DOI] [PubMed] [Google Scholar]
- Guan JC, Jinn TL, Yeh CH, Feng SP, Chen YM, Lin CY. (2004) Characterization of the genomic structures and selective expression profiles of nine class I small heat shock protein genes clustered on two chromosomes in rice (Oryza sativa L.). Plant Mol Biol 56: 795–809 [DOI] [PubMed] [Google Scholar]
- Haralampidis K, Milioni D, Rigas S, Hatzopoulos P. (2002) Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol 129: 1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Vázquez-Pianzola P, Sierra JM, Rivera-Pomar R. (2004) Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA 10: 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr DR, Harris GL. (2004) Close head-to-head juxtaposition of genes favors their coordinate regulation in Drosophila melanogaster. FEBS Lett 572: 147–153 [DOI] [PubMed] [Google Scholar]
- Hofmann NR. (2009) The plasma membrane as first responder to heat stress. Plant Cell 21: 2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Lee U, Vierling E. (2003) Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol 132: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E. (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E. (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27: 25–35 [DOI] [PubMed] [Google Scholar]
- Horváth I, Glatz A, Nakamoto H, Mishkind ML, Munnik T, Saidi Y, Goloubinoff P, Harwood JL, Vigh L. (2012) Heat shock response in photosynthetic organisms: membrane and lipid connections. Prog Lipid Res 51: 208–220 [DOI] [PubMed] [Google Scholar]
- Jagadish SV, Craufurd PQ, Wheeler TR. (2007) High temperature stress and spikelet fertility in rice (Oryza sativa L.). J Exp Bot 58: 1627–1635 [DOI] [PubMed] [Google Scholar]
- Jagadish SV, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. (2010) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J Exp Bot 61: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish SVK, Craufurd PQ, Wheeler TR. (2008) Phenotyping parents of mapping populations of rice for heat tolerance during anthesis. Crop Sci 48: 1140 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston MK, Jacob NP, Brodl MR. (2007) Heat shock-induced changes in lipid and protein metabolism in the endoplasmic reticulum of barley aleurone layers. Plant Cell Physiol 48: 31–41 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Gallie DR, Grover A. (2001) Search for the cellular functions of plant Hsp100/Clp family proteins. Crit Rev Plant Sci 20: 277–295 [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Grover A. (2003) Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol 51: 677–686 [DOI] [PubMed] [Google Scholar]
- Khurana R, Kathuria H, Mukhopadhyay A, Kapoor S, Tyagi AK. (2013) A 286 bp upstream regulatory region of a rice anther-specific gene, OSIPP3, confers pollen-specific expression in Arabidopsis. Biotechnol Lett 35: 455–462 [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Horikoshi T, Katsuyama H, Handa T, Takayanagi K. (1998) A simple and efficient DNA extraction method for plants, especially woody plants. Plant Tissue Culture Biotech 4: 76–80 [Google Scholar]
- Komar AA, Hatzoglou M. (2005) Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem 280: 23425–23428 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. (2007a) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. (2007b) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom N, Ramakrishna W. (2008) Comparative analysis of divergent and convergent gene pairs and their expression patterns in rice, Arabidopsis, and populus. Plant Physiol 147: 1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak S, Tang H. (2000) Regulation of adjacent yeast genes. Trends Genet 16: 109–111 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Vierling E. (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Nagao RT, Key JL. (1994) A soybean 101-kD heat shock protein complements a yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell 6: 1889–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD. (2003) Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res 13: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. (2002) Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet 31: 180–183 [DOI] [PubMed] [Google Scholar]
- Lin MY, Chai KH, Ko SS, Kuang LY, Lur HS, Charng YY. (2014) A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol 164: 2045–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanova ES, Zamchuk LA, Skulachev MV, Ravin NV. (2008) The 5′ untranslated region of the maize alcohol dehydrogenase gene contains an internal ribosome entry site. Gene 420: 11–16 [DOI] [PubMed] [Google Scholar]
- Merret R, Descombin J, Juan YT, Favory JJ, Carpentier MC, Chaparro C, Charng YY, Deragon JM, Bousquet-Antonelli C. (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Reports 5: 1279–1293 [DOI] [PubMed] [Google Scholar]
- Mishkind M, Vermeer JE, Darwish E, Munnik T. (2009) Heat stress activates phospholipase D and triggers PIP accumulation at the plasma membrane and nucleus. Plant J 60: 10–21 [DOI] [PubMed] [Google Scholar]
- Mitra A, Han J, Zhang ZJ, Mitra A. (2009) The intergenic region of Arabidopsis thaliana cab1 and cab2 divergent genes functions as a bidirectional promoter. Planta 229: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A. (2009) Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47: 785–795 [DOI] [PubMed] [Google Scholar]
- Mittal D, Enoki Y, Lavania D, Singh A, Sakurai H, Grover A. (2011) Binding affinities and interactions among different heat shock element types and heat shock factors in rice (Oryza sativa L.). FEBS J 278: 3076–3085 [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Kannan KB, Martínez LM, Segal C. (1999) Characterization of a maize heat-shock protein 101 gene, HSP101, encoding a ClpB/Hsp100 protein homologue. Gene 230: 187–195 [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Martínez LM, Ponce G, Cassab GI, Alagón A, Meeley RB, Ribaut JM, Yang R. (2002) Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. Plant Cell 14: 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Flynn ME, Kaplan G, Racaniello V, Sonenberg N. (1988) Mutational analysis of upstream AUG codons of poliovirus RNA. J Virol 62: 4486–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan VB, D’Silva P. (2009) Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Integr Genomics 9: 433–446 [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. (2010) The heat shock response: life on the verge of death. Mol Cell 40: 253–266 [DOI] [PubMed] [Google Scholar]
- Rogers HJ, Bate N, Combe J, Sullivan J, Sweetman J, Swan C, Lonsdale DM, Twell D. (2001) Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol Biol 45: 577–585 [DOI] [PubMed] [Google Scholar]
- Rubtsova MP, Sizova DV, Dmitriev SE, Ivanov DS, Prassolov VS, Shatsky IN. (2003) Distinctive properties of the 5′-untranslated region of human hsp70 mRNA. J Biol Chem 278: 22350–22356 [DOI] [PubMed] [Google Scholar]
- Ruis H, Schüller C. (1995) Stress signaling in yeast. BioEssays 17: 959–965 [DOI] [PubMed] [Google Scholar]
- Saidi Y, Finka A, Muriset M, Bromberg Z, Weiss YG, Maathuis FJ, Goloubinoff P. (2009) The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 21: 2829–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA, Lindquist S. (1992) Hsp104 is required for tolerance to many forms of stress. EMBO J 11: 2357–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar NK, Kim YK, Grover A. (2009) Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar NK, Kim YK, Grover A. (2014) Coexpression network analysis associated with call of rice seedlings for encountering heat stress. Plant Mol Biol 84: 125–143 [DOI] [PubMed] [Google Scholar]
- Sarkar NK, Kundnani P, Grover A. (2013a) Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 18: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar NK, Thapar U, Kundnani P, Panwar P, Grover A. (2013b) Functional relevance of J-protein family of rice (Oryza sativa). Cell Stress Chaperones 18: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake T, Yoshida S. (1978) High temperature induced sterility in indica rices at flowering. Jpn J Crop Sc 47: 6–17 [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119 [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S, Vierling E. (1994) An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell 6: 1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Grover A. (2010) Plant Hsp100/ClpB-like proteins: poorly-analyzed cousins of yeast ClpB machine. Plant Mol Biol 74: 395–404 [DOI] [PubMed] [Google Scholar]
- Singh A, Mittal D, Lavania D, Agarwal M, Mishra RC, Grover A. (2012) OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB (Hsp100) gene in rice (Oryza sativa L.). Cell Stress Chaperones 17: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Sahi C, Grover A. (2009) Chymotrypsin protease inhibitor gene family in rice: Genomic organization and evidence for the presence of a bidirectional promoter shared between two chymotrypsin protease inhibitor genes. Gene 428: 9–19 [DOI] [PubMed] [Google Scholar]
- Singh A, Singh U, Mittal D, Grover A. (2010) Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes. BMC Genomics 11: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla SL, Pareek A, Kush AK, Grover A. (1998) Distribution patterns of 104 kDa stress-associated protein in rice. Plant Mol Biol 37: 911–919 [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Stoneley M, Bushell M, Willis AE. (2008) Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell 100: 27–38 [DOI] [PubMed] [Google Scholar]
- Sugio A, Dreos R, Aparicio F, Maule AJ. (2009) The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21: 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasseva G, Richard L, Zachowski A. (2004) Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett 566: 115–120 [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. (2004) An abundance of bidirectional promoters in the human genome. Genome Res 14: 62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivinus S, Baulande S, van Zanten M, Campbell F, Topley P, Ellis JH, Dessen P, Coste H. (2001) An element within the 5′ untranslated region of human Hsp70 mRNA which acts as a general enhancer of mRNA translation. Eur J Biochem 268: 1908–1917 [DOI] [PubMed] [Google Scholar]
- Wells DR, Tanguay RL, Le H, Gallie DR. (1998) HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev 12: 3236–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EJ, Bowles DJ. (2004) Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res 14: 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Chen J, Shen B. (2012) The preservation of bidirectional promoter architecture in eukaryotes: what is the driving force? BMC Syst Biol 6(Suppl 1): S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Mizukami Y, Sakurai H. (2005) Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J Biol Chem 280: 11911–11919 [DOI] [PubMed] [Google Scholar]
- Young LW, Cross RH, Byun-McKay SA, Wilen RW, Bonham-Smith PC. (2005) A high- and low-temperature inducible Arabidopsis thaliana HSP101 promoter located in a nonautonomous mutator-like element. Genome 48: 547–555 [DOI] [PubMed] [Google Scholar]
- Young TE, Ling J, Geisler-Lee CJ, Tanguay RL, Caldwell C, Gallie DR. (2001) Developmental and thermal regulation of the maize heat shock protein, HSP101. Plant Physiol 127: 777–791 [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Smith TF. (1998) Yeast “operons”. Microb Comp Genomics 3: 133–140 [DOI] [PubMed] [Google Scholar]
- Zheng G, Tian B, Zhang F, Tao F, Li W. (2011) Plant adaptation to frequent alterations between high and low temperatures: remodelling of membrane lipids and maintenance of unsaturation levels. Plant Cell Environ 34: 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Ye C, Lü H, Chen X, Chai G, Chen J, Wang C. (2006) Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J Plant Res 119: 247–256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.