Genes encoding enolpyruvylshikimate phosphate synthase are tandemly arranged on chromosomes of field-evolved glyphosate-resistant Kochia scoparia.
Abstract
Recent rapid evolution and spread of resistance to the most extensively used herbicide, glyphosate, is a major threat to global crop production. Genetic mechanisms by which weeds evolve resistance to herbicides largely determine the level of resistance and the rate of evolution of resistance. In a previous study, we determined that glyphosate resistance in Kochia scoparia is due to the amplification of the 5-Enolpyruvylshikimate-3-Phosphate Synthase (EPSPS) gene, the enzyme target of glyphosate. Here, we investigated the genomic organization of the amplified EPSPS copies using fluorescence in situ hybridization (FISH) and extended DNA fiber (Fiber FISH) on K. scoparia chromosomes. In both glyphosate-resistant K. scoparia populations tested (GR1 and GR2), FISH results displayed a single and prominent hybridization site of the EPSPS gene localized on the distal end of one pair of homologous metaphase chromosomes compared with a faint hybridization site in glyphosate-susceptible samples (GS1 and GS2). Fiber FISH displayed 10 copies of the EPSPS gene (approximately 5 kb) arranged in tandem configuration approximately 40 to 70 kb apart, with one copy in an inverted orientation in GR2. In agreement with FISH results, segregation of EPSPS copies followed single-locus inheritance in GR1 population. This is the first report of tandem target gene amplification conferring field-evolved herbicide resistance in weed populations.
Glyphosate [N-(phosphonomethyl) Gly] is the most widely used agricultural pesticide globally (Duke and Powles, 2008). Originally, being a nonselective herbicide, its use was limited to vegetation management in noncrop areas; however, introduction of glyphosate-resistant (GR) crops in the late 1990s, coupled with their exceptional adoption, led to accelerated use totaling approximately 128 million ha worldwide in 2012 (James, 2012). GR crop technology has made a significant contribution to global agriculture and the environment, as it not only increased farm income by $32.2 billion (Brookes and Barfoot, 2013), but also moderated the negative environmental impacts of mechanical weed management practices (Gardner and Nelson, 2008; Bonny, 2011). Glyphosate offers a simple, effective, and economic weed management option in GR crops. In addition, it provides immense value in no-till crop production systems by enabling soil and moisture conservation. However, due to intensive glyphosate selection pressure, several weed populations globally have evolved resistance through a variety of mechanisms. Globally, herbicide resistance, in particular the recent proliferation of glyphosate resistance in weed species, is a major crop protection threat; nearly two dozen GR weed species have been reported in the last 15 years (Heap, 2014).
Glyphosate, an aminophosphonic analog of the natural amino acid Gly, nonselectively inhibits 5-Enolpyruvylshikimate-3-Phosphate synthase (EPSPS) in plants, preventing the biosynthesis of the aromatic amino acids Phe, Tyr, and Trp (Steinrücken and Amrhein, 1980), resulting in the death of glyphosate-sensitive individuals. In plants, EPSPS is one of the key enzymes in the shikimate pathway (Herrmann and Weaver, 1999), and glyphosate inhibits EPSPS by binding to EPSPS-shikimate-3-P binary complex forming an EPSPS-shikimate-3-P-glyphosate complex (Alibhai and Stallings, 2001). Bradshaw et al. (1997) hypothesized against the likelihood of weeds evolving resistance to glyphosate, primarily because of its complex biochemical interactions in the shikimate pathway and also due to the absence of known glyphosate metabolism in plants. Nonetheless, several cases of glyphosate resistance, as a result of difference in glyphosate translocation (Preston and Wakelin, 2008) or mutations in the EPSPS, were confirmed (Baerson et al., 2002). More importantly, duplication/amplification of the EPSPS appears to be the basis for glyphosate resistance in several weeds (Sammons and Gaines, 2014). Here, we use duplication to refer to the formation of first repetition of a chromosomal segment and amplification to refer to increase in number of the repetitions (more than two repetitions of a chromosomal segment) under positive selection. The first case of EPSPS amplification as a basis for glyphosate resistance was reported in an Amaranthus palmeri population from GA (Gaines et al., 2010). In this A. palmeri population, there is a massive increase (>100-fold relative to glyphosate-susceptible [GS] plants) in EPSPS copies, and these copies are dispersed throughout the genome (Gaines et al., 2010).
Field-evolved GR Kochia scoparia populations were first reported in western Kansas in 2007 (Heap, 2014). We previously determined that evolution of GR populations of K. scoparia in the U.S. Great Plains is also due to amplification of the EPSPS (A. Wiersma and P. Westra, unpublished data). Unlike in GR A. palmeri, we found relative EPSPS:acetolactate synthase (ALS) copies ranging from three to nine in GR K. scoparia populations. While it quickly became widespread in the region, its presence was reported in another five Great Plains states by 2013 (Heap, 2014). GR K. scoparia populations we tested were 3- to 11-times resistant (population level) to glyphosate compared with a GS population (Godar, 2014), and EPSPS expression positively correlated with genomic EPSPS copy number (A. Wiersma and P. Westra, unpublished data). Here, we reveal the genomic organization of the amplified EPSPS copies in two GR K. scoparia populations, an alternative mechanism of gene amplification to that reported in GR A. palmeri.
RESULTS
Chromosomal Location of the Amplified EPSPS Copies
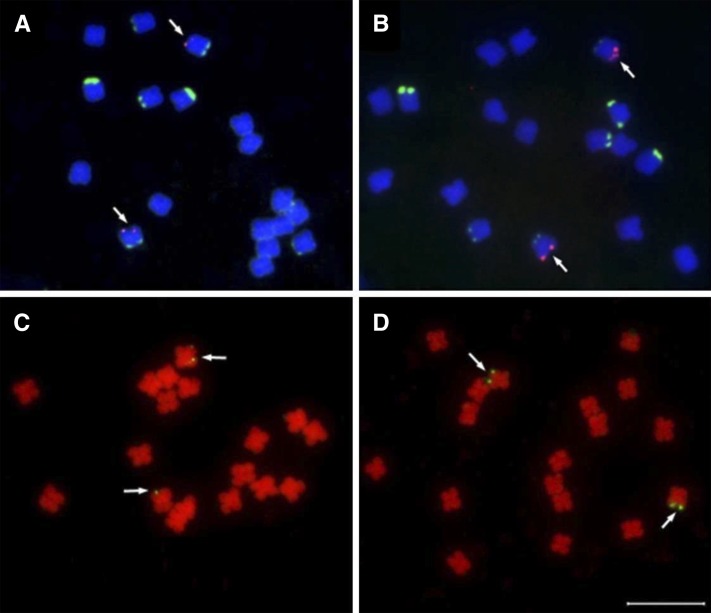
To investigate the location of amplified EPSPS gene copies on GR K. scoparia chromosomes, we used fluorescence in situ hybridization (FISH). Analysis of FISH showed a marked increase in EPSPS signal in GR K. scoparia plants relative to GS plants (Fig. 1). In GS1 and GR1 K. scoparia, three chromosome pairs with nucleolus organizer regions (NORs) were detected, one of which, with a minor NOR signal, had the EPSPS gene on the distal end (Fig. 1, A and B). On metaphase spreads, the EPSPS probe detected much brighter signal on the GR1 chromosome pair relative to the signal on the GS1 chromosome pair (Fig. 1, A and B). Similar EPSPS signals were observed on metaphase chromosomes of GS2 and GR2 plants (Fig. 1, C and D). On prometaphase chromosomes and interphase nuclei, only a faint EPSPS signal was seen on each chromatid (Supplemental Fig. S1, A and C) on GS1 samples, whereas five to seven partially overlapping signals of the EPSPS probe could be distinguished at this location on GR1 samples (Supplemental Fig. S1, C and D).
Figure 1.
FISH mapping of EPSPS gene on chromosome of GS (A and C) and GR (B and D) K. scoparia. A, Three NOR sites on somatic metaphase chromosome pairs of GS1, one with a minor NOR signal showing faint EPSPS signal on the distal end. B, Somatic metaphase spreads of GR1 showing bright EPSPS signal on the same chromosome pair with a minor NOR site. FISH mapping of the EPSPS gene on somatic metaphase chromosomes of GS2 (C) and GR2 (D) K. scoparia. Arrows point to chromosomes with an EPSPS signal. Bar = 10 µm.
High-Resolution Mapping of the EPSPS Cluster
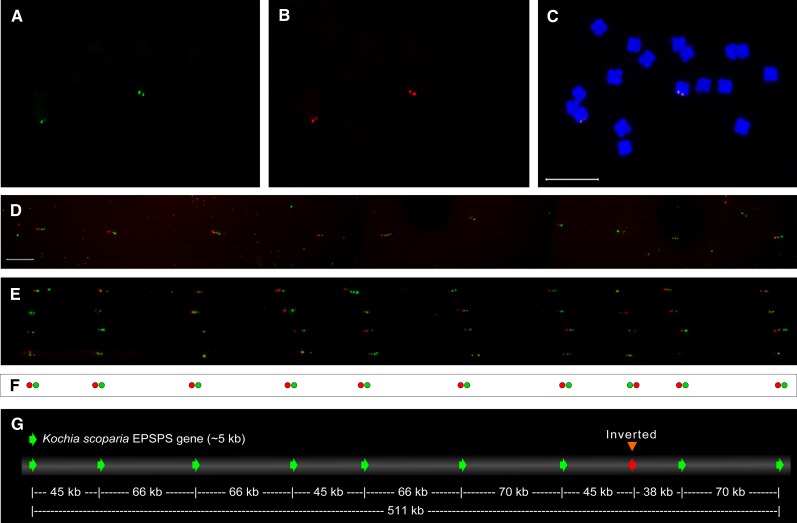
To further explore the arrangement of EPSPS gene copies, we first performed FISH on stretched DNA fiber (Fiber FISH) on GS2 and GR2 plants using one-color probes followed by two-color probes. High-resolution images of Fiber FISH results showed only one EPSPS copy in GS2 plants (Supplemental Fig. S2A). In agreement with the brighter probe hybridization signal in FISH results (Fig. 1D), we observed tandemly configured 10 copies of EPSPS gene on a single DNA fiber of GR2 plants (Supplemental Fig. S2B). On metaphase chromosomes of the GR2 plant, the EPSPS probe hybridized to the same location, giving comparable signal intensity (Fig. 2, A–C) as previously observed in Figure 1C. The two-color probes on a single DNA fiber detected 10 EPSPS copies, one with an inverted EPSPS sequence (Fig. 2, D–F). The total length of the amplified region (measured on seven individual DNA fibers) is approximately 511 ± 26 kb. The EPSPS copies are located approximately 40 to 70 kb apart on a GR2 K. scoparia chromosome (Fig. 2G).
Figure 2.
High-resolution Fiber FISH mapping of EPSPS of GR (GR2) K. scoparia. A to C, Metaphase chromosome spread in two-color FISH showing EPSPS copies clustered at the distal end of homologous chromosomes. D and E, Hybridization of two colored (red and green) EPSPS probes on five different chromosome fibers. F and G represent orientation of EPSPS copies and estimated distance between two adjacent EPSPS copies based on D and E. Red signal (approximately 1.9 kb) and green signal (approximately 2.5 kb) encompass the entire length of EPSPS in K. scoparia. Measurement of the cluster of EPSPS genes was 511.8 ± 26.0 kb (n = 7) in length. Bar = 10 µm.
Inheritance of Glyphosate Resistance
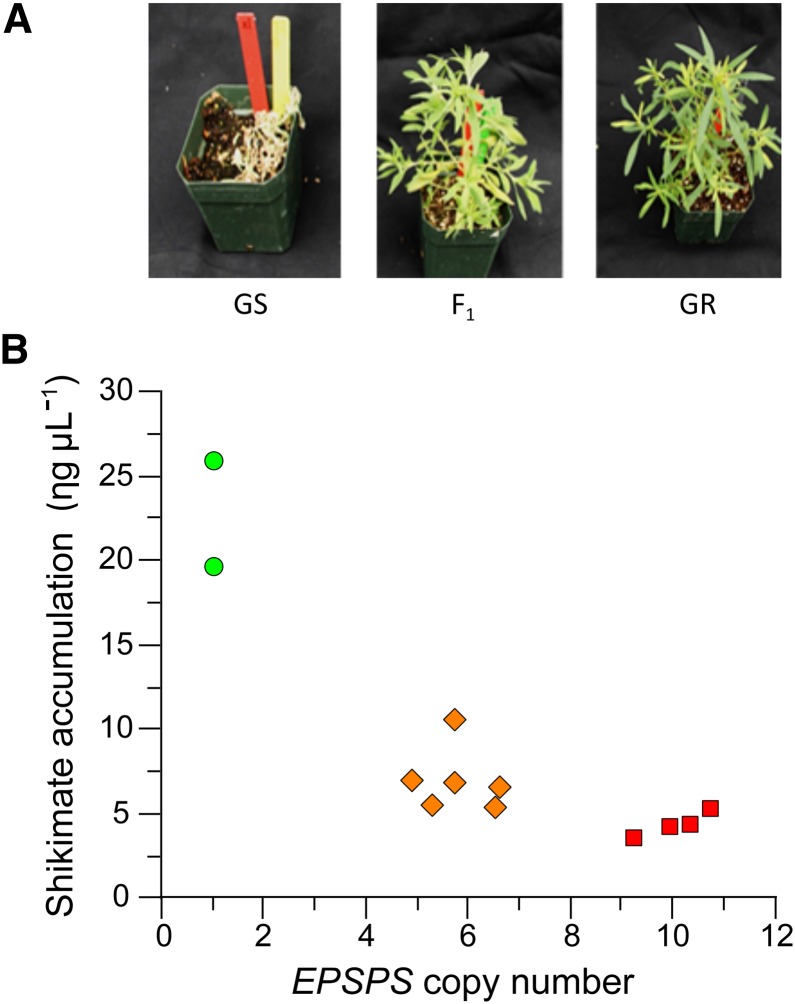
We investigated the inheritance of glyphosate resistance in K. scoparia using a classical genetic approach. We selected nonsegregating GS1 and GR1 lines (see “Materials and Methods”) as parents to generate F1 progeny. F1 seed was successfully generated from reciprocal crosses. F2 seed was produced by self-pollinating F1 plants. In response to a field use rate of glyphosate (868 g acid equivalent [ae] ha–1) application, the GS plants showed stunted growth and eventually died, while GR plants exhibited little or no injury and continued to grow normally (Fig. 3A). As expected for a nuclear-inherited EPSPS gene, F1 plants derived from either GR × GS or GS × GR crosses survived 868 g ae ha–1 glyphosate application (Fig. 3A). A total of 115 F2 plants were evaluated for glyphosate resistance with the same dose (868 g ae ha–1) of glyphosate. F2 progeny segregated as 85 GR to 30 GS (three GR to one GS with χ2 = 0.072), fitting a single-locus inheritance.
Figure 3.
A, Response of GS, F1, and GR plants to a glyphosate rate of 868 g ae ha–1. B, Relationship between EPSPS genomic copy number and shikimate accumulation in two GS (circles), four GR (squares), and six F1 plants (diamonds) of K. scoparia. EPSPS copy number and accumulation of shikimate were determined as described in “Materials and Methods.”
Segregation of EPSPS Copies in F1 and F2 Progeny
The EPSPS copies in parental plants (n = 2, GS1; n = 4, GR1), F1 (n = 6), and F2 progeny (n = 50) were determined using quantitative PCR (qPCR) on genomic DNA. GR1 parental plants possessed nine to 11 copies relative to GS1 plants (Fig. 3B). F1 progeny had approximately five to seven EPSPS copies (Fig. 3B). In F2 progeny, the copy number ranged from one to 13 at a frequency of 24% and 76% with one and four to 13 EPSPS copies, respectively. The EPSPS copy number observed in our genetic populations is in agreement with the inheritance of glyphosate resistance.
We also measured the amount of shikimate accumulated in glyphosate-treated leaf discs to estimate the level of glyphosate resistance. Shikimate accumulation results in plants when EPSPS is inhibited by glyphosate (Amrhein et al., 1980; Herrmann and Weaver, 1999), thus the degree of shikimate accumulation may be used as an indirect measure of a plant’s sensitivity to glyphosate. In this case, if there are higher EPSPS copies, less shikimate accumulation was expected. GR parents accumulated less shikimate in leaf discs compared with GS parents (Fig. 3B). F1 plants accumulated drastically more shikimate than GS parents (P < 0.001) and less shikimate than GR parents (P = 0.064). Overall, shikimate accumulation correlated with the EPSPS copy number (r = –0.841).
K. scoparia Populations Show an Increase in EPSPS Copies and Level of Resistance over Years
We have been collecting field populations of K. scoparia over several years to test whether, in response to glyphosate application (selection event), there is an increase in EPSPS copy number due to recombination and selection at the EPSPS locus. We estimated the EPSPS copies and the level of glyphosate resistance in GR K. scoparia plants from populations that were collected in Kansas in 2007, 2010, and 2012. The results suggest that the GR K. scoparia plants collected in 2007 possessed an average of nine EPSPS copies, while plants from the 2010 and 2012 collection had up to 12 and 16 copies, respectively (Fig. 4). Furthermore, GR K. scoparia plants with nine and 12 copies withstood 1,736 g ae ha–1 but did not survive 3,472 g ae ha–1 glyphosate. However, plants from the 2012 collection survived glyphosate rates up to 5,208 g ae ha–1 (Fig. 4), implying a progression in EPSPS copies and the level of glyphosate resistance from 2007 to 2012. These results indicate that K. scoparia populations with increased EPSPS copies likely arose due to unequal crossing over and may have been favored by glyphosate selection.
Figure 4.
EPSPS genomic copies and level of glyphosate resistance in K. scoparia plants collected in 2007, 2010, and 2012.
DISCUSSION
This study reports the first case of tandem amplification of a target site as a mechanism of naturally evolving resistance to herbicides in plants. Massive amplification of the EPSPS gene randomly dispersed throughout the genome (Gaines et al., 2010), likely mediated by transposable elements (Gaines et al., 2013), has been recently reported in GR A. palmeri. Tandem amplifications of genes that metabolize insecticides have been reported in organophosphate-resistant populations of Culex quinquefasciatus mosquitoes (Paton et al., 2000) and Myzus persicae (Field and Devonshire, 1997). Our results demonstrate the tandem amplification of a target gene itself as a basis for mechanism of herbicide resistance. An intriguing question concerning the resistance mechanism to glyphosate is whether the EPSPS copy number increased in response to a positive selection or whether rare plants with multiple copies existed prior to selection. To date, at least one example of preexistence of the multicopy target gene has been reported, which results in resistance to kinase inhibitors used in lung cancer therapy (Turke et al., 2010).
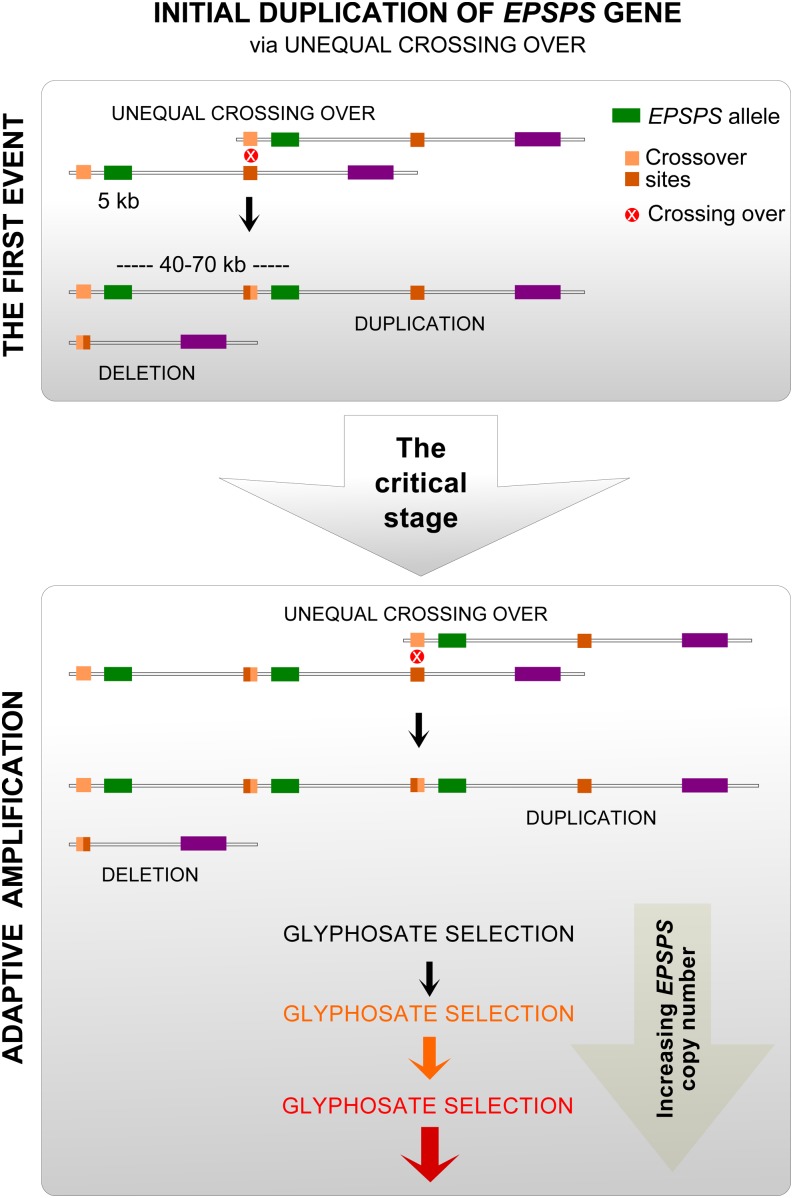
In nature, gene duplication is a common phenomenon and is a precursor for genetic diversity (Wagner et al., 2007). The significance of gene duplication has been comprehensively reviewed (Van de Peer, 2004; Bailey and Eichler, 2006; Conant and Wolfe, 2008; Ponting, 2008). Some duplicated genes confer an immediate adaptive advantage (Perry et al., 2007) and provide a substrate for further amplification under selection, i.e. adaptive amplification. One of the common mechanisms of such amplification is unequal crossing over that takes place between homologs or sister chromatids within the amplified region (van Binsbergen, 2011). The role of unequal meiotic recombination in the formation of many disease resistance gene clusters in crop plants has been documented (Van der Hoorn et al., 2001; Nagy and Bennetzen, 2008; Luo et al., 2011). Continuous variation in EPSPS copy number and a positive correlation between EPSPS expression and the copy number that we have seen (A. Wiersma and P. Westra, unpublished data) suggest that the EPSPS copy number in K. scoparia plants increases through an adaptive process. Furthermore, hybridization of EPSPS probes at distal ends of homologous chromosomes of K. scoparia (Fig. 1) also suggests that an increase in EPSPS copies in GR K. scoparia may have occurred as a result of unequal crossover, as the gene duplication via unequal crossover most likely occurs at the telomere region of chromosomes (Royle et al., 1988; Amarger et al., 1998; Ames et al., 2008).
Here, we illustrate a model for EPSPS amplification via unequal crossover in response to glyphosate selection in K. scoparia (Fig. 5). Survival of the plant that inherits duplicated EPSPS copy (heterozygous for the duplicated copy) from the first duplication event is critical for evolution of resistance to glyphosate, as such an individual will have only a slightly elevated advantage under glyphosate selection. Factors including lower-than-normal use rates of glyphosate, environmental stress (drought and high temperatures), and incomplete spray coverage may result in its survival, thereby allowing it to establish and reproduce one-fourth of the progeny homozygous for the duplicated copies. Sequence homology between the duplicated segments provides a substrate for unequal crossover to happen, leading to EPSPS copy number gain and loss in next-generation progeny. Under continuous selection, plants with higher EPSPS copies will be selected until the return to additional gain in the copy number reaches a plateau. Rate of copy number increase will depend on the interplay between the number of repetitions of sequence homology (copy number) and the return to the gain in additional copy number.
Figure 5.
Illustration of a model suggesting EPSPS amplifications via unequal crossover in response to glyphosate selection and evolution of glyphosate resistance in K. scoparia.
Models for predicting the spread of herbicide resistance in weeds largely rely on underlying genetic mechanisms (single versus polygenic, maternal versus nuclear inherited) and levels of resistance conferred in different states of zygosity. The single-locus inheritance of the EPSPS copies or glyphosate resistance we observed in our classical genetic population is in conformity with tandem arrangement of EPSPS copies. In the context of gene amplification, cytogenetic arrangement of the amplified gene (tandem versus dispersed) and their stability, as well as the magnitude of selection pressure, determine dynamics of the resistance locus (loci), not only at the population, but also at the individual level, unlike in classical genetic models. EPSPS transcript analysis showed that there was no polymorphism in EPSPS transcript sequences, and no EPSPS splice variants were detected in GR K. scoparia (A. Wiersma and P. Westra, unpublished data). Although, initially, most of the EPSPS copies, if not all, will have a complete set of functional motifs and be functionally indistinguishable from the original copy, over time, amplified sequences may diverge and code for new functions. Whether increase in EPSPS gene copies affects fitness of the plants in the absence of selection is unknown; however, a manyfold increase of EPSPS genes in GR A. palmeri does not accrue a measurable fitness cost (Giacomini et al., 2014; Vila-Aiub et al., 2014). These two factors, along with the magnitude of selection pressure, will shape the way for additional increases in the EPSPS copy number.
Our results revealed the tandem genomic organization of amplified EPSPS copies in two GR K. scoparia populations. Nonetheless, the significance of the inverted EPSPS copy in the evolution of glyphosate resistance remains unknown. Although our previous study shows similar levels of EPSPS amplification in other GR K. scoparia populations from the U.S. Great Plains, whether similar amplification patterns exist in those populations remains to be seen. Further investigation of flanking sequences, including breakpoints, will shed additional light on the mechanism of such possibly recurrent rearrangement events that resulted in the evolution of glyphosate resistance in K. scoparia populations.
MATERIALS AND METHODS
Plant Materials
Plants from two GS Kochia scoparia populations, GS1 and GS2, and two GR K. scoparia populations, GR1 and GR2, all from Kansas, were used in this study.
FISH Procedure
Somatic chromosome preparations (using the drop technique), direct probe labeling (by nick translation), and the FISH procedure on GS1 and GR1 plants were performed as described previously (Kato et al., 2004, 2006) with minor modifications. Root tips were collected from young plants and treated in a nitrous oxide gas chamber for 1.5 h, fixed on ice in cold 90% (v/v) acetic acid for 10 min, and washed and stored in 70% (v/v) ethanol at –20°C. For slide preparation, roots were washed in tap water for 10 min and then in KCl buffer for 5 min (75 mm KCl, 7.5 mm EDTA, pH 4). Seven meristems (0.5–1 mm long) were placed in 20 μL of 4% (w/v) cellulase Onozuka R-10 (Tokyo, catalog no. 201069) and 1% (w/v) pectolyase Y23 (Karlan, catalog no. 8006) in KCl buffer and incubated for 43 min at 37°C. Digested meristems were washed for 5 min in ice-cold Tris-EDTA buffer (pH 7.6), with three additional washes in 100% ethanol. Meristems were dispersed with a needle in 20 μL of an ice-cold acetic acid:methanol mix (9:1) and immediately dropped onto three precleaned glass slides placed in a humid chamber. Dried preparations were UV cross linked, soaked in methacarn solution (methanol:chloroform:glacial acetic acid [6:3:1]) for 1 min, dried, and used for hybridization on the same day. For labeling the NOR ribosomal RNA loci, clone pTa71, containing a 9-kb insertion with 18S, 5.8S, and 26S ribosomal RNA wheat (Triticum aestivum) genes and intergenic spacers (Gerlach and Bedbrook, 1979), was used as a probe. Five microliters of probe mixture contained 200 ng of each EPSPS gene PCR product labeled with Texas Red-5-dCTP and 160 ng of pTa71 labeled with Fluorescein-12-deoxyuridine triphosphate (PerkinElmer, catalog nos. NEL413001EA and NEL426001EA). The mixture of probes and the slide preparation were denatured at 100°C separately. The rest of the FISH procedure and washes were performed by using the method described by Kato et al. (2006).
The FISH on the somatic metaphase chromosome of GS2 and GR2 was performed using a procedure as described by Koo et al. (2010). Biotin- and digoxigenin-labeled probes were detected with Alexa Fluor 488 streptavidin antibody (Invitrogen) and rhodamine-conjugated anti-digoxigenin antibody (Roche Diagnostics), respectively.
In both FISH experiments, chromosome preparations were mounted and counterstained with 4',6-diamidino-2-phenylindole solution in Vectashield (Vector Laboratories, catalog nos. H–1200 and H–1300). FISH images were captured with a Zeiss Axioplan 2 microscope using a cooled CCD camera CoolSNAP HQ2 (Photometrics) and AxioVision 4.8 software (Zeiss). The final contrast of the images was processed using Adobe Photoshop CS5 software.
EPSPS FISH Probe Preparation
Sequences of K. scoparia EPSPS mRNA and the Amaranthus palmeri EPSPS gene (Gaines et al., 2010; GenBank accession no. JX56456) were used to develop the PCR primers (Supplemental Fig. S3; Supplemental Table S1). The EPSPS gene was amplified using PSI K. scoparia genomic DNA as a template isolated with Qiagen DNeasy Plant Mini kit (catalog no. 69104). The PCR reaction included JumpStart REDTaq ReadyMix (Sigma, catalog no. P0982), 0.4 μm of each primer, and 0.5 to 4 ng μL–1 template DNA. PCR cycles consisted of initial denaturation at 96°C for 5 min, 35 cycles at 96°C for 30 s, 57°C for 30 s, and 72°C for 4 min, and a final extension of 15 min. PCR products were cut and eluted from agarose gel with a Qiagen Gel Extraction kit (catalog no. 28706) and reamplified using the same primers. PCR products were purified with Invitrogen PCR Purification kit (catalog no. K3100–01) and verified by sequencing (Genewiz). The sequence of the amplified part of K. scoparia EPSPS gene was submitted to the National Center for Biotechnology Information GenBank database with accession number KJ374721. Three PCR products were tested separately by FISH, and products 1 and 3 that showed no background staining on K. scoparia chromosomes were used as a pooled FISH probe.
Fiber FISH Procedure
Young leaf tissues were collected from fast growing plants of GS2 and GR2. Nuclei isolation, DNA fiber preparation, and Fiber FISH were performed following published protocols (Jackson et al., 1998; Koo et al., 2011). Fiber FISH images were captured and processed as previously described in the FISH procedure. The cytological measurements of the Fiber FISH signals were converted into kilobases using a 3 kb μm–1 conversion rate.
EPSPS Fiber FISH Probe Preparation
Sequences of A. palmeri EPSPS gene (GenBank accession no. JX564536) were used to develop the PCR primers (Supplemental Fig. S4) for cloning EPSPS gene from K. scoparia. The probes were amplified with primers (Supplemental Table S1) using GS2 plant genomic DNA as a template. PCR product was cloned in 2.1-TOPO TA vector, and the clones were labeled with either biotin-16-UTP or digoxigenin-11-deoxyuridine triphosphate (Roche Diagnostics) using a standard nick translation reaction.
Creation of Genetic Populations and Their Phenotyping
Plants from K. scoparia populations GS1 and GR1 were grown in a greenhouse. Before flowering, the whole plant was covered with plastic bread bags (33 × 60 cm) with microperforations for self-pollination. Upon plant maturity, seed was harvested separately from individual plants. A subset of selfed progeny seeds were planted, and about 40 plants (8–10 cm tall) from each line were treated with glyphosate (Roundup WeatherMAX, Monsanto Company) at the rate of 868 g ae ha–1 using a chamber sprayer calibrated to deliver 187 L ha–1. None of the GS1 plants survived the glyphosate treatment and were hence identified as a GS line. All GR1 plants survived the glyphosate treatment and were therefore considered for crossing as a GR line. Reciprocal crosses of GS1 and GR1 (R × S represents a GR female pollinated with GS pollen and vice versa for S × R) plants were performed as follows. K. scoparia bears protogynous flowers with stigma being receptive for 1 week before anthesis of the same flower. Therefore, prior to stigma emergence, all the leaves and apical meristems were removed from a few randomly selected branches of GR or GS plants and covered with Lawson 217 pollination bags. After stigma emergence, using a sterile forceps, pollen from the dehisced anthers of GR or GS plants (chosen as male parents) was transferred separately onto the stigmas of the maternal flowers. Immediately after pollination, the flowers were covered with the same pollination bags for 10 d. Mature F1 seed was harvested separately from reciprocal crosses. F1 plants were treated with glyphosate as described above, and all F1 plants from both reciprocal crosses survived a glyphosate rate of 868 g ae ha–1. Randomly selected F1 plants were transplanted into larger pots and self-pollinated (using the plastic bread bags with microperforations as mentioned earlier), and F2 seed was harvested separately. F2 plants were treated with a glyphosate rate of 868 g ae ha–1 as described above.
Shikimate Accumulation Assay
An in vivo measure of shikimate accumulation was determined on parental, F1, and F2 progeny following the procedure developed by Shaner et al. (2005). Six 4-mm leaf discs were collected from a fully expanded young leaf of a single plant when 8 to 10 cm tall. Discs were placed in a 96-well plate with one disc per well containing 100 µm glyphosate solution. Leaf discs were incubated for 16 h under continuous light (µmol 250 m–2 s–1). A shikimate standard curve was used to calculate the shikimate accumulation in ng shikimate µL–1. The experiment was done in triplicate and repeated.
qPCR
EPSPS gene copy number was determined in parental, F1, and F2 plants. Leaf tissue samples were collected in 1.5-mL microcentrifuge tubes and immediately frozen in liquid nitrogen and stored at –20°C. DNA was extracted using the Qiagen DNEasy Plant Mini Kit and quantified using a NanoDrop spectrophotometer. The relative EPSPS gene copy number was determined by qPCR on genomic DNA using ALS as a reference gene. EPSPS forward and reverse primers were 5′-GGCCAAAAGGGCAATCGTGGAG-3′ and 5′-CATTGCCGTTCCCGCGTTTCC-3′, respectively. ALS forward and reverse primers were 5′-ATGCAGACAATGTTGGATAC-3′ and 5′-TCAACCATCGATACGAACAT-3′, respectively. These primers produce products of 102 and 159 bp for EPSPS and ALS, respectively. qPCR was performed using 96-well plates, with each well containing a master mix comprised of 10 µL of iQ SYBR Green Super Mix (BioRad), 1 µL of each corresponding forward and reverse primer, 16 ng of genomic DNA, and 4 µL of deionized water. Each reaction was done in triplicate and was repeated. PCR cycle parameters were set for 95°C for 3 min, denaturing was set at 95°C for 10 s, annealing and extension were set at 60°C for 30 s, and the denaturing/annealing steps were repeated for 39 cycles. Relative quantification of EPSPS was calculated as ∆Ct = CtALS – CtEPSPS, where CtALS and CtEPSPS are the threshold cycles for ALS and EPSPS genes, respectively, and EPSPS copy number was expressed as 2∆Ct (Gaines et al., 2010). The copy number was averaged across replications, and the sd was calculated for each plant sample.
EPSPS Copies and Level of Resistance in K. scoparia Populations Collected over Years
EPSPS copies and the level of resistance to glyphosate were determined in K. scoparia populations collected over years. Seed of GR K. scoparia was collected from fields in 2007, 2010, and 2012. Levels of resistance to glyphosate on 20 to 32 plants from each collection were determined by shikimate accumulation assay and/or treating whole plants with varying rates of glyphosate (0–5,208 g ae ha–1) as previously described. EPSPS copy number of at least six plants from each collection that showed highest levels of resistance was determined using qPCR as previously described.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KJ374721.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. FISH mapping of the EPSPS gene on chromosome of glyphosate-susceptible GS1 and glyphosate-resistant GR1 K. scoparia.
Supplemental Figure S2. High resolution Fiber FISH results showing chromosomal distribution EPSPS copies in GS and GR K. scoparia plants.
Supplemental Figure S3. Part of the K. scoparia EPSPS sequence used for FISH probe production and sequencing.
Supplemental Figure S4. EPSPS structure of A. palmeri and PCR primer positions for K. scoparia EPSPS cloning for FISH and Fiber FISH probe preparation.
Supplemental Table S1. Primers used to develop EPSPS gene FISH and Fiber FISH probes.
Supplementary Material
Acknowledgments
We thank Todd Gaines, Geoff Morris, and Marie Jasieniuk for useful comments on article drafts.
Glossary
- FISH
fluorescence in situ hybridization
- GR
glyphosate-resistant
- EPSPS
5-enolpyruvylshikimate-3-P synthase
- NOR
nucleolus organizer region
- GS
glyphosate-susceptible
- qPCR
quantitative PCR
Footnotes
This work was supported by the Department of Agronomy and the Kansas State Research and Extension (contribution no. 14–359–J), the Kansas Soybean Commission (to M.J.), and the Wheat Genetic Resource Center/Industry and University Cooperative Research Center of the National Science Foundation (grant no. IIP–1338897 to B.S.G).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alibhai MF, Stallings WC. (2001) Closing down on glyphosate inhibition: with a new structure for drug discovery. Proc Natl Acad Sci USA 98: 2944–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarger V, Gauguier D, Yerle M, Apiou F, Pinton P, Giraudeau F, Monfouilloux S, Lathrop M, Dutrillaux B, Buard J, et al. (1998) Analysis of distribution in the human, pig, and rat genomes points toward a general subtelomeric origin of minisatellite structures. Genomics 52: 62–71 [DOI] [PubMed] [Google Scholar]
- Ames D, Murphy N, Helentjaris T, Sun N, Chandler V. (2008) Comparative analyses of human single- and multilocus tandem repeats. Genetics 179: 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrhein N, Deus B, Gehrke P, Steinrücken HC. (1980) The site of the inhibition of shikimate pathway by glyphosate. Plant Physiol 66: 830–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerson SR, Rodriguez DJ, Tran M, Feng Y, Biest NA, Dill GM. (2002) Glyphosate-resistant goosegrass: identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol 129: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. (2006) Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet 7: 552–564 [DOI] [PubMed] [Google Scholar]
- Bonny S. (2011) Herbicide-tolerant transgenic soybean over 15 years of cultivation: pesticide use, weed resistance, and some economic issues: the case of the USA. Sustainability 3: 1302–1322 [Google Scholar]
- Bradshaw LD, Padgette SR, Kimball SL, Wells BH. (1997) Perspectives on glyphosate resistance. Weed Technol 11: 189–198 [Google Scholar]
- Brookes G, Barfoot P. (2013) The global income and production effects of genetically modified (GM) crops 1996-2011. GM Crops Food 4: 74–83 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Duke SO, Powles SB. (2008) Glyphosate: a once-in-a-century herbicide. Pest Manag Sci 64: 319–325 [DOI] [PubMed] [Google Scholar]
- Field LM, Devonshire AL. (1997) Structure and organization of amplicons containing the E4 esterase genes responsible for insecticide resistance in the aphid Myzus persicae (Sulzer). Biochem J 322: 867–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines TA, Wright AA, Molin WT, Lorentz L, Riggins CW, Tranel PJ, Beffa R, Westra P, Powles SB. (2013) Identification of genetic elements associated with EPSPs gene amplification. PLoS ONE 8: e65819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper AS, et al. (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG, Nelson GC. (2008) Herbicides, glyphosate resistance and acute mammalian toxicity: simulating an environmental effect of glyphosate-resistant weeds in the USA. Pest Manag Sci 64: 470–478 [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7: 1869–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini D, Westra P, Ward SM. (2014) Impact of genetic background in fitness cost studies: an example from glyphosate-resistant Palmer amaranth. Weed Sci 62: 29–37 [Google Scholar]
- Godar AS (2014) Glyphosate resistance in kochia. PhD thesis. Kansas State University, Manhattan, Kansas [Google Scholar]
- Heap I (2014) The international survey of herbicide-resistant weeds. http://www.weedscience.com (April 13, 2014)
- Herrmann KM, Weaver LM. (1999) The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 50: 473–503 [DOI] [PubMed] [Google Scholar]
- Jackson SA, Wang ML, Goodman HM, Jiang J. (1998) Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41: 566–572 [PubMed] [Google Scholar]
- James C (2012) 2012 ISAAA Report on Global Status of Biotech/GM Crops. International Service for the Acquisition of Agri-Biotech Applications, Ithaca, New York [Google Scholar]
- Kato A, Albert PS, Vega JM, Birchler JA. (2006) Sensitive FISH signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem 80: 71–78 [DOI] [PubMed] [Google Scholar]
- Kato A, Lamb JC, Birchler JA. (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo DH, Han F, Birchler JA, Jiang J. (2011) Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res 21: 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo DH, Nam YW, Choi D, Bang JW, de Jong H, Hur Y. (2010) Molecular cytogenetic mapping of Cucumis sativus and C. melo using highly repetitive DNA sequences. Chromosome Res 18: 325–336 [DOI] [PubMed] [Google Scholar]
- Luo S, Peng J, Li K, Wang M, Kuang H. (2011) Contrasting evolutionary patterns of the Rp1 resistance gene family in different species of Poaceae. Mol Biol Evol 28: 313–325 [DOI] [PubMed] [Google Scholar]
- Nagy ED, Bennetzen JL. (2008) Pathogen corruption and site-directed recombination at a plant disease resistance gene cluster. Genome Res 18: 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton MG, Karunaratne SH, Giakoumaki E, Roberts N, Hemingway J. (2000) Quantitative analysis of gene amplification in insecticide-resistant Culex mosquitoes. Biochem J 346: 17–24 [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. (2007) Diet and the evolution of human amylase gene copy number variation. Nat Genet 39: 1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. (2008) The functional repertoires of metazoan genomes. Nat Rev Genet 9: 689–698 [DOI] [PubMed] [Google Scholar]
- Preston C, Wakelin AM. (2008) Resistance to glyphosate from altered herbicide translocation patterns. Pest Manag Sci 64: 372–376 [DOI] [PubMed] [Google Scholar]
- Royle NJ, Clarkson RE, Wong Z, Jeffreys AJ. (1988) Clustering of hypervariable minisatellites in the proterminal regions of human autosomes. Genomics 3: 352–360 [DOI] [PubMed] [Google Scholar]
- Sammons RD, Gaines TA. (2014) Glyphosate resistance: state of knowledge. Pest Manag Sci 70: 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner DL, Nadler-Hasser T, Koger CH. (2005) A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci 53: 769–774 [Google Scholar]
- Steinrücken HC, Amrhein N. (1980) The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem Biophys Res Commun 94: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, et al. (2010) Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Binsbergen E. (2011) Origins and breakpoint analyses of copy number variations: up close and personal. Cytogenet Genome Res 135: 271–276 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y. (2004) Computational approaches to unveiling ancient genome duplications. Nat Rev Genet 5: 752–763 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RA, Kruijt M, Roth R, Brandwagt BF, Joosten MH, De Wit PJ. (2001) Intragenic recombination generated two distinct Cf genes that mediate AVR9 recognition in the natural population of Lycopersicon pimpinellifolium. Proc Natl Acad Sci USA 98: 10493–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Aiub MM, Goh SS, Gaines TA, Han H, Busi R, Yu Q, Powles SB. (2014) No fitness cost of glyphosate resistance endowed by massive EPSPS gene amplification in Amaranthus palmeri. Planta 239: 793–801 [DOI] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM. (2007) The road to modularity. Nat Rev Genet 8: 921–931 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.