Fast diversification and versatility of a subfamily of cytochrome P450 enzymes in Brassicaceae has been important in their metabolism of both monoterpenols and herbicides.
Abstract
Comparative genomics analysis unravels lineage-specific bursts of gene duplications related to the emergence of specialized pathways. The CYP76C subfamily of cytochrome P450 enzymes is specific to Brassicaceae. Two of its members were recently associated with monoterpenol metabolism. This prompted us to investigate the CYP76C subfamily genetic and functional diversification. Our study revealed high rates of CYP76C gene duplication and loss in Brassicaceae, suggesting the association of the CYP76C subfamily with species-specific adaptive functions. Gene differential expression and enzyme functional specialization in Arabidopsis thaliana, including metabolism of different monoterpenols and formation of different products, support this hypothesis. In addition to linalool metabolism, CYP76C1, CYP76C2, and CYP76C4 metabolized herbicides belonging to the class of phenylurea. Their ectopic expression in the whole plant conferred herbicide tolerance. CYP76Cs from A. thaliana. thus provide a first example of promiscuous cytochrome P450 enzymes endowing effective metabolism of both natural and xenobiotic compounds. Our data also suggest that the CYP76C gene family provides a suitable genetic background for a quick evolution of herbicide resistance.
Although extensive monoterpenol (especially linalool) oxidative metabolism has been described in many plant species, leading to fragrant and bioactive compounds as diverse as alcohols, aldehydes, acids, and epoxides (Williams et al., 1982; Matich et al., 2003, 2011; Luan et al., 2005, 2006; Ginglinger et al., 2013), pyranoid or furanoid linalool derivatives (Pichersky et al., 1994; Raguso and Pichersky, 1999), and geraniol-derived iridoids and secoiridoids (Dinda et al., 2007a, 2007b, 2011; Tundis et al., 2008), limited information is available on the enzymes generating these oxygenated compounds. Involvement of a cytochrome P450 (P450) enzyme extracted from Vinca rosea (now renamed Catharanthus roseus) in the hydroxylation of geraniol and nerol was suggested as early as 1976 (Madyastha et al., 1976). The first plant P450 gene to be isolated, CYP71A1 from avocado (Persea americana) fruit, was later shown to encode an enzyme with geraniol/nerol epoxidase activity (Hallahan et al., 1992, 1994). To our knowledge, a connection with compounds formed in the fruit has not yet been established. The geraniol 8-hydroxylase (often named geraniol 10-hydroxylase) CYP76B6, involved in the biosynthesis of secoiridoids and monoterpene indole alkaloid anticancer drugs in C. roseus, was found to belong to the CYP76 family in 2001 (Collu et al., 2001). The catalytic function of this enzyme was recently revised, and was shown to include a second oxidation activity, the conversion of 8-hydroxygeraniol into 8-oxogeraniol (Höfer et al., 2013). The same work also revealed a geraniol 8- and 9-hydroxylase activity of CYP76C4 from Arabidopsis thaliana. More recently, another CYP76 enzyme (CYP76A226) from C. roseus was found to metabolize oxidized geraniol derivatives and to have an iridoid oxidase activity, catalyzing the triple oxygenation of cis-trans-nepetalactol into 7-deoxyloganetic acid for the biosynthesis of secoiridoids and terpene indole alkaloids (Miettinen et al., 2014; Salim et al., 2014). Not all CYP76 enzymes seem to be devoted to the metabolism of monoterpenols. In most cases, however, CYP76s seem to be involved in terpenoid metabolism. CYP76Ms from monocots were found to metabolize diterpenoids for the synthesis of antifungal phytocassanes (Swaminathan et al., 2009; Wang et al., 2012; Wu et al., 2013), CYP76AH1 from Salvia miltiorhizza and its ortholog CYP76AH4 from rosemary (Rosmarinus officinalis) were shown to hydroxylate the norditerpene abietatriene in the pathway to labdane-related compounds (Zi and Peters, 2013), whereas CYP76Fs from sandalwood (Santalum album) were found to hydroxylate the sesquiterpenes santalene and bergamotene (Diaz-Chavez et al., 2013). CYP76B1 from Helianthus tuberosus was, however, found to metabolize herbicides belonging to the class of phenylurea (Robineau et al., 1998; Didierjean et al., 2002), but its physiological function was not reported. Other P450s from soybean (Glycine max; CYP71A10; Siminszky et al., 1999) or tobacco (Nicotiana tabacum; CYP71A11 and CYP81B1; Yamada et al., 2000) were also reported to metabolize phenylurea, but their physiological function was not investigated.
A. thaliana ecotype Columbia-0 (Col-0) emits no geraniol and only tiny amounts of linalool, and extensive volatile profiling of different tissues detected only minor amounts of lilac aldehydes (oxygenated linalool derivatives; Rohloff and Bones, 2005). However, ectopic expression of a linalool/nerolidol synthase of strawberry (Fragaria × anannasa cv Elsanta) revealed a potentially efficient oxidative linalool metabolism in A. thaliana rosette leaves (Aharoni et al., 2003). Only recent work started to explore linalool metabolism in A. thaliana, which was found mainly localized in the flowers (Ginglinger et al., 2013). This work demonstrated the existence of two linalool synthases producing different enantiomers, and the concomitant involvement of two P450 enzymes, CYP76C3 and CYP71B31, with predominance of CYP76C3, in linalool oxidation. It also suggested the presence of partially redundant enzymes that may contribute to floral linalool metabolism.
A family of eight CYP76 genes is detected in the A. thaliana genome. We report here an evolutionary and functional analysis of this family. We show that members of the CYP76C subfamily, when successfully expressed in yeast (Saccharomyces cerevisiae), all metabolize monoterpenols with different substrate specificities. Although CYP76Cs seem specific to Brassicaceae, they share common functions with CYP76s from other plants, such as CYP76B1 from H. tuberosus and CYP76B6 from C. roseus. These functions include not only monoterpenol oxidation, but also metabolism and detoxification of herbicides belonging to the class of phenylurea. Because of this property, CYP76Cs can be used simultaneously for monoterpenol oxidation and as selectable markers for plant transformation.
RESULTS
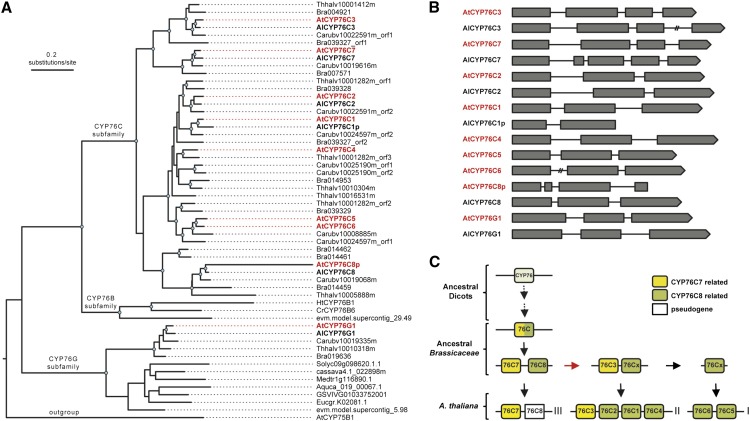
CYP76C Is a Recent P450 Subfamily Specific to Brassicaceae
Eight CYP76 genes have been annotated in the A. thaliana genome (http://www.p450.kvl.dk/p450.shtml). One member belongs to the CYP76G subfamily (CYP76G1), and the seven others fall into the CYP76C subfamily. A BLAST search in other fully sequenced plant genomes (http://www.phytozome.net) indicates that CYP76G1 orthologs are found usually as single copies in dicots (e.g. tomato [Solanum lycopersicum], eucalyptus [Eucalyptus grandis], and papaya [Carica papaya]; Fig. 1A), which suggests that duplicate copies of CYP76G are rapidly purged from the genome. The gene phylogeny (Fig. 1A) shows that CYP76C genes are expanded within Brassicaceae. The timing of this expansion is coincident with the diversification of the family, but did not occur before, because we found no CYP76C copies in Carica papaya, which is representative of an early diverging lineage within the order Brassicales, nor did we find copies in earlier diverging species (i.e. Gossypium raimondii or Theobroma cacao). Thus, the expansion of CYP76C occurred at least 50 million years ago (Beilstein et al., 2010).
Figure 1.
Phylogeny, gene structure, and history of the CYP76 family in Brassicaceae. A, Phylogeny of the CYP76 genes in Brassicaceae. A. thaliana and Arabidopsis lyrata genes are highlighted in bold red and black, respectively. Note that contiguous loci were found, in which cases individual open reading frames (orf) were arbitrarily separated and labeled with the orf tag. Nodes supported with bootstrap values ≥ 85% are marked with blue dots. The tree was rooted with At-CYP75B1. B, Intron-exon map of the CYP76 genes in A. thaliana (red) and A. lyrata (black). C, The likely sequence of duplication events that led to the CYP76C genes found in A. thaliana. The red arrow indicates segmental duplication. Roman numerals indicate the chromosomal location of each gene. Al, A. lyrata; Aquca, Aquilegia coerulea; At, A. thaliana; Bra, Brassica rapa; Carub, Capsella rubella; Cassava; Manihot esculenta; Cr, C. roseus; Eucgr, Eucalyptus grandis; evm.model, Carica papaya; GSVIV, Vitis vinifera; Ht, H. tuberosus; Medtr, Medicago truncatula; Solyc, Solanum lycopersicum; Thhal, Eutrema salsugineum.
The A. thaliana CYP76C genes and a pseudogene (CYP76C8p) are organized in three genomic clusters: CYP76C7 and CYP76C8p on chromosome 3; CYP76C3, CYP76C2, CYP76C1, and CYP76C4 on chromosome 2; and CYP76C5 and CYP76C6 on chromosome 1 (Supplemental Fig. S1A). CYP76C7 and CYP76C3 belong to the same clade and share three common introns, whereas CYP76C8, CYP76C2 CYP76C1, and CYP76C4 belong to a different clade and show only two common introns (Fig. 1, A and B). Based on phylogeny and intron-exon organization, the cluster on chromosome 2 thus most likely derives from a segmental duplication of the cluster formed by CYP76C7 and CYP76C8, possibly as a result of the A. thaliana α whole-genome duplication that occurred during early evolution of Brassicaceae (Bowers et al., 2003), followed by further amplification of the ancestral copy of CYP76C8 to generate CYP76C1, CYP76C2, and CYP76C4 (Fig. 1C). Support for this hypothesis is provided by the analysis of the locus structure in A. lyrata and other Brassicaceae where a copy of the WD-40 repeat family gene is found on the right border and a copy of the PEROXIN11 gene on the left border of both clusters formed by CYP76C7 and CYP76C8 as well as CYP76C3, CYP76C2, CYP76C1, and CYP76C4 (Supplemental Fig. S1B). Loss of CYP76C8 as a pseudogene is recent and only observed in A. thaliana for which no ESTs are reported, and a stop codon is present at position 341 of the protein (i.e. before the heme anchoring Cys in the active site), whereas CYP76C1 is present as a pseudogene in A. lyrata (Supplemental Fig. S1B). The CYP76C5-CYP76C6 tandem present in A. thaliana seems to derive from the dispersion of a tandem duplicate of CYP76C8, followed by a recent duplication event, because only a single homolog is found in other Brassicaceae, associated with the cluster formed by CYP76C3, CYP76C2, CYP76C1, and CYP76C4 (Supplemental Fig. S1B). Overall synteny analysis of the corresponding CYP76 loci in different Brassicaceae (Supplemental Fig. S1B) indicates complex genomic rearrangements with frequent gene duplications and losses or pseudogenizations. The CYP76C subfamily thus radiated in Brassicaceae and shows very high versatility, most likely associated with adaptive functions.
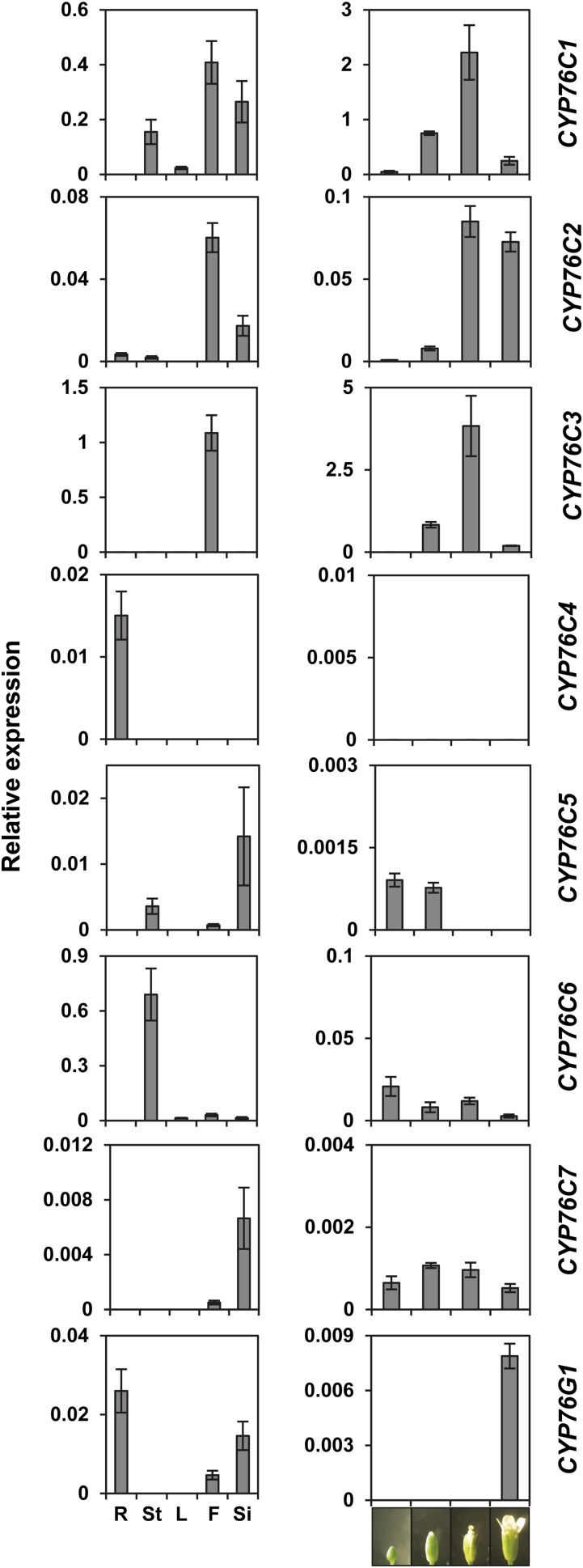
Expression Pattern of the CYP76 Genes in A. thaliana Suggests Limited Functional Redundancy
To evaluate functional specialization of the different members of the CYP76 family in A. thaliana, a quantitative real-time (qRT)-PCR analysis of their expression levels in different organs and floral stages was carried out (Fig. 2). CYP76C1, CYP76C2, and CYP76C3 were mainly expressed in flowers upon anthesis as already reported for CYP76C3 (Ginglinger et al., 2013). CYP76C1 and CYP76C2 were also expressed in siliques as well as at low levels in healthy leaves for CYP76C1, but the expression of CYP76C2 was at least 10 times lower than that of CYP76C1 or CYP76C3. Siliques were the main site of expression of CYP76C5 and CYP76C7, with the expression of the latter being extremely low and thus most likely restricted to very specific tissues. The expression of CYP76C4 was essentially restricted to roots, and was very low. CYP76C6 expression was mainly restricted to the leaves. CYP76C8 expression was investigated in A. lyrata and was the highest in flowers (carpels; Supplemental Fig. S2). CYP76G1 was expressed at very low levels in siliques and roots. Limited functional redundancy of the CYP76 genes is thus expected, except in flowers and siliques.
Figure 2.
Relative CYP76 gene transcripts levels in different plant organs and during flower development in A. thaliana. Evaluation of gene expression in different organs (left) and at different floral stages (right) was carried out by qRT-PCR. The cycle threshold (Ct) values were normalized to the Ct values obtained for four reference genes whose stable expression in A. thaliana tissues is known (Czechowski et al., 2005) and relative expression was calculated with the specific efficiency of each primer pair using the EΔCt method. Results represent the mean ± se of two technical repetitions and five biological replicates for organs, and three for flower stages. F, Flower; L, leaf; R, root; Si, silique; St, stem.
CYP76C Enzymes Are Versatile Monoterpenol Oxidases
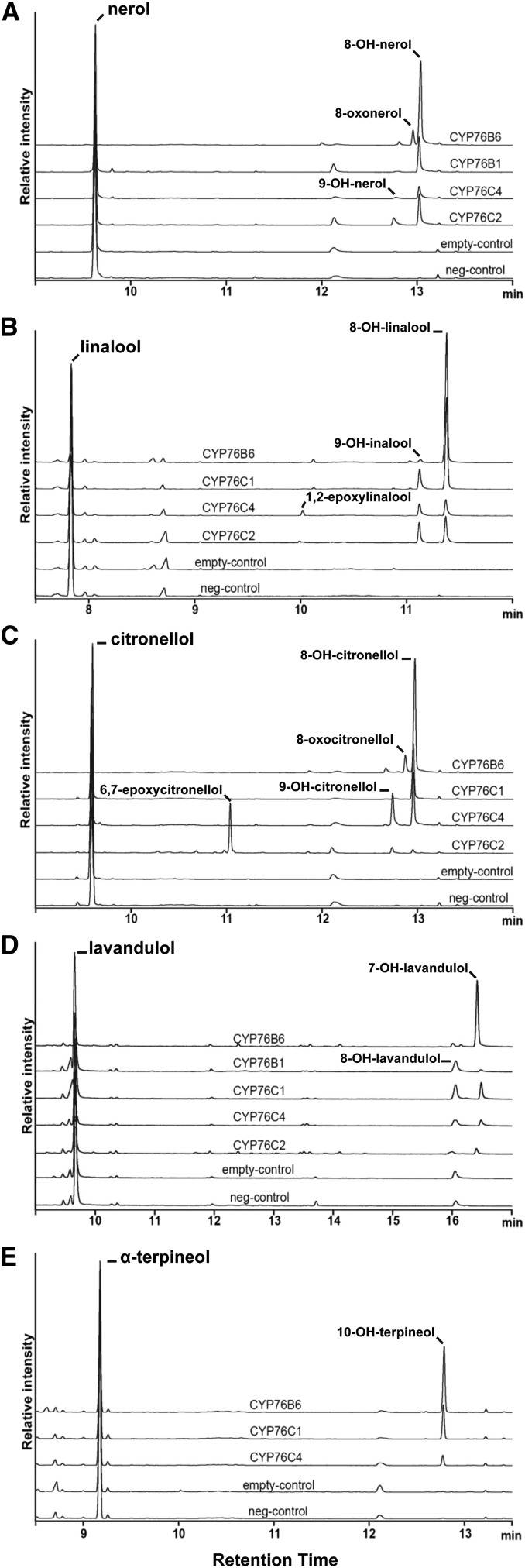
CYP76C3 and CYP76C4 were recently shown to metabolize linalool and geraniol, respectively (Ginglinger et al., 2013; Höfer et al., 2013). To determine whether monoterpenol hydroxylation is a common property of the members of the CYP76 family, a set of monoterpenols and monoterpenes was tested for conversion by CYP76Cs and CYP76G1 enzymes expressed in yeast, and their activities were compared with those of CYP76B6, the geraniol oxidase of the iridoid and terpenoid indole alkaloid pathways of C. roseus (Collu et al., 2001; Höfer et al., 2013), and of CYP76B1 of H. tuberosus (Robineau et al., 1998), the physiological function of which is still unknown. As previously reported (Höfer et al., 2013), expression levels of the A. thaliana CYP76s were low when evaluated from carbon monoxide-bound absorption spectra of the reduced enzymes (Supplemental Fig. S3). They were considered to be significant only for CYP76C1, CYP76C2, and CYP76C4, and were very low and close to the detection limit for CYP76C6, CYP76C7, and CYP76G1. Microsomes prepared from yeast transformed with each of them were tested for activity on 200 μm of five different monoterpenols and four monoterpene olefins (Fig. 3; Table I). No activity was detected with microsomes from yeast expressing CYP76C3, CYP76C5, CYP76C6, CYP76C7, and CYP76G1. Metabolism of geraniol was observed only with CYP74C4 and CYP76B6, confirming previous results (Höfer et al., 2013), and only CYP76B6 further oxidized metabolized 8-hydroxygeraniol. However, nerol was converted by CYP76C2, CYP76C4, CYP76B1, and CYP76B6 into the same major product, most likely 8-hydroxynerol (Fig. 3A; electron-ionization mass spectrum [EI-MS] in Supplemental Fig. S4) and different minor products. Based on mass spectra and data previously reported for geraniol (Höfer et al., 2013), the minor product is expected to be 9-hydroxynerol for CYP76C2 and CYP76C4 (Fig. 3A; EI-MS in Supplemental Fig. S5), and 8-oxonerol for CYP76B6 (Fig. 3A; EI-MS in Supplemental Fig. S6).
Figure 3.
GC-FID chromatograms of the reaction products resulting from the conversion of monoterpenols by the yeast-expressed CYP76C1, CYP76C2, CYP76C4, CYP76B1, and CYP76B6 enzymes. Microsomal membranes from recombinant yeast transformed with the P450 expression vectors or with the empty vector (empty-control) were incubated with 200 μm of substrate for 20 min in the presence of NADPH. No NADPH was added to the negative control (neg-control). Samples corresponding to the major peaks (except for lavandulol and nerol) were analyzed by NMR for compound identification (Supplemental Fig. S9). Identified compounds were then assigned based on their retention time and EI-MS. Mass spectra of the products and references are available in Supplemental Figs. S4 to S17.
Table I. Monoterpenoid metabolism by yeast-expressed CYP76 enzymes.
Microsomal membranes from recombinant yeasts were incubated with 200 μm of substrate for 20 min in the presence of NADPH. Minus signs indicate not metabolized, whereas plus signs indicate formation of minor products that were not quantified. Data are means ± sd of three replicates.
| Substrate |
Enzyme Activity |
||||
|---|---|---|---|---|---|
| CYP76C1 | CYP76C2 | CYP76C4 | CYP76B1 | CYP76B6 | |
| pmol product/min per pmol P450 | |||||
| 8-OH-geraniol | − | − | − | − | 75.9 ± 1.5 |
| Nerol | − | 22.4 ± 1.3 | 6.3 ± 0.5 | 11.7 ± 0.3 | 52.0 ± 1.8 |
| Linalool | 211.7 ± 7.3 | 19.9 ± 0.6 | 6.9 ± 0.1 | − | 35.5 ± 1.1 |
| Citronellol | 147.7 ± 11.2 | 53.4 ± 2.2 | 18.8 ± 1.1 | − | 63.0 ± 0.6 |
| α-Terpineol | 95.2 ± 5.7 | − | 1.8 ± 0.1 | − | 22.5 ± 1.6 |
| Lavandulol | 7.5 ± 0.1 | 4.5 ± 0.2 | 2.4 ± 0.1 | 3.7 ± 0.2 | 118.5 ± 0.3 |
| Limonene | − | − | + | + | + |
| p-Cymene | − | − | + | + | + |
| Camphene | − | − | + | − | − |
| α-Phellandrene | − | − | − | + | + |
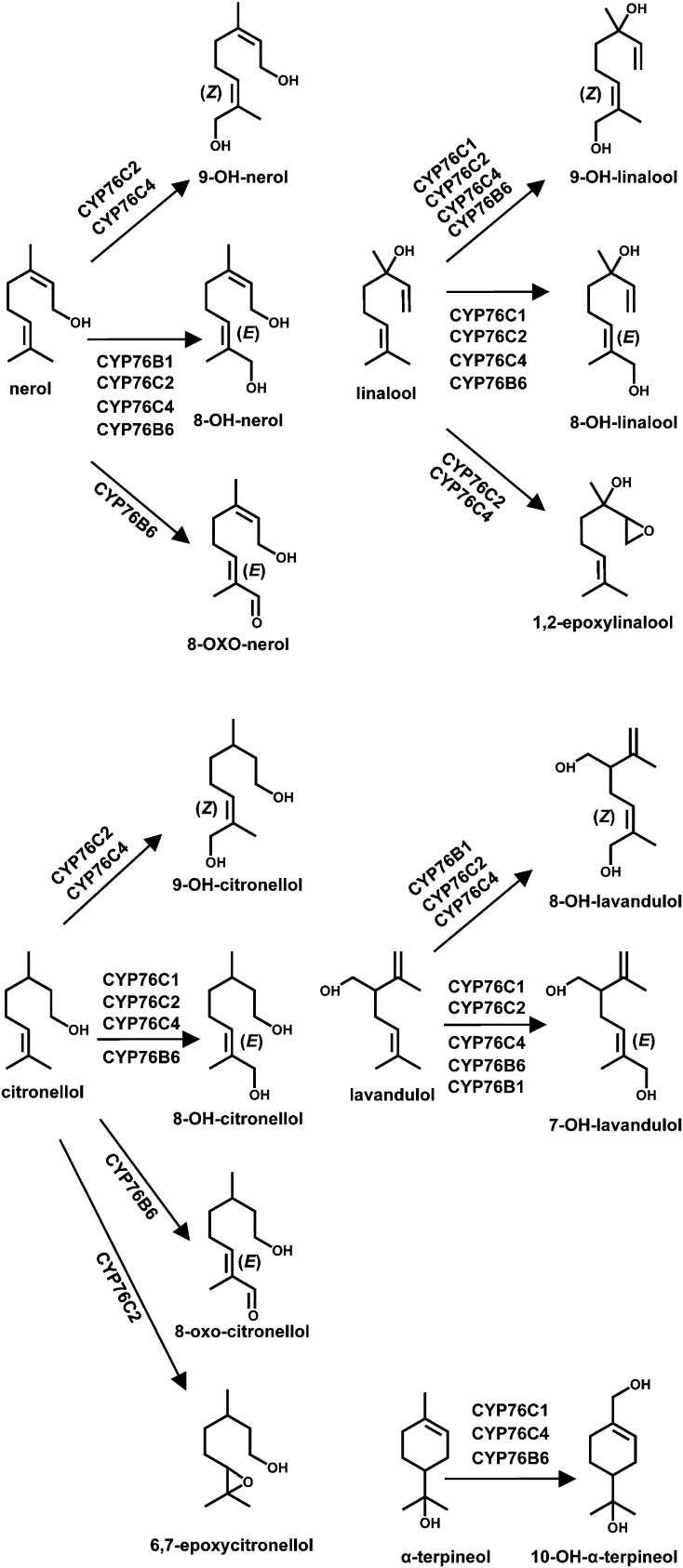
Linalool was found to be metabolized by CYP76C1, CYP76C2, CYP76C4, and CYP76B6 (Fig. 3, B and C). The same products, 8-hydroxylinalool (main) and 9-hydroxylinalool (minor), were obtained from linalool, based on a comparison of retention times and mass spectra with authentic standards and/or NMR validation of the products extracted from upscaled reactions (Fig. 3B; EI-MS in Supplemental Figs. S7 and S8, respectively, and NMR in Supplemental Fig. S9, A and B). CYP76C4 and CYP76C2 additionally formed 1,2-epoxylinalool (Fig. 3B; EI-MS in Supplemental Fig. S10). Metabolism of citronellol by the different enzymes led to several different products (Fig. 3C). In the absence of authentic standards, product structures were assigned by NMR analysis of the products extracted from upscaled reactions (Supplemental Fig. S9, C–E). 8-Hydroxycitronellol (Fig. 3C; EI-MS in Supplemental Fig. S11) appeared as the major product for CYP76C1, CYP76C4, and CYP76B6 and as the sole product for CYP76C1. CYP76C4 also generated 9-hydroxycitronellol as the minor product (Fig. 3C; EI-MS in Supplemental Fig. S12), whereas CYP76B6 rather generated a compound assumed to be 8-oxocitronellol as the minor product (Fig. 3C; EI-MS in Supplemental Fig. S13). CYP76C2, however, catalyzed the formation of a completely different major product (Fig. 3C; EI-MS in Supplemental Fig. S14) with a shorter retention time, which was identified by NMR as 6,7-epoxycitronellol (Supplemental Fig. S9E), and generated very minor amounts of 8- and 9-hydroxylated products. Lavandulol was converted by CYP76B6 into one major product (Fig. 3D), most likely 7-hydroxylavandulol (Fig. 3D; EI-MS in Supplemental Fig. S15), with one minor side product, presumably 8-hydroxylavandulol (Fig. 3D; EI-MS in Supplemental Fig. S16). It was, however, a poor substrate for CYP76C1, CYP76C2, and CYP76C4, which catalyzed the formation of the same two products, most likely 7- and 8-hydroxylavandulol in equal amounts, and for CYP76B1, which formed mostly 7-hydroxylavandulol. The cyclic monoterpenol, α-terpineol, was converted by CYP76B6, CYP76C1, and CYP76C4 into a single product, 10-hydroxy-α-terpineol (Fig. 3E; NMR and EI-MS in Supplemental Figs. S9F and S17, respectively).
Olefinic monoterpenes were very poor substrates and only traces of oxygenated products were obtained with CYP76C4, CYP76B1, or CYP76B6 (Table I). The capacity to oxidize monoterpenols is thus shared by a large number of quite divergent members of the CYP76 family (Fig. 4). Most of them are promiscuous enzymes with regard to monoterpenols, but do not metabolize olefins.
Figure 4.
Summary of the reactions catalyzed by CYP76 enzymes on monoterpenols.
Comparison of the Efficiency of Linalool Metabolism by CYP76Cs from A. thaliana
Evaluation of the catalytic parameters was focused on linalool (Supplemental Fig. S18), the most relevant substrate in A. thaliana. It was carried out using short incubation times and low enzyme concentrations to minimize further conversion of primary products. The catalytic parameters for the different enzymes are summarized in Supplemental Figure S18 and indicate that CYP76C1 is likely to be the most effective linalool oxygenase in A. thaliana.
CYP76Cs Also Metabolize Herbicides Belonging to the Class of Phenylurea
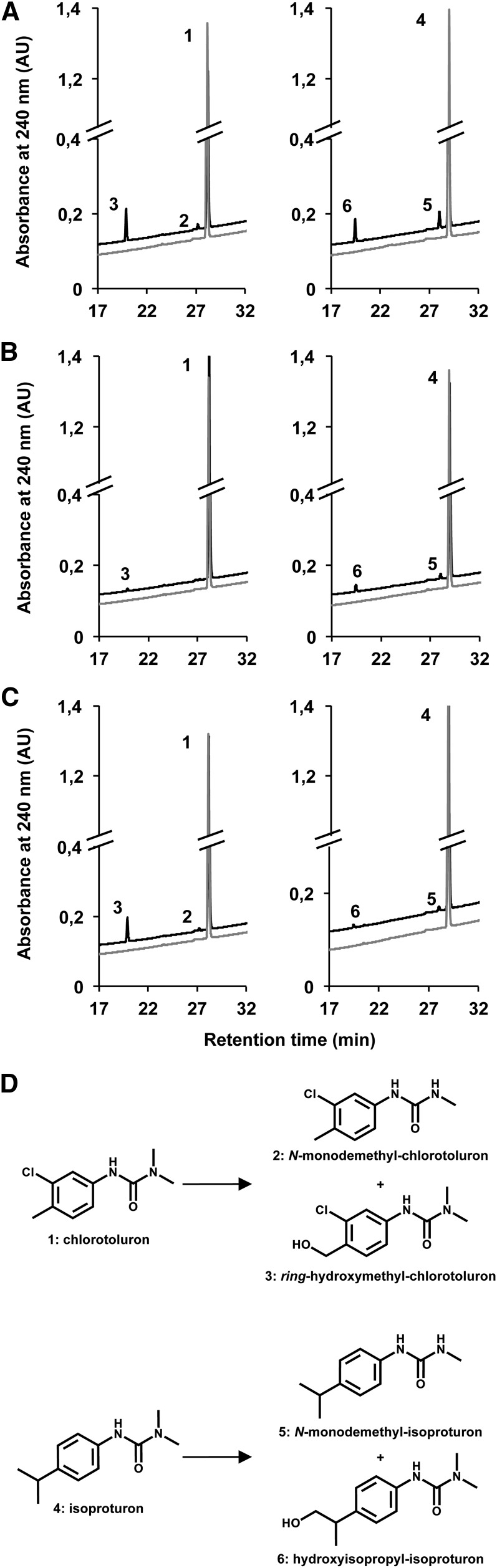
CYP76B1 from H. tuberosus was previously reported (Robineau et al., 1998) to metabolize the PSII inhibitors phenylurea, leading to nonphytotoxic products. As a result, its ectopic expression confers phenylurea resistance and was shown to be effective as a selectable marker for plant transformation (Didierjean et al., 2002). The yeast-expressed CYP76s (all except CYP76C5 and CYP76C6) were thus screened for herbicide metabolism (Supplemental Table S1). The A. thaliana CYP76C enzymes active in vitro on monoterpenols (i.e. CYP76C1, CYP76C2, and CYP74C4) all metabolized a large subset of phenylurea (Supplemental Table S1). CYP76C1 and CYP76C2 metabolized a larger set of compounds. We focused on chlorotoluron and isoproturon, metabolized by all three enzymes, for product determination (Fig. 5). CYP76C1 was the most active, and converted chlorotoluron into ring-methyl-hydroxychlorotoluron as the main product and also produced minor amounts of N-demethyl-chlorotoluron (Fig. 5A; Supplemental Table S2). Isoproturon was similarly converted into isoproturon hydroxylated on the isopropyl side chain and N-demethyl-isoproturon (Fig. 5A; Supplemental Table S2). CYP76C2 and CYP76C4 generated the same compounds in lower amounts (Fig. 5, B and C). Table II compares the catalytic parameters determined with CYP76C1, CYP76C2, and CYP76C4 and shows that the most efficient metabolism was obtained with CYP76C1 and occurs via hydroxylation leading to nonphytotoxic products.
Figure 5.
HPLC-photodiode array chromatograms and phenylurea conversion products of yeast-expressed CYP76C enzymes. Microsomal membranes from recombinant yeasts were incubated with herbicide (400 µm) for 20 min in the presence of NADPH (black), or without NADPH (gray) for negative controls. Products were identified by comparison of retention times and mass spectra with authentic standards. Reference MS data are provided in Supplemental Table S2. A, CYP76C1 + chlorotoluron (left) or isoproturon (right). B, CYP76C2 + chlorotoluron (left) or isoproturon (right). C, CYP76C4 + chlorotoluron (left) or isoproturon (right). D, Phenylurea conversion products of CYP76C1, CYP76C2, and CYP76C4. 1, Chlorotoluron; 2, N-monodemethyl-chlorotoluron; 3, ring-hydroxymethyl-chlorotoluron; 4, isoproturon; 5, N-monodemethyl-isoproturon; 6, hydroxyisopropyl-isoproturon; AU, arbitrary unit.
Table II. Catalytic parameters of phenylurea metabolism by A. thaliana CYP76C enzymes.
Kinetic assays were carried out in a final volume of 200 µL for 20 min in the presence of 1 mm NADPH, 7 pmol of P450, and variable substrate concentrations. Kinetic parameters were deduced from Michaelis-Menten representation. dM-CTU: monodemethyl-chlorotoluron; dM-IPU: monodemethyl-isoproturon; OH-CTU: ring-hydroxymethyl-chlorotuluron; OH-IPU: hydroxyisopropyl-isoproturon. Data are means ± sd of three determinations. Units for catalytic parameters are as follows: Km (μm), kcat (min−1), and kcat/Km (µm−1 min−1). Dash indicates product not formed or in amounts too low for quantification.
| Product | Catalytic Parameter | CYP76C1 | CYP76C2 | CYP76C4 |
|---|---|---|---|---|
| OH-CTU | Km | 589 ± 190 | 249 ± 28 | 96 ± 14 |
| kcat | 61 ± 10 | 1.4 ± 0.2 | 15 ± 3 | |
| kcat/Km | 0.1 | 0.006 | 0.16 | |
| dM-CTU | Km | 135 ± 52 | — | — |
| kcat | 4 ± 0.8 | — | — | |
| kcat/Km | 0.03 | — | — | |
| OH-IPU | Km | 595 ± 98 | 63 ± 4 | 3.8 ± 2 |
| kcat | 38 ± 5 | 2.4 ± 0.9 | 0.9 ± 0.1 | |
| kcat/Km | 0.06 | 0.04 | 0.2 | |
| dM-IPU | Km | 165 ± 67 | 196 ± 112 | — |
| kcat | 12 ± 2 | 1.4 ± 0.7 | — | |
| kcat/Km | 0.07 | 0.007 | — |
Conversely, no metabolism of phenylurea was detected with CYP76B6 from C. roseus, which is the most promiscuous enzyme with monoterpenols. There is thus no systematic correlation of monoterpenols and phenylurea metabolism.
Ectopic Expression of CYP76Cs Confers Resistance to Phenylurea
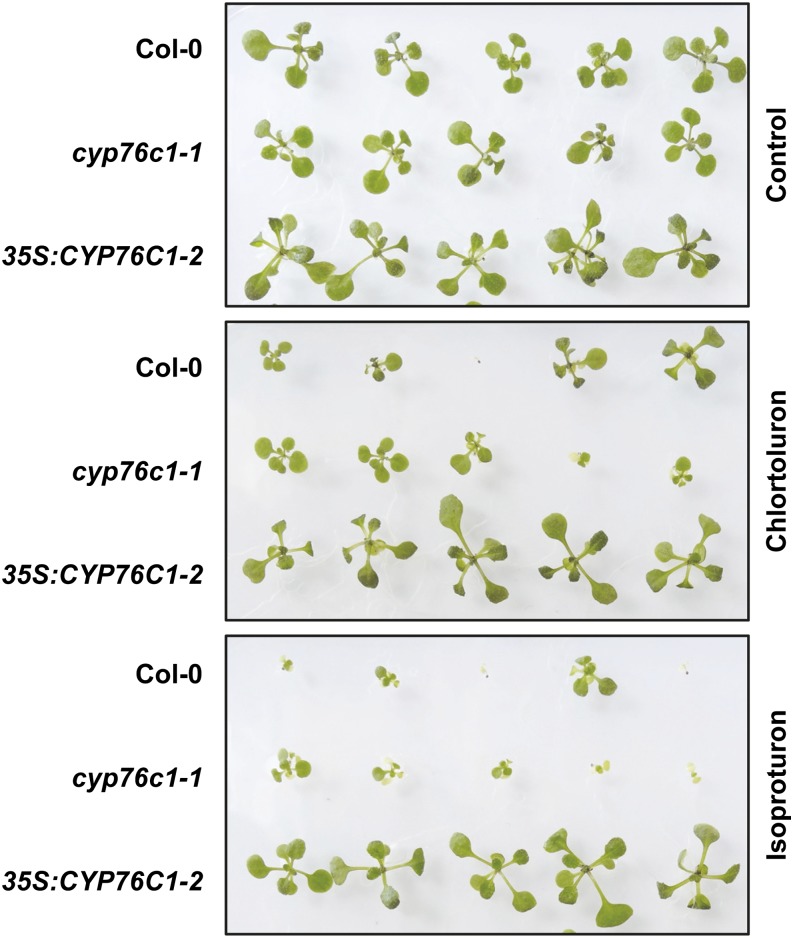
Considering the low and spatially restricted expression of A. thaliana CYP76Cs in roots and leaves, we did not anticipate a significant impact of their current natural expression on plant tolerance to phenylurea. To confirm this hypothesis and to test the influence of gene-increased expression on herbicide resistance, insertion mutants and overexpression lines were isolated for CYP76C1, CYP76C2, and CYP76C4 (Supplemental Fig. S19). Their herbicide tolerance was compared with the wild type. Figure 6 illustrates isoproturon and chlorotoluron tolerance of CYP76C1 insertion and overexpression lines. As anticipated, no significant effect of gene inactivation on herbicide tolerance was observed, independent of the herbicide concentration added to the growth medium. Ectopic overexpression, however, led to a significant gain in herbicide tolerance, the most significant being for isoproturon with all three enzymes (Fig. 6; Supplemental Fig. S20).
Figure 6.
CYP76C1 overexpression confers herbicide tolerance to A. thaliana Col-0, cyp76c1 insertion, or Cauliflower mosaic virus 35S (35S) promoter-driven overexpression lines were grown on Murashige and Skoog medium for 14 d in the presence or absence of 1 μm of chlorotoluron or isoproturon.
DISCUSSION
The CYP76 family of P450 enzymes arose with the emergence of seed plants (Nelson and Werck-Reichhart, 2011) and shows an extensive diversification in monocots and dicots (http://drnelson.uthsc.edu/CytochromeP450.html) with 34 subfamilies named thus far. Based on currently available plant genomes, homologs of the CYP76Cs from A. thaliana are found only in Brassicaceae, but not in papaya. Together with the high frequency of gene duplication and loss observed within Brassicaceae CYP76Cs, this suggests a high versatility, and a role in fast lineage-specific adaptation and plant-herbivore or plant-microbe interaction. A similar trend to high gene duplication is observed in rice (Oryza sativa), in which 29 CYP76 genes were annotated in six subfamilies (Nelson and Werck-Reichhart, 2011). Thus far, the function of only one of these subfamilies is described, being the formation of antifungal diterpenoids phytocassanes (Swaminathan et al., 2009; Wang et al., 2012; Wu et al., 2013). This raises the question of the functional divergence(s) associated with the CYP76 subfamily burst and CYP76Cs duplications in Brassicaceae.
For tandem duplicated genes, the divergence of the expression profile usually occurs at or shortly after gene duplication (Ganko et al., 2007). Clear expression divergences were observed among duplicates such as CYP76C7 (mainly expressed in siliques) and CYP76C3 (flowers), or CYP76C5 (roots), and CYP76C6 (leaves). Large differences were also observed between their respective expression levels, with much higher expression of CYP76C3 and CYP76C6 than of CYP76C7 and CYP76C5. Divergence is even stronger between CYP76C8 and the three duplicates CYP76C1, CYP76C2, and CYP76C4. Whereas CYP76C8 turned into a pseudogene in A. thaliana (but is expressed in flower carpels in A. lyrata), CYP76C1 is the most highly expressed gene of the tandem repeats in chromosome 2, especially in flowers, and CYP76C4 is expressed only at low levels in roots. Overall, expression patterns indicate functional specialization of the different paralogs in A. thaliana, although not excluding some redundancy (e.g. between CP76C3 and CYP76C1). The very low expression of some of them, such as CYP76C7, CYP76C5, or CYP76C4, possibly results from a restricted expression in specific tissues. Considering the high versatility and propensity to gene loss of CYP76Cs, it may also be indicative of ongoing pseudogenization.
If divergence in spatiotemporal expression is a factor that favors gene retention at or just after duplication, it is often followed by a further divergence in expression and in protein sequence and function. We recently reported the activity of CYP76C4 and CYP76C3 in monoterpenol oxidation, the first catalyzing geraniol 8- and 9-hydroxylation (Höfer et al., 2013), and the second the oxygenation of both (3R)- and (3S)-linalool, mainly into 4- and 5-hydroxylinalool, with 8- and 9-hydroxylinalool as minor products (Ginglinger et al., 2013). To further investigate their capacity for monoterpenol oxidation, the whole set of eight CYP76 genes from A. thaliana was expressed in yeast. For most of them, the expression was low if any, possibly reflecting either toxicity for the host or low intrinsic protein stability. However, the three best expressed CYP76Cs (CYP76C1, CYP76C2, and CYP76C4), as well as CYP76B1 from H. tuberosus and CYP76B6 from C. roseus, were all found to metabolize several monoterpenols with different substrate preferences and different efficiencies, and sometimes forming different products. Olefinic monoterpenes were poor substrates for all of them. The activities detected with CYP76C2, CYP76C4, and CYP76B1 were low; however, this might be related to their low levels of expression. Expression of CYP76C2 was previously reported to be strongly activated by bacterial pathogens and in senescent tissues (Godiard et al., 1998). Production of monoterpenols and their oxides is thus far not reported in infected or senescent tissues. However, monoterpenols and their oxides are described for antimicrobial activity (Junker and Tholl, 2013; Radulović et al., 2013). Conversely, CYP76C1 and CYP76B6 very actively catalyzed the oxidation of several tested compounds. CYP76C1 was the most active with linalool, the only monoterpenol for which oxidation products were thus far reported to be emitted by A. thaliana (Rohloff and Bones, 2005) and detected as soluble conjugates (Aharoni et al., 2003; Ginglinger et al., 2013). Moreover, linalool synthases were previously shown to be essentially expressed in A. thaliana flowers, the main site of expression of CYP76C1 (Ginglinger et al., 2013). CYP76C1 therefore appears as a prime candidate to play a significant role in floral linalool metabolism in A. thaliana, which is under current investigation. CYP76C1 also very actively metabolized citronellol and α-terpineol, the latter of which was also detected after volatile profiling of A. thaliana plants (Rohloff and Bones, 2005).
Our preliminary data (Ginglinger et al., 2013; B. Boachon and J. Iglesias, unpublished data) indicate that gene suppression or overexpression of CYP76Cs does not lead to any growth or fertility phenotype. It is thus expected that they play a role in the synthesis of allelochemicals involved in plant-microbe or plant-insect interaction. Identification of the final products resulting from the CYP76C-mediated monoterpenol oxidation is expected to be challenging, because the expression of most of them is low or restricted to very specific tissues available in very small amounts and some of them may use the same substrate (Ginglinger et al., 2013). The primary oxygenated monoterpenols are unlikely to be the final products in the plant and these products, as well as their final degree of oxidation/glycosylation and physicochemical properties, cannot be predicted from published data. However, the differential expression of the CYP76C genes in Brassicaceae predicts that they are unlikely to catalyze successive oxidation steps in a same pathway.
It is interesting to note that the ability to metabolize monoterpenols is not restricted to CYP76Cs from Brassicaceae, but extends to enzymes classified as CYP76Bs from Compositeae and Apocynaceae. CYP76A26 from C. roseus has also been reported to be active with monoterpenols, although its main activity was in iridoid metabolism (Miettinen et al., 2014). Monoterpenol metabolism is thus expected to be a quite common feature of the CYP76 family in dicots. Unexpectedly, CYP76B6, thought to be a specific geraniol oxidase dedicated to the secoiridoid/TIA pathway (Höfer et al., 2013), emerged from the screening as the most promiscuous enzyme with regard to monoterpenols, efficiently metabolizing geraniol, nerol, linalool, citronellol, and lavandulol. CYP76B6 promiscuity thus points to the critical importance of the geraniol synthase for producing the relevant substrate for iridoid and terpene indole alkaloid production (Miettinen et al., 2014) and to the capacity of CYP76B6 and resulting duplicates for evolving multiple functions in different monoterpenols-derived pathways in the plant. In contrast with CYP76Cs from A. thaliana, CYP76B6 essentially formed with all monoterpenols a single hydroxylated derivative and its further oxidation product. The second 9-hydroxylation product observed with the A. thaliana enzymes was not obtained or in tiny amounts. Surprisingly, whereas CYP76B6 acquired an extended capacity to regiospecifically metabolize a large set of monoterpenols, it is completely unable to metabolize phenylurea, which are substrates of the A. thaliana and H. tuberosus enzymes.
Herbicide resistance is a major challenge for modern agriculture (Powles and Yu, 2010). It can result from a mutation at the level of the herbicide target site, from increased metabolism, or from reduced translocation (Powles and Yu, 2010). P450s most often catalyze primary herbicide metabolism and activation, before further processing by conjugation enzymes and storage in the vacuole. Their role in the acquisition of insecticide resistance in insect pests is quite well documented (Ffrench-Constant, 2013), but their part in endowing herbicide resistance and the mechanisms of acquisition of this resistance in weeds are still poorly understood. P450-dependent herbicide metabolism is usually thought to result from the serendipitous docking of herbicides in the active site involved in physiological processes (Powles and Yu, 2010). To our knowledge, CYP76s constitute the first example providing a potential link between the metabolism of physiologically relevant compounds and the metabolism of herbicides and herbicide tolerance. The fast evolution and versatility of the CYP76 family, together with the herbicide tolerance of CYP76C overexpression lines, illustrate how herbicide resistance can be acquired either via gene activation or via gene duplication when those lead to extended or increased gene expression.
CYP76C1, CYP76C2, and CYP76C4, like CYP76B1, metabolize a quite broad set of phenylurea compounds, forming both N-demethylated and ring-methyl(isopropyl)-hydroxylated products. CYP76Cs thus allow herbicide docking in two opposite orientations. Irreversible herbicide detoxification requires either ring-hydroxylation or a double N-dealkylation. Hydroxylation is thus expected to constitute the main CYP76C-dependent detoxification process. In spite of relatively low herbicide turnovers measured in vitro, particularly in the case of CYP76C2, a significant effect on herbicide detoxification is confirmed by the increased herbicide tolerance of overexpressors of all three CYP76C1, CYP76C2, and CYP76C4 enzymes. Natural CYP76C expression of A. thaliana Col-0 does not significantly affect herbicide tolerance. This is not surprising given the restricted tissue-specific expression of each; the amount of enzyme(s) currently present in the wild-type plant is not sufficient to support herbicide resistance. Our data, however, demonstrate the possibility of using the CYP76Cs from A. thaliana to engineer herbicide tolerance. These findings raise the interesting possibility of using genes of the plant’s specialized metabolism as selectable markers for plant transformation. In some cases, the selectable marker and metabolic function could be conveyed by the same gene. These results also point to a potential complex interplay of the metabolism of herbicides with that of specialized plant compounds, and to a possible effect of herbicide treatment and detoxification on plant-insect or plant-pathogen interaction.
MATERIALS AND METHODS
CYP76C Subfamily History and Phylogeny
CYP76 coding sequences from various species were retrieved from Phytozome (http://www.phytozome.org) and GenBank (https://www.ncbi.nlm.nih.gov/genbank/) databases. Coding sequences were translated into amino acid sequences and aligned with MUSCLE (Edgar, 2004) prior to determination of Gblocks (Castresana, 2000) using the SeaView software (http://pbil.univ-lyon1.fr/; Gouy et al., 2010). The corresponding nucleotides Gblocks alignment (Supplemental Data Set S1) was subsequently used for phylogeny reconstruction by maximum likelihood analysis with PhyML 3.0 (Guindon et al., 2010) using the generalized time reversible model (default settings except that the proportion of invariable sites was estimated). Phylogeny consistency was tested by performing 100 bootstrap iterations. The output tree was shaped using the FigTree software (http://tree.bio.ed.ac.uk/software/figtree/). Organization of CYP76C genes in the Arabidopsis thaliana genome was realized according to the chromosome map tool from The Arabidopsis Information Resource (http://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp). Synteny analysis of the CYP76C3 and CYP76C7 loci was realized with the help of the synteny tool on the Phytozome Web site (http://www.phytozome.net/).
Plant Growth
Seeds of A. thaliana Col-0 and Arabidopsis lyrata strain MN47 (Hu et al., 2011) were sown on a standard soil compost mixture. Plants were grown individually in 7-cm pots in growth chambers at 22°C during the 12-h-day period and 19°C during the 12-h-night period under white fluorescent lamps with a photon fluency of 60 μmol m−2 s−1 (rosette leaves) to 90 μmol m−2 s−1 (flower stage). A. lyrata plants were grown individually in 7-cm pots for 5 weeks, before transfer to 12-cm pots containing a standard soil compost mixture completed at 50% (v/v) with sand, and were grown in greenhouses at 24°C during the 16-h-day period and 20°C during the 8-h-night period under a sodium-vapor lamp with a photon fluency of 100 μmol m−2 s−1 to 150 μmol m−2 s−1.
Quantification of Gene Expression
Quantification of gene expression was carried out by qRT-PCR as previously described (Ginglinger et al., 2013). The different organs of A. thaliana and A. lyrata were harvested from five different plants at the flowering stage and were immediately frozen in liquid nitrogen. For the normalization of gene expression in A. lyrata, the orthologs of A. thaliana SAND-like (gene 481666) and TIP41-like (gene 491240) were used after their stable expression was validated among four putative reference genes by the GeNorm (Vandesompele et al., 2002) and NormFinder (Andersen et al., 2004) algorithms and GenEx 4 software (http://genex.gene-quantification.info/). Oligonucleotides used for each gene are provided in Supplemental Table S3. Relative expression was calculated with the specific efficiency of each primer pair using the EΔCt method (Pfaffl, 2001). For A. thaliana, five biological replicates were used for the organs and three were used for the floral stages. For A. lyrata, five biological replicates were used for each tissue.
Generation of Expression Vectors
The generation of the CYP76B1 construct is described in Didierjean et al. (2002). All other constructs are described in Höfer et al. (2013). The yeast (Saccharomyces cerevisiae) and plant expression constructs were generated by PCR amplification from complementary DNA prepared from tissues in which each gene was found to be the most highly expressed. The PCR fragments of CYP76C1, CYP76C2, CYP76C5, CYP76C7, and CYP76B1 were integrated into the yeast expression vector pYeDP60. The constructs for CYP76C3, CYP76C4, CYP76C6, and CYP76B6 were prepared using the Uracil-Specific Excision Reagent (New England Biolabs) cloning technique according to Nour-Eldin et al. (2006) and the PCR fragments were integrated into the yeast expression plasmid pYeDP60u2. For plant expression constructs, CYP76C1 was cloned similarly and integrated in the plant expression vector pCAMBIA2300u. Complementary DNA from CYP76C2 and CYP76C4 was amplified by PCR using specific primers tailed for Gateway cloning technology (Invitrogen) and successively cloned in pDONR 201 and the plant expression vector pB7WG2 (Karimi et al., 2002). Constructs were confirmed by sequencing at each step. Primers used for cloning are provided in Supplemental Table S3.
Heterologous Expression in Yeast
The WAT11 yeast strain was transformed with pYeDP60u2 containing the different P450 sequences as described in Gietz and Schiestl (2007). Yeast cultures were grown and P450 expression was induced as described in Pompon et al. (1996). Cells were harvested by centrifugation and manually broken with glass beads (0.45 mm in diameter) in 50 mm Tris-HCl buffer, pH 7.5, containing 1 mm EDTA and 600 mm sorbitol. The homogenate was centrifuged for 10 min at 10,000g and the resulting supernatant was centrifuged for 1 h at 100,000g. The pellet consisting of microsomal membranes was resuspended in 50 mm Tris-HCl, pH 7.4, 1 mm EDTA, and 30% (v/v) glycerol and stored at −20°C. P450 content of the microsomal preparations was measured by differential spectrophotometry according to Omura and Sato (1964).
Assays for Monoterpenoid Metabolism
A standard enzyme assay using the monoterpenols as substrates was carried out in 100 µL of 20 mm sodium phosphate buffer, pH 7.4, containing varying concentrations of substrate, 600 µm NADPH, and adjusted amounts of P450 enzyme. After addition of NADPH, samples were incubated at 28°C and the reaction was stopped with 10 µL of 1 m HCl and 500 µL of ethyl acetate. Samples were vortexed for 10 s and centrifuged at 4,000g for 2 min. The ethyl acetate phase was transferred to a new vial and the extraction was repeated once. The combined organic phase was dried over anhydrous Na2SO4 (Sigma-Aldrich), concentrated under argon and analyzed by gas chromatography (GC)-flame ionization detection (FID) and GC-mass spectrometry (MS). For the determination of the kinetic parameters of A. thaliana CYP76Cs on linalool, assays were scaled up to a final volume of 400 µL, using R-linalool concentrations ranging from 5 µm to 600 µm, 1 mm NADPH, and about 50 nm P450s. Formation of products was quantified after 4 min of incubation. Kinetic parameters were deduced from Michaelis-Menten representation.
Olefin substrates were incubated in closed 2-mL glass vials for 20 min at 28°C using a thermomixer (Eppendorf) under constant shaking. In a reaction volume of 300 µL, 10% (v/v) of yeast microsomes were diluted in phosphate citrate buffer in the presence of 200 µm of olefin monoterpenes (dissolved in 0.8% [v/v] ethanol) and 1 mm of NADPH. The reaction was quenched on ice and products were extracted with 500 µL of pentane in a thermomixer during 5 min at 20°C. The solvent layer was recovered after centrifugation and analyzed by GC-FID and GC-MS.
GC-FID and GC-MS Analysis
Capillary GC was performed on a Varian 3900 gas chromatograph (Agilent Technologies) equipped with a flame ionization detector and a DB-5 column (30 m, 0.25 mm, and 0.25 µm; Agilent Technologies) with splitless injection at a 250°C injector temperature, and a temperature program of 0.5 min at 50°C, 10°C/min to 320°C, and 5 min at 320°C. Terpenoids were identified based on their retention time and electron-ionization mass spectra (70 eV and mass-to-charge ratio of 50–600) with a PerkinElmer Clarus 680 gas chromatograph coupled to a PerkinElmer Clarus 600T mass spectrometer. Capillary GC-MS was performed as described above. Reference standards of 8-hydroxylinalool was synthesized as previously described (Ginglinger et al., 2013). 1,2-Epoxylinalool was kindly provided by Adam J. Mathich (New Zealand Institute for Plant and Food Research Limited).
NMR Characterization of Products
To generate amounts of products large enough for NMR analysis, the standard enzyme assay was scaled up to a volume of 10 mL containing 400 µm of substrate. After a first incubation for 15 min at 28°C, a second aliquot of P450 enzyme was added and incubated for another 15 min. The reaction was stopped by adding 1 mL of 1 m HCl, vortexing, and cooling on ice. Several upscaled assays were pooled to ensure proper NMR detection of the products. For the extraction of the products, solid-phase extraction columns (Oasis HLB extraction cartridges; Waters) were equilibrated with chloroform, methanol, and water prior to gradual extraction of up to 15 mL of combined samples. After drying, the columns were eluted with CDCl3 and the combined organic phase was dried over anhydrous Na2SO4 and concentrated under argon prior to NMR analysis.
NMR was conducted on a 500-MHz Bruker Avance II spectrometer equipped with a 5-mm dual 13C and 1H cryoprobe with a z-gradient operating at 500.13 MHz for 1H and 125.758 MHz for 13C. A number of different spectra including one-dimensional 1H, 1H to 1H correlation spectroscopy, edited 1H to 13C heteronuclear single-quantum correlation, and 1H to 13C heteronuclear multiple bound correlation were recorded for each sample, adding 1H to 1H nuclear overhauser effect spectroscopy and one-dimensional 13C when required. Pulse sequences were taken from the Bruker library. All experiments were acquired at 293 K with a minimal relaxation delay of 2 s and a mixing time of 600 or 800 ms for nuclear overhauser effect spectroscopy experiments. Coupling constants were assumed to be around 145 Hz and 8 Hz for 1J(13C–1H) and nJ(13C –1H), respectively. Acquisition parameters were adjusted when necessary but typically spectral windows were set to 7 kHz for 1H and 27 or 31 kHz for 13C. For two-dimensional spectra, the data size was at least 2,048 points in the direct dimension and varied between 128 and 256 points in the indirect dimension according to the required resolution.
Assays for Herbicide Metabolism
Screening for herbicide metabolism was carried out in a final volume of 200 µL of 0.1 m sodium phosphate buffer, pH 7.0, containing 400 µm of herbicide in the presence of 1 mm NADPH or without NADPH (control). The assay mixture was equilibrated for 2 min at 27°C before starting the reaction by the addition of microsomal membranes prepared from yeast expressing the different CYP76 genes. After 2 h at 27°C, the reaction was terminated by adding 50 µL of acetonitrile:HCl (99:1), and the reaction medium was analyzed by reverse-phase HPLC on a Purospher 5-µm, 4- × 125-mm endcapped column (Merck). The column was equilibrated in water:acetic acid:acetonitrile (98:1:1) at a flow rate of 0.7 mL/min, and eluted with diode array detection (220–400 nm) using a convex gradient of acetonitrile:methanol (1:1) from 1% to 95% for 32 min, followed by 99% acetonitrile:methanol (1:1) for an additional 6 min. Kinetic assays were conducted in a final volume of 200 µL for 20 min in 0.1 m sodium phosphate, pH 7, containing 1 mm NADPH, 7 pmol of P450, and varying the concentration of substrate. Kinetic parameters were deduced from Michaelis-Menten representation.
Products were characterized by HPLC-MS. The system consisted in a binary solvent delivery pump (SurveyorMS; Thermo-Finnigan) connected to a diode array detector (Surveyor PDA plus; Thermo-Finnigan) and an LTQ mass spectrometer (Thermo Scientific), equipped with an atmospheric pressure ionization interface operated in electrospray ionization (ESI) negative and positive ion modes (ESI− and ESI+, respectively). MS conditions were as follows for ESI+ mode: spray voltage was set at 5 kV; source gases were set (in arbitrary units min−1) for sheath gas, auxiliary gas, and sweep gas at 50, 10, and 10, respectively; capillary temperature was set at 300°C; capillary voltage at 0 V; and tube lens, split lens, and front lens voltages at 60 V, −46 V, and −5.75 V, respectively. For ESI− mode, MS conditions were unchanged except ion optics parameters, which were automatically adapted as follows: capillary voltage at −48 V, and tube lens, split lens, and front lens voltages at −120 V, 34 V, and 4.25 V, respectively. The data were processed using the XCALIBUR software program.
Isolation of Null Mutant and Overexpression Lines
Insertion mutants were selected from SALK lines for CYP76C1 (SALK_010566: cyp76c1-1), CYP76C2 (SALK_037019: cyp76c2-1), and CYP76C4 (SALK_093179: cyp76c4-1) and obtained from the Nottingham Arabidopsis Stock Center (Alonso et al., 2003). Homozygous mutant lines were selected by PCR genotyping on genomic DNA extracted from young leaves using the primers provided in Supplemental Table S3. Absence of transcripts in the insertion lines was assessed by semi-qRT-PCR amplifying the full coding sequence. To generate A. thaliana lines overexpressing CYP76C1, CYP76C2, and CYP76C4, the plant expression vectors harboring each gene were used to transform the Agrobacterium GV3101 strain before transformation of Col-0 plants by floral dip (Clough and Bent, 1998). T1 progeny was screened by germination on glufosinate (BASTA). For each enzyme, two independent T1 BASTA-resistant lines were brought to T3 stable progeny by germination on BASTA to obtain homozygous stable lines. P450 expression was analyzed on T3 lines by qRT-PCR in leaves for CYP76C1- and CYP76C2-overexpressing lines as described above. A primers list is provided in Supplemental Table S3.
Resistance Test to Herbicides
Seeds of Col-0 and of the insertion and overexpression lines of CYP76C1, CYP76C2, and CYP76C4 were sterilized in open 1.5-mL tubes in a glass bottle containing a beaker with 20 mL of bleach (sodium hypochlorite solution). Two mL of 37% fuming HCl was added to the bleach and seeds were sterilized for 4 h. Sterilized seeds were sown on 2.2 g L−1 of Murashige and Skoog medium (Sigma-Aldrich) containing 0.7% agar and 15 g L−1 Suc, adjusted to pH 5.7. After 2 d of stratification at 4°C, plants were grown at 22°C during a 16-h-day period under 70 to 90 µmol m−2 s−1 light and at 20°C during an 8-h-night period. Isoproturon or chlorotoluron was added to the medium at different concentrations for preliminary tests of tolerance. A concentration leading to a clear-cut difference in tolerance is shown.
Sequence data from this article can be found in the Arabidopsis Genome Initiative, GenBank/EMBL, or Phytozome databases under the following accession numbers: At-SAND-like (At2g28390), Al-SAND-like (gene 481666), At-TIP41-like (At4g34270), Al-TIP41-like (491240), At-PP2AA2 (At1g13320), Al-PP2AA2 (gene 936261), At-EXP (At4g26410), At-TUB4 (At5g44340), Al-ACT2 (gene 342019), At-CYP75B1 (At5g07990); At-CYP76C1 (At2g45560), Al-CYP76C1p (gene 871078), At-CYP76C2 (At2g45570), Al-CYP76C2 (gene 346366), At-CYP76C3 (At2g45580), Al-CYP76C3 (gene 322211), At-CYP76C4 (At2g45550), At-CYP76C5 (At1g33730), At-CYP76C6 (At1g33720), At-CYP76C7 (At3g61040), Al-CYP76C7 (gene 486570); At-CYP76C8p (At3g61035), Al-CYP76C8 (gene 867547), At-CYP76G1 (At3g52970), Al-CYP76G1 (gene 348388), Cr-CYP76B6 (AJ251269), and Ht-CYP76B1 (Y10098).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structure of CYP76C loci in Arabidopsis spp. and other Brassicaceae.
Supplemental Figure S2. Expression of the CYP76C paralogs in different organs and floral stages in A. lyrata.
Supplemental Figure S3. Differential carbon monoxide-reduced versus reduced UV-visible absorption spectra of the microsomal membranes prepared from yeasts expressing the CYP76 genes.
Supplemental Figure S4. EI-MS of 8-hydroxynerol.
Supplemental Figure S5. EI-MS of 9-hydroxynerol.
Supplemental Figure S6. EI-MS of 8-oxonerol.
Supplemental Figure S7. EI-MS of 8-hydroxylinalool.
Supplemental Figure S8. EI-MS of 9-hydroxylinalool.
Supplemental Figure S9. NMR characterization of monoterpenol products formed by CYP76C enzymes.
Supplemental Figure S10. EI-MS of 1,2-epoxylinalool.
Supplemental Figure S11. EI-MS of 8-hydroxycitronellol.
Supplemental Figure S12. EI-MS of 9-hydroxycitronellol.
Supplemental Figure S13. EI-MS of 8-oxocitronellol.
Supplemental Figure S14. EI-MS of 6,7-epoxycitronellol.
Supplemental Figure S15. EI-MS of 7-hydroxylavandulol.
Supplemental Figure S16. EI-MS of 8-hydroxylavandulol.
Supplemental Figure S17. EI-MS of 10-hydroxy-α-terpineol.
Supplemental Figure S18. Determination of the catalytic parameters of linalool conversion by CYP76C1, CYP76C2, and CYP76C4.
Supplemental Figure S19. Genotyping of insertion and overexpression lines.
Supplemental Figure S20. CYP76C1, CYP76C2, or CYP76C4 overexpression confers herbicide tolerance to Arabidopsis spp.
Supplemental Table S1. Screening for herbicide metabolism by CYP76 enzymes.
Supplemental Table S2. Retention time, mass, and tandem MS fragmentation patterns for chlorotoluron and isoproturon CYP76C1-dependent products.
Supplemental Table S3. PCR primer list.
Supplemental Data Set S1. Alignment used to generate the tree in Figure 1A.
Supplementary Material
Acknowledgments
We thank Mark Beilstein (University of Arizona) for critical reading of the article.
Glossary
- P450
cytochrome P450
- qRT
real-time quantitative
- EI-MS
electron-ionization mass spectrum
- Col-0
ecotype Columbia-0 of Arabidopsis thaliana
- ESI
electrospray ionization
- GC
gas chromatography
- FID
flame ionization detection
- MS
mass spectrometry
Footnotes
This work was supported by the European Commission Seventh Framework Programme for Research and Technological Development Framework (grant no. KBBE–2007–3–1–01 to the SMARTCELL project), the Centre National de la Recherche Scientifique and the Région Alsace (Bourse de doctorat pour ingénieur to J.-F.G.), the European Fund for Regional Development in the INTERREG IVA Broad Region EU Invests in Your Future programme (to B.B.), the Agence Nationale de la Recherche (grant no. ANR-10–BLAN–1528 to H.R. and D.W.-R. for the PHENOWALL project), and the University of Strasbourg Institute for Advanced Study and the Freiburg Institute for Advanced Studies (funding for the METABEVO project).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aharoni A, Giri AP, Deuerlein S, Griepink F, de Kogel WJ, Verstappen FWA, Verhoeven HA, Jongsma MA, Schwab W, Bouwmeester HJ. (2003) Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250 [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collu G, Unver N, Peltenburg-Looman AMG, van der Heijden R, Verpoorte R, Memelink J. (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508: 215–220 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Chavez ML, Moniodis J, Madilao LL, Jancsik S, Keeling CI, Barbour EL, Ghisalberti EL, Plummer JA, Jones CG, Bohlmann J. (2013) Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS ONE 8: e75053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didierjean L, Gondet L, Perkins R, Lau SMC, Schaller H, O’Keefe DP, Werck-Reichhart D. (2002) Engineering herbicide metabolism in tobacco and Arabidopsis with CYP76B1, a cytochrome P450 enzyme from Jerusalem artichoke. Plant Physiol 130: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinda B, Debnath S, Banik R. (2011) Naturally occurring iridoids and secoiridoids. An updated review, part 4. Chem Pharm Bull (Tokyo) 59: 803–833 [DOI] [PubMed] [Google Scholar]
- Dinda B, Debnath S, Harigaya Y. (2007a) Naturally occurring iridoids. A review, part 1. Chem Pharm Bull (Tokyo) 55: 159–222 [DOI] [PubMed] [Google Scholar]
- Dinda B, Debnath S, Harigaya Y. (2007b) Naturally occurring secoiridoids and bioactivity of naturally occurring iridoids and secoiridoids. A review, part 2. Chem Pharm Bull (Tokyo) 55: 689–728 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant RH. (2013) The molecular genetics of insecticide resistance. Genetics 194: 807–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ. (2007) Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol 24: 2298–2309 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34 [DOI] [PubMed] [Google Scholar]
- Ginglinger JF, Boachon B, Höfer R, Paetz C, Köllner TG, Miesch L, Lugan R, Baltenweck R, Mutterer J, Ullmann P, et al. (2013) Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 25: 4640–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiard L, Sauviac L, Dalbin N, Liaubet L, Callard D, Czernic P, Marco Y. (1998) CYP76C2, an Arabidopsis thaliana cytochrome P450 gene expressed during hypersensitive and developmental cell death. FEBS Lett 438: 245–249 [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hallahan DL, Lau SMC, Harder PA, Smiley DWM, Dawson GW, Pickett JA, Christoffersen RE, O’Keefe DP. (1994) Cytochrome P-450-catalysed monoterpenoid oxidation in catmint (Nepeta racemosa) and avocado (Persea americana); evidence for related enzymes with different activities. Biochim Biophys Acta 1201: 94–100 [DOI] [PubMed] [Google Scholar]
- Hallahan DL, Nugent JHA, Hallahan BJ, Dawson GW, Smiley DW, West JM, Wallsgrove RM. (1992) Interactions of avocado (Persea americana) cytochrome P-450 with monoterpenoids. Plant Physiol 98: 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfer R, Dong L, André F, Ginglinger JF, Lugan R, Gavira C, Grec S, Lang G, Memelink J, Van der Krol S, et al. (2013) Geraniol hydroxylase and hydroxygeraniol oxidase activities of the CYP76 family of cytochrome P450 enzymes and potential for engineering the early steps of the (seco)iridoid pathway. Metab Eng 20: 221–232 [DOI] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker RR, Tholl D. (2013) Volatile organic compound mediated interactions at the plant-microbe interface. J Chem Ecol 39: 810–825 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Luan F, Mosandl A, Degenhardt A, Gubesch M, Wüst M. (2006) Metabolism of linalool and substrate analogs in grape berry mesocarp of Vitis vinifera L. cv. Morio Muscat: demonstration of stereoselective oxygenation and glycosylation. Anal Chim Acta 563: 353–364 [Google Scholar]
- Luan F, Mosandl A, Münch A, Wüst M. (2005) Metabolism of geraniol in grape berry mesocarp of Vitis vinifera L. cv. Scheurebe: demonstration of stereoselective reduction, E/Z-isomerization, oxidation and glycosylation. Phytochemistry 66: 295–303 [DOI] [PubMed] [Google Scholar]
- Madyastha KM, Meehan TD, Coscia CJ. (1976) Characterization of a cytochrome P-450 dependent monoterpene hydroxylase from the higher plant Vinca rosea. Biochemistry 15: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Matich AJ, Comeskey DJ, Bunn BJ, Hunt MB, Rowan DD. (2011) Biosynthesis and enantioselectivity in the production of the lilac compounds in Actinidia arguta flowers. Phytochemistry 72: 579–586 [DOI] [PubMed] [Google Scholar]
- Matich AJ, Young H, Allen JM, Wang MY, Fielder S, McNeilage MA, MacRae EA. (2003) Actinidia arguta: volatile compounds in fruit and flowers. Phytochemistry 63: 285–301 [DOI] [PubMed] [Google Scholar]
- Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, et al. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5: 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Werck-Reichhart D. (2011) A P450-centric view of plant evolution. Plant J 66: 194–211 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MHH, Jensen JK, Halkier BA. (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239: 2370–2378 [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Raguso RA, Lewinsohn E, Croteau R. (1994) Floral scent production in Clarkia (Onagraceae). I. Localization and developmental modulation of monoterpene emission and linalool synthase activity. Plant Physiol 106: 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Powles SB, Yu Q. (2010) Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol 61: 317–347 [DOI] [PubMed] [Google Scholar]
- Radulović NS, Blagojević PD, Stojanović-Radić ZZ, Stojanović NM. (2013) Antimicrobial plant metabolites: structural diversity and mechanism of action. Curr Med Chem 20: 932–952 [PubMed] [Google Scholar]
- Raguso RA, Pichersky E. (1999) New perspectives in pollination biology: floral fragrances. A day in the life of a linalool molecule: chemical communication in a plant-pollinator system. Part 1: linalool biosynthesis in flowering plants. Plant Species Biol 14: 95–120 [Google Scholar]
- Robineau T, Batard Y, Nedelkina S, Cabello-Hurtado F, LeRet M, Sorokine O, Didierjean L, Werck-Reichhart D. (1998) The chemically inducible plant cytochrome P450 CYP76B1 actively metabolizes phenylureas and other xenobiotics. Plant Physiol 118: 1049–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohloff J, Bones AM. (2005) Volatile profiling of Arabidopsis thaliana - putative olfactory compounds in plant communication. Phytochemistry 66: 1941–1955 [DOI] [PubMed] [Google Scholar]
- Salim V, Wiens B, Masada-Atsumi S, Yu F, De Luca V. (2014) 7-deoxyloganetic acid synthase catalyzes a key 3 step oxidation to form 7-deoxyloganetic acid in Catharanthus roseus iridoid biosynthesis. Phytochemistry 101: 23–31 [DOI] [PubMed] [Google Scholar]
- Siminszky B, Corbin FT, Ward ER, Fleischmann TJ, Dewey RE. (1999) Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc Natl Acad Sci USA 96: 1750–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tundis R, Loizzo MR, Menichini F, Statti GA, Menichini F. (2008) Biological and pharmacological activities of iridoids: recent developments. Mini Rev Med Chem 8: 399–420 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: H0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Okada K, Yamazaki K, Wu Y, Swaminathan S, Yamane H, Peters RJ. (2012) Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J Biol Chem 287: 6159–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PJ, Strauss CR, Wilson B, Massy-Westropp RA. (1982) Studies on the hydrolysis of Vitis vinifera monoterpene precursor compounds and model monoterpene β-D-glucosides rationalizing the monoterpene composition of grapes. J Agric Food Chem 30: 1219–1223 [Google Scholar]
- Wu Y, Wang Q, Hillwig ML, Peters RJ. (2013) Picking sides: distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem J 454: 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Kambara Y, Imaishi H, Ohkawa H. (2000) Molecular cloning of novel cytochrome P450 species induced by chemical treatments in cultured tobacco cells. Pestic Biochem Physiol 68: 11–25 [Google Scholar]
- Zi J, Peters RJ. (2013) Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org Biomol Chem 11: 7650–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.