The Usher syndrome type 3–associated protein clarin-1 is functionally important for mechanotransduction channel activity and for proper localization of synaptic components in zebrafish hair cells.
Abstract
Clarin-1 is a four-transmembrane protein expressed by hair cells and photoreceptors. Mutations in its corresponding gene are associated with Usher syndrome type 3, characterized by late-onset and progressive hearing and vision loss in humans. Mice carrying mutations in the clarin-1 gene have hair bundle dysmorphology and a delay in synapse maturation. In this paper, we examined the expression and function of clarin-1 in zebrafish hair cells. We observed protein expression as early as 1 d postfertilization. Knockdown of clarin-1 resulted in inhibition of FM1-43 incorporation, shortening of the kinocilia, and mislocalization of ribeye b clusters. These phenotypes were fully prevented by co-injection with clarin-1 transcript, requiring its C-terminal tail. We also observed an in vivo interaction between clarin-1 and Pcdh15a. Altogether, our results suggest that clarin-1 is functionally important for mechanotransduction channel activity and for proper localization of synaptic components, establishing a critical role for clarin-1 at the apical and basal poles of hair cells.
Introduction
Clarins belong to a large hyperfamily of small integral membrane glycoproteins (4TM) involved in a variety of cellular processes, including receptor trafficking, clustering of signal-associated molecules, and scaffolding functions. These proteins form multimolecular complexes creating an intricate network in which signaling molecules complex together giving origin to subcellular structures known as the tetraspan-enriched microdomains. Thus, by integrating different signaling pathway components, these small transmembrane proteins act as modulators of the signaling cascade and, ultimately, of cell homeostasis (Adato et al., 2002; Charrin et al., 2003; Chen et al., 2003; Xiong et al., 2012).
Mutations in the clarin-1 gene are associated with Usher syndrome type 3, a genetically heterogeneous disorder that affects hearing, balance, and vision in humans (Adato et al., 2002). Amino acid alignments between clarin-1 and different members of the 4TM hyperfamily established a modest degree of similarity with stargazin, a calcium channel subunit involved in expression, mobilization, and clustering of AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors at the synaptic cleft. Since in the inner ear, clarin-1 is not only present at the apical and basal aspects of the hair cells but also in the afferent neurons that innervate them, a similar role in synaptic shaping has been suggested (Adato et al., 2002; Chen et al., 2003; Geng et al., 2009; Zallocchi et al., 2009).
In recent years, significant efforts have been made to characterize clarin-1 function in inner ear. These studies have shown that, similar to other Usher proteins, clarin-1 is necessary for hair bundle and ribbon synapse development and maintenance. Studies in the clarin-1 mutant mouse (Clrn1−/−) showed variable vestibular dysfunction, progressive hearing loss, compromised hair bundle integrity, and a delay in ribbon synapse maturation and neuronal activation (Geng et al., 2009, 2012; Zallocchi et al., 2012b). Expression of the clarin-1 orthologue has also been demonstrated in zebrafish, in which it may be functioning as a scaffold protein between different retinal cell types (Phillips et al., 2013).
Although, the effects of clarin-1 mutations on hair cell function and morphology have been demonstrated by different groups, very little is known regarding clarin-1 function at the molecular level. In this study, we examined clarin-1 expression and function in zebrafish hair cells. We observed expression of clarin-1 between 1 and 7 d postfertilization (dpf). Hair cells from clarin-1 morphants (MOs) show a decrease in mechanotransduction channel activity, defects in vesicular turnover, shorter kinocilia, and mislocalized synaptic markers. Furthermore, we observed an interaction between clarin-1 and the fish orthologue to mouse PCDH15 (protocadherin-15), Pcdh15a. This Usher protein, together with cadherin-23 (CDH23), is a key component of the tip links that gate the mechanotransduction channels in mouse hair cells (Kazmierczak et al., 2007). Absence of clarin-1 in zebrafish hair cells results in altered Pcdh15a distribution. Collectively, our data suggest that clarin-1 may be involved in the processes leading to tip link formation, through vesicle turnover regulation and, ultimately, affecting mechanotransduction channel activity. Cilia and localization of synaptic components may also be affected as a result of an altered vesicle recycling in the absence of clarin-1. In summary, this work underscores novel and critical functions for clarin-1 at both the apical and basal poles of fish hair cells.
Results
Antibody qualification
The clarin-1 antibody developed in our laboratory was qualified by morpholino knockdown and rescued with clarin-1 copy RNA (cRNA) and by exogenous expression of zebrafish tdTomato–clarin-1 in HeLa cells. Immunoblot analyses (Fig. S1) of 2 dpf larvae showed two prominent bands near 30 kD (likely caused by alternative start codon usage, clrn1-001 ENSDART00000149872 and clrn1-201 ENSDART00000062909, and/or to different glycosylation states), consistent with the predicted size for zebrafish clarin-1. Normal expression of clarin-1 was observed at 1 and 2 dpf, but beyond this time point, we were unable to detect clarin-1 by Western blotting (Fig. S1 and not depicted). Clarin-1 is absent from the specific MOs, whereas co-injection with the clarin-1 cRNA restored this expression. Immunohistochemistry analysis of 3 dpf controls shows apical expression of clarin-1 in hair cells from both neuromasts (Fig. S1, A and E) and ears (Fig. S1, I and M). This apical expression is reduced or absent in the clarin-1 MOs (Fig. S1, B, F, J, and N), whereas the co-injection with the cRNA (Fig. S1, C, G, K, and O) restored normal apical expression. These results, together with the immunoreactivity data from HeLa cells exogenously expressing zebrafish clarin-1 (compare Fig. S1, Q–S vs. T–V), demonstrate the specificity of our clarin-1 antibody.
Clarin-1 expression in zebrafish hair cells
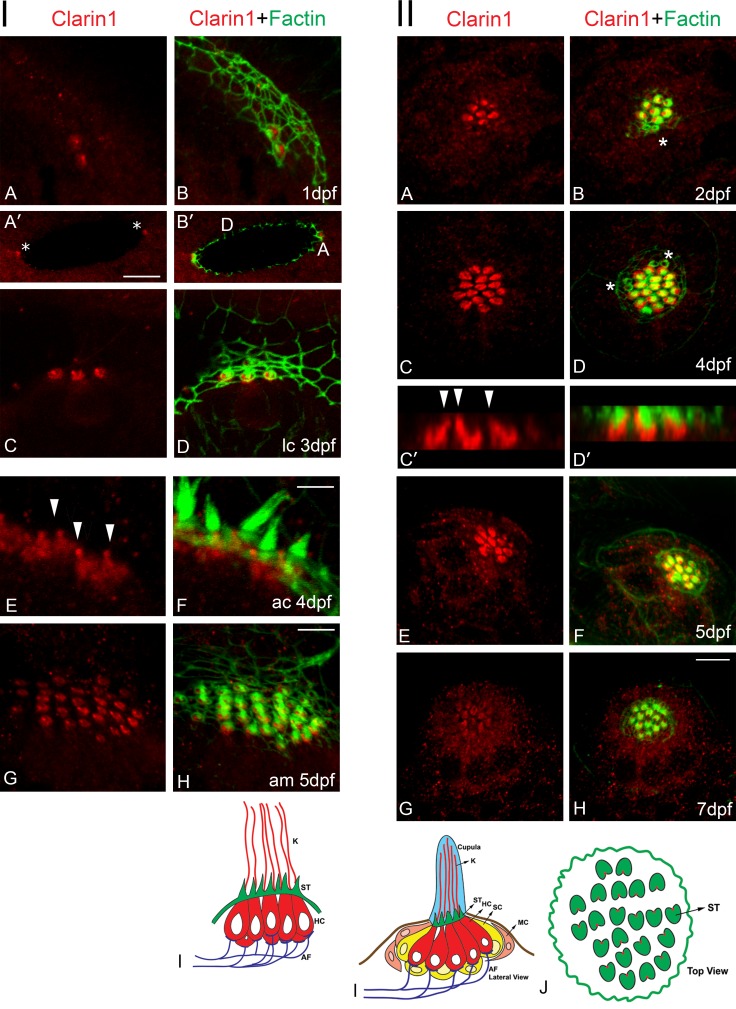
A recent study by Phillips et al. (2013) demonstrated expression of clarin-1 protein at the apical and basal aspect of zebrafish hair cells at 5 dpf. We expanded that work by analyzing its expression between 1 and 7 dpf, using our fusion protein antibody. Immunohistochemistry analyses of whole mount embryos/larvae show apical expression of clarin-1 in hair cells of the ear and lateral line organ (Fig. 1). At 1 dpf, clarin-1 is present in both anterior and posterior macular hair cells (Fig. 1, panel I, A–B′, asterisks). This apical expression persists and extends to the three crista sensory organs until 5 dpf (Fig. 1, panel I, C–H; and not depicted); beyond this time point, we were unable to detect clarin-1 in ear hair cells. Clarin-1 was also observed at the tip of the hair cell bundle starting at 3 dpf; however, its detection was sporadic and variable, likely as a result of epitope masking and/or low antibody sensitivity (not depicted).
Figure 1.
Clarin-1 protein expression in ear and neuromasts. Confocal images of clarin-1 staining. Actin filaments were counterstained with phalloidin (green). Panel I: (A–B′) 1dpf. A, anterior. D, dorsal. Asterisks in A′ denote macular hair cell precursors. (C–D′) 3 dpf lateral crista (lc). (E and F) 4 dpf anterior crista (ac). Arrowheads in E denote point of insertion of the kinocilium. (G and H) 5 dpf anterior macula (am). (I) Cartoon of a lateral view crista sensory organ. Bars: (A, B, C, D, G and H) 7 µm; (A′ and B′) 16 µm; (E and F) 3 µm. Panel II: (A and B) 2 dpf. (C–D′) 4 dpf. C′ and D′ represent z planes of C and D. Arrowheads in C′ denote point of insertion of the kinocilium. (E and F) 5 dpf. (G and H) 7 dpf. Asterisks denote immature sister hair cells. (I and J) Cartoon of the lateral (I) and top (J) views of a neuromast. K, kinocilia; ST, stereocilia; HC, hair cells; SC, supporting cells; MC, mantle cells; AF, afferent fibers. Results are representative images of >10 independent experiments. Bar, 7 µm.
Neuromast hair cells (Fig. 1, panel II) also showed apical expression of clarin-1 starting at 2 dpf but only in mature sensory cells. Immature sister hair cells (determined by the presence of an incipient hair cell bundle; Kindt et al., 2012) were negative for clarin-1 (Fig. 1, panel II, B and D, asterisks). Although weak, we still detected apical expression of clarin-1 at 7 dpf.
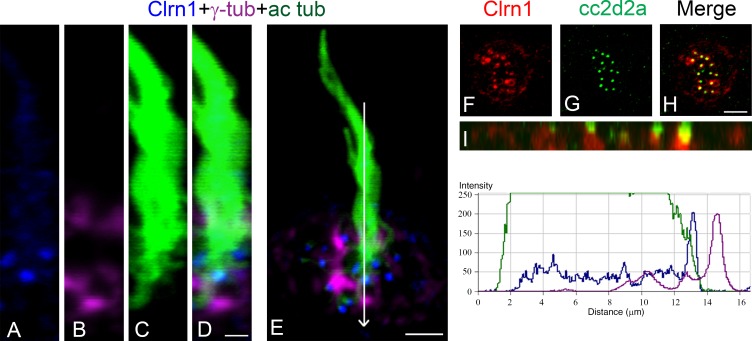
Clarin-1 distribution in ear and neuromast hair cells shows little colocalization with the actin core present at the hair cell bundle and cuticular plate. Interestingly, most of clarin-1 immunostaining is observed basal to the cuticular plate and at the point of insertion of the kinocilium, the fonticulus, where the basal body resides (Fig. 1, panel I [E] and panel II [C′]). Further examination of clarin-1 expression in the cilia compartment (Fig. 2) positions clarin-1 in the transition zone (Ko, 2012; Reiter et al., 2012), the region between the basal body (stained for γ-tubulin) and the axoneme (stained for acetylated tubulin; Fig. 2, A–E). Fluorescence intensity profiles show that most of clarin-1 fluorescence falls between the two ciliary markers. To confirm clarin-1 expression at the ciliary transition zone, 4 dpf neuromasts were immunostained with the transition zone protein Cc2d2a (Fig. 2, F–I; Bachmann-Gagescu et al., 2011; Williams et al., 2011). Partial colocalization was observed between clarin-1 and Cc2d2a, with clarin-1 restricted to the most basal region of the transition zone (Fig. 2 I).
Figure 2.
Clarin-1 is present at the ciliary transition zone. (A–E) Representative images of three independent experiments. 3 dpf neuromast hair cells showing clarin-1 (Clrn1; A), γ-tubulin (γ-tub; B), and acetylated tubulin (ac tub; C). (right) Representative fluorescence intensity profile corresponding to the image presented in E (similar results in the distribution of the fluorescence were observed for three independent experiments). Arrow in E represents the direction of the reading for the fluorescence intensity profile. (F–H) Representative images of three independent experiments. 4 dpf neuromast immunostained for clarin-1 (F) and cc2d2a (G). Cc2d2a stains the transition zone of each hair cell. (I) Z plane view of H showing partial colocalization between these two proteins (yellow). Bars: (A–D) 1 µm; (E) 3 µm; (F–H) 4 µm.
Although clarin-1 is highly enriched at the apical aspect of hair cells (Fig. S2, asterisks), we were still able to detect scattered basal expression in 3 dpf cross sections (Fig. S2, arrowheads). Neuromasts (Fig. S2, A–C) and inner ear sensory organs (Fig. S2, D–L) were dual immunostained with the hair cell marker myosin7a or different synaptic markers, respectively. Macular hair cells showed juxtaposed localization between ZN12 and clarin-1. Similarly, lateral crista immunostaining (Fig. S2, G–L) showed juxtaposed localization between clarin-1 and ribeye b. These results indicate that clarin-1 is also present at the basal aspect of hair cells, near the ribbon synapses.
Characterization of clarin-1 MOs
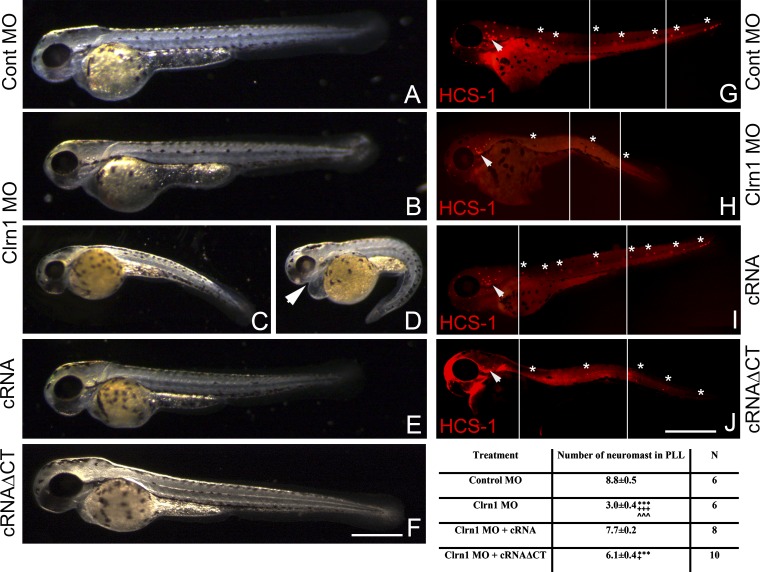
To analyze whether the lack of clarin-1 protein influences hair cell development, one-cell stage eggs were injected with control morpholinos, the clarin-1 morpholino mixture, or the mixture plus clarin-1 cRNA (Fig. 3, A–E and G–I). At 3 dpf, clarin-1 MOs show shortening of the body axis (weak phenotype; Fig. 3 B), ventrally curved body axis, and pericardial edema (mild and severe phenotypes; Fig. 3, C and D, respectively), which are some of the characteristics previously described for ciliary mutants/MOs (Kramer-Zucker et al., 2005; Wilkinson et al., 2009; Austin-Tse et al., 2013). Clarin-1 MOs showed inclined or circle swimming behaviors (Kalueff et al., 2013). The restoration of clarin-1 expression by co-injection with the clarin-1 cRNA (Fig. S1) rescued these morphological defects (Fig. 3 E). The analysis of the lateral line organ by immunostaining with the hair cell marker HCS-1 (hair cell soma-1; Fig. 3, G–I and numerical results), showed a pronounced reduction in the number of neuromasts present in both the anterior and the posterior lateral line (PLL; Fig. 3, G–I, asterisks) upon depletion of clarin-1, even in MOs with a weak or mild phenotype (compare Fig. 3, G vs. H). Again, this effect was reversed when eggs were co-injected with the cRNA (Fig. 3 I and numerical results). Because, during the developmental window under study, we never observed clarin-1 expression in supporting (or mantle) cells (Fig. 1, cartoons), the results imply that clarin-1 is involved in neuromast formation through a role in the regulation of hair cell maturation. To map the protein region involved in this effect, we co-injected clarin-1 MOs with a clarin-1 transcript lacking the cytoplasmic C-terminal tail (cRNAΔCT). This truncated form showed normal apical protein localization in hair cells from both neuromasts and ears (Fig. S1, D, H, L, and P) and restored protein levels (Fig. S1, immunoblot). At 3 dpf, the gross morphology of the larvae injected with this suspension was similar to controls (Fig. 3 F) or showed a weak phenotype, with a slight body curvature (Fig. 3 J). However, the number of neuromasts in these animals was only partially rescued (Fig. 3, J and numerical results), suggesting that the tail domain of clarin-1 is, at least in part, responsible for proper neuromast development.
Figure 3.
Clarin-1 MOs show morphological abnormalities and a reduction in the number of neuromasts. (A–F) Representative images of at least five independent experiments. External phenotype of 3 dpf control (Cont; A), clarin-1 (Clrn1) MOs (weak phenotype [B], mild phenotype [C], and severe phenotype [D]), MOs + cRNA (E), or cRNAΔCT (F) transcripts. Arrowhead in D denotes pericardial edema. (G–J) Representative images of at least six independent experiments. HCS-1 immunostaining of 3-dpf control (G), clarin-1 MOs (H), MOs + cRNA (I), or cRNAΔCT (J). Neuromasts in the PLL are denoted by asterisks. Inner ear is denoted by arrowheads. White lines mark the juxtaposition of the different sections in the composite image. Bars: (A–F) 200 µm; (G–J) 250 µm. Numerical results: number of PLL neuromasts for each treatment. Data are expressed as means ± SEM. ***, P < 0.001 versus control MO. +++, P < 0.001; +, P < 0.05 versus cRNA. ˄˄˄, P < 0.001 versus cRNAΔCT. N, number of independent experiments.
Clarin-1 regulation of mechanotransduction activity and kinocilia elongation
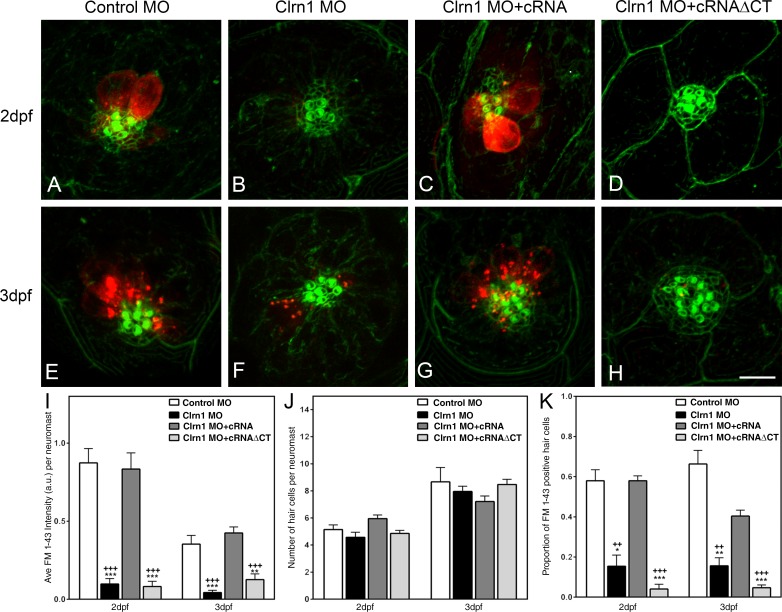
Since clarin-1 knockdown seems to affect hair cell development and because of its apical expression in mature hair cells, we examined whether it might be involved in hair cell function. Injected larvae were assessed for FM1-43 uptake as a way to analyze mechanotransduction channel functionality (Fig. 4; Gale et al., 2001; Meyers et al., 2003; Sidi et al., 2003). We observed a significant decrease of FM1-43 uptake in neuromast hair cells from clarin-1 MOs compared with controls. This impairment in dye uptake did not occur when embryos were co-injected with clarin-1 full-length cRNA (Fig. 4, A–C, E–G, I, and K). Clarin-1 cRNAΔCT did not rescue this phenotype (Fig. 4, D and H). Fluorescence values in clarin-1 cRNAΔCT were similar to those observed for the clarin-1 MOs with almost a complete blockage of FM1-43 uptake, establishing the importance of the clarin-1 C-terminal region for proper mechanotransduction channel activity. Moreover, the reduction in the total fluorescence intensity observed for the clarin-1 MOs and clarin-1 morpholinos + cRNAΔCT (Fig. 4 I) was not caused by a reduction in the number of hair cells per neuromast (Fig. 4 J) but rather in their ability to incorporate the dye (Fig. 4 K), which is consistent with a defect associated with the mechanotransduction machinery.
Figure 4.
FM1-43 uptake experiments. (A–H) Representative images showing dye uptake assessed at 2 dpf (A–D) and 3 dpf (E–H) larvae. (A and E) Control. (B and F) Clarin-1 (Clrn1) MOs. (C and G) MOs + cRNA. (D and H) MOs + cRNAΔCT. Hair cell bundle was counterstained with phalloidin (green). Bar, 7 µm. (I) Semiquantitative analysis of the total fluorescence per neuromast. (J) Number of hair cells per neuromast. (K) Fraction of FM1-43–positive hair cells. Results are expressed as means ± SEM. Number of neuromasts analyzed from independent samples, n > 15. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control MO. ++, P < 0.01; +++, P < 0.001 versus clarin-1 MO + cRNA. a.u., arbitrary unit.
Two closely related pcdh15 genes have been identified in zebrafish (Seiler et al., 2005), with Pcdh15a involved in hearing and vestibular functions presumably through the formation of the tip links that gate the mechanotransduction channels, as is the case in mammalian hair cells (Kazmierczak et al., 2007; Indzhykulian et al., 2013). We generated Pcdh15a MOs and tested them for dye uptake impairment. These experiments (Fig. S3, A–D) showed a reduction in FM1-43 incorporation in the MOs compared with controls. This decrease, qualitatively similar to the one observed in the clarin-1 MOs, provides a positive control for a known component of the mechanotransduction machinery and suggests that clarin-1 might be playing a role in the maturation and/or regulation of mechanotransduction channel activity in the neurosensory epithelia.
Because clarin-1 MOs have a phenotype that resembles ciliary mutants, planar cell polarity (PCP) was also assessed. Neuromast hair cells originate in pairs, with sister cells adopting opposite polarities that can be determined by the position of the kinocilium relative to the actin-rich hair bundle (Kindt et al., 2012; Mirkovic et al., 2012). The results presented in Fig. S4 (A–D) show that clarin-1 is not required for PCP. No obvious differences were observed in the normal orientation of hair bundles from sister hair cells when comparing control and morpholino-injected animals.
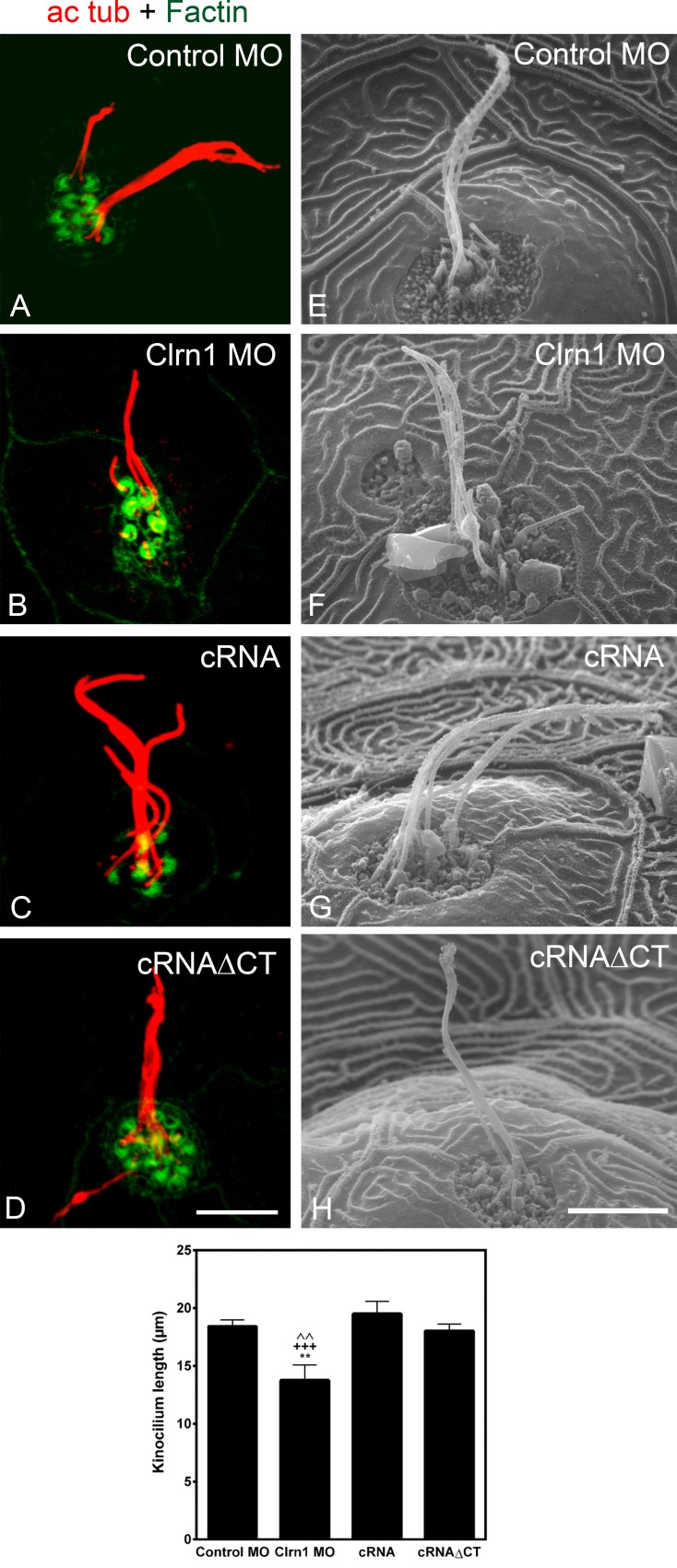
Ultrastructural analysis showed that hair bundles with stereocilia staircase shape developed in the specific clarin-1 MOs; however, further analysis will be needed to completely rule out any abnormalities in hair bundle morphology (Fig. S4, E and F). As clarin-1 is also expressed at the point of insertion of the kinocilium, we examined primary cilia morphology. Immunohistochemistry and scanning EM (SEM) analysis showed a significant reduction of the kinociliary length (Fig. 5, A–H and graph). The C-terminal tail of clarin-1 is not involved in cilia formation/elongation, as the effect was prevented by co-injection with the full or the truncated version of clarin-1 cRNAs (Fig. 5, C, D, G, H, and graph).
Figure 5.
Clarin-1 regulates ciliogenesis. (A–D) Representative images of the kinocilia immunostained for acetylated tubulin (ac tub) in 3 dpf control (A), clarin-1 (Clrn1) MOs (B), MOs + cRNA (C), or cRNAΔCT (D). Hair cell bundle was counterstained with phalloidin. (E–H) Representative images of the ultrastructural analysis of the kinocilia in 3 dpf control (E), clarin-1 MOs (F), MOs + cRNA (G), or cRNAΔCT (H). Bar graph depicts kinocilium lengths for each treatment. Values correspond to the mean length of all the kinocilia per neuromast. Measurements were obtained from confocal (n > 5 independent experiments) and SEM images (n = 6 independent experiments) and expressed as means ± SEM. Total number of neuromasts analyzed, n > 10. Control MO = 18.4 ± 0.5 µm. Clrn1 MO = 13.8 ± 1.3 µm. Clrn1 MO + cRNA = 19.5 ± 1.1 µm. Clrn1 MO + cRNAΔCT = 18.0 ± 0.6 µm. **, P < 0.01 versus control MO. +++, P < 0.001 versus cRNA. ˄˄, P < 0.01 versus cRNAΔCT. Bars: (A–D) 7 µm; (E–H) 5 µm.
Clarin-1 interaction with the mechanotransduction machinery
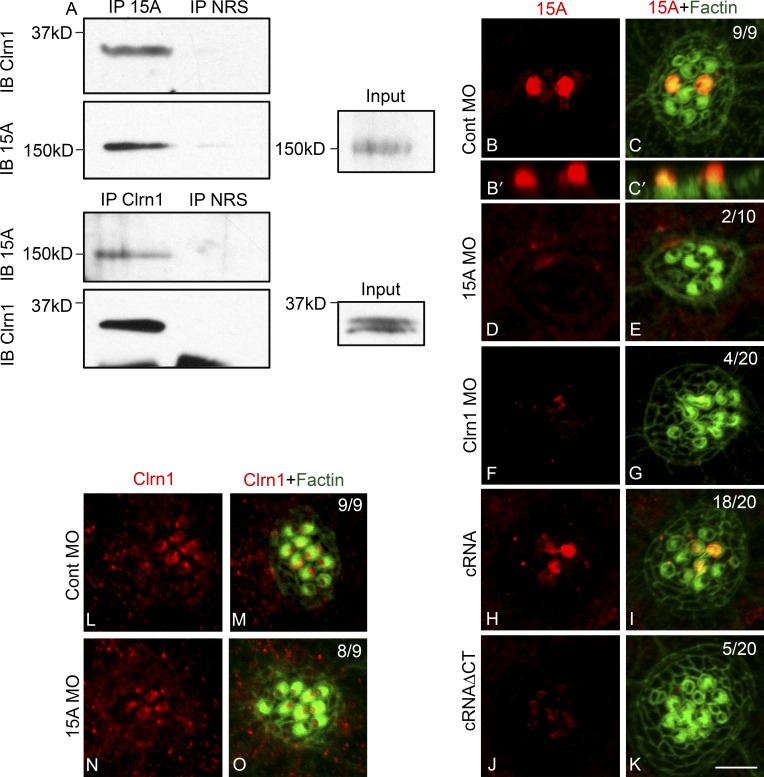
PCDH15 and CDH23 are two major components of the tip links that gate the mechanotransduction channels in mammals (Kazmierczak et al., 2007). Mutations in their corresponding genes are associated with Usher syndrome type I and nonsyndromic hearing loss (Bolz et al., 2001; Ahmed et al., 2003, 2006; Schultz et al., 2011). Several interactions between these and other Usher proteins have been described (Reiners et al., 2006; Zallocchi et al., 2010, 2012a,b; Caberlotto et al., 2011). Because the absence of clarin-1 results in dysfunction of the mechanotransduction machinery, we addressed whether clarin-1, Pcdh15a, and/or Cdh23 were part of a protein complex in fish hair cells (Söllner et al., 2004; Seiler et al., 2005). Reciprocal immunoprecipitation experiments were performed in 3 dpf larvae (Fig. 6 A). Results from these experiments showed a specific interaction between clarin-1 and Pcdh15a. We were able to immunoprecipitate clarin-1 with anti-Pcdh15a (Fig. 6 A, IP 15A) and, vice versa, Pcdh15a with anti–clarin-1 (Fig. 6 A, IP Clrn1). No band was observed when preimmune serum was used as the immunoprecipitating antibody (Fig. 6 A, IP NRS), confirming the specific in vivo interaction between these two Usher proteins. Although immunoblot analysis of 3 dpf larva lysates showed at least three specific bands for Pcdh15a (Fig. S3) only the full length (NCBI Protein database accession no. AAW50923) interacts with clarin-1 under our experimental conditions.
Figure 6.
Clarin-1 regulates Pcdh15a distribution. (A) Representative result of three independent experiments of a reciprocal co-IP in 3 dpf larvae. Protein lysates immunoprecipitated with anti-Pcdh15a (IP 15A), anti–clarin-1 (IP Clrn1), or with normal rabbit serum (IP NRS) and immunoblot for clarin-1 (IB Clrn1) or Pch15A (IB 15A). (inputs) 0.5% Pcdh15a and 3% clarin-1. (B–K) Representative images of 3 dpf neuromasts immunostained for Pcdh15a (15A). (B–C′) Pcdh15a control (Cont). (B′ and C′) Z planes of B and C. (D and E) Pcdh15a MOs. (F and G) Clarin-1 MOs. (H and I) MOs + cRNA. (J and K) MOs + cRNAΔCT. (L–O) 3 dpf neuromasts immunostained for clarin-1. (L and M) Pcdh15a control. (N and O) Pcdh15a MOs. Numbers in dual-color images represent number of neuromasts showing normal Usher protein expression/total of neuromasts analyzed in at least nine independent experiments. Bar, 3 µm.
To further characterize the relationship between Pcdh15a and clarin-1, we analyzed their pattern of expression in the reciprocal MOs (Fig. 6, B–O). Control injected embryos showed Pcdh15a localization in the hair cell bundles (Fig. 6, B–C′). This expression was specific, as pcdh15a MOs lacked apical immunostaining (Fig. 6, D and E). Apical expression was also absent or reduced when clarin-1 protein was depleted from neuromast hair cells (Fig. 6, F and G). Co-injection with the full-length clarin-1 transcript restores Pcdh15a expression (Fig. 6, H and I). In contrast, when MOs were co-injected with the cRNAΔCT, very weak to no expression of Pcdh15a was observed, suggesting that the C-terminal tail of clarin-1 is necessary for proper localization of Pcdh15a in the hair cell bundles (Fig. 6, J and K).
When dependency of clarin-1 expression was analyzed in the Pcdh15a MOs, no effect in clarin-1 distribution was observed (Fig. 6, L–O). These data and the corresponding numerical results (Fig. 6, B–O, top right corner) suggest that clarin-1 association may be required for incorporation of Pcdh15a into the hair cell bundles and that perturbations in clarin-1/Pcdh15a interactions lead, at least in part, to mechanotransduction channel dysfunction. Although clarin-1 appears to be necessary for correct Pcdh15a apical expression, the reverse is not true.
It is worth noting that although Pcdh15a and clarin-1 show apical expression, their distribution patterns are not exactly the same: Pcdh15a is present only in a subpopulation of neuromast hair cells, whereas clarin-1 is present in all mature hair cells (Fig. 1 vs. Figs. 6 and 7). Although the restricted expression of Pcdh15a to a subgroup of hair cells may be caused by epitope masking (Ahmed et al., 2006), the in vivo interaction between these two Usher proteins likely occurs at the base (or the tip) of the hair cell bundle in mature hair cells, where clarin-1 and Pcdh15a colocalize. Similar experiments performed to study a possible clarin-1/Cdh23 association did not show any significant differences compared with controls (not depicted), suggesting that under the experimental conditions used in these experiments, no interaction between clarin-1 and Cdh23 exists.
Figure 7.
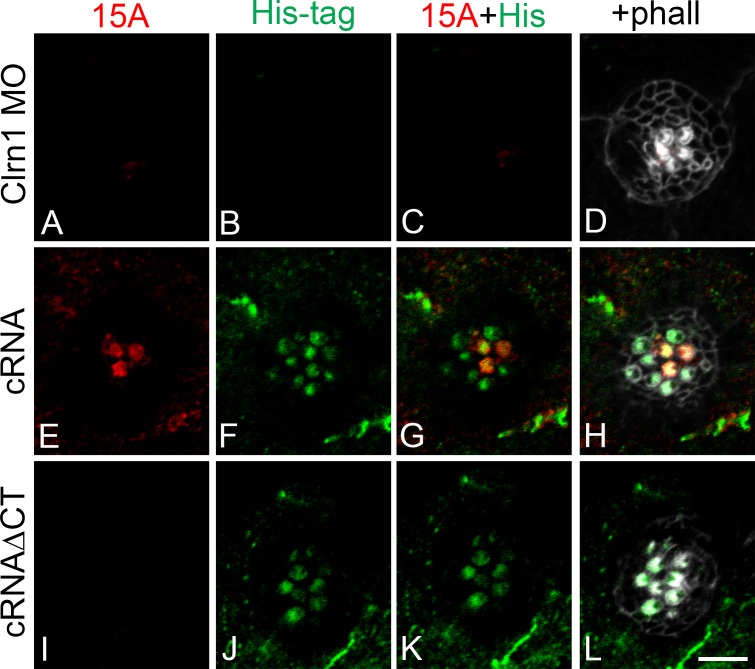
The cytoplasmic tail of clarin-1 is necessary for Pcdh15a apical expression. (A–L) Representative images of 3 dpf neuromasts immunostained for Pcdh15a (15A; A, E, and I), His tag (B, F, and J), and counterstained with phalloidin (phall; gray; D, H, and L). (A–D) Clarin-1 (Clrn1) MOs. (E–H) MOs + cRNA. (I–L) MOs + cRNAΔCT. Bar, 5 µm. Number of independent experiments, n = 5.
To confirm whether absence of Pcdh15a from the hair cell bundle correlates with the absence of clarin-1, we took advantage of the His tag epitope present in the in vitro transcribed clarin-1 cRNAs (Fig. 7). Clarin-1 MOs, lacking clarin-1 apical expression (negative His tag immunostaining), were also negative for Pcdh15a protein expression (Fig. 7, A–D). When eggs were co-injected with the full-length clarin-1 His-tagged transcript (Fig. 7, E–H), expression of both clarin-1 and Pcdh15a was restored to the apical aspect of the hair cells. However, when the cRNAΔCT His-tagged version was used (Fig. 7, I–L), Pcdh15a was absent from the apical aspect of the hair cells in spite of normal apical expression of clarin-1. These results not only demonstrate a dependency for clarin-1 for appropriate apical localization of Pcdh15a but also the requirement of the clarin-1 cytoplasmic C-terminal tail in this process.
Clarin-1 deficiency affects localization of synaptic components
A functional role at the hair cell synapses has been described for several Usher proteins, including synaptic maturation and regulation of calcium channel activity (Kersten et al., 2010; Zallocchi et al., 2012a,b; Gregory et al., 2013). Additionally, floating ribbons have been observed for an Usher fish model, Usher type 1C (Phillips et al., 2011), implicating them in the maturation of the synapses.
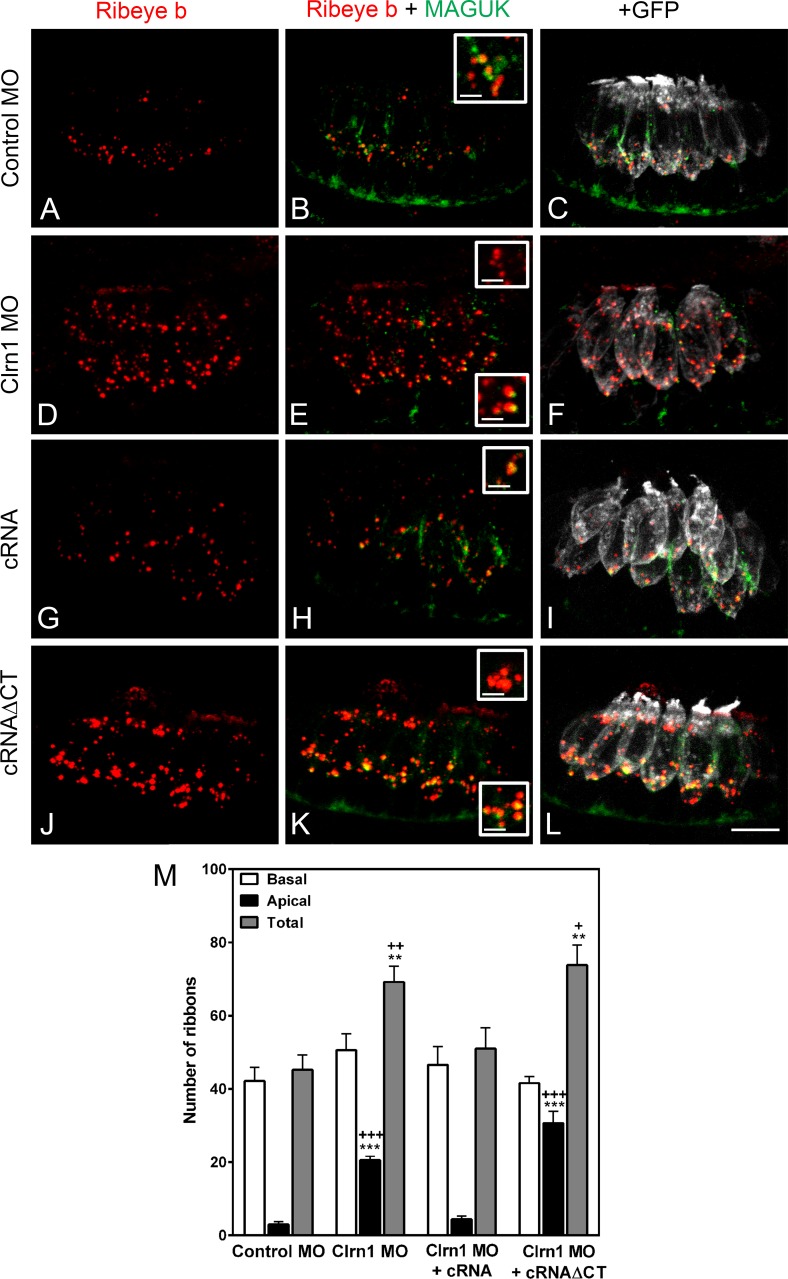
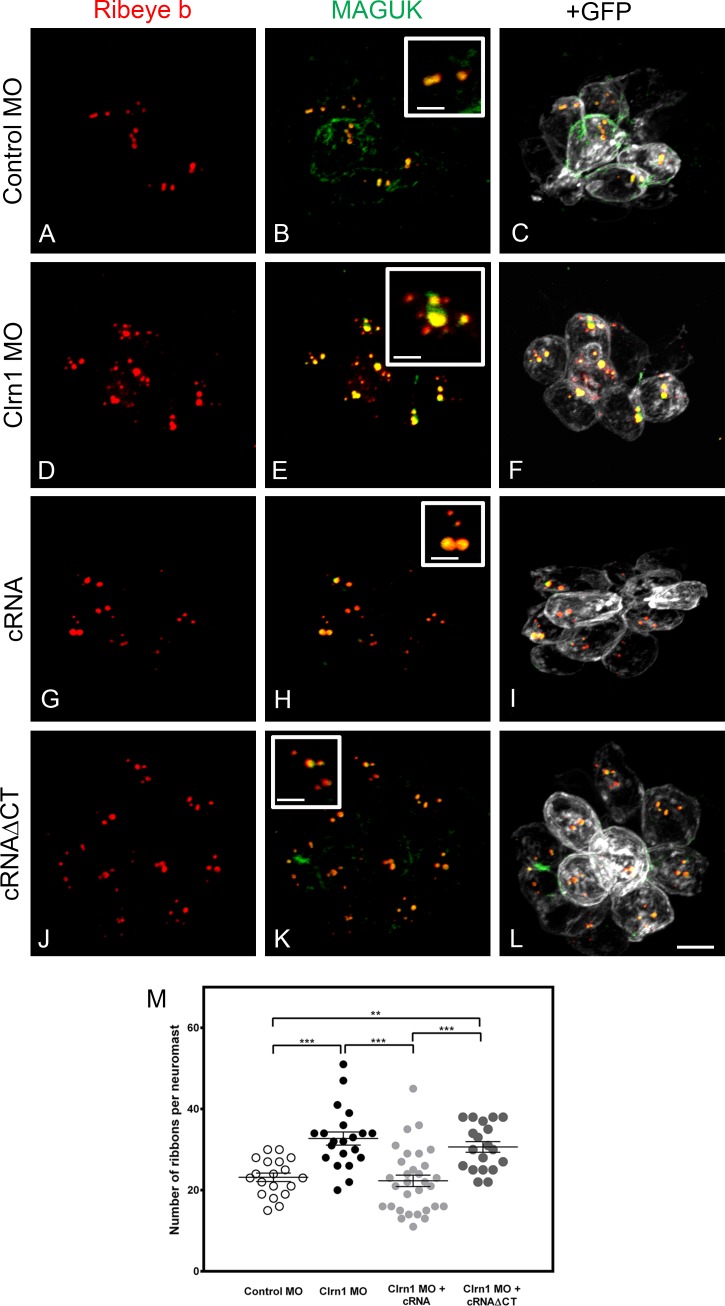
Because clarin-1 can also be detected at the base of the hair cells in mammals and fish (Fig. S2; Zallocchi et al., 2009; Phillips et al., 2013), we analyzed whether its depletion affects distribution of synaptic components. Experiments were performed in a transgenic line expressing a membrane-targeted GFP in ear and neuromast hair cells (Trapani et al., 2009). By using this fish line, we were able not only to identify individual hair cells but also to determine the distribution of the synaptic ribbons along the basolateral plasma membrane. Membrane-associated guanylate kinases (MAGUKs) and ribeye b were used as post- and presynaptic markers, respectively (Figs. 8 and 9). The ribeye b antibody, developed and qualified by our laboratory (Fig. S5), showed the characteristic puncta staining at the base of the hair cells (Sheets et al., 2011) in controls (Fig. S5, A and C) but not in the ribeye b MOs (Fig. S5, B and D). When 2 dpf anterior maculae were analyzed in the clarin-1 MOs, we observed a significant increase in the total number of ribeye b puncta present in the ear hair cells (Fig. 8, A–F and M), with these additional puncta being mislocalized at the apical aspect of the hair cells. Neuromast hair cells were also examined in 3 dpf larvae (Fig. 9, A–F and M). Again, an increase in the number of ribeye b puncta was observed in the clarin-1 MOs. The supernumerary and mislocalized ribeye b expression was not observed when embryos were co-injected with clarin-1 cRNA (Figs. 8 and 9, G–I and M), suggesting that clarin-1 might be playing a role in proper localization of ribeye b. cRNAΔCT did not rescue the synaptic phenotype (Figs. 8 and 9, J–L and M), establishing the importance of the cytoplasmic tail for normal ribeye b localization and, more likely, for synaptic formation. Pre- and postsynaptic boutons, determined by the juxtaposed ribeye b/MAGUK immunofluorescence, did not form in the mislocalized ribeye b, suggesting that those are not functional synapses.
Figure 8.
Clarin-1 MOs show supernumerary boutons and mislocalized expression of ribeye b in ear hair cells. (A–L) Representative experiments of 2 dpf Tg(bnr3c:mGFP) anterior maculae immunostained for ribeye and MAGUK in control MO (A–C), clarin-1 (Clrn1) MOs (D–F), MOs + cRNA (G–I), or cRNAΔCT (J–L). (insets) High magnification of basal and apical ribeye b accumulation. Bars: (main images) 8 µm; (insets) 1.5 µm. (M) Quantification of ribeye b punctate expressed as means ± SEM. Number of anterior macula analyzed, n = 5. **, P < 0.01; ***, P < 0.001 versus control MO. +, P < 0.05; ++, P < 0.01; +++, P < 0.001 versus clarin-1 MO + cRNA.
Figure 9.
Clarin-1 MOs show dysregulation in the number of ribeye b boutons in neuromasts. (A–L) Representative experiments of 3 dpf neuromasts from Tg(bnr3c:mGFP) immunostained for ribeye b and MAGUK in control MOs (A–C), clarin-1 (Clrn1) MOs (D–F), MOs + cRNA (G–I), or cRNAΔCT (J–L). (insets) High magnification of juxtaposed ribeye b/MAGUK boutons. Bars: (main images) 5 µm; (insets) 2 µm. (M) Quantification of ribeye b punctate per neuromast. Results from each independent experiment are represented by open (control animals) or filled (treated animals) circles. Mean for each treatment is depicted as a line with error bars representing SEM. N, number of independent experiments (neuromasts analyzed), n > 14. **, P < 0.01; ***, P < 0.001.
Clarin-1 involvement in vesicle recycling
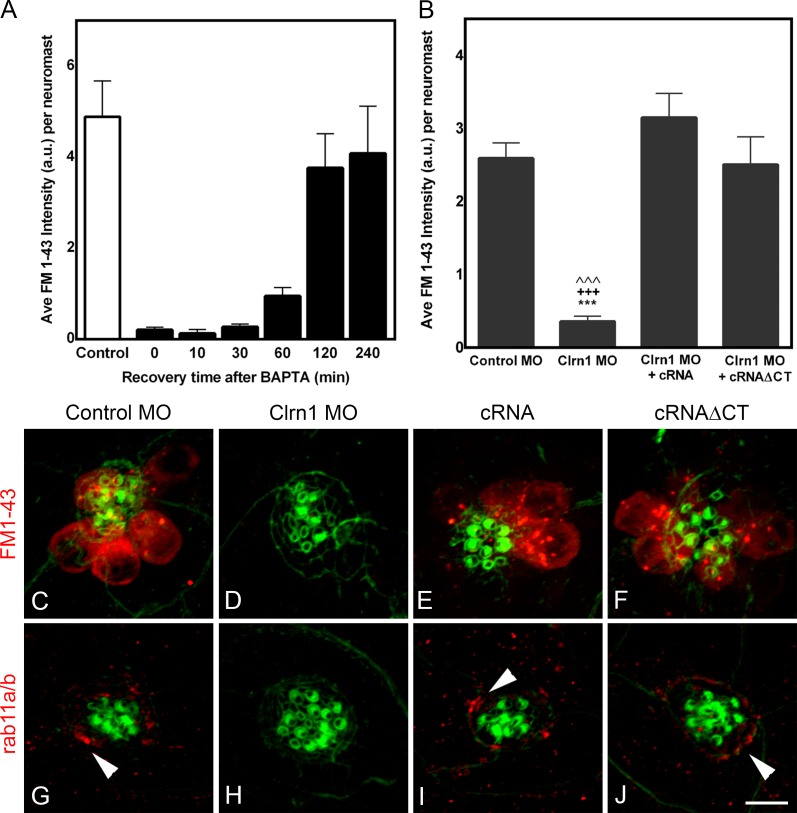
The fact that clarin-1 localized to subcellular regions enriched in vesicles and because several associations with different vesicle markers have been demonstrated (Tian et al., 2009; Zallocchi et al., 2012b), we assessed whether the effects observed in the MOs might be caused by regulation of vesicle recycling by clarin-1. We used two different approaches (Fig. 10). First, we analyzed FM1-43 internalization by neuromast hair cells as an indication of endocytosis. There are two ways FM1-43 can enter the hair cells: (1) through the mechanotransduction channels within a tenth of a second and (2) by endocytosis which requires longer incubations. To block FM1-43 uptake through the mechanotransduction channels, tip links were disrupted by preincubation of the injected animals with BAPTA. This pretreatment allows the specific evaluation of FM1-43 incorporation by endocytosis. BAPTA-treated animals were then incubated for 30 min with FM1-43. The 30-min incubation period was chosen based on previous experiments (Fig. 10 A) in which 3 dpf larvae were preincubated with BAPTA and then with the FM1-43 dye for a very short period of time (20 s), allowing the entrance of the dye through the mechanotransduction channels only. These experiments showed that fish hair cells require ≥2 h to recover from the BAPTA treatment, which implies that after BAPTA and during the first 30 min, dye entry will only occur through endocytosis (Seiler and Nicolson, 1999; Meyers et al., 2003). Results from these experiments (Fig. 10, B–F) showed significant reduction in FM1-43 internalization in the clarin-1 MOs (Fig. 10, B and D) compared with control (Fig. 10, B and C) and cRNA rescued larvae (Fig. 10, B and E). Co-injection with clarin-1 cRNAΔCT also restored FM1-43 incorporation to control levels (Fig. 10, B and F). As a second approach, we analyzed the distribution of the recycling endosomal marker rab11a (Ullrich et al., 1996). Rab11a is mainly expressed at the apical aspect of zebrafish hair cells (Clark et al., 2011), and its involvement in ciliogenesis has been demonstrated (Westlake et al., 2011; Asante et al., 2013). Fig. 10 (G–J) shows representative images of 3 dpf neuromasts immunostained for rab11a/b. Control animals showed expression of rab11a/b in hair cells with an accumulation toward their apical pole (Fig. 10 G, arrowhead). Rab11a/b expression was absent in the clarin-1 MOs but restored when eggs were co-injected with the full or truncated version of clarin-1 (Fig. 10, H vs. I and J). Combined, these results suggest that clarin-1 is involved in hair cell vesicle recycling and that its cytoplasmic tail is not required for this function.
Figure 10.
Clarin-1 is involved in vesicle recycling and endocytosis. (A) Bar graph of fast dye uptake recovery after tip link disruption by BAPTA (means ± SEM; number of independent experiments, n = 3). (B) Semiquantitative analysis of the total fluorescence per neuromast. Means ± SEM. ***, P < 0.001 versus control; +++, P < 0.001 versus clarin-1 (Clrn1) MO + cRNA; ˄˄˄, P < 0.001 versus clarin-1 MO + cRNAΔCT. Number of independent experiments, n > 5. (C–F) Representative images from the semiquantitative analysis in B. Long-term FM1-43 incorporation of 3 dpf larvae preincubated with BAPTA. (G–J) Representative images of at least five independent experiments. Expression of the early endosomal marker rab11a/b. Arrowheads denote apical accumulation of rab11a/b. Bar, 5 µm. a.u., arbitrary unit.
Discussion
In the current study, we provide new lines of evidence suggesting that clarin-1 is playing a role in the assembly and/or proper localization of key components necessary for normal hair cell function. First, we defined clarin-1 subcellular localization in zebrafish hair cells. Clarin-1 localizes to the stereocilia and below the cuticular plate where transport vesicles accumulate (Kachar et al., 1997), in the transition zone ciliary subcompartment that constitutes a docking site for proteins destined for the cilium (Williams et al., 2011; Ko, 2012; Reiter et al., 2012), and at the basal aspect of hair cells where an active process involving vesicle docking, release, and recycling exists (Khimich et al., 2005). Second, absence of clarin-1 leads to defects in endocytosis and the absence of apically localized molecules involved in vesicle recycling. Third, clarin-1 regulates localization of Pcdh15a to the apical aspect of hair cells, contributing to the assembly and/or function of the mechanotransduction apparatus. Fourth, clarin-1 MOs show short kinocilia, suggesting that clarin-1 may be playing a role in ciliogenesis in hair cells. Lastly, clarin-1 is necessary for proper localization of synaptic components, as indicated by the apical accumulation of ribeye b upon clarin-1 depletion. Combined, these results underscore the importance of clarin-1 as a mediator of protein complex formation and/or transport at both the apical and basal aspects of hair cells.
Clarin-1 protein expression was apparent starting at 1 dpf in macular hair cells, the first sensory patches to appear in zebrafish (Tanimoto et al., 2011). This expression persists ≤7 dpf, after which clarin-1 is undetectable from hair cells of both ear and neuromasts. Because clarin-1 transcript levels can be measured beyond this time point (not depicted), this implies that although absent from adult hair cells, clarin-1 may be still present in other tissues, i.e., eye (Phillips et al., 2013), or that there is a down-regulation of the protein that falls below the limit of detection of our antibody preparation.
Clarin-1 MOs showed morphological features associated with ciliopathy mutants/MOs but no differences in PCP. Instead, we observed a decrease in the total number of neuromasts, with hair cells showing dysfunctional mechanotransduction channel activity and short kinocilia. These characteristics of the clarin-1 MOs, i.e., normal hair cell number and PCP, but short kinocilia, coincides with the features observed by Kindt et al. (2012) using ift88 zebrafish mutants. However, in the case of the ift88, they still have normal FM1-43 uptake, suggesting that it is not the abnormal kinocilium per se that is impairing dye uptake and, ultimately, mechanotransduction channel activity in the clarin-1 MOs.
Within the primary cilia, clarin-1 is present at the most basal aspect of the transition zone. This ciliary subcompartment contains molecules involved in the formation of a membrane diffusion barrier that restricts the transport of proteins and lipids to and from the cilia (Fliegauf et al., 2007; Williams et al., 2011; Satish Tammana et al., 2013). The presence of clarin-1 in the transition zone and the fact that clarin-1 MOs are not completely devoid of primary cilia, but instead have significantly shorter kinocilia, an effect that can be rescued by co-injection with clarin-1 transcripts, point to a potential role for clarin-1 as a modulator during late ciliogenesis.
It is worth mentioning that two recent articles have shown a relationship between nasal epithelial cell function and Usher type I and type II proteins in Usher patients (Armengot et al., 2012; Piatti et al., 2014). As for clarin-1, this is the first work presenting evidence that suggests a direct role in ciliogenesis for an Usher protein. Because Usher mouse models show a disruption of the hair cell bundle integrity (Lefèvre et al., 2008), most of the studies related to clarin-1 at the apical aspect of hair cells have been focused on the stereocilia (Geller et al., 2009; Geng et al., 2009, 2012). Its putative role in kinocilium formation, before this work, was unexplored. Given the results presented here, it would be interesting to determine whether clarin-1 plays a role in ciliogenesis in mammalian vestibular and embryonic cochlear hair cells.
Increasing evidence suggests the existence of an Usher interactome. The fact that clarin-1 is not only expressed basally to the hair cell bundle but also at the tip and that mechanotransduction activity was impaired upon clarin-1 depletion lead us to address a possible interaction between clarin-1 and Pcdh15 or Cdh23, the two Usher proteins that are part of the tip links (Kazmierczak et al., 2007). Using a coimmunoprecipitation (co-IP) approach under experimental conditions that preserve secondary interactions (Charrin et al., 2003), we observed a reciprocal association between clarin-1 and Pcdh15a. Further evidence of an in vivo dependency was demonstrated by immunohistochemistry experiments showing abnormal Pcdh15a distribution in clarin-1 MOs. These MOs showed absence or residual expression of Pcdh15a. The residual expression agrees with the published data showing the presence of few tip links in Clrn1−/− mice and supports the notion of clarin-1 acting as a modulator of a process that is already taking place (Geng et al., 2012). In this regard, as a facilitator, clarin-1 may be increasing the efficiency of the process that leads to the final assembly of the mechanotransduction machinery. Reduction of the efficiency in clarin-1 MOs may result in incomplete/delayed complex assembly. Furthermore, this interaction with Pcdh15a is mediated by the C-terminal tail of clarin-1 because MOs expressing a truncated version lacked both Pcdh15a apical localization and FM1-43 uptake capabilities in spite of normal clarin-1 localization. This requirement of an interaction between Pcdh15a and clarin-1 for proper apical protein distribution seems to be unidirectional because clarin-1 expression was normal in the pcdh15a MOs, underscoring the potential importance of clarin-1 for tip link assembly.
It is notable that although clarin-1 knockdown leads to a reduction in Pcdh15a apical localization with the concomitant effect in mechanotransduction channel activity, we did not observe any hair cell bundle dysmorphology. This result contrasts with the published data in the Clrn1−/− mice (Geng et al., 2009, 2012) in which the impairment in dye uptake and the few tip links correlate with hair cell bundle abnormalities. Although speculative, these discrepancies may be explained by a compensatory mechanism through the zebrafish pcdh15a orthologue, pcdh15b. Previous work (Seiler et al., 2005) described expression of the pcdh15b transcript not only in photoreceptor cells but also in inner ear and neuromasts. Because the extracellular domain of both proteins share a high degree of amino acid sequence similarity, it is possible that under conditions in which Pcdh15a is not present, Pcdh15b might be able to help in preserving hair cell bundle integrity. Moreover, under this scenario, we probably will not observe dye uptake, as the intracellular domain of Pcdh15b is highly divergent from Pcdh15a (and mammal PCDH15), which will preclude interactions with other components of the mechanotransduction machinery at the lower tip link density.
Regarding FM1-43 uptake, a modest impairment has already been described for the Clrn1−/− mice (Geng et al., 2012); however, this is the first study demonstrating an in vivo role for clarin-1 in mechanotransduction channel regulation and suggests that clarin-1 has similar functional roles at the apical domains of both fish and rodent hair cells. Given the almost complete blockage of FM1-43 uptake in the clarin-1 MOs and the observation of an interaction between Pchd15a and clarin-1, we were surprised to find that only a subpopulation of the hair cells showed apical expression of Pcdh15a. As we previously mentioned, this discrepancy may be caused by epitope masking (Ahmed et al., 2006) and the necessity of using calcium chelators to expose some of the domains recognized by the Pcdh15a antibodies. An alternative and more likely explanation is that clarin-1 might be affecting the assembly and/or distribution, not only of Pcdh15a but of other key components involved in mechanotransduction function, resulting in a near-complete inhibition of dye uptake.
A direct interaction between PCDH15 and a different tetraspan, TMHS (tetraspan membrane protein of hair cell stereocilia), has recently been described in mouse (Xiong et al., 2012). In that work, the authors demonstrated an obligatory association between TMHS and PCDH15 for normal mechanotransduction activity. Because 4TM members have the tendency to form large aggregates with one another functioning as multimolecular complexes, it is possible that clarin-1 is not acting alone but in conjunction with other members of the same hyperfamily. Elucidating the composition of that putative membrane complex will help clarify the mechanisms through which hair cells regulate mechanotransduction channel function.
In addition to mild stereocilia abnormalities, clarin-1−/− mice have a delay in neuronal maturation and synapse formation (Geng et al., 2009; Zallocchi et al., 2012a). To test whether this is true in zebrafish, we analyzed localization and accumulation of synaptic components in clarin-1 MOs. Results from these experiments showed a defect in ribbon protein localization and number. The additional mislocalized ribeye b boutons did not show juxtaposed staining with postsynaptic markers, suggesting that ribeye b accumulation is not sufficient for synapse formation and that clarin-1 is playing a role in the correct localization of synaptic components. Moreover, the functional domain of clarin-1 that is responsible for this effect mapped within the cytoplasmic tail of the protein.
Similar results, regarding mislocalized ribbons that do not form synapses, were reported in a transgenic zebrafish line overexpressing ribeye b (Sheets et al., 2011). Those results, as well as our findings, suggest that if hair cell presynaptic assembly is altered, either by overexpression of presynaptic components or by depletion of modulators of this assembly, the result is the accumulation and mislocalization of nonfunctional presynaptic components. It is worth mentioning that some of the effects observed in the MOs were irreversible (number of neuromasts and kinocilium length), in that they were not restored after morpholino dilution and clarin-1 reexpression (after 4 dpf), suggesting that there is a specific developmental window during which clarin-1 exerts its effects.
The phenotypes observed in the clarin-1 MOs involving distinct subcellular regions led us to assess whether clarin-1 might be modulating a more general process that ultimately affects these compartments. Because clarin-1 expression correlates with subcellular areas of intense vesicle trafficking and turnover, we studied endocytosis and recycling in neuromast hair cells. Absence of clarin-1 leads to vesicle recycling impairment. Although this effect was rescued when animals were co-injected with the two different versions of clarin-1 transcripts (as well as the kinocilium length and gross animal morphology), Pcdh15a distribution with the concomitant rapid dye uptake and ribeye b localization did not recover. This implies that the truncated form of clarin-1 can still modulate vesicle recycling; however, because the interaction of clarin-1 with different hair cell components occurs through the C-terminal tail, the normal distribution of these molecules in the cRNAΔCT rescued MOs does not occur.
In conclusion, our findings suggest a unified mechanism for clarin-1 action in which, through the modulation of vesicle recycling, clarin-1 facilitates distribution and/or assembly of diverse components necessary for normal hair cell function. These results also reveal an unexpected role for clarin-1 in ciliogenesis and hair cell mechanotransduction channel function through its interaction with Pcdh15a. Future studies exploring the existence of similar mechanisms in photoreceptors may shed light not only on the overall role of clarin-1 in neurosensory cell homeostasis but also in the general processes that are affected in Usher patients.
Materials and methods
Zebrafish strains and husbandry
Zebrafish (Danio rerio), AB strain (catalogue number AB; RRID [Research Resource Identifier]: ZIRC_ZL1; Zebrafish International Resource Center) were grown at 28.5°C under standard conditions. Animal care and husbandry were overseen by the Institutional Animal Care and Use Committee at Boys Town National Research Hospital. Experimental larvae were grown in embryo medium (14 mM NaCl, 0.5 mM KCl, 0.25 mM Na2HPO4, 0.04 mM KH2PO4, 1.3 mM CaCl2, 1 mM MgSO4, and 4 mM NaHCO3, pH 7.2) at 28.5°C with a 10-h/14-h dark/light cycle (Westerfield, 2007). Animals were cryoanaesthetized before the initiation of the experiments (Wilson et al., 2009). The transgenic line Tg(bnr3c:mGFP), AB strain, was provided by T. Nicolson (Oregon Health and Science University, Portland, OR) and has been previously described (Xiao et al., 2005; Trapani et al., 2009).
For 2 dpf experiments in the lateral line organ, only the anterior lateral line neuromasts were analyzed. In the case of 3 dpf larvae of the supraorbital neuromasts, SO2 and SO3 were used for the experiments (Haehnel et al., 2012).
Morpholino and cRNA injections
Mismatch controls, scrambled, and specific morpholinos were designed by and purchased from GeneTools. For the generation of the clarin-1 MOs, morpholinos against the two putative start sites (clrn1-001 ENSDART00000149872 and clrn1-201 ENSDART00000062909; Ensembl database) 5′-GCGCACAGCCTCATCAGTGGAAATC-3′ and 5′-TAACTTGCTTTTGACGGTTAGGCAT-3′ and against intron 2 splice donor site of clarin-1 5′-TGTATTGTTTGCATCTCTTACAGCT-3′ were co-injected into one-cell stage embryos at a concentration of 0.4 mM for each morpholino (total volume of ∼2 nl). This approach proved to efficiently knock down clarin-1 protein between 1 and 3 dpf larvae. For the rescue experiments, a suspension containing the three different clarin-1 morpholinos at a concentration of ∼0.4 mM each plus 100 pg clarin-1 cRNA or clarin-1 cRNAΔCT was co-injected in a final volume of ∼2 nl. Translation-blocking morpholinos against pcdh15a (5′-CCTCCGCATCTTCACTTAATGCCTA-3′) and ribeye b (5′-GAACCGTGCCTCGCACTGCCATCAT-3′) and the corresponding mismatch controls were injected at a final concentration of 0.6 and 1.4 mM, respectively.
Embryos/larvae were maintained in embryo media and life screened at RT using a stereomicroscope (MZ10F; Leica) with a Plan Apochromat 1.0× objective and a magnification of 6.3×. Images were captured with a camera (DFC310 FX; Leica) and the Application Suite V4.0.0 acquisition software (Leica).
Antibodies
The following antibodies were generated against zebrafish proteins under contract with Proteintech: affinity-purified rabbit polyclonal against clarin-1, aa 40–135 (RRID: AB_2307327), rabbit polyclonal against ribeye b, aa 1–231 (RRID: AB_2307328), rabbit polyclonal Pcdh15a, aa 1,777–1,796 (RRID: AB_2307329), and rabbit polyclonal Cdh23, aa 3,195–3,213. Other antibodies used in this study were affinity-purified mouse anti-myosin7A (Soni et al., 2005; Zallocchi et al., 2012a), rabbit anti–clarin-1 DrClrn1 (provided by J. Phillips, University of Oregon, Eugene, OR; Phillips et al., 2013), mouse anti-cc2d2a (provided by R. Bachmann-Gagescu, University of Washington, Seattle, WA; Bachmann-Gagescu et al., 2011), mouse anti–ribeye b (Sheets et al., 2011), rabbit anti-rab11a/b (GeneTex, Inc.), mouse anti-His tag (Thermo Fisher Scientific), mouse anti–acetylated tubulin (T6793; Sigma-Aldrich; RRID: AB_477585), mouse anti–HCS-1 and mouse anti-ZN12 (Developmental Studies Hybridoma Bank, University of Iowa), mouse anti-MAGUK (75–030; Neuromab; RRID: AB_2091920), goat anti–-γ-tubulin (sc-12881; Santa Cruz Biotechnology, Inc.; RRID: AB_641317), and mouse anti-panactin (MAB1501; EMD Millipore; RRID: AB_2223041).
Cloning of the clarin-1 zebrafish transcript
Total RNA was extracted from 3 dpf embryos using TRIZOL reagent and reverse transcribed using the reverse transcription kit (SuperScript III; Life Technologies). Full-length clarin-1 was obtained using 5′-TACTCGAGATGCCTAACCGTCAAAAG-3′ and 5′-CAGAATTCTGTACATGAGATCTGCAGC-3′ as forward and reverse primers, respectively (GenBank accession no. NM_001002671) and the FastStart High Fidelity PCR System (Roche). A plasmid construct was made by inserting the 713-bp fragment downstream the cytomegalovirus promoter into the HindIII–EcoRI sites of the ptdTomato-N1 vector (containing a pUC and SV40 origins of replication; Takara Bio Inc.). The insert was sequence verified by the University of Nebraska Medical Center DNA Sequencing Core Facility.
Cell culture and transfections
HeLa cells were maintained in DMEM/F-12 medium supplemented with 5% of FCS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 0.29 mg/ml l-glutamine (Life Technologies). Subconfluent cells were dissociated with 0.05% trypsin and 0.02% EDTA (Life Technologies) and plated onto poly-l-lysine–coated microscope slides (VWR International) at a ratio of 1:10. Cells were then transfected with 1 µg of the corresponding vector using Lipofectamine 2000 (Invitrogen) and prepared for immunofluorescence analysis.
Clarin-1 cRNA in vitro transcription
Full-length D. rerio clarin-1 was cloned using the TOPO TA cloning kit (Life Technologies) and amplified using a forward primer containing a T7 promoter sequence, 5′-TAATACGACTCACTATAGGATGCCTAACC-3′, and a reverse primer containing a polyadenylation site, 5′-TTTATTTCAGTACATGAGATCTGCAG-3′. For the generation of His tag full-length and ΔCT clarin-1 transcripts, two rounds of PCR were used before cloning them into the TOPO plasmid. Primers for first PCR were full-length and ΔCT forward, 5′-AGGTACCATGCCTAATCGACAGAAACAGG-3′; full-length reverse, 5′-GTGGTGGTACATGAGATCTGCAGCTCC-3′; and ΔCT reverse primer, 5′-GTGCTGGATCCCGGCCAGGCGGATG-3′. Primers for second PCR were full-length and ΔCT forward, 5′-TAATACGACTCACTATAGGTACCATGCCTAATCG-3′; full-length reverse, 5′-CATTATTAGCATCAGTGGTGGTGGTGGTGGTA-3′; and ΔCT reverse primer, 5′-CATTATTAGCATCAGTGGTGGTGGTGGTGCTGGAT-3′.
PCR product was sequence verified and used for in vitro transcription using the mMESSAGE mMACHINE T7 Ultra kit (Life Technologies) according to the manufacturer’s instructions. Quality of the clarin-1 cRNA was verified by electrophoresis in denaturing gels. The wild-type full-length and the His-tagged full-length clarin-1 transcripts were used indistinctly for some of the experiments.
Dye incorporation
Mechanotransduction channel activity assessment.
FM1-43 (fixable version; Life Technologies) uptake was performed in 2 and 3 dpf larvae according to Kindt et al. (2012). In brief, animals were exposed for 20 s to 30 µM (2 dpf) or 3 µM (3 dpf) of FM1-43 in E3 media and washed four times for 5 min each. Animals were PFA fixed and processed for fluorescence analyses. In a set of preliminary results, animals were preexposed to 5 mM BAPTA for 10 min (Kindt et al., 2012) and then to the dye for 5 min; under these conditions, no labeling of the hair cells was observed, confirming that at 20 s the dye entrance is exclusively through the mechanotransduction channels.
Vesicle endocytosis assessment.
Animals were preincubated for 10 min in the presence of BAPTA and then for 30 min with FM1-43. After four washes of 5 min each, animals were PFA fixed and processed for fluorescence analyses.
Fluorescence intensity was estimated using ImageJ version 1.48g obtained from the National Institutes of Health. In brief, Area and Integrated Density of the region of interested were calculated using the corresponding tools. Mean Gray Value was calculated for the background, and the Corrected Total Fluorescence was calculated according to the following equation: Corrected Total Fluorescence = Integrated Density − (Area × Mean Gray Value).
Fluorescence analysis
Whole mount samples.
For ribeye b, MAGUK, and clarin-1 expression analysis, immunohistochemistry was performed in whole larvae according to Mo and Nicolson (2011) with the following modifications: After the 4-h 4% PFA fixation, animals were washed twice for 10 min, permeabilized with cold acetone for 6 min at −20°C, and washed once with distilled water for 5 min and two more times with washing buffer before blocking for 1 h at RT. Primary antibody dilution was 1:500 for ribeye b and MAGUK and 1 µg/250 µl for the affinity-purified anti–clarin-1, in blocking solution. Alexa Fluor 647–, Alexa Fluor 594–, and Alexa Fluor 488–conjugated secondary antibodies were at a dilution 1:1,000. Alexa Fluor 488–conjugated phalloidin was used at a dilution of 1:500 (Life Technologies). Ribeye b punctae were calculated using the Analyze Particles tool of ImageJ with previous transformation to a binary image. Punctae with a size <20 pixels were not included in the final results.
For colocalization analyses of clarin-1 with cc2d2a, fish were processed according to Bachmann-Gagescu et al. (2011). In brief, animals were fixed with 2% trichloroacetic acid in PBS for 3 h at RT, washed several times with PBS, and incubated in blocking solution (1% Triton X-100, 1% BSA, and 1% DMSO in PBS) for 30 min at RT. Anti–clarin-1 and anti-cc2d2a (1:20) were incubated overnight with the same blocking solution. Secondary antibody dilutions were 1:1,000 for Alexa Fluor 594 donkey anti–rabbit and 1:200 for Alexa Fluor 488 donkey anti–mouse.
For any other staining, animals were fixed in 4% PFA overnight at 4°C with rocking. After two washes of 10 min each with PBS containing 0.1% Triton X-100, fish were directly incubated with Alexa Fluor 488–conjugated phalloidin (Life Technologies for F-actin detection in the case of the FM1-43 experiments (dilution at 1:500 in blocking solution: 2.5% BSA, 2.5% FCS, and 0.1% Triton X-100 in PBS) or incubated for ≥1 h at RT with 1% Triton X-100 in PBS to dissolve the otolith. Primary antibodies were added overnight at 4°C with rocking. Dilutions were prepared in blocking solution as follows: anti-Pcdh15a, anti-Cdh23, anti–acetylated tubulin, anti-ZN12, and anti–HCS-1 at 1:500 and anti-His6x and anti–γ-tubulin at 1:100. After three washes of 20 min, Alexa Fluor 594– and/or Alexa Fluor 647–conjugated secondary antibodies were added at a dilution of 1:1,000 in blocking solution and incubated overnight at 4°C with rocking. In all the cases, fish were mounted onto poly-l-lysine–coated microscope slides using 80% glycerol as a mounting medium.
Cross sections.
3 dpf larvae were fixed with 4% PFA and 4% sucrose in PBS overnight at 4°C with rocking. After three washes of 10 min at RT with PBS containing 0.1% Triton X-100 larvae were mounted in Optical Cutting Temperature compound and stored at −80°C. 14-µm cryosections were mounted onto poly-l-lysine–coated microscope slides and air dried for ≥30 min. Sections were incubated with blocking solution (0.1% Triton X-100, 2.5% BSA, and 2.5% FCS in PBS) for 1 h at RT and then overnight with the primary antibodies. Primary antibodies dilutions were as follows: 1 µg/250 µl rabbit affinity-purified anti–clarin-1, mouse affinity-purified anti-myosin7A at 1:250, mouse anti-ZN12 at 1:300, and mouse anti–ribeye b at 1:500. After several washes, samples were incubated with the secondary antibodies Alexa Fluor 647, Alexa Fluor 488, and Alexa Fluor 594, dilution of 1:500 in blocking solution for 1 h at RT. Slides were washed and cover slipped using Vectashield mounting medium containing DAPI to counterstain the nuclei (Vector Laboratories).
Cells.
Transfected HeLa cells were fixed with 4% PFA for 30 min at RT, washed three times for 10 min with PBS, and permeabilized for 7 min with 0.3% Triton X-100 in PBS. After three more washes with PBS, cells were blocked for 1 h at RT with fish gelatin blocking solution (2% FCS and 0.3% fish gelatin in PBS) and incubated overnight with 1 µg/250 µl affinity-purified anti–clarin-1 in fish blocking solution. After several washes and 1-h incubation with the Alexa Fluor 488 goat anti–rabbit secondary antibody (1:500 dilution), slides were coverslipped using Vectashield mounting medium containing DAPI to counterstain the nuclei.
Fluorescence images were captured using a fluorescence microscope (AxioImager.A1; Carl Zeiss), with a 10×, NA 0.3 air objective at RT, equipped with a camera (Flex; SPOT Imaging Solutions) and Basic V4.6 (SPOT Imaging Solutions) as the acquisition software. Confocal images were captured at RT using a confocal laser scanning microscope system (510 Meta; Carl Zeiss) with a built-in camera and a 63×, NA 1.4 oil objective. Z-stack images were acquired using the 63×, NA 1.4 oil objective zoom 2×, with a pinhole auto set at 1 and a sectioning of 0.5 µm. All confocal images were acquired with ZEN 2012 black edition software (Carl Zeiss). Z-stack images are presented as flat z projections. Only linear adjustments were made to brightness and contrast, and the final figures were assembled using Photoshop and Illustrator software (Adobe).
Co-IP
Co-IP experiments were performed according to Zallocchi et al. (2012b) and adapted for zebrafish. In brief, 30 µl of a 50% protein A–Sepharose CL-4B slurry (Sigma-Aldrich) was incubated overnight with 7.5 µl rabbit anti-Pcdh15a, 7 µl rabbit anti–clarin-1 DrClrn1 (Phillips et al., 2013), or normal rabbit serum (immunobeads). The day of the experiment, ∼200 3 dpf larvae were homogenized in co-IP buffer (10 mM Tris/HCl, pH 7.4, 150 mM NaCl, 0.5 mM MgCl2, 0.5 mM CaCl2, and 1% Brij 97) containing protease and phosphatase inhibitors (Thermo Fisher Scientific), and 6 mg protein from this lysate was incubated with the immunobeads overnight at 4°C with rocking. After several washes, co-IPs were resuspended in sample buffer and analyzed by Western blotting.
Western blot
Previous to lysate preparation, embryos/larvae were deyolked according to Link et al. (2006) protocol for deyolking embryos in batches. In brief, 50–100 embryos/larvae were transferred to 200 µl of a 1:2 dilution of Ginzburg Fish Ringer buffer (55 mM NaCl, 1.8 mM KCl, and 1.25 mM NaHCO3), and yolk sac was disrupted by pipetting up and down with a 200-µl tip. Deyolked fish were shaken for 5 min at 1,100 rpm, pelleted, and washed twice with 1 ml of wash buffer (110 mM NaCl, 3.5 mM KCl, 2.7 mM CaCl2, and 10 mM Tris/HCl, pH 8.5). Embryos/larvae were homogenized in 250 µl of lysis buffer (150 mM NaCl, 50 mM Tris, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS, pH 7.4) containing protease inhibitors (Sigma-Aldrich) and cleared by centrifugation, and 30–60 µg protein was used for Western blot analysis. Polyvinyl difluoride membranes were incubated overnight at 4°C in 5% milk blocking solution (0.1% Tween 20, 75 mM NaCl, 5% glycerol, and 5% nonfat dry milk in PBS). Primary antibody dilutions were performed in blocking solution as follows: anti-Pcdh15a at 1:500 and affinity-purified anti–-clarin-1 antibody at 0.2 µg/ml. Incubation with the primary antibodies was performed overnight with rocking. After two washes, membranes were incubated with a goat anti–-rabbit HRP-conjugated secondary antibody for regular Western blots or with HRP-conjugated protein A (GE Healthcare) for co-IP experiments. Dilution for both secondary detection systems was 1:20,000 in milk blocking solution for 1 h at RT. After several washes, membranes were developed using Western blotting substrate (for the co-IPs; ECL 2; Thermo Fisher Scientific) or chemiluminescent substrate (for Western blots; SuperSignal West Femto; Thermo Fisher Scientific).
For panactin immunoblotting, membranes were stripped with Western blot stripping buffer (Restore PLUS; Thermo Fisher Scientific) following the manufacturer’s instructions. Membranes were then blocked in 3% milk blocking solution for 1 h at RT followed by overnight incubation with the primary panactin antibody, dilution of 1:25,000. After two washes, a goat anti–mouse HRP-conjugated secondary antibody was added at a dilution of 1:7,500 for 1 h at RT. Membranes were developed using Western blotting substrate (ECL 2).
Quantification of the specific bands was performed using ImageJ Gel Analysis tool and according to Zallocchi et al. (2012a). In brief, lanes were selected and plotted, and the area of the pick for each band was quantified.
EM
SEM experiments were performed in 3 dpf larvae and according to Kindt et al. (2012). In brief, specimens were fixed with 2.5% glutaraldehyde and 2 mM CaCl2 in 0.1 M cacodylate buffer for 1.5 h at RT. After several washes with 80 mM of cacodylate buffer containing 4 mM CaCl2, samples were postfixed with 50 mM osmium tetroxide. A scanning electron microscope (Quanta 200; FEI; University of Nebraska Medical Center, Electron Microscopy Core Facility) was used to acquire the images, with a beam strength of 30 kV.
Statistical analysis
Numerical data were graphically represented as scattered plots (individual values) or as graph bars (means ± SEM). Statistical analysis was performed using Prism 5 (GraphPad Software). One-way analysis of variance followed by a nonparametric Dunn’s post-test was used to compare the proportion of FM1-43–labeled hair cells. One-way analysis of variance followed by Tukey’s post-test was used to analyze the rest of the experiments.
Online supplemental material
Fig. S1 shows the specificity of the clarin-1 antibody evaluated by Western blotting and immunofluorescence experiments in clarin-1 MOs and HeLa cells exogenously expressing a tdTomato–clarin-1 fusion protein. Fig. S2 shows that clarin-1 is expressed at the basal aspect of hair cells. Fig. S3 shows qualification of the Pcdh15a antibody performed by Western blot experiments in control and pcdh15a MOs. Fig. S4 shows that clarin-1 MOs have normal PCP and hair cell bundle morphology. Fig. S5 shows ribeye b antibody qualification performed by immunohistochemistry in neuromasts and anterior macula hair cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201404016/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201404016.dv.
Supplementary Material
Acknowledgments
We thank J. Taylor and J. Talaska of the Confocal Laser Scanning Microscope Core Facility at University of Nebraska Medical Center (UNMC) for assistance and the Nebraska Research Initiative and the Eppley Cancer Center for support of the Core Facility. We also thank T. Bargar of the Electron Microscope Core Facility at UNMC for SEM assistance. We are grateful to Dr. J. Hudspeth (Howard Hughes Medical Institutes and Laboratory of Sensory Neuroscience, The Rockefeller University) and his research group for their help in establishing the zebrafish laboratory at Boys Town National Research Hospital (BTNRH). We also thank Dr. D. Cosgrove (BTNRH) and Dr. J. Phillips for critically reviewing this manuscript and S. Kennedy for help in figure preparation and art work. The Tg(bnr3c:mGFP) line and the mouse anti–ribeye b were provided by Dr. T. Nicolson. We thank Dr. J. Phillips (for anti–clarin-1 DrClrn1) and Dr. R. Bachmann-Gagescu for anti-cc2d2a.
This work was supported by National Institutes of Health grant 5P20RR018788 and the Tobacco Settlement Fund from the State of Nebraska to M. Zallocchi.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CDH23
- cadherin-23
- co-IP
- coimmunoprecipitation
- cRNA
- copy RNA
- dpf
- day postfertilization
- MAGUK
- membrane-associated guanylate kinase
- MO
- morphant
- PCP
- planar cell polarity
- PLL
- posterior lateral line
- SEM
- scanning EM
References
- Adato A., Vreugde S., Joensuu T., Avidan N., Hamalainen R., Belenkiy O., Olender T., Bonne-Tamir B., Ben-Asher E., Espinos C., et al. . 2002. USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur. J. Hum. Genet. 10:339–350 10.1038/sj.ejhg.5200831 [DOI] [PubMed] [Google Scholar]
- Ahmed Z.M., Riazuddin S., Ahmad J., Bernstein S.L., Guo Y., Sabar M.F., Sieving P., Riazuddin S., Griffith A.J., Friedman T.B., et al. . 2003. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum. Mol. Genet. 12:3215–3223 10.1093/hmg/ddg358 [DOI] [PubMed] [Google Scholar]
- Ahmed Z.M., Goodyear R., Riazuddin S., Lagziel A., Legan P.K., Behra M., Burgess S.M., Lilley K.S., Wilcox E.R., Riazuddin S., et al. . 2006. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci. 26:7022–7034 10.1523/JNEUROSCI.1163-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengot M., Salom D., Diaz-Llopis M., Millan J.M., Milara J., Mata M., and Cortijo J.. 2012. Nasal ciliary beat frequency and beat pattern in retinal ciliopathies. Invest. Ophthalmol. Vis. Sci. 53:2076–2079 10.1167/iovs.11-8666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante D., Maccarthy-Morrogh L., Townley A.K., Weiss M.A., Katayama K., Palmer K.J., Suzuki H., Westlake C.J., and Stephens D.J.. 2013. A role for the Golgi matrix protein giantin in ciliogenesis through control of the localization of dynein-2. J. Cell Sci. 126:5189–5197 10.1242/jcs.131664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin-Tse C., Halbritter J., Zariwala M.A., Gilberti R.M., Gee H.Y., Hellman N., Pathak N., Liu Y., Panizzi J.R., Patel-King R.S., et al. . 2013. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet. 93:672–686 10.1016/j.ajhg.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann-Gagescu R., Phelps I.G., Stearns G., Link B.A., Brockerhoff S.E., Moens C.B., and Doherty D.. 2011. The ciliopathy gene cc2d2a controls zebrafish photoreceptor outer segment development through a role in Rab8-dependent vesicle trafficking. Hum. Mol. Genet. 20:4041–4055 10.1093/hmg/ddr332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz H., von Brederlow B., Ramírez A., Bryda E.C., Kutsche K., Nothwang H.G., Seeliger M., del C-Salcedó Cabrera M., Vila M.C., Molina O.P., et al. . 2001. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet. 27:108–112 10.1038/83667 [DOI] [PubMed] [Google Scholar]
- Caberlotto E., Michel V., de Monvel J.B., and Petit C.. 2011. Coupling of the mechanotransduction machinery and stereocilia F-actin polymerization in the cochlear hair bundles. BioArchitecture. 1:169–174 10.4161/bioa.1.4.17532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S., Manié S., Billard M., Ashman L., Gerlier D., Boucheix C., and Rubinstein E.. 2003. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304:107–112 10.1016/S0006-291X(03)00545-X [DOI] [PubMed] [Google Scholar]
- Chen L., El-Husseini A., Tomita S., Bredt D.S., and Nicoll R.A.. 2003. Stargazin differentially controls the trafficking of α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate and kainate receptors. Mol. Pharmacol. 64:703–706 10.1124/mol.64.3.703 [DOI] [PubMed] [Google Scholar]
- Clark B.S., Winter M., Cohen A.R., and Link B.A.. 2011. Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev. Dyn. 240:2452–2465 10.1002/dvdy.22758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., and Omran H.. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Gale J.E., Marcotti W., Kennedy H.J., Kros C.J., and Richardson G.P.. 2001. FM1-43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J. Neurosci. 21:7013–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller S.F., Guerin K.I., Visel M., Pham A., Lee E.S., Dror A.A., Avraham K.B., Hayashi T., Ray C.A., Reh T.A., et al. . 2009. CLRN1 is nonessential in the mouse retina but is required for cochlear hair cell development. PLoS Genet. 5:e1000607 10.1371/journal.pgen.1000607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R., Geller S.F., Hayashi T., Ray C.A., Reh T.A., Bermingham-McDonogh O., Jones S.M., Wright C.G., Melki S., Imanishi Y., et al. . 2009. Usher syndrome IIIA gene clarin-1 is essential for hair cell function and associated neural activation. Hum. Mol. Genet. 18:2748–2760 10.1093/hmg/ddp210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R., Melki S., Chen D.H., Tian G., Furness D.N., Oshima-Takago T., Neef J., Moser T., Askew C., Horwitz G., et al. . 2012. The mechanosensory structure of the hair cell requires clarin-1, a protein encoded by Usher syndrome III causative gene. J. Neurosci. 32:9485–9498 10.1523/JNEUROSCI.0311-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory F.D., Pangrsic T., Calin-Jageman I.E., Moser T., and Lee A.. 2013. Harmonin enhances voltage-dependent facilitation of Cav1.3 channels and synchronous exocytosis in mouse inner hair cells. J. Physiol. 591:3253–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haehnel M., Taguchi M., and Liao J.C.. 2012. Heterogeneity and dynamics of lateral line afferent innervation during development in zebrafish (Danio rerio). J. Comp. Neurol. 520:1376–1386 10.1002/cne.22798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indzhykulian A.A., Stepanyan R., Nelina A., Spinelli K.J., Ahmed Z.M., Belyantseva I.A., Friedman T.B., Barr-Gillespie P.G., and Frolenkov G.I.. 2013. Molecular remodeling of tip links underlies mechanosensory regeneration in auditory hair cells. PLoS Biol. 11:e1001583 10.1371/journal.pbio.1001583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachar B., Battaglia A., and Fex J.. 1997. Compartmentalized vesicular traffic around the hair cell cuticular plate. Hear. Res. 107:102–112 10.1016/S0378-5955(97)00027-0 [DOI] [PubMed] [Google Scholar]
- Kalueff A.V., Gebhardt M., Stewart A.M., Cachat J.M., Brimmer M., Chawla J.S., Craddock C., Kyzar E.J., Roth A., Landsman S., et al. ; Zebrafish Neuroscience Research Consortium. 2013. Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish. 10:70–86 10.1089/zeb.2012.0861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak P., Sakaguchi H., Tokita J., Wilson-Kubalek E.M., Milligan R.A., Müller U., and Kachar B.. 2007. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 449:87–91 10.1038/nature06091 [DOI] [PubMed] [Google Scholar]
- Kersten F.F., van Wijk E., van Reeuwijk J., van der Zwaag B., Märker T., Peters T.A., Katsanis N., Wolfrum U., Keunen J.E., Roepman R., and Kremer H.. 2010. Association of whirlin with Cav1.3 (α1D) channels in photoreceptors, defining a novel member of the usher protein network. Invest. Ophthalmol. Vis. Sci. 51:2338–2346 10.1167/iovs.09-4650 [DOI] [PubMed] [Google Scholar]
- Khimich D., Nouvian R., Pujol R., Tom Dieck S., Egner A., Gundelfinger E.D., and Moser T.. 2005. Hair cell synaptic ribbons are essential for synchronous auditory signalling. Nature. 434:889–894 10.1038/nature03418 [DOI] [PubMed] [Google Scholar]
- Kindt K.S., Finch G., and Nicolson T.. 2012. Kinocilia mediate mechanosensitivity in developing zebrafish hair cells. Dev. Cell. 23:329–341 10.1016/j.devcel.2012.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H.W.2012. The primary cilium as a multiple cellular signaling scaffold in development and disease. BMB Rep. 45:427–432 10.5483/BMBRep.2012.45.8.167 [DOI] [PubMed] [Google Scholar]
- Kramer-Zucker A.G., Olale F., Haycraft C.J., Yoder B.K., Schier A.F., and Drummond I.A.. 2005. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 132:1907–1921 10.1242/dev.01772 [DOI] [PubMed] [Google Scholar]
- Lefèvre G., Michel V., Weil D., Lepelletier L., Bizard E., Wolfrum U., Hardelin J.P., and Petit C.. 2008. A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development. 135:1427–1437 10.1242/dev.012922 [DOI] [PubMed] [Google Scholar]
- Link V., Shevchenko A., and Heisenberg C.P.. 2006. Proteomics of early zebrafish embryos. BMC Dev. Biol. 6:1 10.1186/1471-213X-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J.R., MacDonald R.B., Duggan A., Lenzi D., Standaert D.G., Corwin J.T., and Corey D.P.. 2003. Lighting up the senses: FM1-43 loading of sensory cells through nonselective ion channels. J. Neurosci. 23:4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic I., Pylawka S., and Hudspeth A.J.. 2012. Rearrangements between differentiating hair cells coordinate planar polarity and the establishment of mirror symmetry in lateral-line neuromasts. Biol. Open. 1:498–505 10.1242/bio.2012570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo W., and Nicolson T.. 2011. Both pre- and postsynaptic activity of Nsf prevents degeneration of hair-cell synapses. PLoS ONE. 6:e27146 10.1371/journal.pone.0027146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.B., Blanco-Sanchez B., Lentz J.J., Tallafuss A., Khanobdee K., Sampath S., Jacobs Z.G., Han P.F., Mishra M., Titus T.A., et al. . 2011. Harmonin (Ush1c) is required in zebrafish Müller glial cells for photoreceptor synaptic development and function. Dis. Model. Mech. 4:786–800 10.1242/dmm.006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.B., Västinsalo H., Wegner J., Clément A., Sankila E.M., and Westerfield M.. 2013. The cone-dominant retina and the inner ear of zebrafish express the ortholog of CLRN1, the causative gene of human Usher syndrome type 3A. Gene Expr. Patterns. 13:473–481 10.1016/j.gep.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti G., De Santi M.M., Brogi M., Castorina P., and Ambrosetti U.. 2014. Emerging ciliopathies: are respiratory cilia compromised in Usher syndrome? Am. J. Otolaryngol. 35:340–346 10.1016/j.amjoto.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Reiners J., Nagel-Wolfrum K., Jürgens K., Märker T., and Wolfrum U.. 2006. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp. Eye Res. 83:97–119 10.1016/j.exer.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Reiter J.F., Blacque O.E., and Leroux M.R.. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13:608–618 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satish Tammana T.V., Tammana D., Diener D.R., and Rosenbaum J.. 2013. Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J. Cell Sci. 126:5018–5029 10.1242/jcs.133439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J.M., Bhatti R., Madeo A.C., Turriff A., Muskett J.A., Zalewski C.K., King K.A., Ahmed Z.M., Riazuddin S., Ahmad N., et al. . 2011. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J. Med. Genet. 48:767–775 10.1136/jmedgenet-2011-100262 [DOI] [PubMed] [Google Scholar]
- Seiler C., and Nicolson T.. 1999. Defective calmodulin-dependent rapid apical endocytosis in zebrafish sensory hair cell mutants. J. Neurobiol. 41:424–434 [DOI] [PubMed] [Google Scholar]
- Seiler C., Finger-Baier K.C., Rinner O., Makhankov Y.V., Schwarz H., Neuhauss S.C., and Nicolson T.. 2005. Duplicated genes with split functions: independent roles of protocadherin15 orthologues in zebrafish hearing and vision. Development. 132:615–623 10.1242/dev.01591 [DOI] [PubMed] [Google Scholar]
- Sheets L., Trapani J.G., Mo W., Obholzer N., and Nicolson T.. 2011. Ribeye is required for presynaptic Ca(V)1.3a channel localization and afferent innervation of sensory hair cells. Development. 138:1309–1319 10.1242/dev.059451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidi S., Friedrich R.W., and Nicolson T.. 2003. NompC TRP channel required for vertebrate sensory hair cell mechanotransduction. Science. 301:96–99 10.1126/science.1084370 [DOI] [PubMed] [Google Scholar]
- Söllner C., Rauch G.J., Siemens J., Geisler R., Schuster S.C., Müller U., and Nicolson T.. Tübingen 2000 Screen Consortium. 2004. Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature. 428:955–959 10.1038/nature02484 [DOI] [PubMed] [Google Scholar]
- Soni L.E., Warren C.M., Bucci C., Orten D.J., and Hasson T.. 2005. The unconventional myosin-VIIa associates with lysosomes. Cell Motil. Cytoskeleton. 62:13–26 10.1002/cm.20080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto M., Ota Y., Inoue M., and Oda Y.. 2011. Origin of inner ear hair cells: morphological and functional differentiation from ciliary cells into hair cells in zebrafish inner ear. J. Neurosci. 31:3784–3794 10.1523/JNEUROSCI.5554-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Zhou Y., Hajkova D., Miyagi M., Dinculescu A., Hauswirth W.W., Palczewski K., Geng R., Alagramam K.N., Isosomppi J., et al. . 2009. Clarin-1, encoded by the Usher Syndrome III causative gene, forms a membranous microdomain: possible role of clarin-1 in organizing the actin cytoskeleton. J. Biol. Chem. 284:18980–18993 10.1074/jbc.M109.003160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani J.G., Obholzer N., Mo W., Brockerhoff S.E., and Nicolson T.. 2009. Synaptojanin1 is required for temporal fidelity of synaptic transmission in hair cells. PLoS Genet. 5:e1000480 10.1371/journal.pgen.1000480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O., Reinsch S., Urbé S., Zerial M., and Parton R.G.. 1996. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135:913–924 10.1083/jcb.135.4.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M.2007. The Zebrafish Book: A guide for the laboratory use of zebrafish (Danio rerio). Fifth edition University of Oregon Press, Eugene, OR. [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. . 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C.J., Carl M., and Harris W.A.. 2009. Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol. 10:17 10.1186/1471-2121-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. . 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192:1023–1041 10.1083/jcb.201012116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.M., Bunte R.M., and Carty A.J.. 2009. Evaluation of rapid cooling and tricaine methanesulfonate (MS222) as methods of euthanasia in zebrafish (Danio rerio). J. Am. Assoc. Lab. Anim. Sci. 48:785–789. [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Roeser T., Staub W., and Baier H.. 2005. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 132:2955–2967 10.1242/dev.01861 [DOI] [PubMed] [Google Scholar]
- Xiong W., Grillet N., Elledge H.M., Wagner T.F., Zhao B., Johnson K.R., Kazmierczak P., and Müller U.. 2012. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell. 151:1283–1295 10.1016/j.cell.2012.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M., Meehan D.T., Delimont D., Askew C., Garige S., Gratton M.A., Rothermund-Franklin C.A., and Cosgrove D.. 2009. Localization and expression of clarin-1, the Clrn1 gene product, in auditory hair cells and photoreceptors. Hear. Res. 255:109–120 10.1016/j.heares.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M., Sisson J.H., and Cosgrove D.. 2010. Biochemical characterization of native Usher protein complexes from a vesicular subfraction of tracheal epithelial cells. Biochemistry. 49:1236–1247 10.1021/bi9020617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M., Meehan D.T., Delimont D., Rutledge J., Gratton M.A., Flannery J., and Cosgrove D.. 2012a. Role for a novel Usher protein complex in hair cell synaptic maturation. PLoS ONE. 7:e30573 10.1371/journal.pone.0030573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M., Delimont D., Meehan D.T., and Cosgrove D.. 2012b. Regulated vesicular trafficking of specific PCDH15 and VLGR1 variants in auditory hair cells. J. Neurosci. 32:13841–13859 10.1523/JNEUROSCI.1242-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.