Xenopus laevis CSF1 renders tadpoles and tadpole macrophages susceptible to the FV3 ranavirus, whereas IL34 increases tadpole and tadpole macrophage resistance.
Keywords: ranavirus, FV3, immunity, CSF-1, IL-34
Abstract
Macrophages are integral to amphibian immunity against RVs, as well as to the infection strategies of these pathogens. Although CSF-1 was considered to be the principal mediator of macrophage development, the IL-34 cytokine, which shares no sequence identity with CSF-1, is now believed to contribute to vertebrate monopoiesis. However, the respective roles of CSF-1- and IL-34-derived macrophages are still poorly understood. To delineate the contribution of these macrophage populations to amphibian immunity against the RV FV3, we identified the Xenopus laevis IL-34 and transcriptionally and functionally compared this cytokine with the previously identified X. laevis CSF-1. The X. laevis CSF-1 and IL-34 displayed strikingly nonoverlapping developmental and tissue-specific gene-expression patterns. Furthermore, only CSF-1 but not IL-34 was up-regulated in the kidneys of FV3-challenged tadpoles. Intriguingly, recombinant forms of these cytokines (rXlCSF-1, rXlIL-34) elicited morphologically distinct tadpole macrophages, and whereas rXlCSF-1 pretreatment decreased the survival of FV3-infected tadpoles, rXlIL-34 administration significantly prolonged FV3-challenged animal survival. Compared with rXlIL-34-elicited macrophages, macrophages derived by rXlCSF-1 were more phagocytic but also significantly more susceptible to in vitro FV3 infections. By contrast, rXlIL-34-derived macrophages exhibited significantly greater in vitro antiranaviral activity and displayed substantially more robust gene expression of the NADPH oxidase components (p67phox, gp91phox) and type I IFN. Moreover, FV3-challenged, rXlIL-34-derived macrophages exhibited several orders of magnitude greater up-regulation of the type I IFN gene expression. This marks the first report of the disparate roles of CSF-1 and IL-34 in vertebrate antiviral immunity.
Introduction
The worldwide decline in approximately one-third (32%) of all amphibian species is posing an imminent threat to the extinction of these organisms [1]. Although the underlining causes behind these die-offs are poorly understood [2, 3], the dramatic increase in the prevalence of RV (family Iridoviridae) infections and resulting mortalities implicates these pathogens as a significant contributing factor to amphibian declines [1–3]. RVs are large, icosahedral dsDNA viruses, the infections of which manifest in systemic diseases, hemorrhaging, and necrotic cell death within multiple afflicted organs [1]. Amphibian tadpoles are generally more susceptible to and succumb from RV infections, whereas mature adults exhibit more natural resistance to these pathogens [4–8]. In contrast to some RV species that appear to be confined to cognate amphibian hosts [9–11], the RV FV3 exhibits an uncanny capacity to traverse species barriers and establish infections to the detriment of new hosts [9, 12–14].
Amphibian macrophage-lineage cells appear to be important to antiranaviral immunity and to the infection strategies of RV pathogens. Recent findings indicate that FV3 infects, persists, and possibly disseminates by infiltrating frog macrophages [15, 16]. These FV3-infected cells display cytosolic pools of assembled viral particles, reminiscent of the HIV-macrophage transmission strategy [17–20]. Indeed, macrophage-lineage cells are integral to homeostasis and immunity of vertebrate species, so it is not surprising that these immune sentinels are often subverted by pathogen-like RVs.
The CSF-1, M-CSF, is considered to be the principal molecule responsible for the development, differentiation, proliferation, and survival of mononuclear phagocyte lineages across all vertebrate species examined to date [21–24]. This growth factor ligates the CSF-1R [25], the cell-surface expression of which is restricted predominantly to committed macrophage precursors and derivative macrophage populations [26, 27]. In a relatively recent development, the IL-34 cytokine, which does not share sequence identity with CSF-1 [28–30], has been demonstrated to bind to and result in the activation of the CSF-1R, contributing to myeloid cell proliferation and differentiation [31–34]. IL-34 appears to be involved in the development and maintenance of tissue macrophages and Langerhans cells [35, 36], as well as osteoclastogenesis [37, 38], maintenance of microglia [35], and the development of mononuclear phagocyte populations responsible for B cell stimulation [39]. However, the precise evolutionary origins and biological necessity for this additional CSF-1R ligand remain to be fully elucidated.
To delineate the respective roles of CSF-1 and IL-34 in amphibian immunity and particularly during FV3 infections, we identified the X. laevis IL-34, produced it in recombinant form and compared tadpole macrophages derived by this growth factor with those elicited by the previously identified X. laevis CSF-1 [40]. This manuscript marks the first report of an amphibian IL-34 and moreover, is the first to demonstrate drastic disparities in the roles of CSF-1- and IL-34-derived macrophages in antiviral defenses.
EXPERIMENTAL PROCEDURES
Animals
Out-bred premetamorphic (stage 54; refs. [41, 42]) tadpoles and metamorphic (stage 64) and adult (2 years old) frogs were obtained from our X. laevis research resource for immunology at the University of Rochester (http://www.urmc.rochester.edu/mbi/resources/xenopus-laevis/). All animals were handled under strict regulations of the laboratory and the University Committee on Animal Research (Approval Number 100577/2003-151).
Identification and in silico analysis of X. laevis IL-34
The identification of the X. laevis CSF-1 was described previously [40]. A partial X. laevis IL-34 cDNA transcript was identified using primers against the predicted Xenopus tropicalis IL-34 sequence (Acc. No.: XM_004914044). Subsequently, RACE PCR was performed in accordance with the manufacturer's directions (Clontech Laboratories, Mountain View, CA, USA) to identify the remainder of the X. laevis cDNA IL-34 transcript (Acc. No.: KJ776754). All primer sequences are available upon request.
Protein-sequence alignments were performed using the Clustal W software (http://www.ebi.ac.uk/clustalw/). Signal-peptide regions were identified using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). Phylogenetic analysis was performed by Clustal X software, using the neighbor-joining method and bootstrapped 10,000 times, with values expressed as percentages.
Production of rXlCSF-1 and rXlIL-34
The production of the rXlCSF-1 has been described previously [40]. The rXlIL-34 was produced by transfecting Sf9 insect cells (Cellfectin II; Invitrogen, Carlsbad, CA, USA) with the pMIB/V5 HisA insect-expression vector (Invitrogen), containing the X. laevis IL-34 sequence, corresponding to the signal peptide-cleaved fragment. Transfected Sf9 supernatants were confirmed to express rXlIL-34, and then positive transfectants were scaled up to 500 mL liquid cultures and grown for 5 days in blasticidin (10 μg/mL)-containing medium. Culture supernatants were dialyzed overnight (16 h) at 4°C against 150 mM sodium phosphate, concentrated against polyethylene glycol flakes (8 kDa) at 4°C, dialyzed overnight at 4°C against 150 mM sodium phosphate, and passage through nickel-nitriloacetic acid agarose columns (Qiagen, Valencia, CA, USA). Columns were washed with 2 × 10 vol high-stringency wash buffer (0.5% Tween 20; 50 mM sodium phosphate; 500 mM sodium chloride; 100 mM imidazole) and 5× with low-stringency wash buffer (as above, but with 40 mM imidazole). The rXlIL-34 was eluted in fractions using 250 mM imidazole. The purity of the eluted fractions was confirmed by silver stain, and the presence of rXlIL-34 was assessed by Western blot using the V5 epitope on rXlIL-34. The rXlIL-34-containing fractions were pooled and the protein concentration determined by the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA, USA). A protease inhibitor cocktail (Roche, Minneapolis, MN, USA) was added to the purified rXlIL-34 and the protein aliquoted and stored at −20°C until use.
The vector control was produced by transfecting Sf9 cells in parallel to the rXlIL-34 production but with an empty expression vector and the methodology described for the generation and isolation of the rXlIL-34.
Cell culture media
The amphibian ASF culture medium used in these studies has been described previously [16]. All cell cultures were established using ASF, supplemented with 10% FBS, 2.5% heat-inactivated X. laevis serum, 20 μg/mL kanamycin, and 100 U/mL penicillin/100 μg/mL streptomycin (Gibco, Life Technologies, Grand Island, NY, USA). APBS has been described previously [16].
Isolation of rXlCSF-1- and rXlIL-34-elicited tadpole macrophages
Tadpoles at developmental stage 54 [41, 42] were i.p. injected with 10 μl of the vector control or 1, 10, 100, or 1000 ng rXlCSF-1 or rXlIL-34 in 10 μl vol. After 1, 3, and 6 days following injections, peritoneal macrophages were collected by lavage with 50 μl vol APBS. Alternatively, tadpoles were injected with a combination of rXlCSF-1 and rXlIL-34 (1 μg each) or 1 μg rXlCSF-1, followed 36 h later by 1 μg rXlIL-34; phagocytes were isolated after 72 h. Cells were enumerated using trypan blue exclusion, cytospun, and stained with Giemsa (Sigma-Aldrich, St. Louis, MO, USA). To acquire sufficient RNA for cDNA synthesis, peritoneal macrophages from two individual tadpoles were pooled, with a total of six pooled fractions (n=6) used per treatment group.
FV3 stocks and animal infections
FHM cells (ATCC No. CCL-42; American Type Culture Collection, Manassas, VA, USA) were maintained in DMEM (Invitrogen), supplemented with 10% FBS (Invitrogen), penicillin (100 U/mL), and streptomycin (100 μg/mL), with 5% CO2 at 30°C. FV3 was grown by a single passage on FHM cells, purified by ultracentrifugation on a 30% sucrose cushion, and quantified by plaque assay on a baby hamster kidney cell monolayer under an overlay of 1% methylcellulose [15].
Tadpoles and adult frogs were infected by i.p. injections with 1 × 104 and 5 × 106 PFUs of FV3, respectively. Three and 6 dpi, animals were euthanized by immersion in 0.5% tricaine methane sulfonate (MS-222) and tissues removed and processed for RNA isolation.
For in vitro FV3 infection studies, tadpoles were i.p. injected with 1 μg rXlCSF-1; rXlIL-34; a combination of both recombinant cytokines (1 μg each); 1 μg rXlCSF-1, followed 36 h later by 1 μg rXlIL-34; or equal volumes of the vector control. Three days later, peritoneal macrophages were isolated by lavage with APBS, and cells were seeded into individual wells of 96-well plates at a density of 1 × 104 cells/well. The derived macrophages were infected at a MOI of 0.5 and cells harvested 2 and 24 h postinfection. To obtain sufficient RNA for cDNA synthesis and qRT-PCR analysis, cells from two individual animals were pooled subsequent to infections and immediately before Trizol RNA extraction. Six such pools were used per treatment group (n=6). For the detection of infectious viral loads by plaque assays, macrophages were not pooled, and cells from six individual tadpoles (n=6) were examined per treatment group by plaque assays, as described above.
For tadpole survival studies, stage 50 tadpoles (10/treatment group; n=10) were injected with 1 μg rXlCSF-1, rXlIL-34, or equal volumes of the vector control and 3 days later, challenged with 1 × 104 PFU of FV3. Tadpoles were checked twice daily, and dead animals were immediately frozen and stored at −20°C for DNA isolation.
Quantitative gene-expression analysis
Total RNA and DNA were extracted from frog tissues and cells using the Trizol reagent following the manufacturer's directions (Invitrogen). All cDNA synthesis was performed using the iScript cDNA synthesis kit, according to manufacturer's directions (Bio-Rad Laboratories) using 500 ng total DNAse-treated (Ambion) RNA.
Relative qRT-PCR gene-expression analyses (CSF-1, IL-34, IL-10, TNF-α, type I IFN, Mx1, p67phox, gp91phox, iNOS, IDO, MHC class I and II, β2m, XNC10, 64R, vDNA Pol II, and MCP) were performed via the ΔΔCT method, with expression examined relative to the GAPDH endogenous control and normalized against the lowest observed expression or vector controls (tissues and peritoneal macrophages, respectively). Absolute qPCR was performed to measure FV3 viral loads in isolated DNA, using a serially diluted standard curve. Briefly, the pGEM-T vector (Promega, Madison, WI, USA), bearing a fragment of the FV3 vDNA Pol II PCR, was amplified in Escherichia coli, quantified, and serially diluted to yield 1010–101 plasmid copies of the vDNA Pol II. These dilutions were used as a standard curve in subsequent absolute qPCR experiments to assess the viral genome transcript copy numbers relative to this standard curve. All experiments were performed using the ABI 7300 real-time PCR system and PerfeCTa SYBR Green FastMix, ROX (Quanta Biosciences, Gaithersburg, MD, USA). Expression analysis was performed using ABI Sequence Detection System software. All primers were validated before use. All primer sequences are available upon request.
Phagocytosis assays
Stage 54 tadpoles were injected with 1 μg rXlCSF-1, rXlIL-34, or equal volumes of the vector control. Three days later, peritoneal macrophages were lavaged with APBS and seeded at a density of 104 cells/well into individual wells of 96-well plates. Cells were incubated overnight (16 h) with 1 μm FITC latex beads (Duke Scientific, Palo Alto, CA, USA). Attached but uningested beads were removed by trypsin treatment and the efficiency of bead removal confirmed by light and fluorescence microscopy. Cells were washed and resuspended in APBS and assessed by flow cytometry using a FACSCanto II (BD Biosciences, San Jose, CA, USA). Data analysis was performed using FlowJo software. A gate excluding FITC-negative cells was used to measure phagocytosis. By back-gating, using the FlowJo software, we established that all phagocytic events were mediated by the large leukocyte populations (forward/side-scatter). We then established a gate to describe these large phagocytes and determined the phagocytic indexes as a measure of the percent of FITC-positive events within this gate for respective vector-, rXlCSF-1-, and rXlIL-34-derived peritoneal leukocyte cultures.
Statistical analysis
Statistical analysis was performed using a one-way ANOVA and Tukey's post hoc test using the VassarStats (http://faculty.vassar.edu/lowry//anova1u.html) statistical program. Statistical analysis of survival data was performed using a Log-rank test (GraphPad Prism 6). Probability level of P < 0.05 was considered significant.
RESULTS
In silico and tissue gene-expression analyses of X. laevis CSF-1 and IL-34
To investigate the respective roles of CSF-1 and IL-34 in amphibian antiviral defenses, we identified the X. laevis IL-34 gene homolog using conventional RACE-PCR and primers designed against the X. tropicalis IL-34 transcript. The derived X. laevis IL-34 cDNA transcript encodes a putative 169-residue protein with a conventional signal sequence and a globular domain, characteristic of IL-34 proteins (data not shown). As is the case with all vertebrate IL-34 genes, the X. laevis IL-34 gene (BLAST-identified on Xenebase) is comprised of six exons (data not shown).
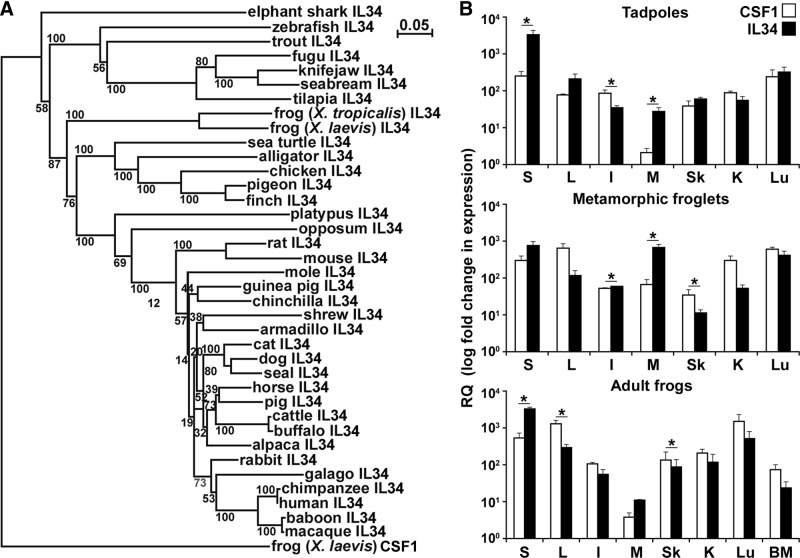
We conducted a phylogenetic comparison of the frog (X. laevis and X. tropicalis) IL-34 with other deduced vertebrate IL-34 protein sequences, using the X. laevis CSF-1 as an outgroup (Fig. 1A). The recently identified elephant shark IL-34 putative protein branched independently from and ancestrally to the other vertebrate IL-34 molecules (Fig. 1A). The fish IL-34 branched as a separate clade from the amphibian IL-34, which also formed a separate clade, ancestral to the reptile, avian, and mammalian IL-34 (Fig. 1A). The reptilian and avian IL-34 sequences formed a separate clade from the marsupial, monotreme, and mammalian IL-34 (Fig. 1A).
Figure 1. Characterization of X. laevis IL-34 (A) phylogeny and (B) CSF-1 and IL-34 quantitative tissue gene expression.
(A) The phylogenetic tree was constructed from multiple deduced protein sequence alignments using the neighbor-joining method and bootstrapped 10,000 times (denoted as percent). (B) Outbred premetamorphic (stage 54) tadpoles and metamorphic (stage 64) and adult (2 years old) frog tissues were assessed. Tissues from three individuals of each stage were examined (n=3). *P < 0.05, statistical difference between indicated groups. Tissues examined: S, spleen; L, liver; I, intestine; M, muscle; Sk, skin; K, kidney; Lu, lung; BM, bone marrow. RQ, Relative quantification.
To discern possible differences in the physiological roles of the X. laevis CSF-1 and IL-34 gene homologs, we compared the tissue gene expression of these two cytokines in tadpoles (stage 54, 2 weeks of age; refs. [41, 42]) and metamorphic (stage 64; 4–5 weeks of age) and adult frogs (2 years old) by qRT-PCR (Fig. 1B). The IL-34 gene expression was particularly prominent in tadpole spleens and was significantly more robust than that of CSF-1 in this organ (Fig. 1B). The splenic IL-34 mRNA levels decreased during metamorphosis but were restored in adult frogs to levels comparable with those detected in tadpoles (Fig. 1B). In contrast, the splenic CSF-1 gene expression marginally increased from tadpoles to metamorphic and adult frogs and remained markedly lower than that of IL-34 at all stages of development (Fig. 1B). Contrary to the splenic expression patterns, the liver CSF-1 transcript levels significantly increased during metamorphosis and remained elevated in adult frogs, whereas the IL-34 liver mRNA levels were modest at all developmental stages examined (Fig. 1B). Whereas the gene expression of CSF-1 and IL-34 increased in the muscle tissues during metamorphosis (derived from regressing tails), the magnitude of this transcriptional increase was significantly greater for the IL-34 gene expression (Fig. 1B). Interestingly, the CSF-1 gene expression in the skin of metamorphic and adult X. laevis was significantly greater than that for IL-34 (Fig. 1B), which is in contrast to the postulated involvement of IL-34 in mammalian Langerhans cell differentiation and maintenance [35]. Notably, MHC class IIhigh Langerhans-like cells have been described in adult Xenopus skin [43]. Whereas CSF-1 gene expression in the kidney was not altered during development, the IL-34 transcript levels were decreased during metamorphosis but were restored in adult frogs to levels comparable with those seen in tadpoles (Fig. 1B). Expression of both genes increased in the lung during development. Finally, CSF-1 and to a lesser extent, IL-34 genes were expressed in the adult bone marrow (Fig. 1B).
rXlCSF-1 and rXlIL-34 elicit the recruitment and differentiation of morphologically distinct mononuclear phagocytes
In accordance with the roles of vertebrate CSF-1 molecules in phagocyte recruitment and differentiation [44, 45], we assessed the capacities of rXlCSF-1 or rXlIL-34 to chemoattract tadpole macrophages upon i.p. injections (Fig. 2). Three days postinjection, both recombinant growth factors resulted in concentration-dependent recruitment of myeloid cells into tadpole peritoneal cavities, with the 100- and 1000-ng doses resulting in significant cell accumulation compared with vector controls (Fig. 2A). The accumulation of cells in response to either recombinant cytokine was much less prominent after 1 and 6 days following cytokine administration (data not shown).
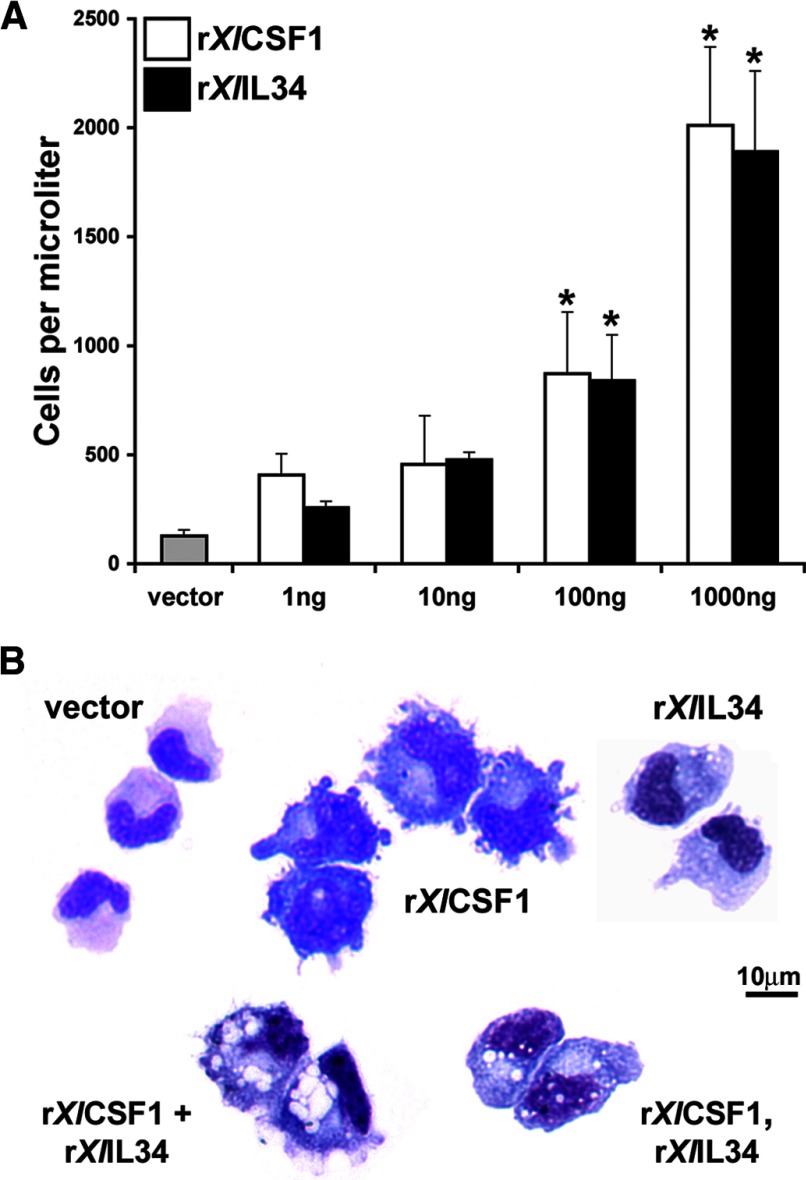
Figure 2. The rXlCSF-1 and rXlIL-34 cytokines elicit morphologically distinct tadpole peritoneal macrophages.
Stage 54 tadpoles were i.p. injected with vector control, 1, 10, 100, or 1000 ng rXlCSF-1 or rXlIL-34. Three days postinjections, peritoneal macrophages were isolated. (A) rXlCSF-1 and rXlIL-34 concentration-dependent elicitation of tadpole peritoneal macrophages. *P < 0.05, statistical difference compared with the vector-treatment group. (B) Giemsa-stained peritoneal phagocytes, isolated from tadpoles, 3 days postinjection, with vector control; 1 μg rXlCSF-1; 1 μg rXlIL-34; a combination of rXlCSF-1 and rXlIL-34 (1 μg each); or 1 μg rXlCSF-1, followed 36 h later by 1 μg rXlIL-34.
We assessed the morphology of recruited cells on Giemsa-stained cytocentrifuged preparations (Fig. 2B). Whereas the peritoneal phagocytes isolated from vector control-injected tadpoles primarily exhibited monocyte and granulocyte morphologies, the cells isolated from rXlCSF-1-treated tadpoles were primarily mononuclear phagocytes that appeared larger, highly vacuolated, and bearing many pseudopodia projections (Fig. 2B). Furthermore, injection of tadpoles with 1, 10, 100, and 1000 ng rXlCSF-1 resulted in, respectively, 14.9 ± 1.5, 51.1 ± 6.5, 72.2 ± 8.1, and 86.0 ± 3.5 mean ± sem percent of total peritoneal leukocytes exhibiting this large, vacuolated, ruffled mononuclear-phagocyte morphology. In contrast, the majority of the cells derived from rXlIL-34-injected animals was predominantly reminiscent of the monocytic fractions seen in the vector controls but with some vacuolation and minimal pseudopod projections (Fig. 2B). Intriguingly, the phagocytes isolated from tadpoles injected with a 1:1 mixture of rXlCSF-1 and rXlIL-34 resembled those derived with rXlCSF-1 alone (Fig. 2B). Conversely, the cells isolated from animals initially injected with rXlCSF-1 and subsequently administered rXlIL-34 36 h later were morphologically more similar to those derived with rXlIL-34 alone (Fig. 2B). The combination and sequential injections of rXlCSF-1 and rXlIL-34 did not result in further increases in the numbers of elicited leukocytes compared with those obtained with the respective individual cytokine treatments (data not shown).
rXlIL-34 and rXlCSF-1 confer distinct effects on tadpole susceptibility to FV3
In light of the critical roles of macrophage-lineage cells during ranaviral infections [15, 16], we examined the effects of preinjecting tadpoles with rXlCSF-1 or rXlIL-34, 3 days before FV3 infection, on their survival (Fig. 3A). Intriguingly, following FV3 infections, tadpoles prestimulated with rXlCSF-1 displayed significantly diminished survival times compared with vector control-treated cohorts, whereas pretreatment with rXlIL-34 significantly increased tadpole survival (Fig. 3A). The mean percent survival for FV3-infected tadpoles pretreated with rXlCSF-1 (53.13%) or rXlIL-34 (75.63%,) was statistically different from those pretreated with vector controls (61.25%). The postmortem tadpole FV3 loads did not significantly differ across the three treatment groups. The tadpole postmortem virus loads, as determined by qPCR, were: 1.07 ± 0.43 × 107 for the vector-control group; 0.74 ± 0.19 × 107 for the XlCSF-1-treatment group; and 0.93 ± 0.22 × 107 for rXlIL-34-treatment group (mean±sem FV3 viral DNA copies/50 ng total DNA).
Figure 3. Tadpoles pretreated with rXlCSF-1 and rXlIL-34 and peritoneal macrophages derived from these animals display distinct susceptibility to FV3.
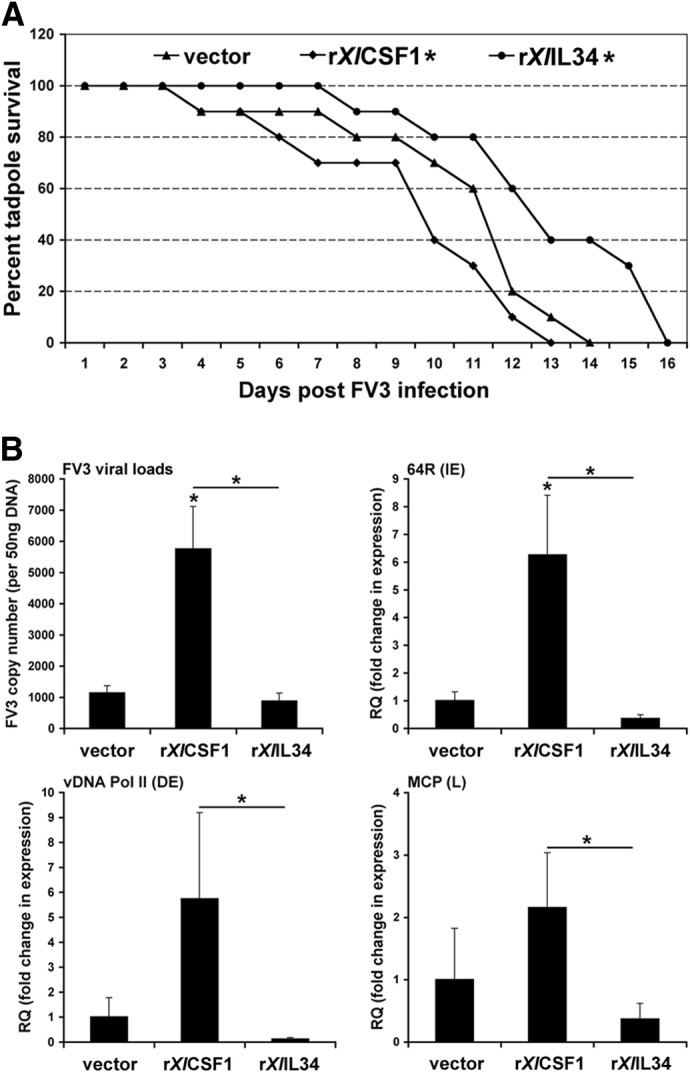
(A) Administration of rXlCSF-1 to tadpoles before FV3 challenge decreases their survival time, whereas preinjection with rXlIL-34 increases tadpole survival. Stage 50 tadpoles (10/treatment group; n=10) were injected with rXlCSF-1, rXlIL-34 (1 μg/tadpole), or equal volume of vector control. Three days postinjection, animals were infected i.p. with 1 × 104 PFU of FV3 (in APBS) or sham infected with equal volumes of APBS. Animal survival was monitored over the course of 16 days. *P < 0.05, statistical difference relative to vector controls. (B) Tadpole rXlCSF-1-pMϕ infected in vitro with FV3 exhibit enhanced FV3 susceptibility, higher viral loads, and increased viral gene expression. Tadpole rXlIL-34-pMϕ and rXlCSF-1-pMϕ were infected at a MOI of 0.5 with FV3 for 24 h, and then viral loads and gene expression were assessed by qPCR. *P < 0.05, statistical difference between indicated groups. IE, Immediate-early; DE, delayed-early; L, late.
rXlCSF-1-derived tadpole macrophages are more susceptible to FV3
We next examined whether peritoneal macrophages, isolated from tadpoles, administered with either vector control, rXlCSF-1 (rXlCSF-1-pMϕ) or rXlIL-34 (rXlIL-34-pMϕ), displayed distinct susceptibilities to FV3 infections in vitro. Accordingly, tadpole peritoneal phagocytes were isolated 3 days after the respective treatments and infected at a 0.5 MOI of FV3. At 24 h postinfection, the FV3 viral loads, as well as the expression of immediate-early (64R), delayed-early (vDNA Pol II), and late (MCP) genes, were assessed by qPCR. Consistent with our observation that rXlCSF-1 rendered tadpoles more susceptible to FV3 (Fig. 3A), rXlCSF-1-pMϕ were also substantially more susceptible to in vitro infections with FV3 compared with the vector control or rXlIL-34-pMϕ (Fig. 3B). The rXlCSF-1-pMϕ exhibited significantly greater FV3 loads, as well as higher expression of the immediate-early 64R gene than the control or rXlIL-34-pMϕ (Fig. 3B). Additionally, rXlCSF-1-pMϕ displayed significantly increased transcript levels of the delayed-early vDNA Pol II and late MCP genes compared with rXlIL-34-pMϕ (Fig. 3B), although it should be noted that MCP was very basally expressed (very high CT values). Notably, we did not observe substantial cell death between and within the different tadpole macrophage cultures 24 h after in vitro FV3 infections. Furthermore, the cell counts of these respective cultures were not significantly altered after 24 h, negating substantial cell proliferation during this time frame (data not shown).
rXlCSF-1-pMϕ exhibit significantly enhanced phagocytic capacities
As FV3 appears to target and gain entry into macrophage-lineage cells as the result of their extensive phagocytic and endocytic potentials, we examined whether differences in the phagocytic capacities of rXlCSF-1-pMϕ and rXlIL-34-pMϕ could account for their differences in FV3 susceptibility. Accordingly, peritoneal phagocytes from tadpoles, pretreated with vector control, rXlCSF-1, or rXlIL-34, were incubated overnight (16 h) with 1 μm FITC latex beads. Phagocytosis of these particles was assessed the following day by flow cytometry (Fig. 4A and B). Compared with vector control and rXlIL-34-pMϕ, the rXlCSF-1-pMϕ ingested significantly more latex beads, resulting in drastic changes in their internal complexities (Fig. 4A). The phagocytes derived from the vector control-treated tadpoles had modest phagocytic capacities, whereas rXlIL-34-pMϕ displayed the least phagocytosis (Fig. 4A and B). Indeed, whereas the phagocytic indexes between the vector control and rXlCSF-1-pMϕ were not statistically different, the rXlCSF-1-pMϕ and rXlIL-34-pMϕ exhibited statistical differences in their respective phagocytic indexes (Fig. 4B).
Figure 4. Tadpole rXlCSF-1-pMϕ are more phagocytic, whereas rXlIL-34-pMϕ are more antiviral.
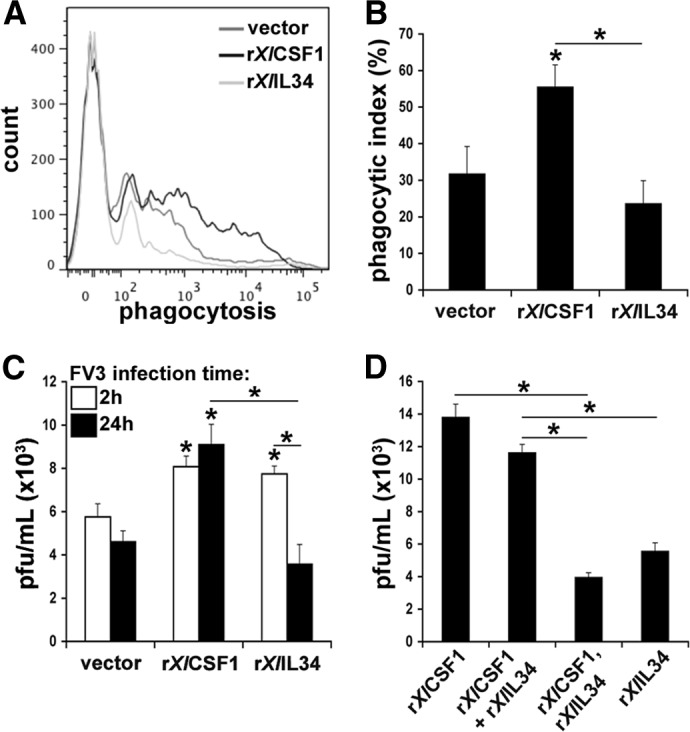
(A) rXlCSF-1-pMϕ, rXlIL-34-pMϕ, and vector control-elicited tadpole peritoneal phagocytes were incubated overnight (16 h) with 1 μm FITC latex beads and analyzed by flow cytometry. Cells from two individuals were pooled immediately before analysis, with six separate pools assessed per treatment group (n=6). (B) Phagocytic indexes of rXlCSF-1-pMϕ, rXlIL-34-pMϕ, and control phagocyte cultures. (C) rXlCSF-1-pMϕ and rXlIL-34-pMϕ exhibit similar FV3 internalization, whereas rXlIL-34-pMϕ eliminate a significant proportion of the virus. rXlCSF-1-pMϕ, rXlIL-34-pMϕ, and control phagocyte cultures were infected in vitro at a MOI of 0.5 and cells harvested for plaque assay analyses at 2 and 24 h. (D) rXlCSF-1 + rXlIL-34-pMϕ are as susceptible to FV3 as rXlCSF-1-pMϕ, whereas macrophages from tadpoles injected with rXlCSF-1 and 36 h later, with rXlIL-34 (rXlCSF-1, rXlIL-34-pMϕ) are significantly more resistant to in vitro FV3 infection. rXlCSF-1-pMϕ, rXlIL-34-pMϕ, rXlCSF-1 + rXlIL-34-pMϕ, and rXlCSF-1, as well as rXlIL-34-pMϕ cultures, were infected in vitro at a MOI of 0.5 for 24 h before harvesting the cells for plaque assay analyses. (B–D) Results are means ± sem. *P < 0.05, significant difference from vector controls; *P < 0.05 (when over bars), significant differences between treatment groups denoted by the bars.
Disparity in FV3 susceptibility of rXlCSF-1-pMϕ and rXlIL-34-pMϕ is not phagocytosis dependent
To evaluate whether the higher phagocytic activity of the rXlCSF-1-pMϕ was responsible for their relatively greater susceptibility to FV3, we pulsed vector-derived phagocytes, rXlCSF-1-pMϕ, and rXlIL-34-pMϕ with FV3 (2 h) or infected them for 24 h before harvesting the cells and determining intracellular FV3-infectious loads by plaque assays (Fig. 4C). According to previous studies [46], 2 h is sufficient for viral entry but not for the generation of virus progeny, whereas 24 h should suffice for several rounds of viral replication. Intriguingly and in contrast to our hypothesis that the rXlCSF-1-pMϕ would exhibit higher FV3 loads as a result of increased phagocytosis of viral particles, rXlCSF-1-pMϕ and rXlIL-34-pMϕ possessed significantly greater FV3 loads at 2 h compared with peritoneal phagocytes from vector-control tadpoles (Fig. 4C). However, whereas the viral loads within the control and rXlCSF-1-pMϕ were not significantly altered at 24 h compared with 2 h postinfection, the rXlIL-34-pMϕ displayed significantly diminished FV3 loads at 24 h (Fig. 4C). This indicates that FV3 does not actively expand within the control peritoneal phagocytes or recombinant cytokine-derived tadpole macrophages and suggests that in addition, the rXlIL-34-pMϕ are capable of reducing a larger proportion of their initial intracellular FV3 loads than the control or rXlCSF-1-pMϕ populations.
Consistent with our observations that macrophages isolated from tadpoles coadministered with both cytokines simultaneously resembled rXlCSF-1-pMϕ (Fig. 2B), in vitro FV3 infection of macrophages derived by combined rXlCSF-1 and rXlIL-34 treatment exhibited similar viral loads to macrophages derived only by rXlCSF-1 (Fig. 4D). Intriguingly, in vitro FV3 infection of macrophages isolated from animals, first injected with rXlCSF-1 and then administered rXlIL-34 36 h later, yielded significantly lower viral loads than those derived by rXlCSF-1, rXlCSF-1 + rXlIL-34, or even rXlIL-34 treatments (Fig. 4D).
Tadpole rXlCSF-1-pMϕ and rXlIL-34-pMϕ exhibit distinct immune gene expression
As our results indicated disparate antiviral roles for the X. laevis CSF-1- and IL-34-derived macrophages, we assessed the immune gene-expression profiles of rXlCSF-1-pMϕ and rXlIL-34-pMϕ to account for these functional differences (Fig. 5). Interestingly, rXlIL-34-pMϕ but not rXlCSF-1-pMϕ exhibited significantly elevated type I IFN gene expression (Fig. 5A), likely accounting for the enhanced antiviral properties of these cells (Figs. 3 and 4). The rXlIL-34-pMϕ and rXlCSF-1-pMϕ had significantly elevated mRNA levels for Mx1 (Fig. 5A), whereas the expression of IL-10 and TNF-α was not significantly different between the different phagocyte populations (Fig. 5A). Furthermore, only the rXlIL-34-pMϕ exhibited significantly up-regulated gene expression of the NADPH oxidase components p67phox and gp91phox, as well as robust, albeit not statistically significant, up-regulation of iNOS (Fig. 5B). Expression of the IDO gene was not significantly different among the examined cell populations. Of an additional note, the rXlIL-34-pMϕ also displayed increased but highly variable expression of MHC class I and II, as well as significantly increased β2m gene expression, whereas expression of these genes was markedly lower in rXlCSF-1-pMϕ (Fig. 5C). The gene expression of the XNC class Ib XNC10 and the CSF-1R was similar across the different phagocyte populations (Fig. 5C, and data not shown).
Figure 5. Quantitative immune gene-expression analysis of rXlCSF-1-pMϕ and rXlIL-34-pMϕ.
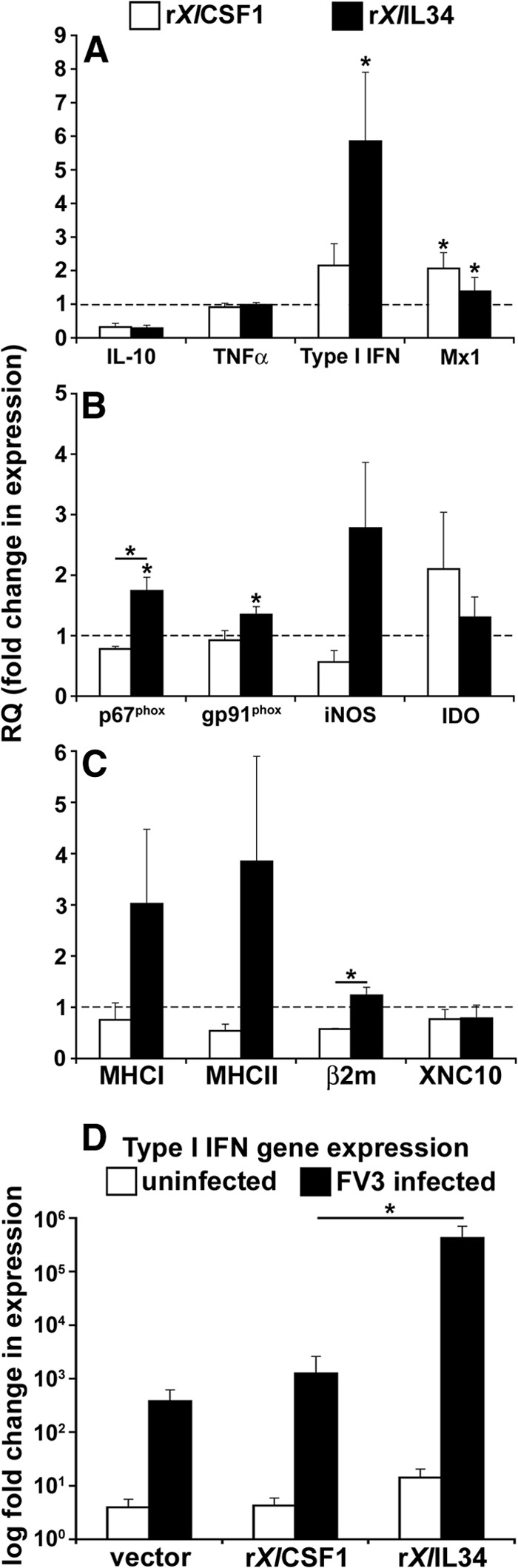
Three days after administration of recombinant cytokines, rXlCSF-1-pMϕ and rXlIL-34-pMϕ, and vector-control peritoneal phagocytes were isolated and assessed by qRT-PCR for immune genes expression of: (A) IL-10, TNF-α, IFN, and Mx1 (Mx1 relative quantification [RQ] values × 10) (B) p67phox, gp91phox, iNOS, and IDO; (C) MHC class I and II, β2m, and XNC10. Gene expression was examined relative to GAPDH control and normalized against respective vector-control gene expression (horizontal dashed lines). (D) Type I IFN gene expression in uninfected and FV3-infected, vector-control leukocytes (vector), rXlCSF-1-pMϕ (rXlCSF1), and rXlIL-34-pMϕ (rXlIL34). All FV3-infected cell populations possessed significantly greater IFN expression than respective uninfected controls. Results are means±sem. *P < 0.05, significant difference from vector controls; *P < 0.05 (when over bars), significant differences between treatment groups denoted by the bars.
To obtain further evidence of the potential antiviral roles of rXlIL-34-pMϕ during FV3 infections, we elicited vector-control leukocytes, rXlCSF-1-pMϕ and rXlIL-34-pMϕ, as before and infected them for 24 h with FV3 before assessing their type I IFN gene expression (Fig. 5D). The FV3-induced increases of type I IFN gene expression were comparable between rXlCSF-1-pMϕ and vector-control cultures, whereas in stark contrast, the FV3-challenged rXlIL-34-pMϕ displayed several logs greater IFN transcript levels (Fig. 5D). This further corroborates the notion that rXlIL-34-pMϕ are more efficient antiviral effectors, at least with respect to FV3.
X. laevis tadpoles and adults exhibit distinct kidney CSF-1 and IL-34 gene-expression profiles during FV3 infections
As our findings suggested that the rXlCSF-1-pMϕ and rXlIL-34-pMϕ conferred distinct roles in amphibian anti-FV3 immune responses, we examined the gene expression of these two cytokines in the kidneys (central site of FV3 replication) of the susceptible tadpoles and resistant adult frogs during FV3 infections (Fig. 6). The mRNA levels for CSF-1 significantly increased in the kidneys of tadpoles and adult frogs at 6 FV3 dpi. In stark contrast, only the adult frogs but not the tadpole X. laevis underwent significant up-regulation of IL-34 kidney gene expression in response to the ranaviral infections (Fig. 6).
Figure 6. Quantitative gene expression of CSF-1 and IL-34 in kidneys of FV3-infected tadpole and adult X. laevis.
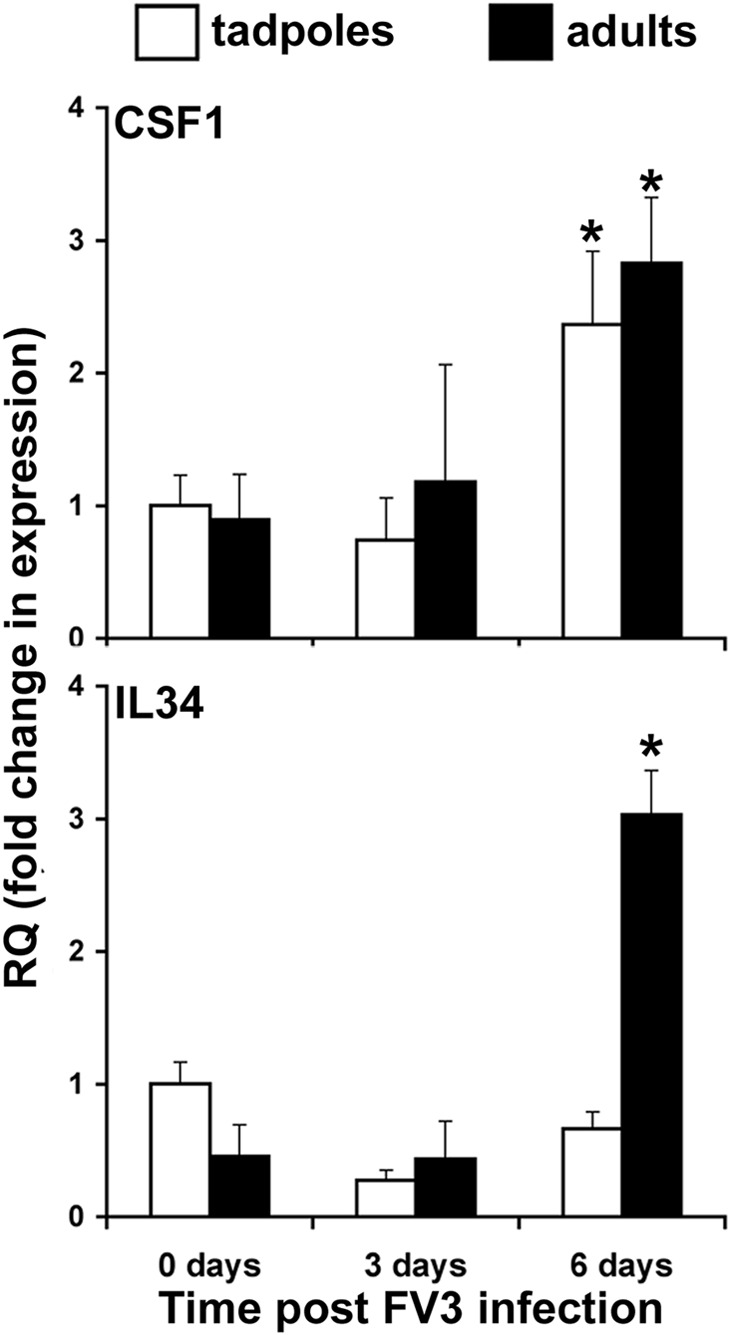
Tadpoles and adult frogs were infected i.p. with 1 × 104 and 5 × 106 PFU of FV3, respectively. At indicated times, animals were euthanized and kidneys collected from six individuals/treatment group (n=6). Gene expression was performed relative to the GAPDH endogenous control and results presented as mean relative quantification ± sem. *P < 0.05, significantly different from the uninfected controls.
DISCUSSION
This is the first account of distinct immunological roles for IL-34 and CSF-1 in vertebrate macrophage antiviral immunity. Notably, structural studies have revealed that although the CSF-1R binds both of these ligands, it does so through distinct surfaces, forming unique receptor conformations and intermolecular interfaces, culminating in unique downstream signaling events [28–30]. These features likely reflect the distinct, functional capacities of phagocytes derived by these respective macrophage growth factors. Considering the notion that IL-34 and CSF-1 drive the differentiation and/or polarization of functionally nonoverlapping macrophage populations largely through the same receptor, the physiological regulation of these events likely occurs, at least in part, through distinct spatiotemporal gene expression of these two growth factors. In fact, this appears to be the case across all vertebrates examined to date. For example, the roles of IL-34 in microglia biology are supported by the robust IL-34 (but not CSF-1) gene expression in mammalian brains during embryogenesis and into adult life [34, 35]. Furthermore, CSF-1-deficient (op/op) mice do not exhibit compensatory IL-34 expression [34]. This suggests that not only are these two genes transcriptionally unrelated, but their respective biological roles are (at least in part) not complementary. Our findings provide further evidence for the nonoverlapping roles of these two cytokines. Indeed, we observed that the X. laevis CSF-1 and IL-34 exhibit strikingly distinct tissue gene-expression patterns across developing animals and during antiviral responses of tadpole and adult frogs. This further corroborates our functional data, which also indicates that these two growth factors have adopted unique roles in macrophage immunity.
Interestingly, the significantly more robust splenic expression of the Xenopus IL-34 coincides with increased immunocompetent properties of tadpole macrophages derived by this growth factor compared with CSF-1. Likewise, the mammalian IL-34 has been recently described for its roles in developing follicular dendritic myeloid cells with B cell-stimulating properties [39]. Considering that the amphibian spleen (in absence of lymph nodes) represents a primary and the main secondary lymphoid tissue, it is possible that the high splenic IL-34 gene expression in X. laevis reflects a similar role. It is equally probable that the CSF-1-derived X. laevis tadpole macrophages will prove to be more effective at dealing with distinct pathogenic challenges. In this regard, the tadpole macrophages induced by this growth factor had significantly increased expression of the antiviral Mx1 gene and extensively more potent phagoctyic capacities, all of which may be effective at eliminating pathogens distinct from RVs. Notably, human CSF-1-derived macrophages are less susceptible to HIV-1 than cells derived by IL-34 [28]. Indeed, CSF-1 stimulation is known to elicit responses of an array of immune genes, including cathepsins, distinct chemokines, FcγRs, and scavenger receptors [47]. While many of the counterpart amphibian genes await functional characterization, their future transcriptional assessment in tadpole CSF1- and IL34-macrophages may lend added perspective to the evolutionary and immunological roles of these phagocyte populations.
Although the X. laevis tadpole IL-34-derived macrophages appear to possess substantially more anti-FV3 activity, we do not believe that CSF-1 stimulation ablates macrophage antiviral efficacies or inhibits type I IFN function. In fact, as described above, human CSF-1-derived macrophages are actually more resistant to HIV-1 than IL-34-Mϕ [28]. It is also noteworthy that following FV3-challenge, the rXlCSF-1-Mϕ up-regulated their gene expression of type I IFN, albeit to a much lower magnitude than rXlIL-34-Mϕ. Additionally, both cytokine-derived macrophage populations expressed comparable levels of the type I IFN-inducible antiviral Mx1 gene, whereas peritoneal macrophages isolated from tadpoles, coinjected with rXlIFN and rXlCSF-1, exhibited commensurable Mx1 expression levels to cells from tadpoles administered rXlIFN alone (data not shown).
The nonoverlapping immune functions of these macrophage growth factors are further substantiated by the observations that overexpression of IL-34, but not of CSF-1, is highly associated with severity of rheumatoid arthritis [48, 49]. Indeed, IL-34 gene expression appears to be linked more intrinsically to inflammatory cues than that of CSF-1, whereas proinflammatory cytokines, such as TNF-α and IL-1β, preferentially up-regulate the transcription of IL-34 through NF-κB-, JNK-, and p44/42 MAPK-mediated pathways [48–50]. Consistent with this notion, the gene expression of the recently identified trout IL-34 is also preferentially up-regulated over CSF-1 in trout cell lines, as well as in primary macrophage cultures following pathogen-associated molecular pattern and proinflammatory cytokine stimulations [51], possibly underlining the evolutionary conservation of the immune contexts of this macrophage growth factor. On this note, we have previously observed that FV3-infected X. laevis tadpoles are inefficient in their kidney proinflammatory (TNF-α, IL-1β) gene expression, as compared to adult frogs [52]. This deficiency may contribute to the susceptibility of tadpoles and may account for the lack of IL-34 gene induction in tadpole kidneys during infections.
In further corroboration with the above, our present findings indicate that whereas CSF-1-derived tadpole macrophages are permissive to FV3, those harvested from IL-34-administered tadpoles are more resistant to infection. Considering that the amphibian kidney is the central site of FV3 replication [15], it is noteworthy that even though tadpoles are capable of up-regulating kidney CSF-1 gene expression, the tadpole kidney IL-34 gene expression remains unchanged during FV3 infections. Thus, the respective susceptibility and resistance of X. laevis tadpoles and adults to FV3 may be defined, at least in part, by effective expression of IL-34 and the presence of macrophage populations derived by this growth factor. Furthermore, we previously demonstrated that whereas the X. laevis type I IFN is a potent antiviral mediator, X. laevis tadpoles do not sufficiently increase the kidney expression of this gene during FV3 infections [5]. Interestingly, our present findings indicate that rXlIL-34-pMϕ but not rXlCSF-1-pMϕ display significantly up-regulated type I IFN transcript levels. Thus, tadpole susceptibility to FV3 may be marked by their inability to generate and/or increase the numbers of IL-34-induced macrophages within infected kidneys, where these animals would suffer from the absence of a crucial type I IFN-producing cell population at the central virally targeted organ. Moreover, in the absence of IL-34-derived macrophages, FV3 would encounter only the CSF-1 macrophages, which according to our findings, would enhance the tadpole susceptibility to this virus.
Our observations indicate that although FV3 undergoes active expression of select genes within infected macrophages, the virus does not significantly expand within these cells. Rather, it appears that roughly the amount of infectious FV3 particles that have successfully penetrated macrophage hosts is maintained within these cells, possibly reflecting an FV3 dissemination strategy. In support of this, we observed that the FV3 MCP gene is poorly expressed in infected macrophages, suggesting inefficient assembly of new virions within these cells. Indeed, FV3 infections result in considerable apoptotic death of epithelial cells, such as the A6 kidney cell line, but much less so frog phagocytes. In corroboration with this and irrespective of treatment, we did not observe substantial death of tadpole phagocytes over the 24 h in vitro infection period. We also did not observe notable increases in cell counts that would indicate macrophage proliferation. Furthermore, we did not detect increased viral particles in CSF1- and IL34-derived macrophages over 24 h, suggesting that FV3 does not rely on the proliferative state of these cell populations. In this regard, CSF-1- and IL-34-derived macrophages appear to be equally permissive to the initial FV3 entry, but the latter population markedly decreases its intracellular viral loads, possibly as the result of increased autocrine type I IFN stimulation and reactive oxygen intermediate production (up-regulated NADPH oxidase components).
It is interesting that even though the pretreatment of tadpoles with rXlCSF-1 significantly reduced their mean survival following FV3 challenge and that rXlIL-34 conferred significantly prolonged tadpole survival, irrespectively, all animal-treatment groups examined had similar postmortem FV3 loads. Likewise, we recently demonstrated that although X. laevis tadpoles stimulated with type I IFN before FV3 inoculation exhibited several logs lower viral loads, they nonetheless incurred extensive infection-related tissue damage and eventually (although significantly later) succumbed to this viral infection [5]. This suggests that tadpole susceptibility reflects detrimental pathologies that may be induced even by low FV3 titers. With respect to our present findings, the prolonged (IL-34-stimulated) tadpole survival would likely reflect the delayed viral dissemination and concomitant tissue damage. Where Xenopus macrophages serve as vectors for RV dissemination, the decreased and increased FV3 loads within respective IL-34 and CSF-1 macrophages likely account for the delayed and expedited tadpole mortality, whereas the similar postmortem loads possibly mark the eventuality of the spread of this highly infectious virus.
It is intriguing that whereas tadpole macrophages derived by the combination of rXlCSF-1 and rXlIL-34 resembled the rXlCSF-1-Mϕ morphologically and with respect to FV3 susceptibility, the macrophages elicited by injecting tadpoles first with CSF-1 and later with IL-34 resembled the rXlIL-34-Mϕ in these respects. Notably, compared with the mammalian CSF-1, IL-34 possesses significantly lower affinity for the CSF-1R [34], so a similar relationship between the amphibian cytokines and CSF-1R may account for why the combination treatments resulted in cells resembling rXlCSF-1-Mϕ. Conversely, with sequential treatment, CSF-1 may be recruiting and initiating tadpole macrophage differentiation, whereas the later IL-34 administration is plausibly, functionally polarizing these cells toward rXlIL-34-Mϕ. At this time, it is difficult to speculate on why the sequential rXlCSF-1, rXlIL-34 treatment resulted in cells that were even more resistant to in vitro FV3 challenges than the rXlIL-34-Mϕ. Possibly, CSF-1 may function as a more global macrophage growth factor, whereas IL-34 may confer further immune-related roles in functionally polarizing the CSF-1-differentiated macrophages. Such a physiological strategy would manifest in the results described here, where in vivo CSF-1-differentiated, IL-34-polarized macrophages would possess more effective antiviral properties than cells derived with IL-34 alone.
The current limitation of X. laevis-specific reagents prevents more selective characterization of peritoneal phagocyte subsets based on specific cell-surface markers. Nevertheless, we feel that our present findings are of pivotal importance, as they clearly underline very disparate roles for amphibian CSF-1 and IL-34 in macrophage antiviral immunity. Furthermore, although the purity of the rXlIL-34-pMϕ and rXlCSF-1-pMϕ cultures is not to the degree as would be permitted by the availability of Xenopus macrophage-specific antibodies, these cultures did express very similar levels of the CSF-1R gene and were morphologically comprised of primarily mononuclear phagocytes (>80%).
IL-34 orthologs have not only been found across all classes of vertebrates [24, 51, 53, 54], but it would also seem that this gene actually predates CSF-1, as an IL-34 but no CSF-1 homolog has been recently identified in elasmobranchs [55]. At this time, it is difficult to speculate whether our findings pertaining to the functional differences between X. laevis IL-34- and CSF-1-derived tadpole macrophages extend to evolutionarily and physiologically distinct vertebrate species or reflect unique physiological and immunological strategies on the part of amphibians. Garnering insight into the respective biological roles of these macrophage growth factors across multiple, physiologically distinct species will be intriguing from an evolutionary perspective and may well reveal an added parameter to vertebrate antiviral-immune defenses.
ACKNOWLEDGMENTS
This work was supported by the U.S. National Institutes of Health (R24-AI-059830) and National Science Foundation (IOB-074271) grants to J.R. L.G. was supported by a National Sciences and Engineering Research Council of Canada Postdoctoral Fellowship and a Life Sciences Research Fellowship from the Howard Hughes Medical Institute.
We thank Tina Martin for animal husbandry, Dr. Eva-Stina Eldhom for her critical review of this manuscript, and Dr. Yuko Ota for kindly providing us with the elephant shark IL-34 sequence. This manuscript was improved by the insightful comments of two anonymous reviewers.
Footnotes
- β2m
- β2-microglobulin
- APBS
- amphibian PBS
- ASF
- amphibian serum free medium
- CT
- comparative threshold
- dpi
- days postinfection
- FHM
- fathead minnow
- FV3
- frog virus 3
- MCP
- major capsid protein
- MOI
- multiplicity of infection
- Pol II
- polymerase II
- qRT-PCR
- quantitative RT-PCR
- RV
- ranavirus
- rXlCSF-1
- recombinant Xenopus laevis CSF-1
- rXlIFN
- recombinant Xenopus laevis interferon
- rXlCSF-1-pMϕ
- recombinant Xenopus laevis CSF-1-derived peritoneal macrophages
- rXlIL-34
- recombinant Xenopus laevis IL-34
- rXlIL-34-pMϕ
- recombinant Xenopus laevis IL-34-derived peritoneal macrophages
- vDNA Pol II
- viral DNA polymerase II
- XNC
- Xenopus nonclassical
AUTHORSHIP
L.G. and J.R. designed and planned the studies, analyzed the data, wrote the manuscript, and prepared the figures. L.G. performed the experiments.
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1. Chinchar V. G. (2002) Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch. Virol. 147, 447–470. [DOI] [PubMed] [Google Scholar]
- 2. Williams T., Barbosa-Solomieu V., Chinchar V. G. (2005) A decade of advances in iridovirus research. Adv. Virus Res. 65, 173–248. [DOI] [PubMed] [Google Scholar]
- 3. Chinchar V. G., Hyatt A., Miyazaki T., Williams T. (2009) Family Iridoviridae: poor viral relations no longer. Curr. Top. Microbiol. Immunol. 328, 123–170. [DOI] [PubMed] [Google Scholar]
- 4. Bayley A. E., Hill B. J., Feist S. W. (2013) Susceptibility of the European common frog Rana temporaria to a panel of ranavirus isolates from fish and amphibian hosts. Dis. Aquat. Organ. 103, 171–183. [DOI] [PubMed] [Google Scholar]
- 5. Grayfer L., De Jesus Andino F., Robert J. (2014) The amphibian (Xenopus laevis) type I interferon response to frog virus 3: new insight into ranavirus pathogenicity. J. Virol. 88, 5766–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoverman J. T., Gray M. J., Miller D. L. (2010) Anuran susceptibilities to ranaviruses: role of species identity, exposure route, and a novel virus isolate. Dis. Aquat. Organ. 89, 97–107. [DOI] [PubMed] [Google Scholar]
- 7. Landsberg J. H., Kiryu Y., Tabuchi M., Waltzek T. B., Enge K. M., Reintjes-Tolen S., Preston A., Pessier A. P. (2013) Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Dis. Aquat. Organ. 105, 89–99. [DOI] [PubMed] [Google Scholar]
- 8. Reeve B. C., Crespi E. J., Whipps C. M., Brunner J. L. (2013) Natural stressors and ranavirus susceptibility in larval wood frogs (Rana sylvatica). Ecohealth 10, 190–200. [DOI] [PubMed] [Google Scholar]
- 9. Jancovich J. K., Bremont M., Touchman J. W., Jacobs B. L. (2010) Evidence for multiple recent host species shifts among the ranaviruses (family Iridoviridae). J. Virol. 84, 2636–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jancovich J. K., Davids E. W., Seiler A., Jacobs B. L., Collins J. P. (2001) Transmission of the Ambystoma tigrinum virus to alternative hosts. Dis. Aquat. Organ. 46, 159–163. [DOI] [PubMed] [Google Scholar]
- 11. Speare R., Smith J. R. (1992) An iridovirus-like agent isolated from the ornate burrowing frog Limnodynastes ornatus in northern Australia. Dis. Aquat. Organ. 14, 51–57. [Google Scholar]
- 12. Becker J. A., Tweedie A., Gilligan D., Asmus M., Whittington R. J. (2003) Experimental infection of Australian freshwater fish with epizootic haematopoietic necrosis virus (EHNV). J. Aquat. Anim. Health 25, 66–76. [DOI] [PubMed] [Google Scholar]
- 13. Haislip N. A., Gray M. J., Hoverman J. T., Miller D. L. (2011) Development and disease: how susceptibility to an emerging pathogen changes through anuran development. PLoS One 6, e22307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hyatt A. D., Gould A. R., Zupanovic Z., Cunningham A. A., Hengstberger S., Whittington R. J., Kattenbelt J., Coupar B. E. (2000) Comparative studies of piscine and amphibian iridoviruses. Arch. Virol. 145, 301–331. [DOI] [PubMed] [Google Scholar]
- 15. Morales H. D., Abramowitz L., Gertz J., Sowa J., Vogel A., Robert J. (2010) Innate immune responses and permissiveness to ranavirus infection of peritoneal leukocytes in the frog Xenopus laevis. J. Virol. 84, 4912–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robert J., Abramowitz L., Gantress J., Morales H. D. (2007) Xenopus laevis: a possible vector of ranavirus infection? J. Wildl. Dis. 43, 645–652. [DOI] [PubMed] [Google Scholar]
- 17. Coiras M., Lopez-Huertas M. R., Perez-Olmeda M., Alcami J. (2009) Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 7, 798–812. [DOI] [PubMed] [Google Scholar]
- 18. Goodenow M. M., Rose S. L., Tuttle D. L., Sleasman J. W. (2003) HIV-1 fitness and macrophages. J. Leukoc. Biol. 74, 657–666. [DOI] [PubMed] [Google Scholar]
- 19. Gousset K., Ablan S.D., Coren L.V., Ono A., Soheilian F., Nagashima K., Ott D.E., Freed E. O. (2008) Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog. 4, e1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Groot F., Welsch S., Sattentau Q. J. (2008) Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111, 4660–4663. [DOI] [PubMed] [Google Scholar]
- 21. Wang T., Hanington P. C., Belosevic M., Secombes C. J. (2008) Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J. Immunol. 181, 3310–3322. [DOI] [PubMed] [Google Scholar]
- 22. Pixley F. J., Stanley E. R. (2004) CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 14, 628–638. [DOI] [PubMed] [Google Scholar]
- 23. Hanington P. C., Wang T., Secombes C. J., Belosevic M. (2007) Growth factors of lower vertebrates: characterization of goldfish (Carassius auratus L.) macrophage colony-stimulating factor-1. J. Biol. Chem. 282, 31865–31872. [DOI] [PubMed] [Google Scholar]
- 24. Garceau V., Smith J., Paton I. R., Davey M., Fares M. A., Sester D. P., Burt D. W., Hume D. A. (2010) Pivotal Advance: Avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J. Leukoc. Biol. 87, 753–764. [DOI] [PubMed] [Google Scholar]
- 25. Dai X. M., Ryan G. R., Hapel A. J., Dominguez M. G., Russell R. G., Kapp S., Sylvestre V., Stanley E. R. (2002) Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120. [DOI] [PubMed] [Google Scholar]
- 26. Lichanska A. M., Browne C. M., Henkel G. W., Murphy K. M., Ostrowski M. C., McKercher S. R., Maki R. A., Hume D. A. (1999) Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU. 1. Blood 94, 127–138. [PubMed] [Google Scholar]
- 27. Guilbert L. J., Stanley E. R. (1980) Specific interaction of murine colony-stimulating factor with mononuclear phagocytic cells. J. Cell Biol. 85, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chihara T., Suzu S., Hassan R., Chutiwitoonchai N., Hiyoshi M., Motoyoshi K., Kimura F., Okada S. (2010) IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 17, 1917–1927. [DOI] [PubMed] [Google Scholar]
- 29. Liu H., Leo C., Chen X., Wong B. R., Williams L. T., Lin H., He X. (2014) The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1. Biochim. Biophys. Acta 1824, 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma X., Lin W. Y., Chen Y., Stawicki S., Mukhyala K., Wu Y., Martin F., Bazan J. F., Starovasnik M. A. (2012) Structural basis for the dual recognition of helical cytokines IL-34 and CSF-1 by CSF-1R. Structure 20, 676–687. [DOI] [PubMed] [Google Scholar]
- 31. Belosevic M., Hanington P. C., Barreda D. R. (2006) Development of goldfish macrophages in vitro. Fish Shellfish Immunol. 20, 152–171. [DOI] [PubMed] [Google Scholar]
- 32. Droin N., Solary E. (2010) Editorial: CSF-1R, CSF-1, and IL-34, a “menage a trois” conserved across vertebrates. J. Leukoc. Biol. 87, 745–747. [DOI] [PubMed] [Google Scholar]
- 33. Lin H., Lee E., Hestir K., Leo C., Huang M., Bosch E., Halenbeck R., Wu G., Zhou A., Behrens D., Hollenbaugh D., Linnemann T., Qin M., Wong J., Chu K., Doberstein S. K., Williams L. T. (2008) Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 320, 807–811. [DOI] [PubMed] [Google Scholar]
- 34. Wei S., Nandi S., Chitu V., Yeung Y. G., Yu W., Huang M., Williams L. T., Lin H., Stanley E. R. (2010) Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J. Leukoc. Biol. 88, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greter M., Lelios I., Pelczar P., Hoeffel G., Price J., Leboeuf M., Kundig T. M., Frei K., Ginhoux F., Merad M., Becher B. (2012) Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y., Colonna M. (2014) Interkeukin-34, a cytokine crucial for the differentiation and maintenance of tissue resident macrophages and Langerhans cells. Eur. J. Immunol. 44, 1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baud'huin M., Renault R., Charrier C., Riet A., Moreau A., Brion R., Gouin F., Duplomb L., Heymann D. (2010) Interleukin-34 is expressed by giant cell tumours of bone and plays a key role in RANKL-induced osteoclastogenesis. J. Pathol. 221, 77–86. [DOI] [PubMed] [Google Scholar]
- 38. Chen Z., Buki K., Vaaraniemi J., Gu G., Vaananen H. K. (2011) The critical role of IL-34 in osteoclastogenesis. PLoS One 6, e18689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamane F., Nishikawa Y., Matsui K., Asakura M., Iwasaki E., Watanabe K., Tanimoto H., Sano H., Fujiwara Y., Stanley E. R., Kanayama N., Mabbott N. A., Magari M., Ohmori H. (2014) CSF-1 receptor-mediated differentiation of a new type of monocytic cell with B cell-stimulating activity: its selective dependence on IL-34. J. Leukoc. Biol. 95, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grayfer L., Robert J. (2013) Colony-stimulating factor-1-responsive macrophage precursors reside in the amphibian (Xenopus laevis) bone marrow rather than the hematopoietic subcapsular liver. J. Innate Immun. 5, 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thors F., de Kort E. J., Nieuwenhuys R. (1982) On the development of the spinal cord of the clawed frog, Xenopus laevis. I. Morphogenesis and histogenesis. Anat. Embryol. (Berl). 164, 427–441. [DOI] [PubMed] [Google Scholar]
- 42. Thors F., de Kort E. J., Nieuwenhuys R. (1982) On the development of the spinal cord of the clawed frog, Xenopus laevis. II. Experimental analysis of differentiation and migration. Anat. Embryol. (Berl). 164, 443–454. [DOI] [PubMed] [Google Scholar]
- 43. Ramanayake T., Simon D. A., Frelinger J. G., Lord E. M., Robert J. (2007) In vivo study of T-cell responses to skin alloantigens in Xenopus using a novel whole-mount immunohistology method. Transplantation 83, 159–166. [DOI] [PubMed] [Google Scholar]
- 44. Barreda D. R., Hanington P. C., Belosevic M. (2004) Regulation of myeloid development and function by colony stimulating factors. Dev. Comp. Immunol. 28, 509–554. [DOI] [PubMed] [Google Scholar]
- 45. Grayfer L., Hanington P. C., Belosevic M. (2009) Macrophage colony-stimulating factor (CSF-1) induces pro-inflammatory gene expression and enhances antimicrobial responses of goldfish (Carassius auratus L.) macrophages. Fish Shellfish Immunol. 26, 406–413. [DOI] [PubMed] [Google Scholar]
- 46. Majji S., Thodima V., Sample R., Whitley D., Deng Y., Mao J., Chinchar V. G. (2009) Transcriptome analysis of frog virus 3, the type species of the genus ranavirus, family Iridoviridae. Virology 391, 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beck A. H., Espinosa I., Edris B., Li R., Montgomery K., Zhu S., Varma S., Marinelli R. J., van de Rijn M., West R. B. (2009) The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin. Cancer Res. 15, 778–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chemel M., Le Goff B., Brion R., Cozic C., Berreur M., Amiaud J., Bougras G., Touchais S., Blanchard F., Heymann M. F., Berthelot J. M., Verrecchia F., Heymann D. (2012) Interleukin 34 expression is associated with synovitis severity in rheumatoid arthritis patients. Ann. Rheum. Dis. 71, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hwang S. J., Choi B., Kang S. S., Chang J. H., Kim Y. G., Chung Y. H., Sohn D. H., So M. W., Lee C. K., Robinson W. H., Chang E. J. (2012) Interleukin-34 produced by human fibroblast-like synovial cells in rheumatoid arthritis supports osteoclastogenesis. Arthritis Res. Ther. 14, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eda H., Shimada H., Beidler D. R., Monahan J. B. (2011) Proinflammatory cytokines, IL-1beta and TNF-alpha, induce expression of interleukin-34 mRNA via JNK- and p44/42 MAPK-NF-kappaB pathway but not p38 pathway in osteoblasts. Rheumatol. Int. 31, 1525–1530. [DOI] [PubMed] [Google Scholar]
- 51. Wang T., Kono T., Monte M. M., Kuse H., Costa M. M., Korenaga H., Maehr T., Husain M., Sakai M., Secombes C. J. (2013) Identification of IL-34 in teleost fish: differential expression of rainbow trout IL-34, MCSF-1 and MCSF2, ligands of the MCSF receptor. Mol. Immunol. 53, 398–409. [DOI] [PubMed] [Google Scholar]
- 52. De Jesus Andino F., Chen G., Li Z., Grayfer L., Robert J. (2012) Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus frog-virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology 432, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gow D. J., Garceau V., Kapetanovic R., Sester D. P., Fici G. J., Shelly J. A., Wilson T. L., Hume D. A. (2012) Cloning and expression of porcine colony stimulating factor-1 (CSF-1) and colony stimulating factor-1 receptor (CSF-1R) and analysis of the species specificity of stimulation by CSF-1 and interleukin 34. Cytokine 60, 793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gow D. J., Garceau V., Pridans C., Gow A. G., Simpson K. E., Gunn-Moore D., Hume D. A. (2013) Cloning and expression of feline colony stimulating factor receptor (CSF-1R) and analysis of the species specificity of stimulation by colony stimulating factor-1 (CSF-1) and interleukin-34 (IL-34). Cytokine 61, 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Venkatesh B., Lee A. P., Ravi V., Maurya A. K., Lian M. M., Swann J. B., Ohta Y., Flajnik M. F., Sutoh Y., Kasahara M., Hoon S., Gangu V., Roy S. W., Irimia M., Korzh V., Kondrychyn I., Lim Z. W., Tay B. H., Tohari S., Kong K. W., Ho S., Lorente-Galdos B., Quilez J., Marques-Bonet T., Raney B. J., Ingham P. W., Tay A., Hillier L. W., Minx P., Boehm T., Wilson R. K., Brenner S., Warren W. C. (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505, 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]