Genome function is regulated dynamically in part by chromatin, which consists of the histones, non-histone proteins and RNA molecules that package DNA. Studies in C. elegans and D. melanogaster have contributed significantly to our understanding of molecular mechanisms of genome function in humans, and revealed conservation of chromatin components and mechanisms1–3. Nevertheless, the three organisms have prominent differences in genome size, chromosome architecture, and gene organization. On human and fly chromosomes, for instance, pericentric heterochromatin flanks single centromeres, whereas worm chromosomes have dispersed heterochromatin-like regions enriched in the distal chromosomal ‘arms,’ and centromeres distributed along their lengths4,5. To systematically investigate chromatin organization and associated gene regulation across species, we generated and analyzed a large collection of genome-wide chromatin datasets from cell lines and developmental stages in worm, fly and human. Here we present over 800 new datasets from our ENCODE and modENCODE consortia, bringing the total to over 1400. Comparison of combinatorial patterns of histone modifications, nuclear lamina-associated domains, organization of large-scale topological domains, chromatin environment at promoters and enhancers, nucleosome positioning, and DNA replication patterns reveals many conserved features of chromatin organization among the three organisms. We also find significant differences, notably in the composition and locations of repressive chromatin. These datasets and analyses provide a rich resource for comparative and species-specific investigations of chromatin composition, organization, and function.
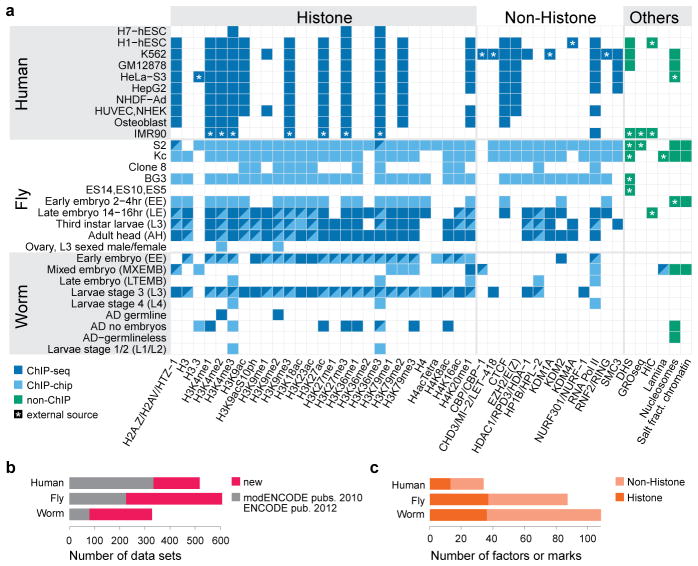
We used chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) or microarray hybridization (ChIP-chip) to generate profiles of core histones, histone variants, histone modifications, and chromatin-associated proteins (Fig. 1, Supplementary Fig. 1, Supplementary Tables 1, 2). Additional data include DNase I hypersensitivity sites in fly and human cells, and nucleosome occupancy maps in all three organisms. Compared to our initial publications1–3, this represents a tripling of available fly and worm datasets and a substantial increase in human datasets (Fig. 1b,c). Uniform quality standards for experimental protocols, antibody validation, and data processing were used throughout the projects6. All data are freely available at modMine (http://intermine.modencode.org), the project data portal (http://data.modencode.org), the ENCODE Data Coordination Center (http://genome.ucsc.edu/ENCODE), or our database and web application (http://encode-x.med.harvard.edu/data_sets/chromatin/) with faceted browsing that allows users to choose tracks for visualization or download. Detailed analyses of related transcriptome and transcription factor data are presented in accompanying papers7,8.
Fig. 1. Dataset overview.
a, Histone modifications, chromosomal proteins, and other profiles mapped in at least two species (see Supplementary Fig. 1 for full dataset and Supplementary Table 1 for detailed descriptions). Different protein names for orthologs are separated by slash. (see Supplementary Table 2). b, Number of all datasets generated by this and previous consortia publications1–3 (new: 815; old: 638). Each dataset corresponds to a replicate-merged normalized profile of a histone, histone variant, histone modification, non-histone chromosomal protein, nucleosome, or salt-fractionated nucleosome. c, Number of unique histone marks or non-histone chromosomal proteins profiled.
We performed systematic cross-species comparisons of chromatin composition and organization, focusing on targets profiled in at least two organisms (Fig. 1). Sample types utilized are human cell lines H1-hESC, GM12878 and K562; fly late embryos (LE), third instar larvae (L3) and cell lines S2, Kc, BG3; and worm early embryos (EE) and stage 3 larvae (L3). Our conclusions are summarized in Extended Data Table 1.
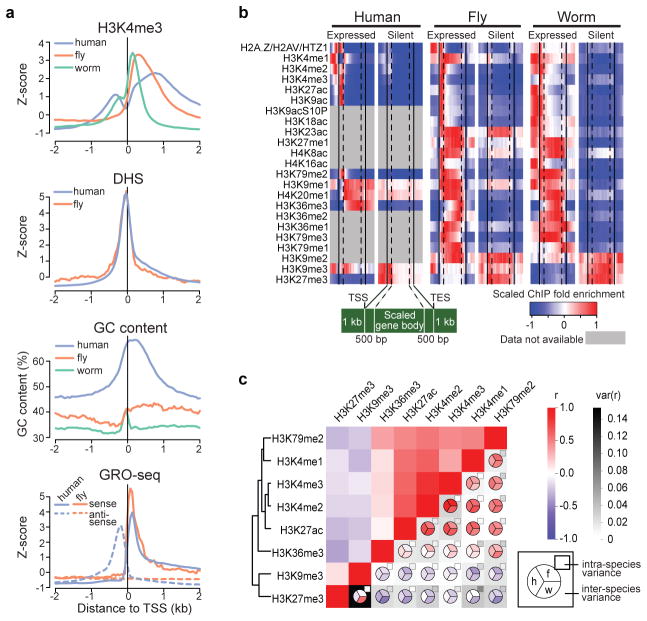
Not surprisingly, the three species show many common chromatin features. Most of the genome in each species is covered by at least one histone modification (Supplementary Fig. 2), and modification patterns are similar around promoters, gene bodies, enhancers, and other chromosomal elements (Supplementary Figs. 3 –12). Nucleosome occupancy patterns around protein-coding genes and enhancers are also largely similar across species, although we observed subtle differences in H3K4me3 enrichment patterns around transcription start sites (TSSs) (Extended Data Fig. 1a, Supplementary Figs. 12–14). The configuration and composition of large-scale features such as lamina-associated domains (LADs) are similar (Supplementary Figs. 15 –17). LADs in human and fly are associated with late replication and H3K27me3 enrichment, suggesting a repressive chromatin environment (Supplementary Fig. 18). Finally, DNA structural features associated with nucleosome positioning are strongly conserved (Supplementary Figs. 19, 20).
Although patterns of histone modifications across active and silent genes are largely similar in all three species9, there are some notable differences (Extended Data Fig. 1b). For example, H3K23ac is enriched at promoters of expressed genes in worm, but is enriched across gene bodies of both expressed and silent genes in fly. H4K20me1 is enriched on both expressed and silent genes in human but only on expressed genes in fly and worm (Extended Data Fig. 1b). Enrichment of H3K36me3 in genes expressed with stage- or tissue-specificity is lower than in genes expressed broadly, possibly because profiling was done on mixed tissues (Supplementary Figs. 21–23; see Supplementary Methods). While the co-occurrence of pairs of histone modifications are largely similar across the three species, there are clearly some species-specific patterns (Extended Data Fig. 1c, Supplementary Figs. 24, 25).
Previous studies showed that in human9,10 and fly1,11 prevalent combinations of marks or ‘chromatin states’ correlate with functional features such as promoters, enhancers, transcribed regions, Polycomb-associated domains, and heterochromatin. ‘Chromatin state maps’ provide a concise and systematic annotation of the genome. To compare chromatin states across the three organisms, we developed and applied a novel hierarchical non-parametric machine learning method called hiHMM (see Supplementary Methods) to generate chromatin state maps from eight histone marks mapped in common, and compared the results with published methods (Fig. 2; Supplementary Figs. 26–28). We find that combinatorial patterns of histone modifications are largely conserved. Based on correlations with functional elements (Supplementary Figs. 29–32), we categorized the 16 states into six groups: promoter (state 1), enhancer (states 2–3), gene body (states 4–9), Polycomb-repressed (states 10–11), heterochromatin (states 12–13), and weak or low signal (states 14–16).
Fig. 2. Shared and organism-specific chromatin states.
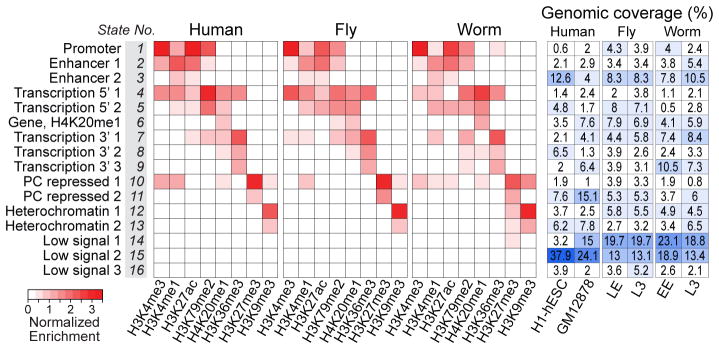
16 chromatin states derived by joint segmentation using hiHMM (hierarchical HMM; see Supplementary Methods) based on enrichment patterns of 8 histone marks. The genomic coverage of each state in each cell type or developmental stage is also shown (see Supplementary Figs. 26–32 for detailed analysis of the states). States are named for putative functional characteristics.
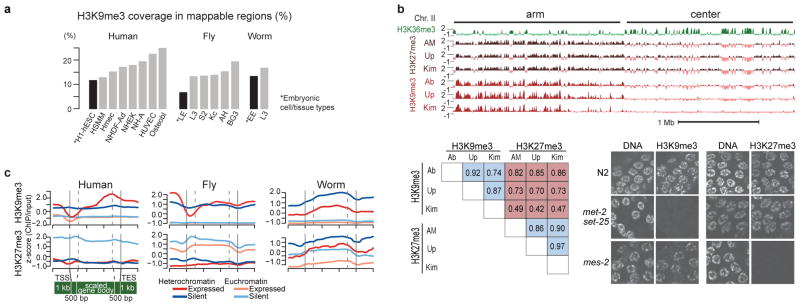
Heterochromatin is a classically defined and distinct chromosomal domain with important roles in genome organization, genome stability, chromosome inheritance, and gene regulation. It is typically enriched for H3K9me312, which we used as a proxy for identifying heterochromatic domains (Fig. 3a, Supplementary Figs. 33, 34). As expected, the majority of the H3K9me3-enriched domains in human and fly are concentrated in the pericentromeric regions (as well as other specific domains, such as the Y chromosome and fly 4th chromosome), whereas in worm they are distributed throughout the distal chromosomal ‘arms’11,13,14 (Fig. 3a). In all three organisms, we find that more of the genome is associated with H3K9me3 in differentiated cells/tissues compared to embryonic cells/tissues (Extended Data Fig. 2a). We also observe large cell-type-specific blocks of H3K9me3 in human and fly11,14,15 (Supplementary Fig. 35). These results suggest a molecular basis for the classical concept of “facultative heterochromatin” formation to silence blocks of genes as cells specialize.
Fig. 3. Genome-wide organization of heterochromatin.
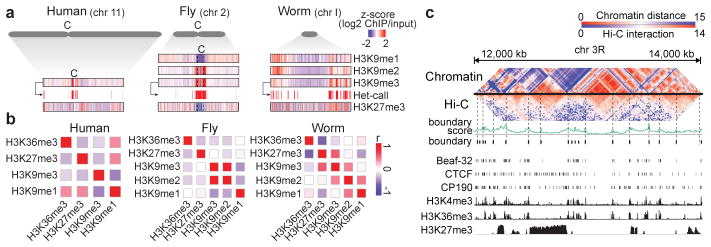
a, Enrichment profiles of H3K9me1/me2/me3 and H3K27me3 and identification of heterochromatin domains based on H3K9me3 (illustrated for human H1-hESC, fly L3, and worm L3). For fly chr2, 2L, 2LHet, 2RHet and 2R are concatenated (dashed lines); C: centromere, Het: heterochromatin. b, Genome-wide correlation among H3K9me1/me2/me3, H3K27me3, and H3K36me3 in human K562 cells, fly L3, and worm L3; no H3K9me2 profile is available for human. c, Comparison of Hi-C-based and chromatin-based topological domains in fly LE. Heatmaps of similarity matrices for histone modification and Hi-C interaction frequencies are juxtaposed (see Supplementary Fig. 40).
Two distinct types of transcriptionally-repressed chromatin have been described. As discussed above, classical ‘heterochromatin’ is generally concentrated in specific chromosomal regions and enriched for H3K9me3 and also H3K9me212. In contrast, Polycomb-associated silenced domains, involved in cell-type-specific silencing of developmentally regulated genes11,14, are scattered across the genome and enriched for H3K27me3. We found that the organization and composition of these two types of transcriptionally silent domains differ across species. First, human, fly, and worm display significant differences in H3K9 methylation patterns. H3K9me2 shows a stronger correlation with H3K9me3 in fly than in worm (r= 0.89 vs. r= 0.40, respectively), whereas H3K9me2 is well correlated with H3K9me1 in worm but not in fly (r= 0.44 vs. r= −0.32, respectively) (Fig. 3b). These findings suggest potential differences in heterochromatin in the three organisms (see below). Second, the chromatin state maps reveal two distinct types of Polycomb-associated repressed regions: strong H3K27me3 accompanied by marks for active genes or enhancers (Fig. 2, state 10; perhaps due to mixed tissues for fly and worm), and strong H3K27me3 without active marks (state 11) (see also Supplementary Fig. 31). Third, we observe a worm-specific association of H3K9me3 and H3K27me3. These two marks are enriched together in states 12 and 13 in worm but not in human and fly. This unexpected strong association between H3K9me3 and H3K27me3 in worm (observed with several validated antibodies; Extended Data Fig. 2b) suggests a species-specific difference in the organization of silent chromatin.
We also compared the patterns of histone modifications on expressed and silent genes in euchromatin and heterochromatin (Extended Data Fig. 2c, Supplementary Fig. 36). We previously reported prominent depletion of H3K9me3 at TSSs and high levels of H3K9me3 in the gene bodies of expressed genes located in fly heterochromatin14, and now find a similar pattern in human (Extended Data Fig. 2c, Supplementary Fig. 36). In these two species, H3K9me3 is highly enriched in the body of both expressed and silent genes in heterochromatic regions. In contrast, expressed genes in worm heterochromatin have lower H3K9me3 enrichment across gene bodies compared to silent genes (Extended Data Fig. 2c, Supplementary Figs. 36, 37). There are also conspicuous differences in the patterns of H3K27me3 in the three organisms. In human and fly, H3K27me3 is highly associated with silent genes in euchromatic regions, but not with silent genes in heterochromatic regions. In contrast, consistent with the worm-specific association between H3K27me3 and H3K9me3, we observe high levels of H3K27me3 on silent genes in worm heterochromatin, while silent euchromatic genes show modest enrichment of H3K27me3 (Extended Data Fig. 2c, Supplementary Fig. 36).
Our results suggest three distinct types of repressed chromatin (Extended Data Fig. 3). The first contains H3K27me3 with little or no H3K9me3 (human and fly states 10 and 11 and worm state 11), corresponding to developmentally regulated Polycomb-silenced domains in human and fly, and likely in worm as well. The second is enriched for H3K9me3 and lacks H3K27me3 (human and fly states 12 and 13), corresponding to constitutive, predominantly pericentric heterochromatin in human and fly, which is essentially absent from the worm genome. The third contains both H3K9me3 and H3K27me3 and occurs predominantly in worm (worm states 10, 12, and 13). Co-occurrence of these marks is consistent with the observation that H3K9me3 and H3K27me3 are both required for silencing of heterochromatic transgenes in worms16. H3K9me3 and H3K27me3 may reside on the same or adjacent nucleosomes in individual cells17,18; alternatively the two marks may occur in different cell types in the embryos and larvae analyzed here. Further studies are needed to resolve this and determine the functional consequences of the overlapping distributions of H3K9me3 and H3K27me3 observed in worm.
Genome-wide chromatin conformation capture (Hi-C) assays have revealed prominent topological domains in human19 and fly20,21. While their boundaries are enriched for insulator elements and active genes19,20 (Supplementary Fig. 38), the interiors generally contain a relatively uniform chromatin state - active, Polycomb-repressed, heterochromatin, or low signal22 (Supplementary Fig. 39). We found that chromatin state similarity between neighboring regions correlates with chromatin interaction domains determined by Hi-C (Fig. 3c, Supplementary Fig. 40, Supplementary Methods). This suggests that topological domains can be largely predicted by chromatin marks when Hi-C data are not available (Supplementary Figs. 41, 42).
C. elegans and D. melanogaster have been used extensively for understanding human gene function, development, and disease. Our analyses of chromatin architecture and the large public resource we have generated provide a blueprint for interpreting experimental results in these model systems, extending their relevance to human biology. They also provide a foundation for researchers to investigate how diverse genome functions are regulated in the context of chromatin structure.
Methods
For full details of Methods, see Supplementary Information.
Extended Data
Extended Data Fig. 1. Chromatin features at TSSs and gene bodies and co-occurrence of histone modifications.
a, Comparative analysis of promoter architecture at Transcription Start Sites (TSSs). From the top, H3K4me3 (human GM12878, fly L3, and worm L3), DNase I hypersensitivity sites (DHS), GC content, and nascent transcript (GRO-seq in human IMR90 and fly S2 cells). Human promoters, and to a lesser extent worm promoters (as defined using recently published capRNA-seq data23), exhibit a bimodal enrichment for H3K4me3 and other active marks around TSSs. In contrast, fly promoters clearly exhibit a unimodal distribution of active marks, downstream of TSSs. Since genes that have a neighboring gene within 1 kb of a TSS or TES (Transcription End Site) were removed from this analysis, any bimodal histone modification pattern cannot be attributed to nearby genes. This difference is also not explained by chromatin accessibility determined by DNase I hypersensitivity (DHS), or by fluctuations in GC content around the TSSs, although the GC profiles are highly variable across species. b, Average gene body profiles of histone modifications on protein-coding genes in human GM12878, fly L3, and worm L3. c, Genome-wide correlations between histone modifications show intra- and inter-species similarities and differences. Upper left half: pairwise correlations between marks in each genome, averaged across all three species. Lower right half: pairwise correlations, averaged over cell types and developmental stages, within each species (pie chart), with inter-species variance (grey-scale background) and intra-species variance (grey-scale small rectangles) of correlation coefficients for human (h), fly (f), and worm (w). Modifications enriched within or near actively transcribed genes are consistently correlated with each other in all three organisms. In contrast, we found a major difference in the co-occurrence pattern of two key repressive chromatin marks (black cell in bottom left): H3K27me3 (related to Polycomb (Pc)-mediated silencing) and H3K9me3 (related to heterochromatin). These two marks are strongly correlated at both developmental stages analyzed in worm, whereas their correlation is low in human (r = −0.24 ~ −0.06) and fly (r = −0.03 ~ −0.1).
Extended Data Fig. 2. Histone modifications in heterochromatin.
a, Genomic coverage of H3K9me3 in multiple cell types and developmental stages. Embryonic cell lines/stages are marked with an asterisk and a black bar. b, Evidence that overlapping H3K9me3 and H3K27me3 ChIP signals in worm are not due to antibody cross-reactivity. ChIP-chip experiments were performed from early embryo (EE) extracts with three different H3K9me3 antibodies (from Abcam, Upstate, and H. Kimura) and three different H3K27me3 antibodies (from Active Motif, Upstate, and H. Kimura). The H3K9me3 antibodies show similar enrichment profiles (upper panel) and high genome-wide correlation coefficients (lower left). The same is true for H3K27me3 antibodies. There is significant overlap between the H3K9me3 and H3K27me3 ChIP signal, especially on chromosome arms, resulting in relatively high genome-wide correlation coefficients (Extended Data Fig. 1c). The Abcam and Upstate H3K9me3 antibodies showed low level cross-reactivity with H3K27me3 on dot blots24, and the Abcam H3K9me3 ChIP signal overlapped with H3K27me3 on chromosome centers. The Kimura monoclonal antibodies against H3K9me3 and H3K27me3 showed the least overlap and smallest genome-wide correlation. In ELISA assays using histone H3 peptides containing different modifications, each Kimura H3K9me3 or H3K27me3 antibody recognized the modified tail against which it was raised and did not cross-react with the other modified tail25,26, providing support for their specificity. Specificity of the Kimura antibodies was further analyzed by immunostaining germlines from wild type, met-2 set-25 mutants (which lack H3K9 HMT activity16), and mes-2 mutants (which lack H3K27 HMT activity27) in the lower right panel. Staining with anti-HK9me3 was robust in wild type and in mes-2, but undetectable in met-2 set-25. Staining with anti-HK27me3 was robust in wild type and in met-2 set-25, but undetectable in mes-2. Finally, we note that the laboratories that analyzed H3K9me3 and H3K27me3 in other systems used Abcam H3K9me3 (for human and fly) and Upstate H3K27me3 (for human), and in these cases observed non-overlapping distributions. Chandra et al. also reported non-overlapping distributions of H3K9me3 and H3K27me3 in human fibroblast cells using the Kimura antibodies26. The overlapping distributions that we observe in worms using any of those antibodies suggest that H3K9me3 and H3K27me3 occupy overlapping regions in worms. Those overlapping regions may exist in individual cells or in different cell sub-populations in embryo and L3 preparations. c, Average gene body profiles of H3K9me3 and H3K27me3 on expressed and silent genes in euchromatin and heterochromatin in human K562 cells, fly L3, and worm L3.
Extended Data Fig. 3. Organization of silent domains.
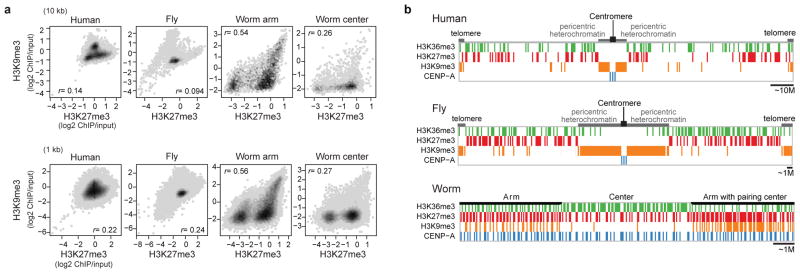
a, The correlation of H3K27me3 and H3K9me3 enrichment for human K562 (left most), fly L3 (second left), and worm EE chromosome arms (second right) and centers (right most) with a 10 kb bin (upper) and a 1 kb bin (lower). The density was calculated as a frequency of bins that fall in the area in the scatter plot (darker grey at a higher frequency). r indicates Pearson correlation coefficients between binned H3K27me3 fold enrichment (log2) and H3K9me3 fold enrichment (log2). Worm chromosome arms have a distinctly high correlation between H3K27me3 and H3K9me3. The lower correlation in worm chromosome centers is due to the overall absence of H3K9me3 in these regions. b, Schematic diagrams of the distributions of silent domains along the chromosomes in human (H1-hESC), fly (S2), and worm (EE). In human and fly, the majority of the H3K9me3-enriched domains are located in the pericentric regions (as well as telomeres), while the H3K27me3-enriched domains are distributed along the chromosome arms. H3K27me3-enriched domains are negatively correlated with H3K36me3-enriched domains, although in human, there is some overlap of H3K27me3 and H3K36me3 in bivalent domains. CENP-A resides at the centromere. In contrast, in worm the majority of H3K9me3-enriched domains are located in the arms, while H3K27me3-enriched domains are distributed throughout the arms and centers of the chromosomes and are anti-correlated with H3K36me3-enriched domains. In arms and centers, domains that are permissive for CENP-A incorporation generally reside within H3K27me3-enriched domains.
Extended Data Table 1.
Summary of key shared and organism-specific chromatin features in human, fly, and worm.
| Chromatin features | Human | Fly | Worm | Figures |
|---|---|---|---|---|
|
Promoters
| ||||
| H3K4me3 enrichment pattern around TSS | Bimodal peak | Unimodal peak* | Weak bimodal peak | ED1a,b,S12 |
| Well positioned +1 nucleosome at expressed genes | Yes | Yes | Yes | S13 |
|
| ||||
|
Gene bodies
| ||||
| Lower H3K36me3 in specifically expressed genes | Yes | Yes | Yes | S21–S23 |
|
| ||||
|
Enhancers
| ||||
| High H3K27ac sites are closer to expressed genes | Yes | Yes | Yes | S5–6 |
| Higher nucleosome turnover at high H3K27ac sites | Yes | Yes | ND | S7 |
|
| ||||
|
Nucleosome positioning
| ||||
| 10-bp periodicity profile | Yes | Yes | Yes | S19a |
| Positioning signal in genome | Weak | Weak | Less weak | S19b |
|
| ||||
|
LADs
| ||||
| Histone modification in short LADs | H3K27me3 | H3K27me3 | H3K27me3 | S17 |
| Histone modification in long LADs | H3K9me3 internal, H3K27me3 borders | ND | H3K9me3 +H3K27me3 | S15 |
| Associated with late replication in S-phase | Yes | Yes | ND | S18 |
|
| ||||
|
Genome-wide correlation
| ||||
| Correlation between H3K27me3 and H3K9me3 | Low | Low | High (in arms) | ED1c,ED3a |
|
| ||||
|
Chromatin state maps
| ||||
| Similar marks and genomic features at each state | Yes | Yes | Yes | 2,S29–32 |
|
| ||||
|
Silent domains: constitutive heterochromatin
| ||||
| Composition | H3K9me3 | H3K9me3 | H3K9me3 +H3K27me3 | 2,ED3b |
| Predominant location | Pericentric+chrY | Pericentric+chr4/Y | Arms | 3a,ED3b |
| Depletion of H3K9me3 at TSS of expressed genes | Yes | Yes | Weak | ED2c |
|
| ||||
|
Silent domains: Polycomb-associated
| ||||
| Composition | H3K27me3 | H3K27me3 | H3K27me3 | 2 |
| Predominant location | Arms | Arms+Chr4 | Arms+Centers | 3a,ED3b |
|
| ||||
|
Topological domains
| ||||
| Active promoters enriched at boundaries | Yes | Yes | ND | S38 |
| Similar chromatin states are enriched in each domain | Yes | Yes | ND | S39 |
Unimodal peak enriched downstream of TSS
ND: No Data
Supplementary Material
Acknowledgments
This project is mainly funded by NHGRI U01HG004258 (GHK, SCRE, MIK, PJP, VP), U01HG004270 (JDL, JA, AFD, XSL, SS), U01HG004279 (DMM), U54HG004570 (BEB) and U01HG004695 (WSN). It is also supported by NHBIB 5RL9EB008539 (JWKH), NHGRI K99HG006259 (MMH), NIGMS fellowships (SCJP, ENL), NIH U54CA121852 (TDT), NSF 1122374 (DSD), National Natural Science Foundation of China 31028011 (XSL), MEST Korea MHW-2013-HI13C2164 (JHK), NRF-2012-0000994 (K-AS), and Wellcome Trust 54523 (JA). We thank David Acevedo and Cameron Kennedy for technical assistance.
Footnotes
Author Contributions
Lead data analysis team: JWKH, YLJ, TL, BHA, SL, K-AS, MYT, SCJP, AK, EB, SSH, AR. Lead data production team: KI, AM, AA, TG, NCR, TAE, AAA, DA. (Ordered alphabetically) Data analysis team: JAB, DSD, XD, FF, NG, PH, MMH, PVK, NK, ENL, MWL, RP, NS, CW, HX; Data production team: SKB, QBC, RA-JC, YD, ACD, CBE, SE, JMG, DH, MH, TEJ, PK-Z, CVK, SAL, IL, XL, HNP, AP, BQ, PS, YBS, AV, CMW. NIH scientific project management: EAF, PJG, MJP. The role of the NIH Project Management Group was limited to coordination and scientific management of the modENCODE and ENCODE consortia. Paper writing: JWKH, YLJ, TL, BHA, SL, K-AS, MYT, SCJP, SSH, AR, KI, TDT, MK, DMM, SS, SCRE, XSL, JDL, JA, GHK, and PJP. Group leaders for data analysis or production: REK, JHK, BEB, AFD, VP, MIK, WSN, TDT, MK, DMM, SS, SCRE, JA, XSL, GHK, JDL, and PJP. Overall project management, and corresponding authors: DMM, SS, SCRE, XSL, JDL, JA, GHK, and PJP.
Completing Financial Interests
The authors declare no competing financial interests.
References
- 1.The modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gassmann R, et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484:534–7. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerstein MB, et al. Comparative analysis of the transcriptome across distant species. Nature. doi: 10.1038/nature13424. in submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyle AP, et al. Comparative analysis of regulatory information and circuits across distant species. Nature. doi: 10.1038/nature13668. in submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic acids research. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, et al. Broad Chromosomal domains of histone modification patterns in C. elegans. Genome Research. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riddle NC, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome research. 2011;21:147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towbin BD, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 17.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen RA, et al. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res. 2013;23:1339–1347. doi: 10.1101/gr.153668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egelhofer TA, et al. An assessment of histone-modification antibody quality. Nature Structural & Molecular Biology. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi-Takanaka Y, et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic acids research. 2011;39:6475–6488. doi: 10.1093/nar/gkr343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra T, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Molecular cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Current biology: CB. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.