Abstract
Apoptosis has essential roles in a variety of cellular and developmental processes. Although the pathway is well studied, how the activities of individual components in the pathway are regulated is less understood. In Drosophila, a key component in apoptosis is Drosophila inhibitor of apoptosis protein 1 (DIAP1), which is required to prevent caspase activation. Here, we demonstrate that Drosophila CG42593 (ubr3), encoding the homolog of mammalian UBR3, has an essential role in regulating the apoptosis pathway. We show that loss of ubr3 activity causes caspase-dependent apoptosis in Drosophila eye and wing discs. Our genetic epistasis analyses show that the apoptosis induced by loss of ubr3 can be suppressed by loss of initiator caspase Drosophila Nedd2-like caspase (Dronc), or by ectopic expression of the apoptosis inhibitor p35, but cannot be rescued by overexpression of DIAP1. Importantly, we show that the activity of Ubr3 in the apoptosis pathway is not dependent on its Ring-domain, which is required for its E3 ligase activity. Furthermore, we find that through the UBR-box domain, Ubr3 physically interacts with the neo-epitope of DIAP1 that is exposed after caspase-mediated cleavage. This interaction promotes the recruitment and ubiquitination of substrate caspases by DIAP1. Together, our data indicate that Ubr3 interacts with DIAP1 and positively regulates DIAP1 activity, possibly by maintaining its active conformation in the apoptosis pathway.
Morphogenesis in multicellular organisms is a process with a balanced control of cell proliferation and cell death. To maintain this homeostasis, superfluous or unwanted cells are usually removed promptly via programmed cell death or apoptosis.1, 2 Compelling evidence has shown that dysregulation of apoptosis results in a variety of diseases, such as cancer, neurodegenerative disorders and autoimmune diseases.3, 4, 5, 6 The apoptotic machinery is conserved from invertebrates to vertebrates. Drosophila has been used as an excellent model to study apoptosis because of its advantages in genetic manipulation. A crucial step in apoptosis is the cascade activation of initiator and effector caspases that eventually causes cell death. Under normal circumstances, the activities of caspases are kept in check by a conserved family of anti-apoptotic proteins termed inhibitor of apoptosis proteins (IAPs). The Drosophila genome encodes four IAPs, including Drosophila inhibitor of apoptosis protein 1 (DIAP1), DIAP2, DBruce and Deterin.7, 8, 9, 10 Among these four proteins, DIAP1 is stringently required to prevent caspase activation.11, 12 Although the requirement of DIAP1 in the apoptosis pathway is well documented, it is unclear how the activity of DIAP1 is regulated during development.
The covalent attachment of ubiquitin to proteins is a crucial regulatory mechanism in many developmental and physiological processes.13 Ubiquitination is a catalytic cascade involving ubiquitin-activating (E1), ubiquitin-conjugating (E2) and ubiquitin-ligating (E3) enzymes.14 The E3 proteins that specifically recognize a distinctive set of substrates for ubiquitination are an exceptionally large family.15 The RING domain of DIAP1 is an E3 ligase that inactivates caspases mainly through ubiquitination.16 Previous studies have shown that the anti-apoptotic activity of DIAP1 is negatively regulated by three pro-apoptotic proteins called Reaper, head involution defective (Hid) and Grim (RHG).2, 17 These proteins negatively regulate DIAP1 function through distinct mechanisms, either by disrupting interactions between DIAP1 and the initiator caspase Drosophila Nedd2-like caspase (Dronc), or by promoting the ubiquitination-dependent degradation of DIAP1.18, 19 In addition to regulation by RHG, DIAP1 has been considered a substrate of the N-end rule pathway. Ditzel et al.20 first discovered that DIAP1 can be cleaved by effector caspases at its NH2 terminus, exposing a binding motif for UBR-box-containing E3 ligases and subsequently be degraded by the N-end rule-mediated degradation. Later, Yokokura et al.21 reported that the N-end rule pathway does not have a major role in the turnover of N-terminally truncated DIAP1. More recently, Ditzel et al.22 reported that the UBR-binding motif of DIAP1 is essential for its anti-apoptotic function, indicating UBR family proteins regulate apoptosis via ways other than destroying DIAP1. Although these reports highlight the importance of UBR-box-containing E3 ligases in regulating DIAP1, it is currently unknown which UBR family member is involved in apoptosis.
The mammalian genome encodes seven evolutionarily conserved members of UBR E3 ligases (UBR1–UBR7).23, 24 All of the UBR E3 ligases share a ∼70-amino-acid UBR-box and can function as N-recognins, which are involved in the N-end rule pathway of the ubiquitination system.25 Among them, UBR1, UBR2 and UBR3 are subfamily members containing both a UBR-box and a Ring domain, which is required for its E3 ligase activity.26 Data from mouse models indicate that UBR1 and UBR2 are involved in the regulation of apoptosis in spermatocytes, skeletal muscle and cardiovascular development with partial redundancy.27, 28, 29 UBR3 was first characterized in mice as an E3 ligase involved in the regulation of olfactory and other sensory systems.30 Recently, human UBR3 was also found to be required for genome stability by regulating the essential DNA repair protein APE1.31 As UBR3 does not bind to the known N-end rule substrates of UBR1 and UBR2,30 it is currently unknown whether UBR3 is involved in the apoptosis pathway. Here, we have generated Drosophila ubr3 mutant and characterized its role in development. Our data suggest that Ubr3 is involved in the apoptosis pathway by regulating the activity of DIAP1 during development.
Results
Disruption of ubr3 results in impaired eye and wing growth
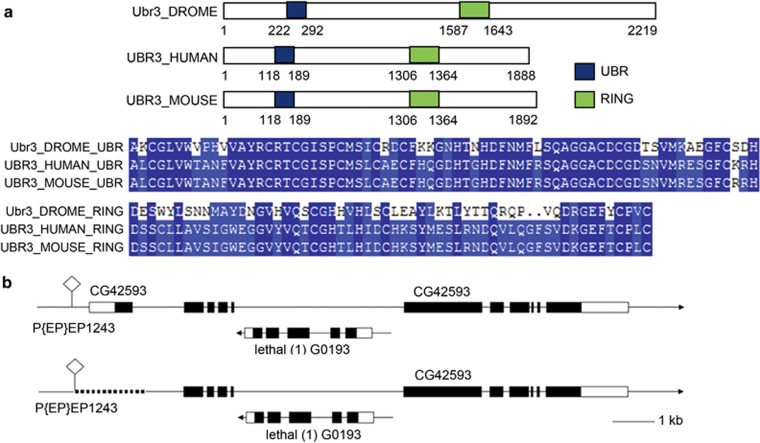
The establishment of a genome-wide RNA interference (RNAi) library has facilitated genetic screening for genes affecting particular pathways or biological processes in Drosophila. To identify E3 ligases that are involved in regulating eye development, we carried out a systematic RNAi screen by selectively knocking down individual E3 ligases in the developing eye of Drosophila. One of the candidates identified from the screen was CG42953, which is the Drosophila homolog of UBR3 (ubr3). The Drosophila CG42593 mRNA is predicted to encode a protein of 2219 amino acids with a theoretical molecular weight of 244 kDa. Domain analysis and BlastP search shows that the amino-acid sequence of the UBR-box domain of CG42593 is 39% identical and 51% similar to the human UBR3 (NP_742067.3) (Figure 1a), indicating that CG42593 encodes the homolog of mammalian UBR3.
Figure 1.
Genomic and protein structures of Ubr3. (a) UBR-box domain of Ubr3 is highly conserved among different species. (b) Generation of a null allele by P-element-mediated imprecise excision. The dotted line indicates the deleted region, which includes the start codon and portion of N-terminal region
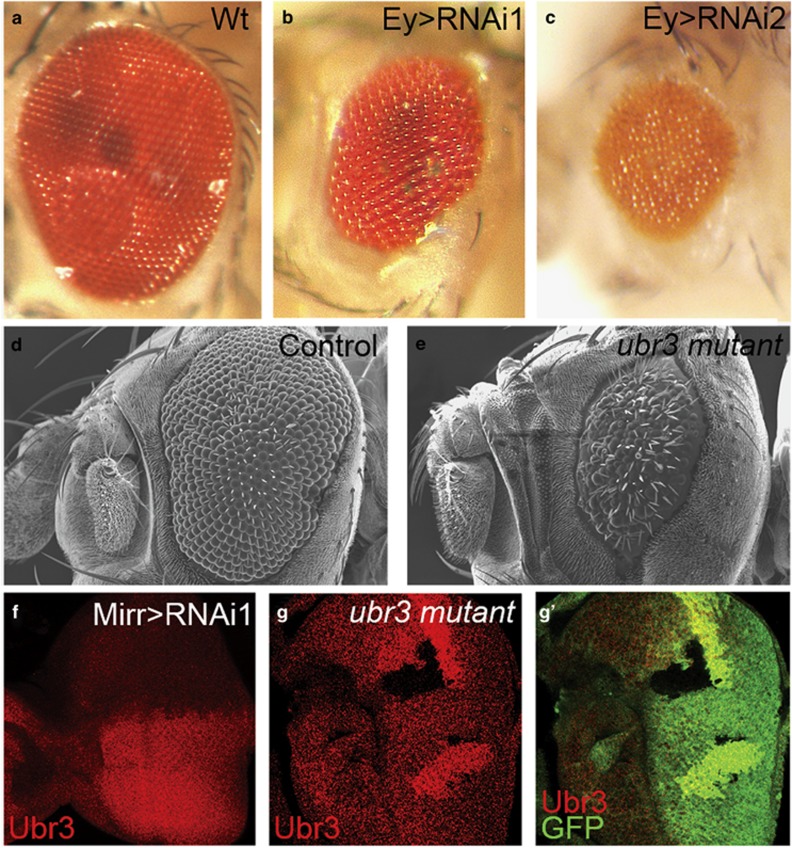
As shown in Figures 2b and c, eyeless-Gal4-induced ubr3 knock-down resulted in rough and smaller adult eyes. Similarly, knock-down of ubr3 by RNAi in the wing also impaired wing development (Supplementary Figure S1). To monitor the RNAi-mediated reduction of Ubr3, we generated an Ubr3-specific antibody and examined the protein levels by immunostaining. We observed that the expression of Ubr3 is greatly reduced in the dorsal compartment of the eye imaginal disc because of Mirror-Gal4-driven expression of ubr3 RNAi (Figure 2f). To further examine the role(s) of Ubr3 in eye development, we have generated a null allele of ubr3 by P-element-mediated imprecise excision (Figure 1b). Although the transcription of the gene CG2206, which lies within ubr3, is not affected by this allele (Supplementary Figure S1c), immunostaining indicates that expression of Ubr3 is completely absent in mutant clones (Figures 2g and g'). As the null allele of ubr3 is homozygous lethal, the adult eye phenotypes of this mutant were examined by virtue of the EGUF/hid method.32 Consistent with the RNAi results, we observed strong eye defects including eye roughness and small eye size (Figure 2e).
Figure 2.
Knockdown and knockout of ubr3 impaired the development of eye. All pictures are oriented anterior left, dorsal up. (a–c) Compared with wild type (a), RNAi-mediated knockdown of ubr3 driven by Ey-Gal4 (b and c) induces growth defect in eyes. (d and e) Scanning electron microscope (SEM) analyses of adult eyes in ubr3 knockout mutant. Eye phenotypes were generated by virtue of EGUF/hid method.26 (d) FRT19A line was used as a control. (f) Ubr3 staining is greatly reduced in the dorsal compartment of the eye imaginal disc expressing ubr3 RNAi driven by Mirr-Gal4. (g-g') Ubr3 protein is absent in ubr3 mutant clones. Genotypes: (b) w, UAS-ubr3 RNAi 1/Ey-Gal4, (c) w,UAS-ubr3 RNAi 2/Ey-Gal4, (d) FRT19A/yw,GMR-Hid, FRT19A;+/Ey-Gal4, UAS-flp, (e) w,ubr31-FRT19A/ yw,GMR-Hid, FRT19A;+/Ey-Gal4, UAS-flp, (f) w,+/UAS-ubr3 RNAi 1; +/Mirr-Gal4, (g) w, ubr31-FRT19A /hsFLP, Ubi-mRFP FRT19A
Loss of ubr3 in eye and wing discs induces apoptosis
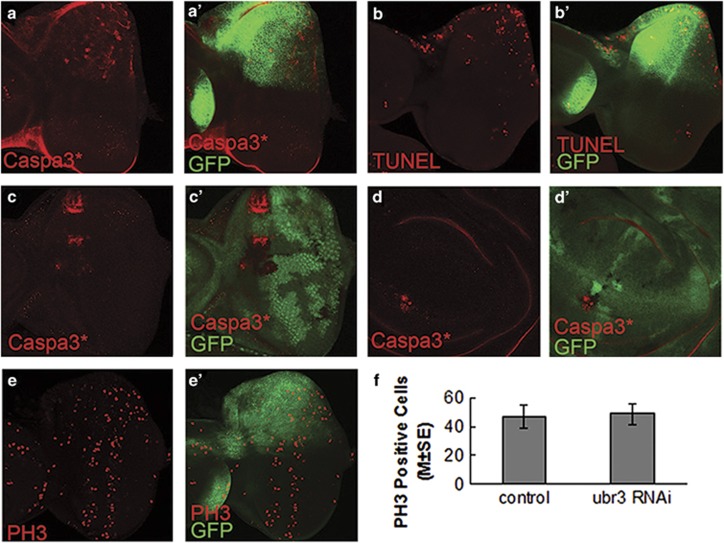
To determine whether the aforementioned eye defects were caused by apoptosis, we used multiple methods to monitor the apoptosis in the eye and wing imaginal discs of third instar larvae upon ubr3 mutation and knockdown. We found that the levels of both activated caspases (cleaved caspase-3) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) were significantly increased when ubr3 is knocked down by RNAi in the dorsal half of the eye disc (Figures 3a and b). We further performed mosaic clonal analysis using the ubr3 mutant in both eye and wing discs. Consistently, activation of caspase-3 was also observed in cells mutant for ubr3 (Figures 3c and d). To exclude the possibility that the eye phenotypes may also arise from defects in cell proliferation, we examined the mitosis status by staining with the mitotic marker phospho-histone H3 (PH3). As shown in Figures 3e and e' and f, cell proliferation in ubr3 RNAi was not affected. Collectively, these data argue that Ubr3 is involved in the control of apoptosis rather than cell proliferation in Drosophila.
Figure 3.
Loss of ubr3 activity induces apoptosis in eye imaginal discs. (a-b') ubr3 was knocked-down by Mirr-Gal4>RNAi in the dorsal compartment of eye disc. Dorsal is up as marked by GFP. Apoptosis was detected by activated caspase-3 (cleaved caspase-3 (Caspa3*)) staining (a-a') and TUNEL assay (b-b'). (c-d') In ubr3 mutant clones (GFP negative), Caspa3* is increased in eye disc (c) and wing disc (d). (e-e') Eye discs labeled with mitosis marker PH3. Knockdown of ubr3 does not affect PH3 levels. (f) Quantification of PH3-positive cells. There is no statistical difference between the dorsal compartment (ubr3 RNAi) and ventral compartment (control) of the eye discs, as analyzed by paired t-test (P=0.506 (two-tailed), n=9). Genotypes: (a, b and e) w;+/UAS-ubr3 RNAi 1; +/Mirr-Gal4
Ubr3 acts upstream of Dronc, and downstream of or in parallel to DIAP1
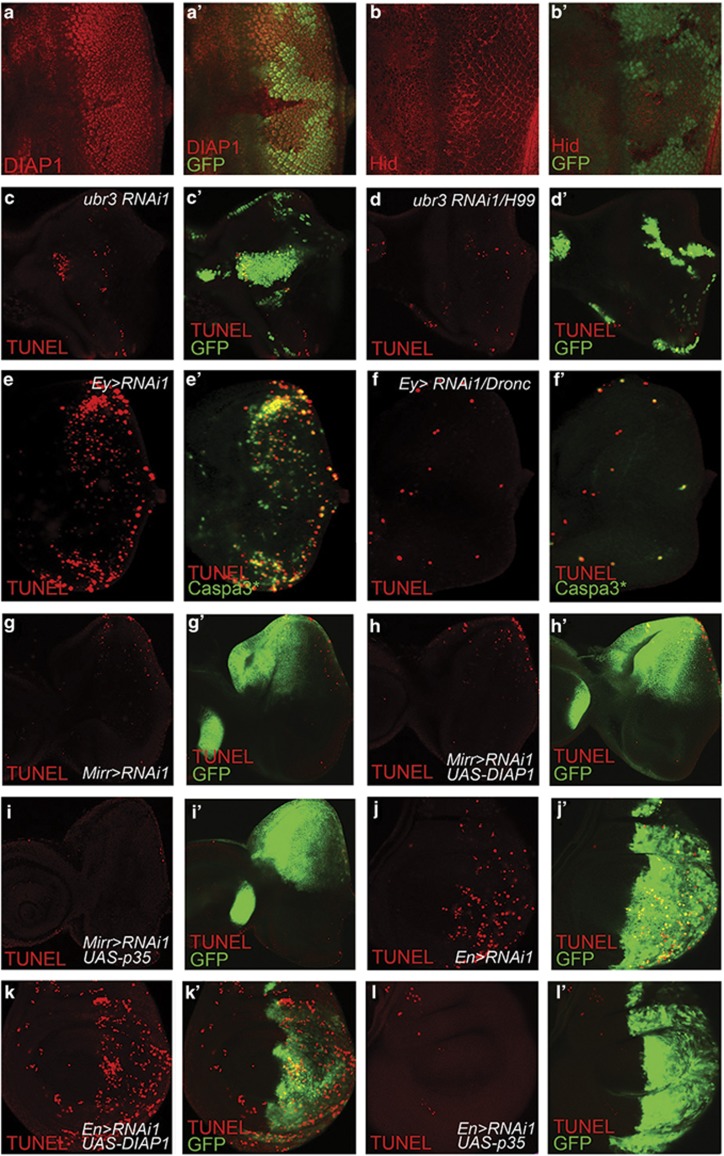
A variety of evidence supports the view that DIAP1 is the primary suppressor of apoptosis in Drosophila.11 Previous reports have shown that DIAP1 can be ubiquitinated by UBR family proteins (also called N-recognins) and degraded by proteasome.33 To test this possibility, first we examined the DIAP1 protein levels in ubr3 mutant cells using a DIAP1-specific antibody (Figures 4a and a''), whose specificity is verified in Supplementary Figures S2a-a''. The fact that protein levels of DIAP1 were slightly reduced only in some but not all ubr3 mutant cells excludes the possibility that Ubr3 targets DIAP1 for degradation. Although the change in DIAP1 levels may result from caspase activation in apoptotic cells, these data imply that Ubr3 do not directly control DIAP1 stability. Next, we checked the expression of Hid, one major pro-apoptotic proteins upstream of DIAP1. The increase in Hid protein levels is subtle if any in ubr3 mosaic clones (Figure 4b). Then, we asked whether the change in RHG complex is accounted for the apoptosis induced by ubr3 knockdown. We found that in both eye discs and wing discs (Figures 4c and d' and Supplementary Figures S2c-e), ubr3 knockdown still induced significant cell death in clones of cells homozygous for H99, a deletion that removes rpr, hid and grim, arguing that ubr3 regulates apoptosis not mainly through RHG proteins.
Figure 4.
Ubr3 functions in parallel to or downstream of DIAP1. (a-a') Protein levels of DIAP1 are slightly reduced in some ubr3 mutant cells. (b-b') The pro-apoptotic factor hid is slightly increased in cells mutant for ubr3. (c-d') In the eye discs, loss of RHG proteins fails to block the cell death induced by Ubr3 knockdown. Ubr3 RNAi is expressed in control clones (c-c') or clones homozygous for H99 (d-d'). All clones are positively marked by GFP. (e-f') Ey>ubr3 RNAi-caused cell death in the eye discs is markedly suppressed by Dronc mutation. ubr3 RNAi is expressed throughout the eye discs in wild-type background (e-e') or Dronc homozygous mutant background (f-f'). (g-i') Eye discs. (g-g') Mirr>ubr3 RNAi induces apoptosis. (h-h') Overexpression of wild-type DIAP1 fails to suppress apoptosis caused by Mirr>ubr3 RNAi. (i-i') p35 overexpression is able to block the apoptosis caused by Mirr>ubr3 RNAi. (j-l') In wing discs, cell death induced by En>ubr3 RNAi was examined when DIAP1 (k-k') or p35 (l-l') was co-expressed. Genotypes: (c) hsFLP, tubGal4, UAS-GFP; UAS-RNAi1/+ FRT80B/ tubGal80 FRT80B, (d) hsFLP, tubGal4, UAS-GFP; RNAi1/+ H99 FRT80B/ tubGal80 FRT80B, (e) w; UAS-ubr3 RNAi1/ Ey-Gal4, (f) w; Ey-Gal4/ubr3 RNAi1; DroncI29, (g) w;+/UAS-ubr3 RNAi 1; +/Mirr-Gal4, (h) w;+/UAS-ubr3 RNAi 1; UAS-DIAP1/Mirr-Gal4, (i) w;+/UAS-ubr3 RNAi 1; UAS-p35/Mirr-Gal4, (j) w; UAS-ubr3 RNAi 1/En-Gal4, (k) w; UAS-ubr3 RNAi 1/En-Gal4,+/ UAS-DIAP1, (l) w; UAS-ubr3 RNAi 1/En-Gal4,+/ UAS-p35
We performed further genetic epistasis analysis to place ubr3 in the apoptosis pathway. In response to upstream death stimuli, the initiator caspase Dronc is auto-activated as a component of the apoptosome, and further activates effector caspases, including DrICE and DCP-1.34 In this experiment, we knocked down ubr3 in a Dronc mutant background and found that the cell death induced by ubr3 RNAi was massively suppressed in the absence of Dronc (Figures 4e and f). This result argues that Ubr3 acts upstream of Dronc in the apoptosis pathway. Next, we asked whether overexpression of DIAP1 is able to suppress ubr3 RNAi-induced apoptosis. Although overexpression of DIAP1 can suppress GMR>rpr-induced apoptosis (Supplementary Figure S3), it fails to block ubr3 RNAi-induced cell death in both eye discs and wing discs (Figures 4h and h' and k-k'). In contrast, overexpression of baculovirus pancaspase inhibitor p35, which serves as an effective apoptosis inhibitor in Drosophila,35 blocked the apoptosis in both cases (Figures 4i and i' and l-l'). We also verified the apoptosis levels with acridine orange staining in eye discs (Supplementary Figure S4).
Taken together, these results indicate that Ubr3 regulates apoptosis mainly in parallel to or downstream of DIAP1, but upstream of Dronc.
The UBR-box domain rather than the Ring domain is essential for Ubr3's anti-apoptotic function
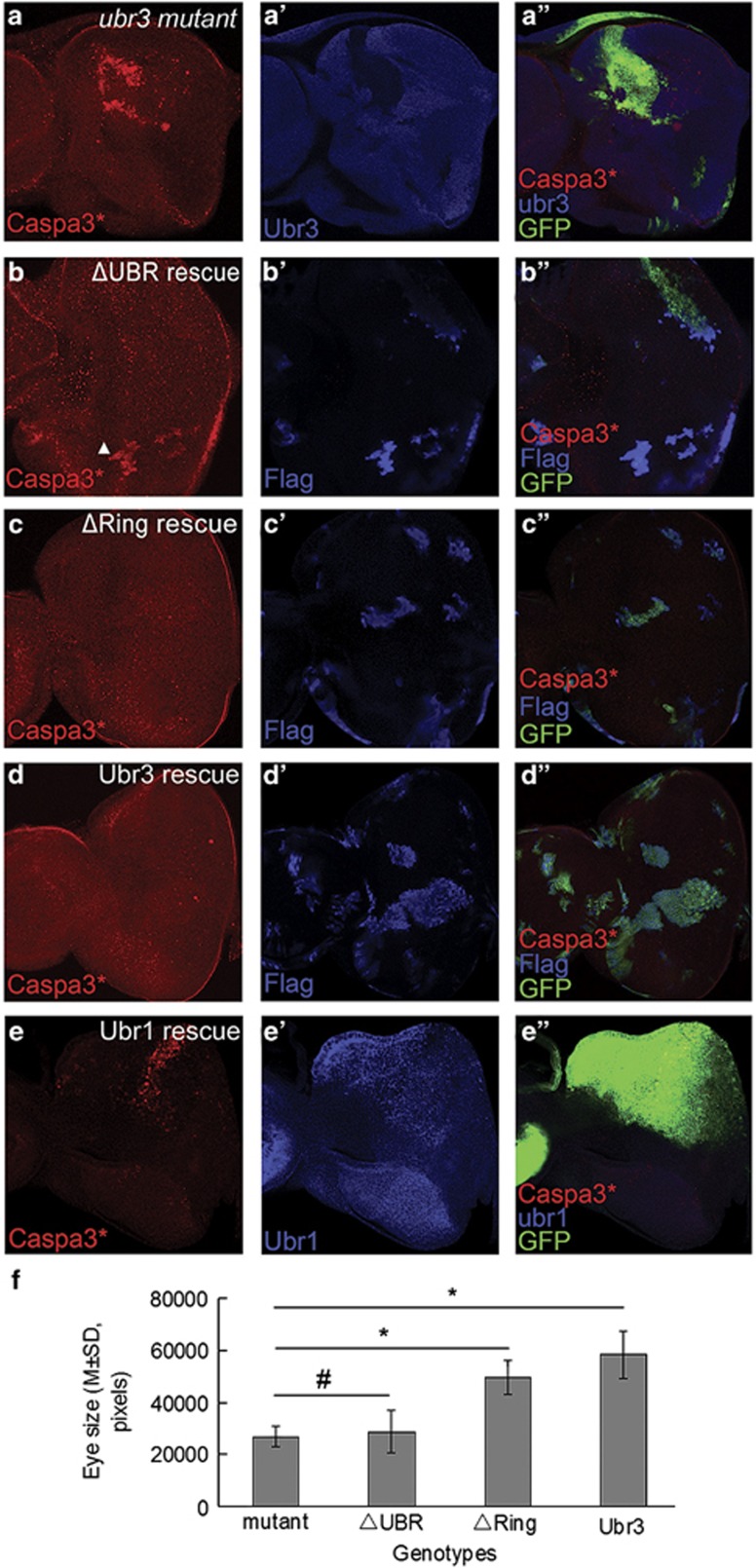
Ubr3 contains two conserved domains, a UBR-box domain involved in protein–protein interaction and a Ring domain required for its E3 ligase activity.23, 31 To further elucidate the molecular mechanism underlying Ubr3's function in apoptosis, we dissected the role(s) of each domain by genetic rescue experiments using the MARCM system.36 We generated transgenic flies expressing Gal4-inducible wild-type Ubr3 or two deletion mutants, Ubr3ΔUBR (UBR-box domain deleted) and Ubr3ΔRing (Ring domain deleted). Expression of Ubr3ΔRing or wild-type Ubr3 fully blocked the ectopic apoptosis in ubr3 mutant eye imaginal disc cells, as indicated by the absence of activated caspase-3 staining (Figures 5c and c'' and 5d and d''). In contrast, ectopic expression of Ubr3ΔUBR failed to suppress or only partially suppressed caspase-3 activation (Figures 5b and b''). Similar results were observed in wing discs (Supplementary Figure S5). In addition to caspase activation, we also examined the effect of different Ubr3 variants in restoring the eye size of ubr3 mutants. As shown before, by virtue of the EGUF/hid method, we were able to generate adult eyes composed of ubr3 mutant cells. Compared with the small and rough eyes of ubr3 mutants, expression of Ubr3ΔRing and wild-type Ubr3 both greatly restored the eye size (Figures 5f, P<0.05), while Ubr3ΔUBR showed little effect (Figures 5f, P>0.05, n=8, one-way ANOVA with LSD post hoc). Representative pictures of each genotype are shown in Supplementary Figures S5. These data suggest that the UBR-box domain is required for Ubr3's anti-apoptotic activity. However, the Ring domain is not essential, suggesting that Ubr3 may not act as an E3 ligase in this process.
Figure 5.
Genetic rescue of apoptosis in ubr3 mutant by overexpression of different Ubr3 constructs and Ubr1. (a-d'') MARCM clone analysis (GFP as positive labeling marker). (a-a'') Caspase-3 is activated in ubr3 mutant clones. (b-d'') Caspase-3 activation is rescued by expression of Ubr3ΔRing or Ubr3 (WT). Ubr3ΔUBR expression partially suppresses caspase-3 activation. (e-e''') Overexpression of Ubr1 fails to inhibit caspase-3 activation in ubr3 deficient cells. (f) Statistical analysis of the sizes of ubr3 mutant eyes rescued by expression of different Ubr3 variants (one-way ANOVA with LSD post hoc, *denotes P<0.05, #denotes P>0.05). Ubr3ΔRing and Ubr3 show rescuing effects, but not Ubr3ΔUBR. Genotypes: (a) ubr31-FRT19A/w, hsflp, Tub-Gal80 FRT19A; +/Act-Gal4,UAS-GFP, (b) ubr31-FRT19A/w, hsflp, Tub-Gal80 FRT19A; +/Act-Gal4, UAS-GFP; +/UAS-ubr3ΔUBR, (c) ubr31-FRT19A/w, hsflp, Tub-Gal80 FRT19A; +/Act-Gal4, UAS-GFP; +/UAS-ubr3ΔRing, (d) ubr31-FRT19A/w, hsflp, Tub-Gal80 FRT19A; +/Act-Gal4, UAS-GFP; +/UAS-ubr3, (e) w;+/UAS-ubr3 RNAi 1; UAS-ubr1/Mirr-Gal4
Among UBR family members in Drosophila, Ubr1 (CG9086) and Ubr3 (CG42593) have similar domain organizations, as both contain a Ring domain and a UBR-box domain. Interestingly, when we overexpressed Ubr1 in the ubr3 RNAi background, no rescue of apoptosis was observed (Figures 5e and e''). Consistently, no significant difference was found in adult eye sizes between the two groups (Supplementary Figures S5 j–l). These results suggest that Ubr1 and Ubr3 have distinct roles in Drosophila, despite the high similarities in protein structure.
Ubr3 binds to DIAP1 and promotes DIAP1-dependent caspases ubiquitination
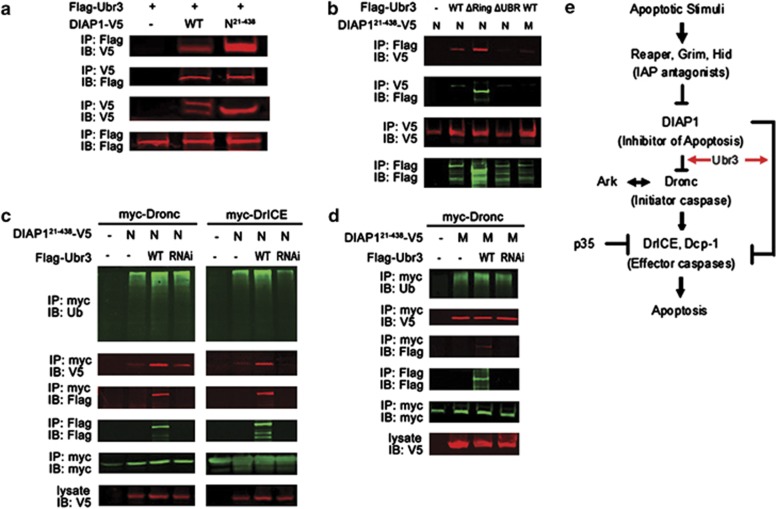
As reported by Ditzel et al.,22 isolated UBR domain can recognize the neo-epitope of DIAP1 exposed after caspase-mediated cleavage. We have found that UBR3 regulates apoptosis in a UBR-box-dependent manner downstream of or in parallel to DIAP1. To further characterize the interaction between Ubr3 and DIAP1, we performed co-immunoprecipitation (co-IP) assays in cultured S2 cells. First, we confirmed that Ubr3 binds to the N-terminally cleaved form of DIAP1 (N-DIAP121-438, the low band in lane 2), which exposes the binding motif for the UBR-box domain. However, Ubr3 fails to interact with full-length DIAP1 (top band in lane2) (Figure 6a). Second, the physical interaction between Ubr3 and DIAP1 relies on the UBR-box domain and the neo-epitope of DIAP1. Either deletion of the UBR-box domain of Ubr3 or replacement of N21 to M21 in N-DIAP121-438 can destroy this interaction (Figure 6b). However, UBR3ΔRing can interact effectively with DIAP1, consistent with the in vivo findings that UBR3ΔRing is a functional protein, but UBR3ΔUBR is not (Figure 5). Similarly, we found that N-DIAP121-438 instead of M-DIAP121-438 can pull down endogenous Ubr3 (Supplementary Figure S6).
Figure 6.
DIAP1 binds to Ubr3 and promotes ubiquitination of DrICE and Dronc. (a) Co-IP assays with Ubr3 and DIAP1. S2 cells were co-transfected with V5-tagged wild-type DIAP1 or DIAP121–438 and Flag-tagged Ubr3. Co-precipitated proteins were analyzed by immunoblotting (IB) with the indicated antibodies. The wild-type DIAP1 exhibits two bands, the full length (top band) and the N-terminally cleaved one (low band). The interaction of Ubr3 with the cleaved DIAP1 (DIAP121–438) but not the full-length DIAP1 can be detected. (b) Co-IPs were performed as in (a) with N- and M-DIAP121–438 and the Ubr3 variants. (c) Effects of Ubr3 co-expression or knockdown on DIAP1-mediated caspase ubiquitination. S2 cells were co-transfected with myc-tagged Dronc/DrICE together with N-DIAP121-438 and constructs encoding Flag-tagged Ubr3 or Ubr3 shRNA. Half of the cells were lysed under denaturing conditions using hot lysis protocol and ubiquitinated caspases were pulled down by myc affinity gel followed by immunoblotting with anti-ubiquitin antibody. The other half of cells were processed for co-IP assays as in (a). (d) Ubr3 fails to affect M-DIAP121-438's catalytic function in Dronc ubiquitination. Experiments were performed similar to (c). (e) Model of Ubr3's function in apoptosis. Ubr3 helps block apoptosis mainly through its interaction with DIAP1. This interaction promotes the binding of downstream caspases to DIAP1 and facilitates subsequent ubiquitination reactions
It has been shown that M-DIAP121-438, which cannot be recognized by UBR family proteins, has reduced activities in catalyzing caspase ubiquitination and in inhibiting apoptosis.22 Here, we further explored how the interaction of DIAP1 with Ubr3 affects DIAP1's activities. As shown in Figure 6c, expression of N-DIAP121–438 in S2 cells readily ubiquitinated the downstream caspases Dronc and DrICE. Introduction of ectopic Ubr3 further enhanced N-DIAP121-438-induced caspase ubiquitination, while ubr3 knockdown significantly reduced DrICE ubiquitination (Figure 6c, the efficacy of Ubr3 RNAi is verified in Supplementary Figure S7), suggesting that Ubr3 regulates the anti-apoptotic activities of DIAP1. Although DIAP1 and Dronc/DrICE interact weakly with each other in normal cells, Ubr3 is found to be able to enhance their interaction possibly by forming a tertiary complex as shown by the co-IP results (Figure 6c). The interaction of Ubr3 with DIAP1 is important for the capability of Ubr3 in controlling DIAP1's activities, as Ubr3 fails to affect M-DIAP121-438's catalytic function in Dronc ubiquitination (Figure 6d). In support of this view, Ubr3ΔUBR, which fails to interact with DIAP1, has markedly reduced activity on DIAP1-induced DrICE ubiquitination (Supplementary Figure S8). Altogether, these results indicate that the presence of Ubr3 is required to maximize DIAP1's E3 ligase activities on substrate caspases.
Discussion
In this study, we used the developing Drosophila eye and wing as in vivo models to analyze the function of ubr3 at the molecular and cellular levels. We showed that loss of ubr3 causes massive cell death in Drosophila. Our detailed genetic and molecular analyses led us to conclude that Ubr3 controls the apoptosis pathway by regulating DIAP1 activity.
The Drosophila genome encodes six UBR-box-containing E3 ligases, and the functional details of each UBR protein remain elusive. Although DIAP1 has been shown as a substrate for N-end rule-mediated degradation and could interact with isolated UBR domains of multiple UBR family members, no single UBR protein has ever been identified in the regulation of DIAP1.20, 21 We for the first time characterized Ubr3, a highly conserved UBR protein, as an anti-apoptotic factor, which controls the function rather than the stability of DIAP1. Interestingly, the role of Ubr3 in apoptosis pathway cannot be replaced by other UBR proteins such as Ubr1. Since N-end rule degradation pathway has been implicated in the regulation of DIAP1, it is possible that this process involves other UBR family members. More studies are needed to fully elucidate the role of UBR family proteins in the apoptosis pathway.
Our genetic epistasis experiments place Ubr3 in parallel to or downstream of DIAP1 in the apoptosis pathway. First, loss of RHG proteins fails to suppress cell death because of ubr3 knockdown, arguing that RHG complex is upstream of Ubr3 in the apoptotic pathway. Although Hid levels are slightly elevated in cells mutant for ubr3, it is probably a secondary effect because of the reduced activities of DIAP1. Interestingly, RHG proteins have been shown to be also ubiquitinated by DIAP1.37 It is possible that ubr3 is also important for this activity of DIAP1. Second, Dronc mutation or ectopic expression of p35 can effectively suppress apoptosis induced by the loss of ubr3 activity. Third, overexpression of DIAP1 fails to inhibit loss-of-ubr3-induced apoptosis. Finally, we show that Ubr3 interacts with DIAP1 and promotes DIAP1-mediated caspase ubiquitination. On the basis of these data, we propose a model in which Ubr3 interacts with DIAP1 to regulate its activities in the apoptosis pathway.
How does Ubr3 regulate DIAP1's activity? Previously, Ditzel et al.22 showed that the isolated UBR-box domains of UBR1, -3, -5, -6 and -7 can all bind to DIAP1. By using the M-DIAP121-438 mutant, which fails to interact with UBR proteins, they also showed that the UBR-DIAP1 interaction is required for DIAP1's anti-apoptotic activity. However, the molecular nature of the involving UBR protein(s) was unclear. Our studies presented here resolved this issue by identifying Ubr3 as an essential protein required for DIAP1 activity. Moreover, we showed that overexpression of Ubr1, a homolog of Ubr3, fails to rescue the apoptosis defect in ubr3 mutant cells. This result shows that the activity of Ubr3 in the regulation of DIAP1 is highly specific, and strongly argues against the previous view that significant redundancy exists among UBR proteins in apoptotic regulation. It is important to mention that ectopic expression of an E3-defective Ubr3 can effectively suppress caspase-3 activation in ubr3 mutant cells, suggesting that Ubr3 does not act as an E3 ligase in regulating DIAP1 activity. Interestingly, we notice that Ring domain deletion renders Ubr3 more stable in cultured cells, implying a potential role of Ubr3 in its auto-regulation through E3 ligase activity. Finally, we observed that Ubr3 forms a complex together with DIAP1 and caspases. Although Ubr3 regulates the ubiquitination levels of caspases, it also affects the interaction between caspases and their E3 ligase, DIAP1 (Figure 6d). Together, these data lead to a model in which Ubr3 promotes the interaction of DIAP1 with caspases, and therefore facilitates the subsequent ubiquitinating reaction (Figure 6e). Interestingly, recent data suggest that DIAP1 rests in an ‘inactive' conformation whose activation requires caspase-mediated cleavage.22 As Ubr3 interacts with DIAP1 and caspases, the binding of Ubr3 to DIAP1 may also help to maintain DIAP1 in the active conformation, which is essential for its activity. Further biochemical and molecular experiments are needed to explore the detailed mechanisms underlying the regulation of DIAP1's anti-apoptotic activity by Ubr3.
Materials and Methods
Drosophila stocks, genetics
Upstream activating sequence (UAS)-p35, UAS-DIAP1, actin-Gal4, MS1096-Gal4, Eyeless-Gal4, mirror-Gal4, yw FRT101, yw neoFRT19A, yw Ubi-green fluorescent protein (GFP) FRT101, heat shock-flippase (hs-FLP), UAS-CD8::GFP, w hsflp, Tub-Gal80 FRT19A and GMR-Hid, yw FRT19A; eyeless-Gal4, UAS-FLP lines were obtained from Bloomington Stock Center (Indiana University, Bloomington, IN, USA). MARCM clone tool lines yw, hsFLP; tubGAL4 UASGFP; FRT80B tubGal80 and yw; sp/Cyo; FRT80B/TM6B were received from Dr. Lei Zhang (SIBCB, Shanghai, China). DroncI29 is described previously.34 H99 is a deletion of hid, rpr and grim. A null allele of ubr31 was generated by P-element-mediated imprecise excision from p (EP) EP1243, which deletes 1555 bp including the start codon and the first exon. ubr3 RNAi lines v106993 (RNAi1) and v22901 (RNAi2) were obtained from Vienna Drosophila RNAi Center (VDRC), Vienna, Austria.
Mosaic clones of ubr3 mutant were generated by the FLP-FRT method and induced in first/second-instar larvae, as described previously.38 Flies were raised on standard cornmeal/yeast/molasses/agar media at 25 °C.
Constructs and transgenic lines
Ubr3 cDNA was generated by synthetic method from ESTs and exons, and details are available on request. pUAST-3Flag-Ubr3 was constructed by cloning the full-length ubr3 cDNA into pUAST vector with XhoI and XbaI. Forward primer: 5′-ACGCTCGAGATGGACGAGGATGACAACCTG-3′ reverse primer: 5′-ACGTCTAGACAAGAGATCCCGGTGAAACACCCATG-3′. pUAST-3Flag-Ubr3ΔUBR construct has a deletion of 222-292aa, and pUAST-3Flag-Ubr3ΔRing construct has a deletion of 1515–2219aa. YFP-based DIAP1 construct is a modified version of the ubiquitin fusion construct described previously by Lévy et al.39 Briefly, YFP was used as a reference protein to link to the N terminus of ubiquitin and the C terminus of ubiquitin was then linked to DIAP1. After cleavage of the tripartite fusion protein at the C terminus of ubiquitin, equimolar amounts of the unmodified DIAP1 and the reference protein will be produced. The YFP-Ubiquitin-DIAP1 construct was generated by overlapping PCR and cloned into pUAST vector with C-terminal V5 tag (details are available on request). Three different DIAP1 variants were generated by the same method: YFP-Ubiquitin-DIAP1-V5, YFP-Ubiquitin-N-DIAP121–438-V5 and YFP-Ubiquitin-M-DIAP121–438-V5, referred to as DIAP1(-V5), N-DIAP121–438 (-V5) and M-DIAP121–438 (-V5), respectively, in the text. DIAP1(-V5) is a wild-type DIAP1 protein. N-DIAP121–438 (-V5) is a truncated DIAP1, from which the first 20 N-terminal amino acids are removed. In M-DIAP121–438 (-V5), the N21 was replaced by M21.
RNAi-mediated knockdown of ubr3 in S2 cells was achieved by transfection of a short hairpin RNA transgene in pWALIUM20 knockdown vector.40 A gene-specific 21-bp oligonucleotide (5′-GCACATTTGTGATGCACTACA-3′) was selected and inserted into the vector.
Eye size quantification and statistical analysis
All groups were crossed at the same time and raised under identical circumstances. All the pictures were taken on female flies at the same magnification, using AxioCam (Zeiss, Cambridge, UK). The areas of the eyes were measured by a colleague who is unaware of the genotypes, using Photoshop software (Adobe Systems Inc., San Jose, CA, USA). Data were analyzed with SPSS15.0 (IBM Corp., Chicago, IL, USA), using one-way ANOVA with LSD post hoc tests. All data were presented as means±S.D., and P<0.05 was considered statistically significant.
Apoptosis assays
For TUNEL assay, dissected larval and pupal tissues were fixed for 20 min at RT in 4% formaldehyde in PBST, washed in PBST (0.3% Triton X-100) and permeabilized by incubation in 100 mM Na citrate/0.1% Triton X-100 at 65 °C for 30 min. Samples were rinsed in PBST and TUNEL was performed using a kit from Roche (Indianapolis, IN, USA). Tissues were mounted for confocal analysis (Zeiss LSM780).
Immunostaining
Dissection, fixation and immunostaining of imaginal discs were performed as described previously.41 Guinea pig anti-Ubr3 primary antibody was produced against amino acids 1831–2072. Guinea pig anti-Ubr1 primary antibody was targeted against amino acids 1485–1816. Primary antibodies used for the immunostainings were rabbit anti-cleaved caspase-3 (Cell Signaling Technology, Danvers, MA, USA), Mouse anti-PH3 (Abcam, Cambridge, MA, USA), rabbit anti-Hid (d-300) (Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti Flag (Sigma, St. Louis, MO, USA) and rabbit anti-GFP Alexa Fluor 488 (Molecular Probes, Eugene, OR, USA). Mouse anti-DIAP1 (1:5) was generously provided by Dr. Bruce A Hay (California Institute of Technology, Pasadena, CA, USA). The primary antibodies were detected by fluorescent-conjugated secondary antibodies from Jackson ImmunoResearch (West Grove, PA, USA).
Immunoprecipitation and western blotting
Drosophila S2 cells were maintained at 25 °C in Hyclone SFX-Insect cell culture medium (SH30278.02, Thermo Scientific, Waltham, MA, USA). For co-IP experiments, Drosophila S2 cells were transfected in 60 mm dishes with 2 μg of total DNA, using Effectene (Qiagen, Hilden, Germany). Forty-eight hours after transfection, cells were harvested and lysed in 1% Trition X-100 lysis buffer: 1% Triton X-100, 20 mM Tris (pH 7.5), 1 mM EDTA, 150 mM NaCl, protease inhibitor cocktail (Roche, added right before use). Co-IP and western blot experiments were performed as described.38 For ubiquitination assays, transfected cells were treated with 50 μM MG-132 (Sigma) for 6 h before harvest. Forty-eight hours after transfection, cells were harvested and lysed by hot lysis protocol. Generally, cells were boiled at 100 °C in hot lysis buffer I (1% SDS, 50 mM NaF and 1 mM EDTA) followed by extensive shearing and mixing. After centrifugation and fivefolds dilution with hot lysis buffer II (25 mM Tris-HCl, pH 7.5, 1.25% Triton X-100, 125 mM NaCl and 50 mM NaF, protease inhibitors and a final concentration of 50 μM MG132 were added just before use), the supernatant was collected as cell lysate. The following antibody-bound beads were used for immunoprecipitation: EZview Red Anti-FLAG M2 Affinity Gel (Sigma), EZview Red Anti-c-Myc Affinity Gel (Sigma), Anti-V5 Agarose Affinity Gel (Sigma). The primary antibodies used for western blotting were rabbit anti-V5 (Sigma), mouse anti-V5 (Invitrogen, Carlsbad, CA, USA), mouse anti-Ub (P4D1, Santa Cruz Biotechnology), mouse anti-Flag (Sigma) and mouse anti-myc (Sigma).
Acknowledgments
We thank Dr. Bruce A Hay (Caltech, USA) for providing the anti-DIAP1 antibody and Dr. Meier for sending us various tub>DIAP1 transgenic lines and the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing the pVALIUM20 vector. Many lines are obtained from the Bloomington Stock Center and the Vienna Drosophila RNAi Center. This work is supported by grants the National Basic Research Program of China (2011CB943901 and 2011CB943902), Research Foundation for Advanced Talents of Wenzhou Medical University (QTJ08012) and Wenzhou Medical University research grant (XNK07005), National Natural Science Foundation of China (31030049), NIH grants (2R01 GM063891 and 1R01GM087517) and Strategic Priority Research Program of the Chinese Academy of Sciences Grant (XDA01010101). YF is supported by Marie Curie Career Integration Grant 630846 from the EU FP7 Program and the Birmingham Fellowship, University of Birmingham, UK. AB is supported by Grant R01 GM068016 from the National Institute of General Medical Sciences (NIGMS).
Glossary
- IAPs
inhibitor of apoptosis proteins
- DIAP1
Drosophila inhibitor of apoptosis protein 1
- Dronc
Drosophila Nedd2-like caspase
- En
engrailed
- Ey
eyeless
- Mirr
mirror
- FLP
flippase
- FRT
flippase recombination target
- UAS
upstream activating sequence
- GFP
green fluorescent protein
- Hid
head involution defective
- Hs
heat shock
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Baehrecke
Supplementary Material
References
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke EH. How death shapes life during development. Nat Rev Mol Cell Biol. 2002;3:779–787. doi: 10.1038/nrm931. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Flavell RA. Apoptosis genes and autoimmunity. Curr Opin Immunol. 2000;12:719–724. doi: 10.1016/s0952-7915(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Vucic D, Miller LK. The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 1998;440:243–248. doi: 10.1016/s0014-5793(98)01465-3. [DOI] [PubMed] [Google Scholar]
- Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ, et al. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 2007;5:e251. doi: 10.1371/journal.pbio.0050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Jones D, Zhou L, Steller H, Deterin ChuY. a new inhibitor of apoptosis from Drosophila melanogaster. J Biol Chem. 2000;275:22157–22165. doi: 10.1074/jbc.M000369200. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ashwell JD. IAPs: what's in a name. Mol Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem. 2002;277:49644–49650. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A. The role of ubiquitylation for the control of cell death in Drosophila. Cell Death Differ. 2010;17:61–67. doi: 10.1038/cdd.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule and regulation of apoptosis. Nat Cell Biol. 2003;5:373–376. doi: 10.1038/ncb0503-373. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P, et al. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem. 2004;279:52603–52612. doi: 10.1074/jbc.M409691200. [DOI] [PubMed] [Google Scholar]
- Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, et al. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell. 2008;32:540–553. doi: 10.1016/j.molcel.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Sriram SM, Kim BY, Kwon YT. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol. 2011;12:735–747. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol Cell Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, Kwon YT. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci USA. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Sohr R, Xia Z, Hellweg R, Hortnagl H, Varshavsky A, et al. Biochemical and genetic studies of UBR3, a ubiquitin ligase with a function in olfactory and other sensory systems. J Biol Chem. 2007;282:18510–18520. doi: 10.1074/jbc.M701894200. [DOI] [PubMed] [Google Scholar]
- Meisenberg C, Tait PS, Dianova II, Wright K, Edelmann MJ, Ternette N, et al. Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability. Nucleic Acids Res. 2012;40:701–711. doi: 10.1093/nar/gkr744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14:861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Olson MR, Holley CL, Yoo SJ, Huh JR, Hay BA, Kornbluth S. Reaper is regulated by IAP-mediated ubiquitination. J Biol Chem. 2003;278:4028–4034. doi: 10.1074/jbc.M209734200. [DOI] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Standley HJ, Lin X, Houston DW, Heasman J. pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Development. 2002;129:4089–4101. doi: 10.1242/dev.129.17.4089. [DOI] [PubMed] [Google Scholar]
- Levy F, Johnsson N, Rumenapf T, Varshavsky A. Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci USA. 1996;93:4907–4912. doi: 10.1073/pnas.93.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, et al. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev Cell. 2008;14:120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.