Abstract
DNA damage-regulated autophagy modulator protein 1 (DRAM1), a multi-pass membrane lysosomal protein, is reportedly a tumor protein p53 (TP53) target gene involved in autophagy. During cerebral ischemia/reperfusion (I/R) injury, DRAM1 protein expression is increased, and autophagy is activated. However, the functional significance of DRAM1 and the relationship between DRAM1 and autophagy in brain I/R remains uncertain. The aim of this study is to investigate whether DRAM1 mediates autophagy activation in cerebral I/R injury and to explore its possible effects and mechanisms. We adopt the oxygen-glucose deprivation and reperfusion (OGD/R) Neuro-2a cell model to mimic cerebral I/R conditions in vitro, and RNA interference is used to knock down DRAM1 expression in this model. Cell viability assay is performed using the LIVE/DEAD viability/cytotoxicity kit. Cell phenotypic changes are analyzed through Western blot assays. Autophagy flux is monitored through the tandem red fluorescent protein–Green fluorescent protein–microtubule associated protein 1 light chain 3 (RFP–GFP–LC3) construct. The expression levels of DRAM1 and microtubule associated protein 1 light chain 3II/I (LC3II/I) are strongly up-regulated in Neuro-2a cells after OGD/R treatment and peaked at the 12 h reperfusion time point. The autophagy-specific inhibitor 3-Methyladenine (3-MA) inhibits the expression of DRAM1 and LC3II/I and exacerbates OGD/R-induced cell injury. Furthermore, DRAM1 knockdown aggravates OGD/R-induced cell injury and significantly blocks autophagy through decreasing autophagosome-lysosome fusion. In conclusion, our data demonstrate that DRAM1 knockdown in Neuro-2a cells inhibits autophagy by blocking autophagosome-lysosome fusion and exacerbated OGD/R-induced cell injury. Thus, DRAM1 might constitute a new therapeutic target for I/R diseases.
Keywords: DNA damage-regulated autophagy modulator protein 1 (DRAM1), oxygen-glucose deprivation and reperfusion (OGD/R), cerebral ischemia and reperfusion (I/R) injury, autophagy
1. Introduction
Cerebral homeostasis and function rely on an adequate supply of oxygenated blood. Stroke, often caused by hypoxic ischemic encephalopathy and acute cerebrovascular incidents, interrupts cerebral blood supply and results in high mortality and long-term disability worldwide [1]. The goal of stroke treatment is to restore the blood supply as rapidly as possible; however, reperfusion may aggravate cerebral injury. The prevention and treatment of cerebral ischemia/reperfusion (I/R)-induced injury have been considered pivotal strategies for stroke intervention. Multiple pathophysiology processes may be involved in cerebral I/R injury, including neutrophil infiltration, post-ischemic hyperperfusion, and impairment of the blood-brain barrier [2]. However, the mechanisms involved in these processes are complex and remain uncertain.
Autophagy is a basic catabolic mechanism involving intracellular components and damaged organelle degradation that maintains normal organelle function and nutrient restoration [3], and it is an evolutionarily conserved mechanism from yeast to Homo sapiens [4]. Generally, autophagy is activated by many physiological and pathological conditions. A double-membrane-bound autophagosome forms and travels through the cytoplasm of the cell to the lysosome, and the two organelles fuse into an autophagolysosome and degrade in the lysosome. The degradation products can be reused by the cell for metabolism [5]. During cerebral I/R injury, autophagy is activated, and the regulation of autophagy affects the results of the brain I/R injury. However, its effects on cerebral I/R are controversial. On the one hand, some investigators [6,7,8,9] have shown that cerebral ischemia induced autophagy-like cell death, and inhibition of autophagy prevents neuron death after ischemic injury. On the other hand, other investigators [10,11,12,13,14] have shown that autophagy activation is involved in neuroprotection in brain I/R injury. Autophagy has been considered a double-edged sword with pro-survival or pro-death potential in cerebral I/R injury [15].
DNA damage-regulated autophagy modulator protein 1 (DRAM1) is a multi-pass membrane lysosomal protein that has shown a high degree of conservation throughout evolution and has been reported as a tumor protein p53 (TP53) target gene involved in autophagy and apoptosis [16]. During cerebral I/R injury, DRAM1 expression is altered along with the ratio of microtubule associated protein 1 light chain 3II/I (LC3II/I) [17]; however, whether DRAM1 regulates autophagy during brain I/R is uncertain. Given that DRAM1 is mainly located in the lysosomal membrane and that lysosomes play pivotal roles in the autophagic pathway, we hypothesized that DRAM1 may play a role in autophagy through the lysosomes during cerebral I/R injury. To investigate this hypothesis, the oxygen glucose deprivation and reperfusion (OGD/R) model was used to mimic ischemia/reperfusion conditions in vitro. We found that OGD/R induced DRAM1-dependent stimulation of autophagy in Neuro-2a cells. Knockdown of DRAM1 inhibited autophagy flux by decreasing autophagosome-lysosome fusion.
2. Results and Discussion
2.1. Oxygen Glucose Deprivation and Reperfusion (OGD/R) Treatment Increases DNA Damage-Regulated Autophagy Modulator Protein 1 (DRAM1) Expression and Induces Autophagy Activation
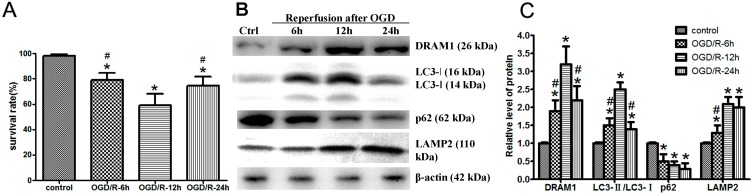
OGD/R-induced Neuro-2a cell injury acts as a model to mimic cerebral I/R injury in vitro and results in neuronal insult [18,19]. This model could mimic extracellular condition in cerebral ischemia and subsequently reperfusion, which is used as a common model for research in vitro. First, we assessed Neuro-2a cell survival rate after OGD/R. The cell death assay showed 20.98% of the cells died after a 4 h OGD and reperfusion for 6 h. This cell death percentage increased with reperfusion time to 40.25% after 12 h and then decreased to 25.33% after 24 h (Figure 1A), which indicated the most serious injury after OGD/R occurred at 12 h, and then cells recovered and proliferated.
Figure 1.
Oxygen glucose deprivation and reperfusion (OGD/R) treatment increases DNA damage-regulated autophagy modulator protein 1 (DRAM1) expression and induces autophagy activation. Neuro-2a cells were subjected to 4 h OGD and harvested after 6, 12, and 24 h. (A) Cell viability was assessed using the LIVE/DEAD viability/cytotoxicity kit. Neuro-2a cells were exposed to OGD for 4 h followed by reperfusion for the indicated times; (B) Relative protein expression levels. At the indicated reperfusion time points, Neuro-2a cells were collected, and protein was extracted to assess DRAM1, microtubule associated protein 1 light chain 3 (LC3), lysosomal-associated membrane protein 2 (LAMP2), and ubiquitin-binding protein (p62) expression levels through Western blot analysis. β-Actin was used as an internal control; and (C) The quantitative results from (B). β-Actin was used as an internal control (* p < 0.05 vs. control, # p < 0.05 vs. ODG/R 12 h group, n = 6).
In ischemic cerebral model, autophagy is activated, followed by the rapid elimination of autophagic flux [20]. However, little is known regarding autophagy in the reperfusion phase after cerebral ischemia. OGD/R-induced changes in DRAM1 and certain autophagic pathway proteins in Neuro-2a cells were determined from 6 to 24 h after OGD/R treatment. The ratio of LC3II/I significantly increased and peaked at 12 h after reperfusion (Figure 1B,C), which suggested the presence of autophagy during reperfusion [21]. OGD/R treatment induced a significant increase in DRAM1 protein levels, with a peak at 12 h after reperfusion (Figure 1B,C). Lysosomal-associated membrane protein 2 (LAMP2), a single-span lysosomal membrane protein, which functions as a lysosomal membrane receptor of autophagy, was also investigated [22]. It has been reported that LAMP2 dysfunction results in inefficient autophagosome clearance [23]. OGD/R increased LAMP2 protein levels compared with the control group (Figure 1B,C). Ubiquitin-binding protein (p62), a substrate of autophagy, binds to LC3 and decreases during autophagy [24,25]. We found that p62 levels decreased in OGD/R-induced Neuro-2a cell injury. These data suggested that autophagy was activated in Neuro-2a cells after OGD/R treatment.
2.2. Inhibition of Autophagy Exacerbates OGD/R-Induced Neuro-2a Cell Injury
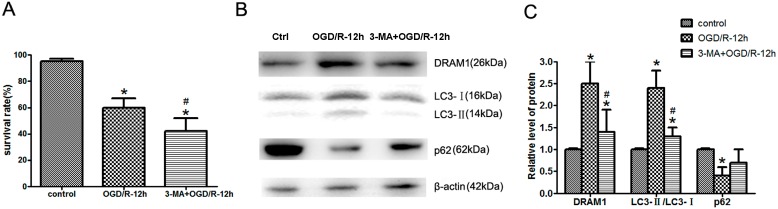
To identify the role of autophagy activation in OGD/R-induced Neuro-2a cell injury, we used the autophagy-specific inhibitor 3-Methyladenine (3-MA) to suppress autophagy activation. We found that the increases in DRAM1 expression and the LC3II/I ratio in the Neuro-2a cells treated with OGD/R were significantly inhibited after co-treatment with 3-MA, and the p62 decline was also attenuated (Figure 2B,C). In addition, the survival rate of Neuro-2a cells was markedly reduced after 3-MA treatment with OGD/R (Figure 2A), which contrasts with the ischemic cerebral model [26], indicating that autophagy inhibition exacerbated OGD/R-induced Neuro-2a cell injury and that autophagy may play a protective role in OGD/R-mediated Neuro-2a cell injury [14].
Figure 2.
Inhibition of autophagy exacerbates OGD/R-induced Neuro-2a cell injury. Neuro-2a cells were pretreated with the autophagy-specific inhibitor 3-Methyladenine (3-MA) and then subjected to 4 h OGD and 12 h reperfusion. (A) Cell viability was assessed using a LIVE/DEAD viability/cytotoxicity kit; (B) Relative protein expression levels. At the OGD/R 12 h time point, Neuro-2a cells were collected, and protein was extracted to assess DRAM1, LC3, and p62 expression levels through Western blot analysis. β-Actin was used as an internal control; and (C) The quantitative results from (B). β-Actin was used as an internal control (* p < 0.05 vs. control, # p < 0.05 vs. ODG/R 12 h group, n = 6).
2.3. DRAM1 Knockdown Inhibits Autophagy Activation and Aggravates OGD/R-Induced Neuro-2a Cell Injury
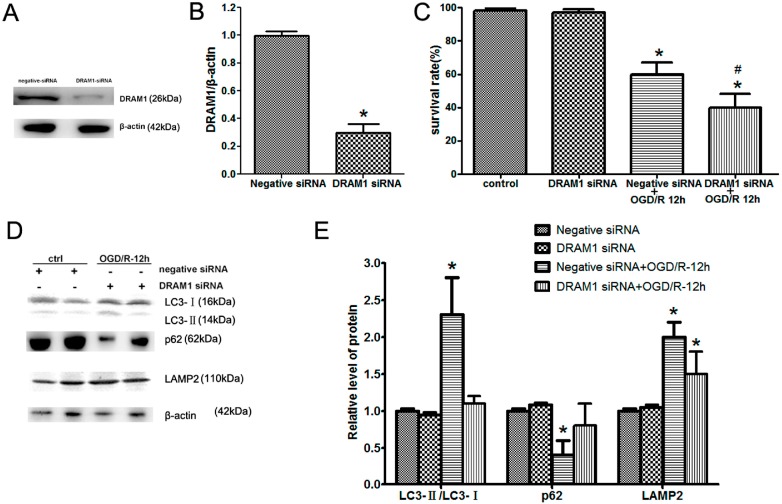
DRAM1 expression was significantly up-regulated in OGD/R-induced Neuro-2a cells, peaking at 12 h of reperfusion along with the LC3II/I ratio. Meanwhile, 3-MA treatment reduced DRAM1 expression and LC3II/I ratio. These data suggest that the DRAM1 expression pattern and autophagy were regulated in the in the in vitro model. DRAM1 encodes a 238-amino acid protein and is highly conserved in a number of species, including humans, mice, zebra fish, Drosophila, and Caenorhabditis elegans (C. elegans) [27,28]. A previous report showed that DRAM1 has a potential tumor-suppressive function and is required for p53-induced autophagy [29]. Although we have demonstrated a protective role of autophagy and increased DRAM1 expression during OGD/R-induced Neuro-2a cell injury, it is unknown how autophagy and DRAM1 protect against cell injury and death. To confirm the links among DRAM1 expression, autophagy activation, and OGD/R-induced Neuro-2a cell injury, we used a siRNA specifically targeting DRAM1. We found that DRAM1 expression in Neuro-2a cells was significantly inhibited after DRAM1 siRNA treatment (Figure 3A,B). The inhibition of DRAM protein production had no effect on the LC3II/I ratio or the levels of p62 and LAMP2 of Neuro-2a cells under normal conditions and resulted in a significant decrease in the LC3II/I ratio and an increase of p62 in OGD/R-induced Neuro-2a cells (Figure 3D,E). Galavotti et al. also documented that DRAM1 knockdown inhibits p62 targeting to autophagosomes and reduces its degradation [30]. However, DRAM1 silencing had little effect on LAMP2 expression in OGD/R-induced Neuro-2a cells. LAMP2 is a single-span lysosomal membrane protein [31], and knockout of the entire gene results in abnormal lysosomal biogenesis and inefficient autophagosome clearance [32]. We hypothesized that LAMP2 might show a compensatory increase after DRAM1 knockdown. Given that DRAM1 and LAMP2 are both lysosomal membrane proteins, they likely exert a similar effect on the autophagy-lysosome pathway. Furthermore, the survival rate of Neuro-2a cells was markedly reduced after OGD/R treatment in the absence of DRAM1 (Figure 3C). Our results indicated that siRNAs specific for DRAM1 inhibited autophagy activation and aggravated OGD/R-induced Neuro-2a cell injury. However, there was little effect on LAMP2 expression after DRAM1 knockdown.
Figure 3.
DRAM1 knockdown inhibits autophagy activation and aggravates OGD/R-induced Neuro-2a cell injury. Neuro-2a cells were transfected with negative siRNA or DRAM1 siRNA and then subjected to 4 h OGD and 12 h reperfusion. (A) Neuro-2a cells were transfected with negative siRNA or DRAM1 siRNA; (B) The quantitative results from (A); (C) Cell viability was assessed using the LIVE/DEAD viability/cytotoxicity kit; (D) Relative protein expression levels. At the indicated pretreatment, Neuro-2a cells were collected, and protein was extracted to assess LC3, p62, and LAMP2 expression through Western blot analysis. β-Actin was used as an internal control; and (E) The quantitative results from (D). β-Actin was used as an internal control (* p < 0.05 vs. control, # p < 0.05 vs. ODG/R 12 h group, n = 6).
2.4. DRAM1 Knockdown Blocks Autophagy by Decreasing Autophagosome-Lysosome Fusion
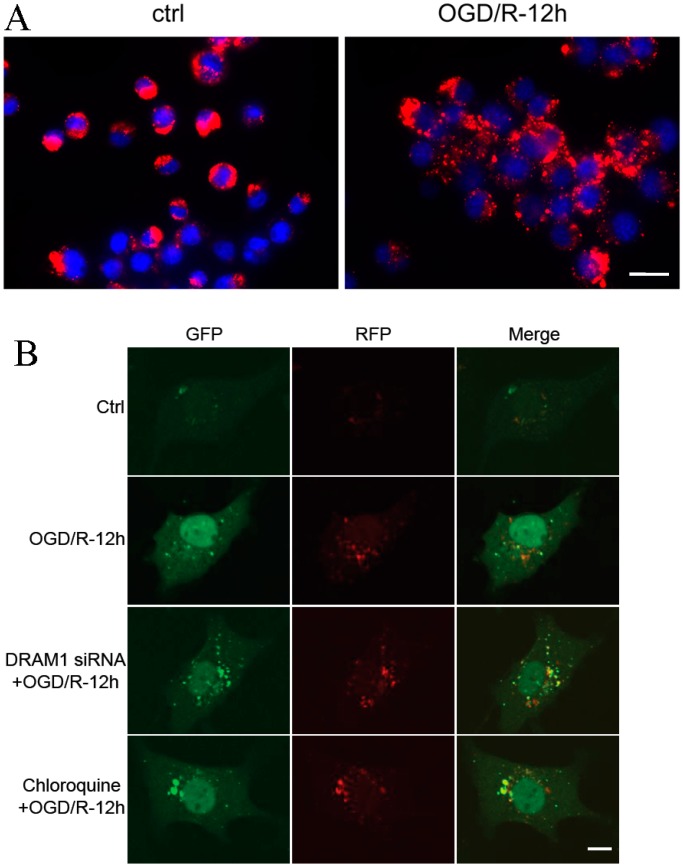
To investigate the mechanism by which DRAM1 regulates autophagy, we investigated the effects of DRAM1 on autophagosome-lysosome fusion because of its subcellular location. Lysosomes are rich in hydrolytic enzymes and are responsible for the degradation of intracellular materials captured by autophagy. LysoTracker Red was used to stain lysosomes. OGD/R treatment increased the number of LysoTracker-labeled lysosomes (Figure 4A). Neuro-2a cells were transfected with a red fluorescent protein–Green fluorescent protein–microtubule associated protein 1 light chain 3 (RFP–GFP–LC3) expression vector to monitor autophagy [33]. Increases in both autophagosomes and autolysosomes (as indicated by RFP–GFP–LC3) [34] were considered an “intact” flux in vivo. After autophagosome formation, autophagosome turnover is mostly determined by the process of autophagosome-lysosome fusion and the degradation of autophagy contents by lysosomal enzymes [35]. Due to differences in acid tolerance, dual fluorescent protein expression vectors were used to observe the autophagy process. When autophagosomes fuse with lysosomes, GFP disappears within the lysosomal environment and RFP remains. Thus, different fluorescence signals may indicate different steps in autophagy. At 12 h after OGD/R, the red LC3 puncta had increased compared with the control group; however, the yellow LC3 puncta were less because the autophagosome clearance was smooth. DRAM1 knockdown increased the yellow puncta (Figure 4B), which suggest that the accumulated autophagosomes in OGD/R-induced Neuro-2a cells are not attributable to inhibited autophagic flux [9]. Pretreatment with chloroquine resulted in a phenomenon similar to DRAM1 knockdown. Chloroquine raises the lysosomal pH and inhibits the fusion of autophagosomes and lysosomes, thus preventing the maturation of autophagosomes into autolysosomes and blocking a late step of autophagy [36]. Lysosome destabilization is induced by DRAM1 in virus-cell interactions, and knocking down DRAM expression with specific siRNAs inhibits autophagy and lysosomal membrane permeabilization [37]. Zhang et al. reported that DRAM1 regulates autophagy in a 3-Nitropropionic Acid (3-NP) treatment A549 cell model through promoting lysosomal acidification and activating lysosomal enzymes [38]. Lysosomal pH and permeability might be involved in DRAM1-mediated autophagy. Furthermore, some reports have documented that mitochondria are also involved in DRAM1-mediated autophagy. A mitochondrial inhibitor upregulates DRAM1 expression, and mitochondrial protein synthesis inhibition induces autophagy through the DRAM1 pathway [39]. Autophagy, at least partially, contributes to the neurodegeneration induced by mitochondria dysfunction [40]. DRAM1-mediated autophagy may be a complex process regulated by multi-organelles. These results suggested that DRAM1 knockdown might block autophagy through decreasing autophagosome-lysosome fusion, and the specific mechanism of DRAM1 on autophagosome-lysosome fusion requires further study in the future to fully understand the role of DRAM1 in autophagy.
Figure 4.
DRAM1 knockdown blocks autophagy by decreasing autophagosome-lysosome infusion. (A) After OGD/R 12 h treatment, Neuro-2a cells were incubated with LysoTracker Red (50 mM) for 30 min; and (B) After DRAM1 siRNA or chloroquine pretreatment and red fluorescent protein–Green fluorescent protein–microtubule associated protein 1 light chain 3 (RFP–GFP–LC3) reporter plasmid transfection, Neuro-2a cells were subjected to OGD/R 12 h and then observed with confocal microscope. Scale bar = 15 µm.
3. Experimental Section
3.1. Cell Culture
The mouse neuroblastoma cell line Neuro-2a was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 100 μg/mL streptomycin (Gibco, Grand Island, NY, USA) and 100 U/mL penicillin (Gibco, Grand Island, NY, USA). The cells were maintained at 37 °C in a saturated-humidity atmosphere containing 95% air/5% CO2.
3.2. Transfection with the Red Fluorescent Protein–Green Fluorescent Protein–Microtubule Associated Protein 1 Light Chain 3 (RFP–GFP–LC3) Expression Vector and DRAM1 Knockdown
Neuro-2a cells were transfected with the GFP–RFP–LC3 expression vector or DRAM1 siRNA using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. Empty vector controls or negative siRNA controls were used for each transfection. The GFP–RFP–LC3 reporter plasmid (ptfLC3; Addgene, Cambridge, MA, USA) has previously been described [34]. Small interfering RNAs (siRNA) targeting DRAM1 mRNA (5'-CCACGATGTATACAAGATA-3', Invitrogen, Waltham, MA, USA) and a negative siRNA (5'-TAAGGCTATGAAGAGATAC-3', Invitrogen, Waltham, MA, USA) have been described and validated previously [16,41]. The cells were subjected to treatment 48 h after transfection.
3.3. Oxygen-Glucose Deprivation (OGD) and Reperfusion
The OGD and reperfusion model was established as described previously [42]. DMEM medium was removed, and the Neuro-2a cells were washed twice with oxygen and glucose-free Hank’s Balanced Salt Solution (HBSS, pH 7.4, Gibco, Grand Island, NY, USA). Then, the medium was replaced with oxygen and glucose-free HBSS, and the cells were transferred to an anaerobic chamber containing 95% N2 and 5% CO2 at 37 °C. After 4 h, the cells were maintained in DMEM with 10% FBS and returned to normal conditions for 0, 6, 12 or 24 h.
3.4. Cell Viability Assay
The cell viability assay was performed using the LIVE/DEAD viability/cytotoxicity kit for mammalian cells (Invitrogen, Waltham, MA, USA). Neuro-2a cells were cultured in 96-well plates for 24–48 h until acceptable cell density levels were reached. The cells were rinsed twice with D-PBS (Gibco, Grand Island, NY, USA) to remove serum esterases and then incubated for 30 min at room temperature in a mixture of 2 μM calcein AM (Invitrogen, Waltham, MA, USA) and 4 μM ethidium homodimer-1 (Invitrogen, Waltham, MA, USA). The live (calcein AM-labeled, green) and dead (ethidium homodimer-1-labeled, red) cells were visualized on a fluorescence microscope (Olympus IX 71, Tokyo, Japan) and automatically counted using ImageJ software (National Institutes of Health, Maryland, MD, USA).
3.5. Lysosome Staining and GFP–RFP–Fluorescence Observation
The LysoTracker Red and Hoechst stains were diluted in DMEM. The cells were cultured in 37 °C medium containing 50 nM LysoTracker Red (Sigma-Aldrich, St. Louis, MO, USA) and 10 μg/mL Hoechst (Sigma-Aldrich, St. Louis, MO, USA) for 30 min. The cells were observed and analyzed using a fluorescence microscope. After GFP–RFP–LC3 reporter plasmid transfection and different treatment, the cells were observed and imaged with a confocal microscope (Leica Camera AG, Solms, Germany), more than 30 cells were calculated.
3.6. Western Blot Analyzes
Western blotting was performed as previously described [43]. The cells were rinsed twice with ice-cold PBS (Beyotime, Jiangsu, China) and harvested with RIPA lysis buffer (Beyotime, Jiangsu, China). Total protein was quantified and separated on a 10%–15% SDS/polyacrylamide gel (SDS–PAGE, Beyotime, Jiangsu, China) and transferred to nitrocellulose membranes (Millipore, Massachusetts, MA, USA). The membranes were blocked in TBS–Tween buffer containing 20 mM Tris-HCl (Beyotime, Jiangsu, China, 150 mM NaCl, 5% nonfat milk, and 0.05% Tween-20 (pH 7.5) (Beyotime, Jiangsu, China) for 1 h at room temperature and probed with specific antibodies against human LC3 (1:400; Abcam, Cambridge, UK), DRAM1 (1:1000; Abcam, Cambridge, UK), LAMP2 (1:400; Abcam, Cambridge, UK), p62 (1:500; Abcam, Cambridge, UK), and β-actin (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4 °C overnight. The protein levels were normalized to β-actin. The membranes were rinsed five times (5 min each) with PBS-T (Beyotime, Jiangsu, China) and then incubated with an appropriate secondary antibody (diluted 1:10,000 in PBS, 0.01% Tween-20; Thermo Scientific, Maryland, MA, USA) for 1 h, followed with rinsing five times (5 min each) in PBS-T. Immunoreactivity was detected with enhanced chemoluminescent autoradiography (eECL Western Blot Kit, Cwbiotech, Beijing, China) according to the manufacturer’s instructions. The protein expression levels were quantitatively analyzed using ImageJ software.
3.7. Statistical Analyzes
All results were expressed as means ± SDs. The differences between multiple groups were computed using a one-way analysis of variance (ANOVA) followed by Tukey’s test. A value of p < 0.05 was considered significant.
4. Conclusions
In conclusion, we demonstrated that DRAM1 regulated autophagy in an OGD/R-induced mouse Neuro-2a cell injury model. DRAM1 was initially described as regulating autophagy in cancer; here, we extended its role in the context of cerebral I/R. Our study suggested that DRAM1 expression patterns and autophagic pathways were regulated in OGD/R-induced Neuro-2a cell injury. Inhibition of autophagy exacerbated OGD/R-induced Neuro-2a cell injury, which involved the effects of DRAM1 and autophagosome-lysosome infusion. These findings may provide experimental and therapeutic options for the treatment of a broad range of pathologic disorders associated with cerebral ischemia reperfusion injury.
Author Contributions
Mengqiang Yu and Yugang Jiang conceived and designed the experiments; Mengqiang Yu, Qingliang Feng, Yi’an Ouyang and Jie Gan performed the experiments; Mengqiang Yu and Yugang Jiang analyzed the data and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Donnan G.A., Fisher M., Macleod M., Davis S.M. Stroke. Lancet. 2008;371:1612–1623. doi: 10.1016/S0140-6736(08)60694-7. [DOI] [PubMed] [Google Scholar]
- 2.Pan J., Konstas A.A., Bateman B., Ortolano G.A., Pile-Spellman J. Reperfusion injury following cerebral ischemia: Pathophysiology, MR imaging, and potential therapies. Neuroradiology. 2007;49:93–102. doi: 10.1007/s00234-006-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchiyama Y., Koike M., Shibata M. Autophagic neuron death in neonatal brain ischemia/hypoxia. Autophagy. 2008;4:404–408. doi: 10.4161/auto.5598. [DOI] [PubMed] [Google Scholar]
- 4.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z., Klionsky D.J. Eaten alive: A history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui D., Wang L., Qi A., Zhou Q., Zhang X., Jiang W. Propofol prevents autophagic cell death following oxygen and glucose deprivation in PC12 cells and cerebral ischemia-reperfusion injury in rats. PLoS One. 2012;7:e35324. doi: 10.1371/journal.pone.0035324. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Xu F., Li J., Ni W., Shen Y.W., Zhang X.P. Peroxisome proliferator-activated receptor-γ agonist 15d-prostaglandin J2 mediates neuronal autophagy after cerebral ischemia-reperfusion injury. PLoS One. 2013;8:e55080. doi: 10.1371/journal.pone.0055080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koike M., Shibata M., Tadakoshi M., Gotoh K., Komatsu M., Waguri S., Kawahara N., Kuida K., Nagata S., Kominami E., et al. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am. J. Pathol. 2008;172:454–469. doi: 10.2353/ajpath.2008.070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen Y.D., Sheng R., Zhang LS., Han R., Zhang X., Zhang X.D., Han F., Fukunaga K., Qin Z.H. Neuronal injury in rat model of permanent focal cerebral ischemia is associated with activation of autophagic and lysosomal pathways. Autophagy. 2008;4:762–769. doi: 10.4161/auto.6412. [DOI] [PubMed] [Google Scholar]
- 10.Liu K., Sun Y., Gu Z., Shi N., Zhang T., Sun X. Mitophagy in ischaemia/reperfusion induced cerebral injury. Neurochem. Res. 2013;38:1295–1300. doi: 10.1007/s11064-013-1033-0. [DOI] [PubMed] [Google Scholar]
- 11.Su F., Zhang P., Jiang Z.W., Peng D.Q., Gao L.Y., Liu S.Q., Qian L.B., Ye Z.G., Xia Q. Expression and function of autophagy after ischemia/reperfusion in rats hippocampus neuron. Chin. J. Appl. Physiol. 2011;27:187–191. [PubMed] [Google Scholar]
- 12.Carloni S., Buonocore G., Balduini W. Protective role of autophagy in neonatal hypoxia–ischemia induced brain injury. Neurobiol. Dis. 2008;32:329–339. doi: 10.1016/j.nbd.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Wang P., Guan Y.F., Du H., Zhai Q.W., Su D.F., Miao C.Y. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemic stroke. Autophagy. 2012;8:77–87. doi: 10.4161/auto.8.1.18274. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X., Yan H., Yuan Y., Gao J., Shen Z., Cheng Y., Shen Y., Wang R.R., Wang X., Hu W.W., et al. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013;9:1321–1333. doi: 10.4161/auto.25132. [DOI] [PubMed] [Google Scholar]
- 15.Wei K., Wang P., Miao C.Y. A double-edged sword with therapeutic potential: An updated role of autophagy in ischemic cerebral injury. CNS Neurosci. Ther. 2012;18:879–886. doi: 10.1111/cns.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crighton D., Wilkinson S., O’Prey J., Syed N., Smith P., Harrison P.R., Gasco M., Garrone O., Crook T., Ryan K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi N., Qipshidze N., Munjal C., Vacek J.C., Metreveli N., Givvimani S., Tyagi S.C. Tetrahydrocurcumin ameliorates homocysteinylated cytochrome-c mediated autophagy in hyperhomocysteinemia mice after cerebral ischemia. J. Mol. Neurosci. 2012;47:128–138. doi: 10.1007/s12031-011-9695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contartese A., Valoti M., Corelli F., Pasquini S., Mugnaini C., Pessina F., Aldinucci C., Sgaragli G., Frosini M. A novel CB2 agonist, COR167, potently protects rat brain cortical slices against OGD and reperfusion injury. Pharmacol. Res. 2012;66:555–563. doi: 10.1016/j.phrs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L.P., Ji C., Lu P.H., Li C., Xu B., Gao H. Oxygen glucose deprivation (OGD)/re-oxygenation-induced in vitro neuronal cell death involves mitochondrial cyclophilin-D/P53 signaling axis. Neurochem. Res. 2013;38:705–713. doi: 10.1007/s11064-013-0968-5. [DOI] [PubMed] [Google Scholar]
- 20.Rami A., Langhagen A., Steiger S. Focal cerebral ischemia induces upregulation of Beclin 1 and autophagy-like cell death. Neurobiol. Dis. 2008;29:132–141. doi: 10.1016/j.nbd.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Adhami F., Schloemer A., Kuan C.Y. The roles of autophagy in cerebral ischemia. Autophagy. 2007;3:42–44. doi: 10.4161/auto.3412. [DOI] [PubMed] [Google Scholar]
- 22.Eskelinen E.L., Illert A.L., Tanaka Y., Schwarzmann G., Blanz J., von Figura.K., Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol. Biol. Cell. 2002;13:3355–3368. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eskelinen E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Asp. Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shvets E., Elazar Z. Autophagy-independent incorporation of GFP–LC3 into protein aggregates is dependent on its interaction with p62/SQSTM1. Autophagy. 2008;4:1054–1056. doi: 10.4161/auto.6823. [DOI] [PubMed] [Google Scholar]
- 26.Xing S., Zhang Y., Li J., Zhang J., Li Y., Dang C., Li C., Fan Y., Yu J., Pei Z., et al. Beclin 1 knockdown inhibits autophagic activation and prevents the secondary neurodegenerative damage in the ipsilateral thalamus following focal cerebral infarction. Autophagy. 2012;8:63–76. doi: 10.4161/auto.8.1.18217. [DOI] [PubMed] [Google Scholar]
- 27.Kerley-Hamilton J.S., Pike A.M., Hutchinson J.A., Freemantle S.J., Spinella M.J. The direct p53 target gene, FLJ11259/DRAM, is a member of a novel family of transmembrane proteins. Biochim. Biophys. Acta. 2007;1769:209–219. doi: 10.1016/j.bbaexp.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Prey J., Skommer J., Wilkinson S., Ryan K.M. Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy. Cell Cycle. 2009;8:2260–2265. doi: 10.4161/cc.8.14.9050. [DOI] [PubMed] [Google Scholar]
- 29.Crighton D., Wilkinson S., Ryan K.M. DRAM links autophagy to p53 and programmed cell death. Autophagy. 2007;3:72–74. doi: 10.4161/auto.3438. [DOI] [PubMed] [Google Scholar]
- 30.Galavotti S., Bartesaghi S., Faccenda D., Shaked-Rabi M., Sanzone S., McEvoy A., Dinsdale D., Condorelli F., Brandner S., Campanella M., et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 31.Eskelinen E.L., Cuervo A.M., Taylor M.R., Nishino I., Blum J.S., Dice J.F., Sandoval I.V., Lippincott-Schwartz J., August J.T., Saftig P. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic. 2005;6:1058–1061. doi: 10.1111/j.1600-0854.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- 32.Hariharan N., Zhai P., Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid. Redox Signal. 2011;14:2179–2190. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka Y., Guhde G., Suter A., Eskelinen E.L., Hartmann D., Lullmann-Rauch R., Janssen P.M., Blanz J., von Figura K., Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 35.Ni H.M., Bockus A., Wozniak A.L., Jones K., Weinman S., Yin X.M., Ding W.X. Dissecting the dynamic turnover of GFP–LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klionsky D.J., Baehrecke E.H., Brumell J.H., Chu C.T., Codogno P., Cuervo A.M., Debnath J., Deretic V., Elazar Z., Eskelinen E.L., et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6:438–448. doi: 10.4161/auto.6.4.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laforge M., Limou S., Harper F., Casartelli N., Rodrigues V., Silvestre R., Haloui H., Zagury J.F., Senik A., Estaquier J. DRAM triggers lysosomal membrane permeabilization and cell death in CD4+ T cells infected with HIV. PLoS Pathog. 2013;9:e1003328. doi: 10.1371/journal.ppat.1003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X.D., Qi L., Wu J.C., Qin Z.H. DRAM1 regulates autophagy flux through lysosomes. PLoS One. 2013;8:e63245. doi: 10.1371/journal.pone.0063245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X., Le L., Fan Y., Lv L., Zhang J. Autophagy is induced through the ROS–TP53–DRAM1 pathway in response to mitochondrial protein synthesis inhibition. Autophagy. 2012;8:1071–1084. doi: 10.4161/auto.20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X.D., Wang Y., Wang Y., Zhang X., Han R., Wu J.C., Liang Z.Q., Gu Z.L., Han F., Fukunaga K., et al. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy. 2009;5:339–350. doi: 10.4161/auto.5.3.8174. [DOI] [PubMed] [Google Scholar]
- 41.Bensaad K., Cheung E.C., Vousden K.H. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi Z., Ma X., Cui G., Li M., Li F. Cinnamtannin B-1 regulates cell proliferation of spinal cord astrocytes and protects the cell fromoxygen-glucose-serum deprivation/reoxygenation-induced apoptosis. Int. J. Mol. Sci. 2013;14:15827–15837. doi: 10.3390/ijms140815827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bendix I., Schulze C., von Haefen C., Gellhaus A., Endesfelder S., Heumann R., Felderhoff-Mueser U., Sifringer M. Erythropoietin modulates autophagy signaling in the developing rat brain in an in vivo model of oxygen-toxicity. Int. J. Mol. Sci. 2012;13:12939–12951. doi: 10.3390/ijms131012939. [DOI] [PMC free article] [PubMed] [Google Scholar]