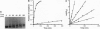Abstract
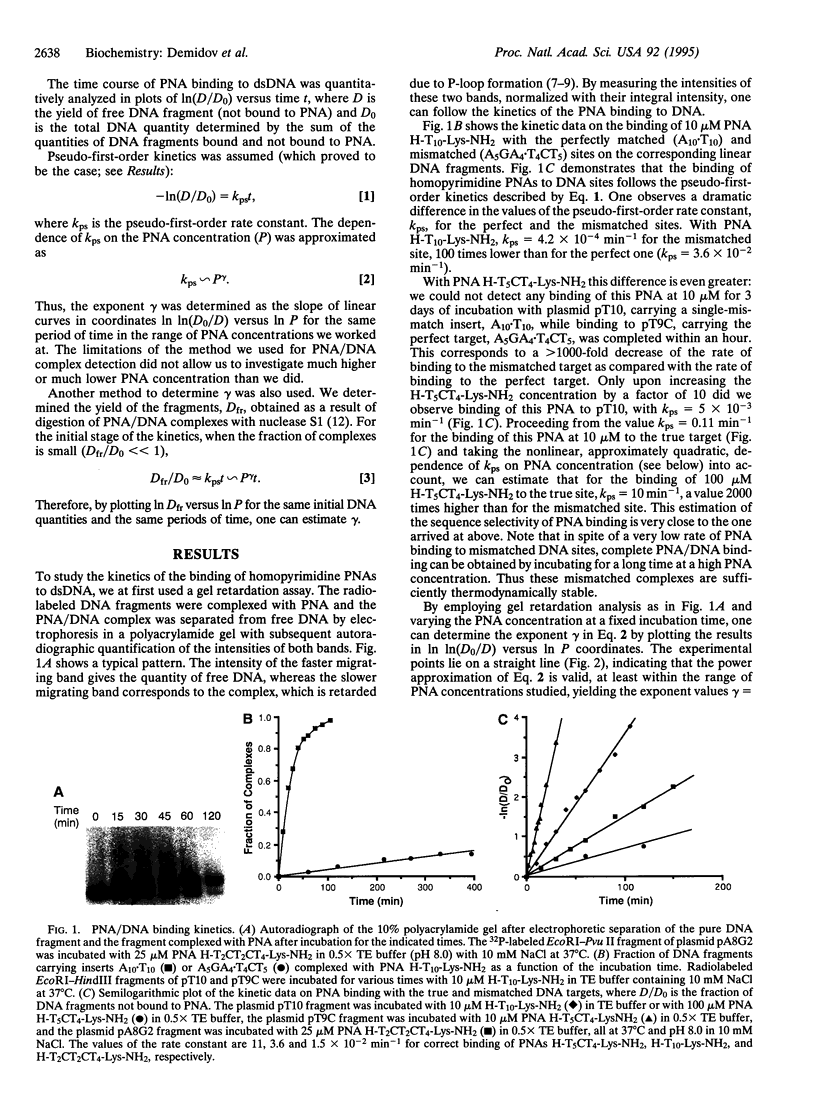
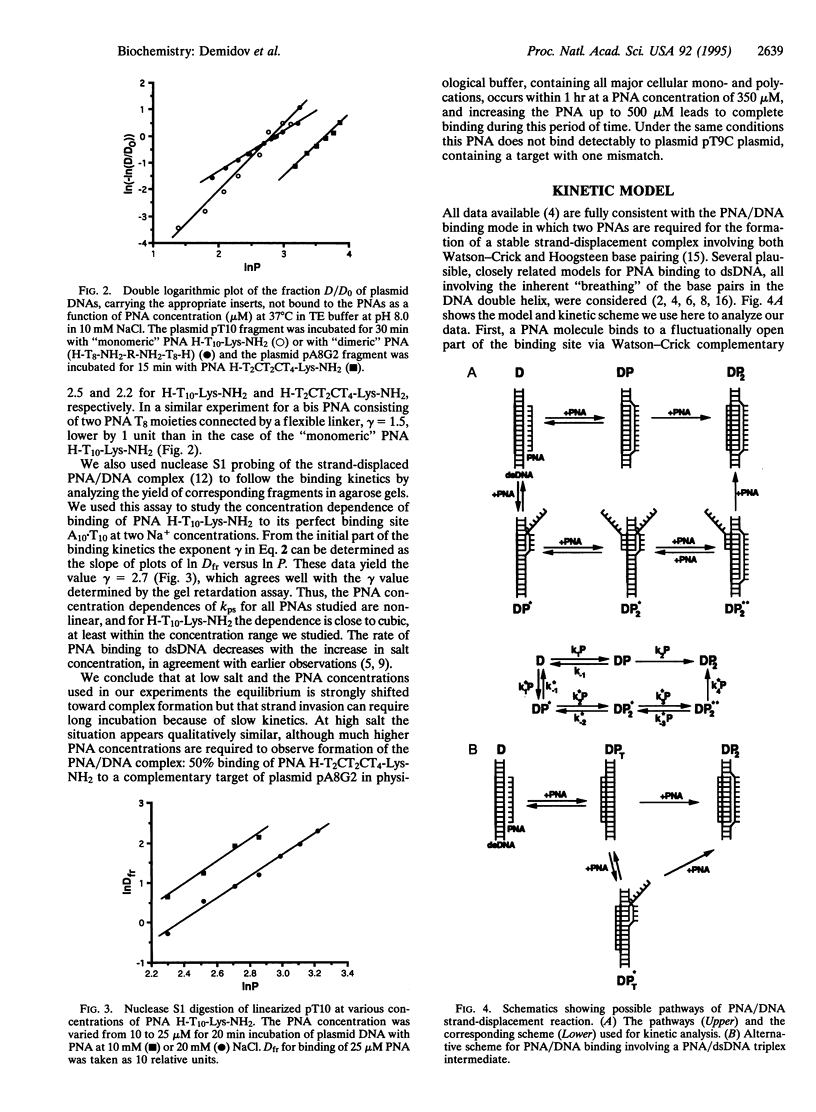
To elucidate the mechanism of recognition of double-stranded DNA (dsDNA) by homopyrimidine polyamide ("peptide") nucleic acid (PNA) leading to the strand-displacement, the kinetics of the sequence-specific PNA/DNA binding have been studied. The binding was monitored with time by the gel retardation and nuclease S1 cleavage assays. The experimental kinetic curves obey pseudo-first-order kinetics and the dependence of the pseudo-first-order rate constant, kps, on PNA concentration, P, obeys a power law kps approximately P gamma with 2 < gamma < 3. The kps values for binding of decamer PNA to dsDNA target sites with one mismatch are hundreds of times slower than for the correct site. A detailed kinetic scheme for PNA/DNA binding is proposed that includes two major steps of the reaction of strand invasion: (i) a transient partial opening of the PNA binding site on dsDNA and incorporation of one PNA molecule with the formation of an intermediate PNA/DNA duplex and (ii) formation of a very stable PNA2/DNA triplex. A simple theoretical treatment of the proposed kinetic scheme is performed. The interpretation of our experimental data in the framework of the proposed kinetic scheme leads to the following conclusions. The sequence specificity of the recognition is essentially provided at the "search" step of the process, which consists in the highly reversible transient formation of duplex between one PNA molecule and the complementary strand of duplex DNA while the other DNA strand is displaced. This search step is followed by virtually irreversible "locking" step via PNA2/DNA triplex formation. The proposed mechanism explains how the binding of homopyrimidine PNA to dsDNA meets two apparently mutually contradictory features: high sequence specificity of binding and remarkable stability of both correct and mismatched PNA/DNA complexes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almarsson O., Bruice T. C., Kerr J., Zuckermann R. N. Molecular mechanics calculations of the structures of polyamide nucleic acid DNA duplexes and triple helical hybrids. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7518–7522. doi: 10.1073/pnas.90.16.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anshelevich V. V., Vologodskii A. V., Lukashin A. V., Frank-Kamenetskii M. D. Slow relaxational processes in the melting of linear biopolymers: a theory and its application to nucleic acids. Biopolymers. 1984 Jan;23(1):39–58. doi: 10.1002/bip.360230105. [DOI] [PubMed] [Google Scholar]
- Cherny D. Y., Belotserkovskii B. P., Frank-Kamenetskii M. D., Egholm M., Buchardt O., Berg R. H., Nielsen P. E. DNA unwinding upon strand-displacement binding of a thymine-substituted polyamide to double-stranded DNA. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1667–1670. doi: 10.1073/pnas.90.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M. E., Crothers D. M., Doty P. Relaxation kinetics of dimer formation by self complementary oligonucleotides. J Mol Biol. 1971 Dec 14;62(2):383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- Demidov V. V., Cherny D. I., Kurakin A. V., Yavnilovich M. V., Malkov V. A., Frank-Kamenetskii M. D., Sönnichsen S. H., Nielsen P. E. Electron microscopy mapping of oligopurine tracts in duplex DNA by peptide nucleic acid targeting. Nucleic Acids Res. 1994 Dec 11;22(24):5218–5222. doi: 10.1093/nar/22.24.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov V. V., Potaman V. N., Frank-Kamenetskii M. D., Egholm M., Buchard O., Sönnichsen S. H., Nielsen P. E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 1994 Sep 15;48(6):1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Demidov V., Frank-Kamenetskii M. D., Egholm M., Buchardt O., Nielsen P. E. Sequence selective double strand DNA cleavage by peptide nucleic acid (PNA) targeting using nuclease S1. Nucleic Acids Res. 1993 May 11;21(9):2103–2107. doi: 10.1093/nar/21.9.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egholm M., Buchardt O., Christensen L., Behrens C., Freier S. M., Driver D. A., Berg R. H., Kim S. K., Norden B., Nielsen P. E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993 Oct 7;365(6446):566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Peffer N. J., Bisi J. E., Thomson S. A., Cadilla R., Josey J. A., Ricca D. J., Hassman C. F., Bonham M. A., Au K. G. Antisense and antigene properties of peptide nucleic acids. Science. 1992 Nov 27;258(5087):1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- Møllegaard N. E., Buchardt O., Egholm M., Nielsen P. E. Peptide nucleic acid.DNA strand displacement loops as artificial transcription promoters. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3892–3895. doi: 10.1073/pnas.91.9.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Peptide nucleic acids (PNAs): potential antisense and anti-gene agents. Anticancer Drug Des. 1993 Feb;8(1):53–63. [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Sequence specific inhibition of DNA restriction enzyme cleavage by PNA. Nucleic Acids Res. 1993 Jan 25;21(2):197–200. doi: 10.1093/nar/21.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991 Dec 6;254(5037):1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Buchardt O. Evidence for (PNA)2/DNA triplex structure upon binding of PNA to dsDNA by strand displacement. J Mol Recognit. 1994 Sep;7(3):165–170. doi: 10.1002/jmr.300070303. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Buchardt O. Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjug Chem. 1994 Jan-Feb;5(1):3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Buchardt O. Sequence-specific transcription arrest by peptide nucleic acid bound to the DNA template strand. Gene. 1994 Nov 4;149(1):139–145. doi: 10.1016/0378-1119(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Peffer N. J., Hanvey J. C., Bisi J. E., Thomson S. A., Hassman C. F., Noble S. A., Babiss L. E. Strand-invasion of duplex DNA by peptide nucleic acid oligomers. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10648–10652. doi: 10.1073/pnas.90.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibanyenda N., De Bruin S. H., Haasnoot C. A., van der Marel G. A., van Boom J. H., Hilbers C. W. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G) . d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur J Biochem. 1984 Feb 15;139(1):19–27. doi: 10.1111/j.1432-1033.1984.tb07970.x. [DOI] [PubMed] [Google Scholar]
- Wittung P., Nielsen P. E., Buchardt O., Egholm M., Nordén B. DNA-like double helix formed by peptide nucleic acid. Nature. 1994 Apr 7;368(6471):561–563. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]