Abstract
The present study aimed to examine the changes in mesenteric lymph during the development of sepsis and to identify the distinct proteins involved, as targets for further study. The sepsis animal model was constructed by cecal ligation and puncture (CLP). The mesenteric lymph was collected from 28 adult male Sprague-Dawley rats, which were randomly divided into the following four groups (n=7 per group): CLP-6 h, CLP-24 h, sham-6 h and sham-24 h groups. Capillary high performance liquid chromatography-tandem mass spectrometry was performed to analyze the proteome in mesenteric lymph. A comprehensive bioinformatic analysis was then conducted to investigate the distinct proteins. Compared with the sham group, 158 distinct proteins were identified in the lymph samples from the CLP group. Five of these proteins associated with the same lipid metabolism pathway were selected, apolipoprotein E (ApoE), annexin A1 (Anxa1), neutrophil gelatinase-associated lipocalin (NGAL), S100a8 and S100a9. The expression of ApoE, Anxa1, NGAL, S100a8 and S100a9 were all elevated in the progression of sepsis. The five proteins were reported to be closely associated with disease development and may be a potential target for the diagnosis and treatment of sepsis. In conclusion, identifying proteome changes in mesenteric lymph provides a novel perspective to understand the pathological mechanisms underlying sepsis.
Keywords: lymph, proteome, sepsis, cecal ligation and puncture, bioinformation
Introduction
Sepsis, a systemic inflammatory response syndrome, is a leading cause of mortality in critically ill patients (1). Sepsis resulting from severe infection is frequently associated with confusion, metabolic acidosis, low blood pressure, decreased systemic vascular resistance and blood coagulation dysfunctions (2). Severe sepsis may lead to organ dysfunction and septic shock (3). Although the recommendations of the treatment targeting for sepsis are updated every four years (4–6), the mortality rate resulting from sepsis remains high, at between 30 and 50% (7).
To decrease the mortality rate and identify an effective treatment for sepsis, numerous studies have focused on identifying the pathogenic mechanisms that underlie sepsis. Disease-conditioned mesenteric lymph, but not portal blood, carries high levels of gut-derived factors leading to organ injury (8,9). As outlined in previous studies, protein fractions in the lymph are partly responsible for gut-origin sepsis (10,11). In addition, certain proteins in the lymph promote the progression of sepsis by acting as carriers of biologically active molecules or effector molecules. With advances in modern technology, proteomic analysis in lymph has provided a number of breakthroughs in examining the mechanisms underlying this critical disease (12–15).
Previously, investigations concerning sepsis have primarily focused on the proteome in the plasma (16,17), heart (1), liver (18) and skeletal muscle (19); however, there is little information regarding the role of lymph proteins. The experimental sepsis model induced by cecal ligation and puncture (CLP) has been widely used to reproduce numerous pathophysiological features of clinical sepsis (20,21). The significant changes in the levels of plasma proteins in mice with CLP-induced sepsis were reported to be during the late stage of septic development (24 h following surgery) (17). Therefore, in the present study, a sepsis model was constructed by CLP and the lymph was collected at 6 h and 24 h following surgery, which represented the early and the late stages of the acute phase, respectively. The aim was to examine the changes in rodent proteome mesenteric lymph in response to sepsis, in order to contribute to the current understanding of the pathogenesis of sepsis progression.
Materials and methods
Animals
A total of 28 adult male Sprague-Dawley rats (Slaccas Laboratory Inc., Shanghai, China), weighing 250–300 g, were housed at 25°C and subjected to 12 h light/dark cycle treatment conditions. All of the animals were fed a standard rodent diet (Slaccas Laboratory Inc.) with access to water ad libitum. All of the animals received humane care in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals. Approval was obtained from the Institutional Animal Care and Use Committee at the Second Military Medical University (Shanghai, China).
Experimental model
The animals were randomly divided into the following four groups (n=7 per group): CLP-6 h, sham-6 h, CLP-24 h and sham-24 h groups. In the CLP groups, the sepsis models were constructed by CLP as previously described (22). Briefly, general anesthetic was induced by intraperitoneal injection of pentobarbital (40 mg/kg; Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) and CLP was performed with a single in-and-out puncture (20 gage needle; Yangzhou Great Wall Medical Equipment Factory, Yangzhou, China) at the middle part of the cecum. To ensure successful puncture, droplets of feces were extruded from the penetration holes. The rats in the sham groups underwent cecum exenteration under general anesthetic instead of CLP. After surgery, all the rats were injected with 100 g/5 ml of normal saline subcutaneously. Following recovery from the anesthesia, the rats received free access to food and water ad libitum.
Lymph collection
Prior to the CLP/sham surgery, the thoracic duct was cannulated with micro-urethane tubing (inner diameter, 0.635 mm; outer diameter, 1.02 mm; American Health & Medical Supply International Corp. Co., Ltd., New York, NY, USA) and externalized through a 14-gage angiocatheter (Hospira, Inc., Lake Forest, IL, USA) in the left flank of all the experimental animals. At 6 h and 24 h following CLP/sham surgery, the lymph samples were collected from all groups. Lymph collection was conducted by thoracic duct drainage according to a modified method that has been previously described (23). In brief, the rats were re-anesthetized with intraperitoneal injection of 40 mg/kg pentobarbital. The lymph samples were collected directly into sterile ice-chilled vacuum EDTA tubes (Shanghai Kehua Bio-engineering Co., Ltd., Shanghai; China) for 60 min. One sample from the seven rats within one group was centrifuged at 4°C (1,700 × g) for 15 min and immediately stored at −80°C for further analysis.
Proteomic analysis
Proteomic analysis, according to a previous description (24), and bioinformatic analysis were performed by Sensichip Infotech Co., Ltd. (Shanghai, China). The bioinformatics analysis was used to analyze the molecular functions of differentially expressed genes of the identified proteins. All of the chemical reagents, software and equipment were provided by this company. The experimental procedures were performed according to standardized manufacturer’s instructions.
Major protein depletion
The lymph samples were defrosted, diluted and filtered through a 0.22-μm filter membrane (AB SCIEX, Framingham, MA, USA). Next, three major abundant proteins (albumin, IgG and transferrin) were depleted by the Multiple Affinity Removal Column (M-3; Agilent Technologies Co., Ltd., Santa Clara, CA, USA). When the samples had been individually depleted, quantitative analysis of the lymph samples was performed with the Dc protein assay reagent (Bio-Rad, Hercules, CA, USA). The samples were then desalted and concentrated by ultrafiltration with a Vivaspin 4 centrifugal concentrator and 5-kDa polyethersulfone filter (Sartorius AG, Göttingen, Germany). The target proteins were detected by 12.5% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) electrophoresis.
Capillary high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS)-based proteomics
The proteins from each sample (150 μg) were dissolved in 200 μl UA buffer (8 M urea in 150 mM Tris-HCl; pH 8.0). The solution was centrifuged (14,000 × g, 30 min) twice and the supernate was abandoned. The protein sample was then incubated with 100 μl iodoacetamide in the dark at room temperature for 30 min. Following centrifugation (14,000 × g, 20 min), the sample was mixed with 100 μl UA buffer followed by reduplicate centrifugation (14,000 × g, 20 min). The protein sample underwent reduction (blending with 100 μl dissolution buffer and reduplicate centrifugation at 14000 × g for 20 min) and was digested in 40 μl trypsin buffer (2 μg trypsin in 40 μl dissolution buffer) at 37°C for 16–18 h. Following centrifugation (14,000 × g, 10 min) in a new collection tube, the peptides were then eluted and quantified at optical density (OD)280 in the Unicam SP 600 spectrophotometer (Pye Unicam Ltd., Cambridge, UK)
The sample (50 μg) was labeled with iTRAQ reagents (iTRAQ Reagent-4 plex Multiplex kit; AB SCIEX) according to the manufacturer’s instructions. Samples from CLP-6 h, CLP-24 h, sham-6 h and sham-24 h groups were labeled with 114, 115, 116 and 117 iTRAQ isobaric tags, respectively. The labeled materials were then combined and pre-fractionated on a polysulfoethyl 4.6×100-mm column (5 μm, 200 Å; PolyLC Inc., Columbia, MD, USA) using an AKTA Purifier 100 (GE Healthcare, Little Chalfont, UK). Approximately 30 effluent fractions were collected and merged into ten fractions according to the strong cation exchange chromatogram. The fractions were freeze-dried and desalted using a C18 cartridge (Sigma, St. Louis, MO, USA).
Subsequently, HPLC analysis was performed. The samples were added to the sample column (2 cm × 100 μm, 5 μm-C18; EASY-Column Capillary Column; AB Sciex, Waltham, MA, USA) automatically and were then separated through an analytical column (75 μm × 100 mm, 3 μm-C18; EASY-Column Capillary Column; Thermo Fisher Scientific). The HPLC Easy nLC gradient between buffer A (0.1% formic acid in water) and buffer B (0.1% formic acid in acetonitrile) was formed at 250 μl/min as follows: Increasing to 35% B from 0–100 min, increasing to 100% B by 108 min and held at 100% until 120 min. The sample effluent was directed into the ion spray source of a Q-Exactive mass spectrometer (Thermo Finnigan, San Jose, CA, USA) scanning from 300 to 1800 m/z for 120 min.
Sequence database searches
The sequence database searches and quantitative analysis were performed by Mascott 2.2 software (Matrix Science Ltd., Boston, MA, USA), Proteome Discoverer 1.3 and Xcalibur softwares (Thermo Fisher Scientific).
The data were searched against the ipi.RAT.v3.87.fasta database (sequence no. 39925) using Mascot 2.2 software with the search parameters as follows: Type of search, MS/MS ion search; enzyme, trypsin; mass values, monoisotopic; max missed cleavages, 2; fixed modifications, carbamidomethyl (C), iTRAQ4plex (N-term), iTRAQ4plex (K); variable modifications, oxidation (M), iTRAQ4plex (Y); peptide mass tolerance, ±20 ppm; and fragment mass tolerance, 0.1 Da. Peptide false discovery rates ≤0.01 and a peptide Mascot score >20 were defined as significant.
Data analysis
Quantitative analysis was conducted using Proteome Discoverer 1.3 software according to the peak intensity of labeled peptides. For different samples labeled with different tags, the peptide quantification in one sample was calculated as the ratio of the signal intensity of one tag compared with other the tags. The median of the calculated ratios was considered as the result of protein quantitation. Each median of the signal ratio was normalized to eliminate human sample loading error. Differences in the relative abundance were calculated as differences in the log peak areas and reported as fold changes between two groups. Differences among the groups were analyzed by Student’s t-test. P<0.05 was considered to indicate a statistically significant difference. Differentially expressed proteins were screened by the t-test (P<0.05) and fold change (fold change>1.5) method.
Bioinformatics
In the gene ontology (GO) analysis, differentially expressed genes (DEGs) encoding the 158 identified proteins were mapped to the GO database using the GSEABase software package on the R statistics platform (http://www.r-project.org/). DEGs were classified according to three ontologies: Biological processes, cellular components and molecular functions.
In the pathway analysis, DEGs were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (www.bioconductor.org) by GenMAPP v2.1 (Gladstone Institutes, San Francisco, CA, USA) and the enrichment degree of each gene was counted in different pathways.
In the network analysis, the interactions between DEGs were analyzed by downloading the pathway data from the KEGG, MIPS (http://mips.helmholtz-muenchen.de/proj/ppi/) and PubMed databases (25) and KEGGSOAP software package (www.bioconductor.org). Inter-correlations between DEGs were analyzed using the co-citation algorithm.
Finally, the results of the three types of analyses were integrated into a network of protein-protein correlations with comprehensive consideration. The network was illustrated graphically using Medusa software (Informer Technologies, Inc., Bilbao, Spain).
Results
Sepsis model
Following induction of the sepsis model, the rats in the sham and CLP groups presented physiological changes of different degrees.
At 6 h following surgery, the rats in the sham-6 h group presented with normal physical ability. The mental state (assessed by observation of movements of the animal) and weight loss of rats was marginally lower than prior to surgery. The rats were responsive, and actively feeding and drinking. No piloerection or abdominal distension were observed and the rats had dry granular feces, soft stools and a clean anus. At 24 h, the rats in the sham-24 h presented with a similar physiological status.
However, rats in the CLP-6h group exhibited poor mental state, decreased coat glossiness, piloerection, abdominal distension, and a fairly sensitive response to stimuli (stimuloation by a needle in the tail). The rats stayed in groups, did not eat and only about half of them were actively drinking water. The abdominal incisions of the rats were red with a small quantity of exudate and stools were soft or loose. In the CLP-24 h group, the rats were more sluggish and also stayed in groups. The rats had evident abdominal distention, decreased eating and drinking habits and secretion from their eyes. The rats responded slowly in spite of intense stimulation. Feces were observed on the anuses and tails of the rats. The perianal hairs of the rats were wet and the feces were primarily loose stools.
The mortality rate was 60% in the CLP groups following ten days post-operation.
Sample preparation
After the highly-abundant protein was depleted, the concentrations of the remaining lymph protein in the CLP-6, CLP-24, sham-6 and sham-24 groups were 1.52, 1.54, 1.51 and 2.09 μg/μl, respectively. By quantitative analysis, the relative concentrations of peptides were estimated by absorbance at 280 nm (OD2801.1=1 μg/μl). The OD280 of the samples were 1.31 (CLP-6), 1.63 (CLP-24), 1.44 (sham-6) and 1.39 (sham-24), respectively. The depletion rate of the highly-abundant protein was relatively high, while the concentration of the target proteins was low.
Lymph proteomics
The abundances of 984 proteins identified in the rat lymph samples were compared between the CLP-6 and sham-6; CLP-24 and sham-24; CLP-6 and CLP-24; and sham-6 and sham-24 groups. A total of 158 distinct proteins with a fold change ≥1.5; P<0.05 between the different groups were identified in the present study. Compared with the sham-6 h group, 30 of the 158 proteins were significantly increased in the CLP-6 h group (Table I). These proteins were listed according to their molecular function (using the freely available protein database UniProt; http://www.uniprot.org/) and there were shown to be 13 binding proteins, two structural proteins, three enzymes, two protease inhibitors, one cytokine, one transcription factor, five uncharacterized proteins and three proteins with unclear function. An additional uncharacterized protein, Pfkp (fold=0.28, P=0.0016), was found to decrease significantly (fold change≤0.3, P<0.05) in the CLP-6 group.
Table I.
Upregulated proteins in the lymph of the cecal ligation and puncture-6 h group.
| Gene name | Description | Fold change | P-value |
|---|---|---|---|
| Sbp | Uncharacterized protein | 4.29 | 0.0000 |
| Scgb2a1 | Secretoglobin family 2A member 2 | 4.19 | 0.0000 |
| S100a9 | S100-A9 protein | 3.73 | 0.0000 |
| Ngp | Uncharacterized protein | 3.37 | 0.0000 |
| S100a8 | S100-A8 protein | 3.04 | 0.0000 |
| Retnlg | Resistin-like molecule gamma | 2.83 | 0.0000 |
| Psbpc2 | Prostatic steroid-binding protein C2 | 2.71 | 0.0000 |
| Psbpc1 | Prostatic steroid-binding protein C1 | 2.51 | 0.0000 |
| Klk1c9 | Submandibular glandular kallikrein-9 | 2.48 | 0.0000 |
| Tceb2 | Transcription elongation factor B polypeptide 2 | 2.38 | 0.0000 |
| LOC298109 | Uncharacterized protein | 2.07 | 0.0000 |
| Lmnb1 | Lamin-B1 | 2.06 | 0.0000 |
| Igfbp1 | Insulin-like growth factor-binding protein 1 | 2.00 | 0.0002 |
| Lcn2 | Neutrophil gelatinase-associated lipocalin | 1.97 | 0.0002 |
| Fh1 | Isoform Mitochondrial of Fumarate hydratase, mitochondrial | 1.96 | 0.0002 |
| Pigr | Polymeric immunoglobulin receptor | 1.90 | 0.0002 |
| Clca1 | Uncharacterized protein | 1.90 | 0.0002 |
| Tsku | Tsukushin | 1.89 | 0.0002 |
| LOC679994 | Histone H3.1 | 1.80 | 0.0004 |
| GF20391-like isoform 1 | 1.76 | 0.0010 | |
| Mpst | 3-Mercaptopyruvate sulfurtransferase | 1.71 | 0.0041 |
| Andpro | Cystatin-related protein 1 | 1.69 | 0.0056 |
| Myh6 | Myosin-6 | 1.64 | 0.0074 |
| Mmrn1 | Uncharacterized protein | 1.63 | 0.0072 |
| Anxa1 | Annexin A1 | 1.61 | 0.0103 |
| Nono | Non-POU domain-containing octamer-binding protein | 1.59 | 0.0123 |
| ApoE | Apolipoprotein E | 1.53 | 0.0265 |
| Serpinb1a | Leukocyte elastase inhibitor A | 1.52 | 0.0301 |
| Chi3l1 | Chitinase-3-like protein 1 | 1.51 | 0.0310 |
| Ccdc48 | EF-hand domain-containing protein ENSP00000381169 homolog | 1.50 | 0.0311 |
Compared with the sham-24 group, 20 proteins were confirmed to increase significantly (fold change≥1.5; P<0.05) in abundance of the CLP-24 group (Table II) and included six binding proteins, two enzymes, one protease inhibitor, one signal transducer protein and ten uncharacterized proteins. By contrast, two proteins were identified to decrease significantly (fold change≤0.3; P<0.05) in the CLP-24h group compared with the sham-24 group: Svs4 (fold=0.25, P=0.0016) and one uncharacterized protein, Svs6 (fold=0.27; P=0.0016).
Table II.
Upregulated proteins in the lymph of the cecal ligation and puncture-24 h group.
| Gene name | Description | Fold change | P-value |
|---|---|---|---|
| Siae | Sialate O-acetylesterase | 3.60 | 0.0000 |
| Lcn2 | Neutrophil gelatinase-associated lipocalin | 2.30 | 0.0000 |
| Mgp | Matrix Gla protein | 2.05 | 0.0001 |
| Ldlr | Uncharacterized protein | 1.81 | 0.0004 |
| Ngp | Uncharacterized protein | 1.81 | 0.0004 |
| RGD1565682 | Uncharacterized protein | 1.81 | 0.0004 |
| Ptgr1 | Prostaglandin reductase 1 | 1.80 | 0.0004 |
| Sbp | Uncharacterized protein | 1.78 | 0.0014 |
| Tnnc1 | Cardiac troponin C | 1.70 | 0.0037 |
| LOC299282 | Uncharacterized protein | 1.70 | 0.0033 |
| Vcan | Uncharacterized protein (Fragment) | 1.67 | 0.0057 |
| Myh6 | Myosin-6 | 1.64 | 0.0075 |
| Sema6d | Uncharacterized protein | 1.64 | 0.0074 |
| Il1r2 | Interleukin-1 receptor type 2 | 1.62 | 0.0102 |
| Andpro | Cystatin-related protein 1 | 1.60 | 0.0114 |
| Slc4a1 | Uncharacterized protein | 1.57 | 0.0148 |
| Fgl2 | Fibrinogen-like 2 | 1.56 | 0.0190 |
| Mpo | Uncharacterized protein | 1.54 | 0.0218 |
| Chi3l1 | Chitinase-3-like protein 1 | 1.52 | 0.0276 |
| Itpa | Uncharacterized protein | 1.51 | 0.0296 |
Bioinformatics
The 158 proteins identified in the CLP and sham groups were further analyzed for functional and biological relevance. Using GO analysis, the proteins were analyzed based of their involvement in biological processes, cellular components and molecular functions (Figs. 1–3 and cross-referenced to Tables III–V).
Figure 1.
Biological process analysis. The chart shows the biological processes that the 158 proteins are involved in.
Figure 3.
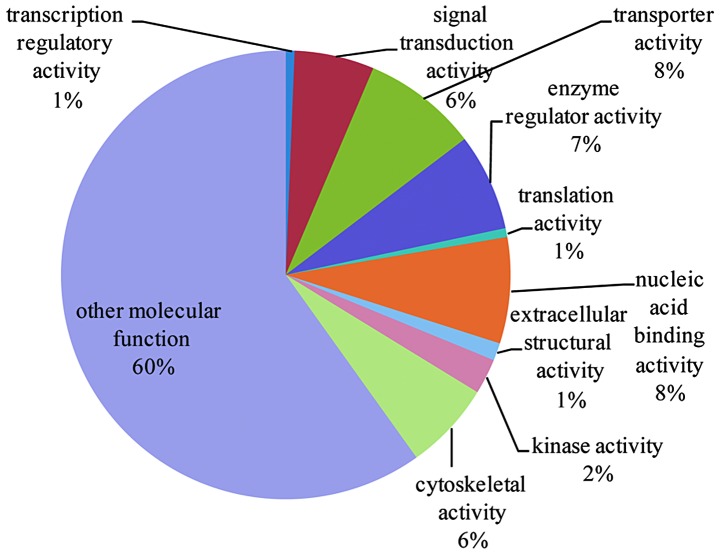
Molecular function analysis. The chart shows the function the 158 proteins are involved in.
Table III.
Biological process analysis.
| Term | Gene count | P-value | Gene list |
|---|---|---|---|
| Cell cycle and proliferation | 5 | 0.912015 | Agt, Rcc2, Anxa1, Hprt1, Ak1 |
| Transport | 27 | 0.002065 | ApoE, Hnrnpa1, Slc4a1, Agt, Fabp5, Anxa1, Ptx3, Ttr, Apof, Ldlr, Hpx, Fabp4, Mpst, Abp1, Fabp3, Lcn2, Clca1, Hba-a2, Fabp6, Myh6, Lbp, Tnnc1, Col1a1, Mb, Atp7a, Orm1, Atp5a1 |
| Stress response | 20 | 3.25E-05 | F13a1, ApoE, Saa4, Thbs1, Agt, Ngp, Kng1, Ddb1, Nono, Mpo, Hpx, Fabp4, Fn1, Hspa1l, Serpina3n, Lbp, Hspa5, Serping1, Mb, Orm1 |
| Developmental processes | 23 | 0.069737 | ApoE, Neb, Thbs1, Agt, Hist1h1b, Vcan, Mgp, Acan, Timp1, Hspa1l, Hba-a2, Myh6, Sema6d, Tpm1, Tpi1, Tagln, Hprt1, Pdlim3, Col1a1, Mb, Atp7a, Hexb, Atp5a1 |
| RNA metabolism | 4 | 0.999991 | Hnrnpa1, Nono, Fabp4, Tceb2 |
| DNA metabolism | 2 | 0.74583 | Ddb1, Nono |
| Protein metabolism | 14 | 0.86033 | F13a1, Pgc, Dpm1, Ddb1, Serpinb1a, Acy1, Hpx, Rps28, Tceb2, Serping1, Hp, Rpl8, Atp7a, sHexb |
| Other metabolic processes | 40 | 2.44E-07 | Aox1, ApoE, Agt, Fabp5, Dpm1, Ptx3, Acy1, Pgm1, Ttr, Apof, Car3, Ptgr1, Ldlr, Acan, Chi3l1, Car1, Mpo, Hpx, Fabp4, Abp1, Fabp3, Pfkp, Fabp6, Fh1, Pgam2, Lbp, Tpi1, Pgls, Pla1a, Ldha, Ckm, Hprt1, Ak1, Itpa, Eno3, Gstm1, Atp7a, Hexb, S100a9, Atp5a1 |
| Cell organization and biogenesis | 19 | 0.034714 | ApoE, Neb, Agt, Hist1h1b, Rcc2, Hist1h1d, Ptx3, Ldlr, Acan, Fn1, Myh6, Lbp, Hprt1, Hist1h1a, Pdlim3, Atp7a, Hexb, S100a9, Hist1h2ai |
| Signal transduction | 6 | 1 | Fgl2, Agt, Anxa1, Fgl1, Hpx, Hspa5 |
| Cell adhesion | 7 | 0.103657 | Thbs1, Agt, Vcan, Acan, Fn1, Col1a1, Col6a2 |
| Death | 4 | 0.788118 | Fgl2, Agt, Hprt1, Atp7a |
| Other biological processes | 46 | 0.332232 | ApoE, Pgc, Mrfap1, Agt, Ngp, Kng1, Rcc2, Sbp, Gemin4, Ptx3, A2m, Mgp, Apof, Tpm2, Ldlr, Glrx3, Hpx, Fabp4, Igfbp1, Hspa1l, Retnlg, Serpina3n, Lcn2, Cald1, Fh1, Myh6, Sema6d, Tpm1, Lbp, Ldha, Myom2, Hprt1, Hist1h1a, Serping1, Tnnc1, Semg1, Col1a1, Rpl8, Mb, Atp7a, Orm1, S100a8, Hexb, S100a9, Mybpc1, Prg4 |
Table V.
Molecular function analysis.
| Term | Gene count | P-value | Gene list |
|---|---|---|---|
| Transcription regulatory activity | 1 | 0.999674 | Fabp4 |
| Signal transduction activity | 9 | 0.999986 | Fgl2, Agt, Ttr, Ldlr, Fgl1, Lbp, Pigr, Il1r2, Prg4 |
| Transporter activity | 13 | 0.030194 | ApoE, Slc4a1, Fabp5, Fabp4, Fabp3, Lcn2, Clca1, Hba-a2, Fabp6, Mb, Atp7a, Orm1, Atp5a1 |
| Enzyme regulator activity | 11 | 0.006352 | ApoE, Agt, Kng1, Anxa1, Serpinb1a, A2m, Timp1, Fn1, Serpina3n, Serping1, Slpi |
| Translation activity | 1 | 0.285497 | Hspa5 |
| Nucleic acid binding activity | 12 | 0.962945 | Hnrnpa1, Hist1h1b, Dpm1, Ddb1, Serbp1, Tsn, Hist1h1d, Nono, Hmgn2, Hist1h1a, Rpl8, Hist1h2ai |
| Extracellular structural activity | 2 | 0.013584 | Col1a2, Col1a1 |
| Kinase activity | 4 | 0.84497 | Pfkp, Ckm, Ckb, Ak1 |
| Cytoskeletal activity | 10 | 0.002977 | Tpm2, Tnni2, Tpm3, Cald1, Myh6, Tpm1, Myom2, Pdlim3, Tnnc1, Mybpc1 |
| Other molecular function | 94 | 0.222994 | Aox1, F13a1, ApoE, Pgc, Neb, Hnrnpa1, Mrfap1, Thbs1, Slc4a1, Lmnb1, Ngp, Fabp5, Hist1h1b, Dpm1, Siae, Vcan, Anxa1, Sbp, Gemin4, Tsn, Hist1h1d, Ptx3, A2m, Acy1, Mgp, Pgm1, Tnnc2, Apof, Tpm2, Car3, Ptgr1, Ldlr, Acan, Glrx3, Chi3l1, Nono, Car1, Mpo, Hpx, Fabp4, Mpst, Fn1, Abp1, Igfbp1, Fabp3, Hspa1l, Col1a2, Retnlg, Rps28, Pfkp, Lcn2, Chad, Tceb2, S100a11, Hba-a2, Ostf1, Fabp6, Fh1, Myh6, Sema6d, Pgam2, Lbp, Tpi1, Trim72, Pgls, Hspa5, Pla1a, Ldha, Ckm, Tsku, Hprt1, Ckb, Pdlim3, Ak1, Itpa, Serping1, Eno3, Tnnc1, Semg1, Gstm1, Hp, Col1a1, Rpl8, Mb, Atp7a, Orm1, Slpi, S100a8, Hexb, Col6a2, S100a9, Atp5a1, Limd2, Prg4 |
Following pathway analysis, the 158 proteins were shown to be mainly involved in extracellular matrix-receptor interactions (count=7; P<0.01), focal adhesions (count=7; P<0.05), and complement and coagulation cascades (count=6; P<0.01; Table VI).
Table VI.
Proteins in signaling pathway.
| Title | Gene count | P-value | Genes |
|---|---|---|---|
| ECM-receptor interaction | 7 | 5.98E-05 | Thbs1,Col6a3,Fn1,Col1a2,Chad,Col1a1,Col6a2 |
| Complement and coagulation cascades | 6 | 0.000165 | F13a1,Kng1,C4bpb,A2m,C4bpa,Serping1 |
| Focal adhesion | 7 | 0.010068 | Thbs1,Col6a3,Fn1,Col1a2,Chad,Col1a1,Col6a2 |
| Cardiac muscle contraction | 5 | 0.002137 | Tpm2,Tpm3,Myh6,Tpm1,Tnnc1 |
| PPAR signaling pathway | 4 | 0.009466 | Fabp5,Fabp4,Fabp3,Fabp6 |
| Renin-angiotensin system | 1 | 0.187962 | Agt |
| Ribosome | 2 | 0.271977 | Rps28,Rpl8 |
| Antigen processing and presentation | 2 | 0.290114 | Hspa1l,Hspa5 |
| Nucleotide excision repair | 1 | 0.417476 | Ddb1 |
| Ubiquitin mediated proteolysis | 2 | 0.484216 | Ddb1,Tceb2 |
| Calcium signaling pathway | 2 | 0.643433 | Tnnc2,Tnnc1 |
| TGF-β signaling pathway | 1 | 0.653817 | Thbs1 |
| Hematopoietic cell lineage | 1 | 0.658099 | Il1r2 |
| Endocytosis | 2 | 0.664494 | Ldlr,Hspa1l |
| Toll-like receptor signaling pathway | 1 | 0.712848 | Lbp |
| Vascular smooth muscle contraction | 1 | 0.755917 | Cald1 |
| Lysosome | 1 | 0.758953 | Hexb |
| Axon guidance | 1 | 0.79776 | Sema6d |
| Tight junction | 1 | 0.805238 | Myh6 |
| Cell adhesion molecules | 1 | 0.80767 | Vcan |
| MAPK signaling pathway | 2 | 0.848604 | Hspa1l,Il1r2 |
| Regulation of actin cytoskeleton | 1 | 0.930221 | Fn1 |
| Cytokine-cytokine receptor interaction | 1 | 0.96282 | Il1r2 |
| Olfactory transduction | 1 | 0.993046 | Clca1 |
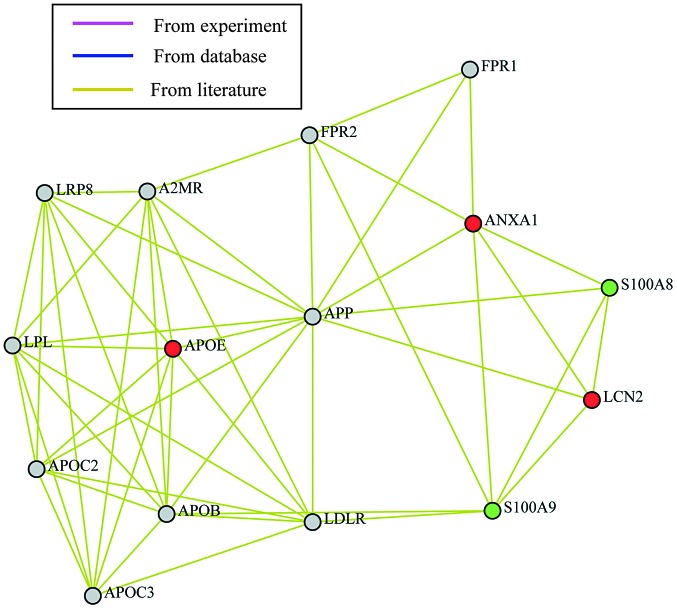
The gene symbols were mapped on a protein-protein interaction network (Fig. 4). The closest connection was identified between hub Fn1 and A2m genes. However, no significant fold-change was found in the Fn1 gene and A2m-gene associated protein expression. This network only shows the peripheral interactions between the differentially expressed proteins, but not the close interactions between individual proteins. Therefore, further analysis of the time-course data was performed. Comprehensively considering the fold change (>1.5 or <0.5) and a P-value <0.05, the biological effects of the distinct proteins and involvement of proteins in the induction of inflammation and sepsis, five proteins were selected for further gene network analysis, including apolipoprotein E (ApoE), annexin A1 (Anxa1), neutrophil gelatinase-associated lipocalin (NGAL), S100a8 and S100a9 (Fig. 5). Compared with the sham-6 h group, Anxa1 and ApoE were upregulated in the CLP-6 h group. There was no significant difference in the expression of Anxa1 and ApoE between the CLP-24 h and CLP-6 h groups. S100a8 and S100a9 were overexpressed at 6 h following CLP compared with the sham-6 h group, while downregulated 24 h after CLP. In addition, the expression of NGAL was detected to be elevated in the CLP-6 h (compared with sham-6 h) and CLP-24 h groups (compared with CLP-6 h group). All of the five proteins were found to be involved in the same lipid metabolism-associated network, with amyloidogenic glycoprotein as the hub. The alterations in the relative abundance of these five proteins between the CLP-6 h and CLP-24 h groups are demonstrated in Fig. 6. Among the proteins, NGAL expression revealed a significant increase from CLP-6 h to CLP-24 h.
Figure 4.
Differentially expressed protein network analysis (1), using gene symbols. The protein-protein interaction network of differentially expressed proteins; the gene with the highest connectivity in network connectivity analysis are termed the hub genes, e.g. Fn1 and A2m. Red, hub gene; blue, general gene.
Figure 5.
Differentially expressed protein network analysis (2), using gene symbols. It is observed that the five selected proteins: ApoE, Anxa1, NGAL (Lcn2), S100a8 and S100a9 are involved in the same lipid metabolism-associated network, with amyloidogenic glycoprotein (APP) as the hub. ApoE, apolipoprotein E; Anxa1, annexin A1; NGAL, neutrophil gelatinase-associated lipocalin. Red, upregulated genes (CLP24 vs. CLP-6); green, downregulated genes (CLP-24 vs. CLP-6).
Figure 6.

Relative abundance of five selected proteins at 6 h and 24 h following CLP. Of the five selected proteins, the relative abundance of S100a9 exhibited the greatest fold change 6 h following CLP (fold change, 3.7; P<0.0001), followed by S100a8 (fold change, 3; P<0.0001). The relative abundance of ApoE and Anxa1 were almost unchanged from 6 to 24 h following CLP. The levels of S100a8/S100a9 progressively decreased, while NGAL increased rapidly. ApoE, apolipoprotein E; Anxa1, annexin A1; NGAL, neutrophil gelatinase-associated lipocalin.
Discussion
Although numerous investigations concerning sepsis have been conducted, the detailed and precise mechanism of sepsis remains unclear. The present study provided a novel prospective to aid in the elucidation of the mechanism of sepsis by determining proteome changes in the mesenteric lymph. The results demonstrated that a total of 158 proteins in the mesenteric lymph, which were found to exist in all of the groups, were identified to be significantly differentially expressed (fold change ≥1.5; P<0.05). The 158 proteins demonstrated abundant differences between the CLP and sham groups as well as the CLP-6 h and CLP-24 h groups. Following analysis of the 158 proteins by bioinformatic methods, five proteins were investigated as targets: ApoE, Anxa1, NGAL, S100a8 and S100a9.
ApoE, the ligand of the low density lipoprotein receptor, has a vital role in lipid metabolism and is involved in the pathogenesis of critical diseases (26,27). It has been suggested that ApoE polymorphisms are associated with severe sepsis in surgical patients (28). In a previous study, the majority of patients with severe sepsis did not have the APOɛ3 allele. The APOɛ3 allele significantly protected carriers against the risk of severe sepsis (29). Although the role of ApoE in sepsis was not clear, ApoE was determined to be associated with a variety of different factors. In sepsis models induced by CLP, ApoE reversed the increase of hepatic T cell apoptosis and necrosis, and promoted Th1 cytokine levels (30). In addition, ApoE−/− mice demonstrated notably higher levels of proinflammatory cytokines in the serum, compared with wild-type mice (31,32). ApoE inhibited the lipopolysaccharide (LPS)-induced increase of tumor necrosis factor-α, interleukin (IL)-1β and IL-6 in serum and decreased the mortality rates of mice with sepsis (32). By contrast, another study suggested that ApoE increased the mortality rates of rats with sepsis (30). Seven days following CLP, rats injected with recombinant ApoE3 had a mortality rate of 82.14 and 100% at doses of 114 μg/kg or 1.6 mg/kg, respectively, while none of the saline-injected rats died (30). The results demonstrated that ApoE was involved in multiple biological processes, including transport, stress responses, cell organization and biogenesis. ApoE was also involved in transporter and enzyme regulator activity. At 6 h following CLP, ApoE was indicated to be significantly upregulated in the lymph, but not in the CLP-24 h group. Therefore, it is suggested that ApoE demonstrated protective effects in the early stages of sepsis, but that this effect was reversed in the later stages.
Anxa1, an annexin, is known to be an endogenous anti-inflammatory mediator (33). The expression of annexins (AnxA1–AnxA7, AnxA9 and AnxA11) has been demonstrated to be regulated by pro- and anti-inflammatory stimulation (34). Furthermore, pro- and anti-inflammatory responses were revealed to be present throughout the entire process of sepsis progression (35). Anxa1 decreased the degree of the acute inflammatory response (36) and exerted active effects on the suppression of acute inflammation by preventing the adhesion and migration of neutrophils, adjusting the production of pro-inflammatory and anti-inflammatory cytokines and promoting neutrophil apoptosis (37,38). There was evidence that endogenous Anxa1 exhibited a protective role in the cerebral microcirculation of sepsis (39). However, it was reported that the Anxa1 plasma level decreased in patients with sepsis, as compared with control patients (35). In the present study, Anxa1 was overexpressed in the lymph of the CLP-6 h group (fold=1.61, P=0.0103) compared with that in the sham-6 h group. It indicated that Anxa1 had a key role in sepsis progression and that the expression of Anxa1 may be different in the lymph and plasma of patients. Bioinformatics analysis revealed that Anxa1 was mainly involved in enzyme regulator activity, signal transduction and cell adhesion, which corresponded with previous studies.
S100a8 and S100a9 as members of the S100 protein family, are released from neutrophils and activate phagocytes during sepsis (40). S100a8 and S100a9 most commonly emerge as heterodimers and have a prominent role in the pathogenesis of various inflammatory diseases (41).
S100A8 and/or S100A9 have been reported to possess multiple important biological activities, including mediating neutrophil adhesion to fibronectin (42), inducing neutrophil chemotaxis (43) and mediating apoptosis (44). It has also been demonstrated that S100A8 and S100A9 enhanced inflammatory responses by mediating nuclear factor-κB activation and inducing cytokine secretion, including IL-6, IL-8 and IL-1β (41). During the early phase of sepsis, excessive release of proinflammatory cytokines and chemokines reflect hyperreactive immune responses and result in hyporeactive immune responses in the later phase along with intractable shock, refractory infection, multiple organ failure and even mortality (45). As outlined in a previous study, the S100a8/S100a9 plasma levels were significantly elevated in patients with severe sepsis (46). In abdominal sepsis patients, the S100a8/S100a9 levels in the abdominal fluid were >15-fold that in the plasma, indicating that S100a8/S100a9 expression was induced primarily at the site of infection during sepsis (46). The present study indicated that the expression of S100a8 and S100a9 were significantly elevated in the lymph during early stages of sepsis (CLP-6h) but decreased in later stages (CLP-24h). As S100 proteins have been defined as clinical markers of inflammation in a previous study (47), S100a8 and S100a9 levels in the lymph may be a marker for the diagnosis of early stage sepsis.
NGAL, a lipocalin, is overexpressed in acute kidney injury due to ischemia, toxic factors and sepsis (48,49). In patients with sepsis, the overexpression of NGAL attributed to the response of the kidneys to sepsis. NGAL expression was found to be elevated in the serum of patients with sepsis (50). The concentration of urinary NGAL was also considered to be an indicator of early acute kidney injury in patients with sepsis (49). Furthermore, NGAL levels in the urine were reported to be correlated with the plasma or serum levels (51). In the present study, NGAL levels in the lymph increased progressively and reached a peak at 24 h following CLP. The NGAL levels in the plasma and urine have been widely adopted as an early predictor of acute kidney injury (52–54). NGAL had a critical role in sepsis progression and further studies are required to determine its clinical utility as a diagnostic marker for sepsis.
In conclusion, the present study demonstrated that the expression of five proteins (ApoE, Anxa1, NGAL, protein S100a8 and S100a9) was significantly elevated in the progression of sepsis. All five proteins appeared to have vital roles in critical disease development and may therefore be potential targets for the treatment and diagnosis of sepsis. The present study provides a novel perspective to aid in the understanding of the pathological mechanism of sepsis.
Figure 2.
Cellular component analysis. The chart shows the cellular components that the 158 proteins are involved in.
Table IV.
Cellular component analysis.
| Term | Gene count | P-value | Gene list |
|---|---|---|---|
| Extracellular matrix | 8 | 0.00073 | Vcan, Acan, Timp1, Fn1, Col1a2, Chad, Col1a1, Col6a2 |
| Non-structural extracellular | 43 | 0 | F13a1, ApoE, Fgl2, Pgc, Saa4, Thbs1, Agt, Svs6, Kng1, Vcan, Svs4, Col6a3, Sbp, Ptx3, A2m, Mgp, Ttr, Apof, Ldlr, Acan, Chi3l1, Fgl1, Hpx, Timp1, Fn1, Abp1, Igfbp1, Col1a2, Serpina3n, Lcn2, Chad, Lbp, Pla1a, Pigr, Tsku, Serping1, Mmrn1, Hp, Col1a1, Orm1, Slpi, Col6a2, Prg4 |
| Cytosol | 3 | 0.430499 | Pfkp, Ldha, Eno3 |
| Cytoskeleton | 12 | 0.028496 | Neb, Slc4a1, Lmnb1, Rcc2, Tpm2, Tpm3, Cald1, Myh6, Tpm1, Myom2, Pdlim3, Mybpc1 |
| Nucleus | 19 | 0.988576 | Hnrnpa1, Mrfap1, Lmnb1, Hist1h1b, Rcc2, Dpm1, Anxa1, Ddb1, Serbp1, Gemin4, Tsn, Hist1h1d, Nono, Fabp4, Hmgn2, Tceb2, S100a11, Hist1h1a, Hist1h2ai |
| Plasma membrane | 9 | 0.99651 | Slc4a1, Col6a3, Anxa1, Fn1, Clca1, Pigr, Ak1, Atp7a, Col6a2 |
| Other membranes | 18 | 1 | Slc4a1, Lmnb1, Dpm1, Col6a3, Anxa1, Ldlr, Mpst, Fn1, Clca1, Cald1, Sema6d, Pigr, Il1r2, Ak1, Atp7a, Hexb, Col6a2, Atp5a1 |
| Translational apparatus | 2 | 0.39956 | Rps28, Rpl8 |
| ER/Golgi | 3 | 0.993138 | Dpm1, Hspa5, Atp7a |
| Mitochondrion | 6 | 0.652947 | Mpo, Mpst, Fh1, Ckb, Ak1, Atp5a1 |
| Other cytoplasmic organelle | 4 | 0.38439 | Siae, Ldlr, Mpo, Hexb |
| Other cellular component | 57 | 0.025085 | Aox1, F13a1, Hnrnpa1, Mrfap1, Ngp, Fabp5, Hist1h1b, Rcc2, Dpm1, Siae, Anxa1, Ddb1, Serbp1, Serpinb1a, Tsn, Hist1h1d, Acy1, Apof, Tpm2, Car3, Ptgr1, Glrx3, Tpm3, Car1, Fabp4, Mpst, Hmgn2, Fabp3, Retnlg, Rps28, Pfkp, S100a11, Ostf1, Fabp6, Fh1, Myh6, Sema6d, Tpm1, Trim72, Tagln, Hspa5, Ldha, Ckm, Hprt1, Hist1h1a, Ckb, Pdlim3, Ak1, Itpa, Eno3, Semg1, Gstm1, Col1a1, Rpl8, Atp7a, Hexb, Hist1h2ai |
Acknowledgements
This study was supported by the Shanghai Key Discipline Construction of Public Health Funds (Shanghai, China) (grant no. O8GWZX1103). The authors would like to thank Shanghai Sensichip Co., Ltd. (Shanghai, China) for the proteomic and bioinformatics analysis and Dr. Karen Bysouth who provided editing services on behalf of Elixigen Co. (Shanghai, China).
References
- 1.Hinkelbein J, Kalenka A, Schubert C, Peterka A, Feldmann J, Robert E. Proteome and metabolome alterations in heart and liver indicate compromised energy production during sepsis. Protein Pept Lett. 2010;17:18–31. doi: 10.2174/092986610789909520. [DOI] [PubMed] [Google Scholar]
- 2.Mckean SC, Ross JJ, Dressler DD, Brotman DJ, Ginsberg J. Principles and practice of hospital medicine. McGraw-Hill; 2012. [Google Scholar]
- 3.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 4.Dellinger RP, Carlet JM, Masur H, et al. Surviving Sepsis Campaign guidelines for management of severe sepsis and septic shock. Intensive Care Med. 2004;30:536–555. doi: 10.1007/s00134-004-2210-z. [DOI] [PubMed] [Google Scholar]
- 5.Dellinger RP, Levy MM, Carlet JM, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Intensive Care Med. 2008;34:17–60. doi: 10.1007/s00134-007-0934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okazaki Y, Matsukawa A. Pathophysiology of sepsis and recent patents on the diagnosis, treatment and prophylaxis for sepsis. Recent Pat Inflamm Allergy Drug. 2009;3:26–32. doi: 10.2174/187221309787158416. [DOI] [PubMed] [Google Scholar]
- 8.Magnotti LJ, Upperman JS, Xu DZ, Lu Q, Deitch EA. Gut-derived mesenteric lymph but not portal blood increases endothelial cell permeability and promotes lung injury after hemorrhagic shock. Ann Surg. 1998;228:518–527. doi: 10.1097/00000658-199810000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittal A1, Phillips AR, Middleditch M, et al. The proteome of mesenteric lymph during acute pancreatitis and implications for treatment. JOP. 2009;10:130–142. [PubMed] [Google Scholar]
- 10.Dayal SD, Haskó G, Lu Q, et al. Trauma/hemorrhagic shock mesenteric lymph upregulates adhesion molecule expression and IL-6 production in human umbilical vein endothelial cells. Shock. 2002;17:491–495. doi: 10.1097/00024382-200206000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Moore EE. Claude H. Organ, Jr. memorial lecture: splanchnic hypoperfusion provokes acute lung injury via a 5-lipoxygenase-dependent mechanism. Am J Surg. 2010;200:681–689. doi: 10.1016/j.amjsurg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leak LV, Liotta LA, Krutzsch H, et al. Proteomic analysis of lymph. Proteomics. 2004;4:753–765. doi: 10.1002/pmic.200300573. [DOI] [PubMed] [Google Scholar]
- 13.Mittal A, Middleditch M, Ruggiero K, et al. The proteome of rodent mesenteric lymph. Am J Physiol Gastrointest Liver Physiol. 2008;295:G895–G903. doi: 10.1152/ajpgi.90378.2008. [DOI] [PubMed] [Google Scholar]
- 14.Mittal A, Phillips A, Middleditch M, et al. The proteome of mesenteric lymph during acute pancreatitis and implications for treatment. JOP. 2009;10:130–142. [PubMed] [Google Scholar]
- 15.Clement CC, Aphkhazava D, Nieves E, et al. Protein expression profiles of human lymph and plasma mapped by 2D-DIGE and 1D SDS-PAGE coupled with nanoLC-ESI-MS/MS bottom-up proteomics. J Proteomics. 2012;78:172–187. doi: 10.1016/j.jprot.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mcdunn JE, Townsend RR, Cobb JP. The murine plasma protein response to polymicrobial intra-abdominal sepsis. Proteomics Clin Appl. 2007;1:373–386. doi: 10.1002/prca.200600494. [DOI] [PubMed] [Google Scholar]
- 17.Ren Y, Wang J, Xia J, et al. The alterations of mouse plasma proteins during septic development. J Proteome Res. 2007;6:2812–2821. doi: 10.1021/pr070047k. [DOI] [PubMed] [Google Scholar]
- 18.Dear JW, Leelahavanichkul A, Aponte A, et al. Liver proteomics for therapeutic drug discovery: inhibition of the cyclophilin receptor CD147 attenuates sepsis-induced acute renal failure. Crit Care Med. 2007;35:2319–2328. doi: 10.1097/01.ccm.0000281858.44387.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan X, Berthiaume F, Yarmush D, Yarmush ML. Proteomic analysis of altered protein expression in skeletal muscle of rats in a hypermetabolic state induced by burn sepsis. Biochem J. 2006;397:149–158. doi: 10.1042/BJ20051710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otero-Antón E, González-Quintela A, López-Soto A, López-Ben S, Llovo J, Pérez L. Cecal ligation and puncture as a model of sepsis in the rat: influence of the puncture size on mortality, bacteremia, endotoxemia and tumor necrosis factor alpha levels. Eur Surg Res. 2001;33:77–79. doi: 10.1159/000049698. [DOI] [PubMed] [Google Scholar]
- 21.Freise H, Brückner U, Spiegel H. Animal models of sepsis. J Invest Surg. 2001;14:195–212. doi: 10.1080/089419301750420232. [DOI] [PubMed] [Google Scholar]
- 22.Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Wang J, Li Y, Lin ZF. A modified rat model for cannulation and collection of thoracic duct lymph. Lymphology. 2011;44:82–84. [PubMed] [Google Scholar]
- 24.Mittal A, Middleditch M, Ruggiero K, et al. Changes in the mesenteric lymph proteome induced by hemorrhagic shock. Shock. 2010;34:140–149. doi: 10.1097/SHK.0b013e3181cd8631. [DOI] [PubMed] [Google Scholar]
- 25.Neil AL, Christensen H. Australian school-based prevention and early intervention programs for anxiety and depression: a systematic review. Med J Aust. 2007;186:305–308. doi: 10.5694/j.1326-5377.2007.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 26.Bonomini F, Filippini F, Hayek T, et al. Apolipoprotein E and its role in aging and survival. Exp Gerontol. 2010;45:149–157. doi: 10.1016/j.exger.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Seshasai RK, Katz R, de Boer IH, et al. Apolipoprotein E and kidney function in older adults. Clin Nephrol. 2012;78:174–180. doi: 10.5414/CN107427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moretti EW, Morris RW, Podgoreanu M, et al. APOE polymorphism is associated with risk of severe sepsis in surgical patients. Crit Care Med. 2005;33:2521–2526. doi: 10.1097/01.ccm.0000186368.96146.fb. [DOI] [PubMed] [Google Scholar]
- 29.Moretti EW, Morris RW, Podgoreanu M, et al. Perioperative Genetics and Safety Outcomes Study (PEGASUS) Investigative Team: APOE polymorphism is associated with risk of severe sepsis in surgical patients. Crit Care Med. 2005;33:2521–2526. doi: 10.1097/01.ccm.0000186368.96146.fb. [DOI] [PubMed] [Google Scholar]
- 30.Kattan OM, Kasravi FB, Elford EL, Schell MT, Harris HW. Apolipoprotein E-mediated immune regulation in sepsis. J Immunol. 2008;181:1399–1408. doi: 10.4049/jimmunol.181.2.1399. [DOI] [PubMed] [Google Scholar]
- 31.de Bont N, Netea MG, Demacker PN, et al. Apolipoprotein E knock-out mice are highly susceptible to endotoxemia and Klebsiella pneumoniae infection. J Lipid Res. 1999;40:680–685. [PubMed] [Google Scholar]
- 32.Van Oosten M, Rensen PC, Van Amersfoort ES, et al. Apolipoprotein E protects against bacterial lipopolysaccharide-induced lethality. A new therapeutic approach to treat gram-negative sepsis. J Biol Chem. 2001;276:8820–8824. doi: 10.1074/jbc.M009915200. [DOI] [PubMed] [Google Scholar]
- 33.Kamal AM, Flower RJ, Perretti M. An overview of the effects of annexin 1 on cells involved in the inflammatory process. Mem Instit Oswaldo Cruz. 2005;100(Suppl 1):39–47. doi: 10.1590/s0074-02762005000900008. [DOI] [PubMed] [Google Scholar]
- 34.Kreft S. Annexinabhängige modulation der immunantwort und hämostase. Universität zu Köln; 2013. [Google Scholar]
- 35.Tsai WH, Shih CH, Yu YB, Hsu HC. Plasma levels in sepsis patients of annexin A1, lipoxin A4, macrophage inflammatory protein-3a, and neutrophil gelatinase-associated lipocalin. J Chin Med Assoc. 2013;76:486–490. doi: 10.1016/j.jcma.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Flower RJ. Eleventh Gaddum memorial lecture. Lipocortin and the mechanism of action of the glucocorticoids. Br J Pharmacol. 1988;94:987–1015. doi: 10.1111/j.1476-5381.1988.tb11614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vago JP, Nogueira CR, Tavares LP, et al. Annexin A1 modulates natural and glucocorticoid-induced resolution of inflammation by enhancing neutrophil apoptosis. J Leukoc Biol. 2012;92:249–258. doi: 10.1189/jlb.0112008. [DOI] [PubMed] [Google Scholar]
- 38.Perretti M. Editorial: to resolve or not to resolve: Annexin A1 pushes resolution on track. J Leukoc Biol. 2012;92:245–247. doi: 10.1189/jlb.0312128. [DOI] [PubMed] [Google Scholar]
- 39.Gavins FN, Hughes EL, Buss NA, et al. Leukocyte recruitment in the brain in sepsis: involvement of the annexin 1-FPR2/ALX anti-inflammatory system. FASEB J. 2012;26:4977–4989. doi: 10.1096/fj.12-205971. [DOI] [PubMed] [Google Scholar]
- 40.Nacken W, Roth J, Sorg C, Kerkhoff C. S100A9/S100A8: Myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc Res Tech. 2003;60:569–580. doi: 10.1002/jemt.10299. [DOI] [PubMed] [Google Scholar]
- 41.Simard J-C, Cesaro A, Chapeton-Montes J, et al. S100A8 and S100A9 induce cytokine expression and regulate the NLRP3 inflammasome via ROS-dependent activation of NF-κB(1.) PLoS One. 2013;8:e72138. doi: 10.1371/journal.pone.0072138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anceriz N, Vandal K, Tessier PA. S100A9 mediates neutrophil adhesion to fibronectin through activation of β2 integrins. Biochem Biophys Res Commun. 2007;354:84–89. doi: 10.1016/j.bbrc.2006.12.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryckman C, Vandal K, Rouleau P, et al. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 44.Yui S, Nakatani Y, Mikami M. Calprotectin (S100A8/S100A9), an inflammatory protein complex from neutrophils with a broad apoptosis-inducing activity. Biol Pharm Bull. 2003;26:753–760. doi: 10.1248/bpb.26.753. [DOI] [PubMed] [Google Scholar]
- 45.Riedemann NC, Guo RF, Ward PA. Novel strategies for the treatment of sepsis. Nat Med. 2003;9:517–524. doi: 10.1038/nm0503-517. [DOI] [PubMed] [Google Scholar]
- 46.van Zoelen MA, Vogl T, Foell D, et al. Expression and role of myeloid-related protein-14 in clinical and experimental sepsis. Am J Respir Crit Care Med. 2009;180:1098–1106. doi: 10.1164/rccm.200810-1552OC. [DOI] [PubMed] [Google Scholar]
- 47.Foell D, Frosch M, Sorg C, Roth J. Phagocyte-specific calcium-binding S100 proteins as clinical laboratory markers of inflammation. Clin Chim Acta. 2004;344:37–51. doi: 10.1016/j.cccn.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 48.Mishra J, Dent C, Tarabishi R, et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet. 2005;365:1231–1238. doi: 10.1016/S0140-6736(05)74811-X. [DOI] [PubMed] [Google Scholar]
- 49.Bagshaw SM, Bennett M, Haase M, et al. Plasma and urine neutrophil gelatinase-associated lipocalin in septic versus non-septic acute kidney injury in critical illness. Intensive Care Med. 2010;36:452–461. doi: 10.1007/s00134-009-1724-9. [DOI] [PubMed] [Google Scholar]
- 50.Parravicini E, Nemerofsky SL, Michelson KA, et al. Urinary neutrophil gelatinase-associated lipocalin is a promising biomarker for late onset culture-positive sepsis in very low birth weight infants. Pediatr Res. 2010;67:636–640. doi: 10.1203/PDR.0b013e3181da75c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uttenthal L. NGAL: a marker molecule for the distressed kidney. Clin Lab Int. 2005;29:39–41. [Google Scholar]
- 52.Gupta A, Berg DT, Gerlitz B, et al. Role of protein C in renal dysfunction after polymicrobial sepsis. J Am Soc Nephrol. 2007;18:860–867. doi: 10.1681/ASN.2006101167. [DOI] [PubMed] [Google Scholar]
- 53.Peng ZY, Wang H-Z, Srisawat N, et al. Bactericidal antibiotics temporarily increase inflammation and worsen acute kidney injury in experimental sepsis. Crit Care Med. 2012;40:538–543. doi: 10.1097/CCM.0b013e31822f0d2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seija M, Baccino C, Nin N, et al. Role of peroxynitrite in sepsis-induced acute kidney injury in an experimental model of sepsis in rats. Shock. 2012;38:403–410. doi: 10.1097/SHK.0b013e31826660f2. [DOI] [PubMed] [Google Scholar]