Abstract
The aim of this study was to identify the molecular events that distinguish serrated colorectal carcinoma (SCRC) from conventional colorectal carcinoma (CCRC) through differential gene expression, pathway and protein-protein interaction (PPI) network analysis. The GSE4045 and GSE8671 microarray datasets were downloaded from the Gene Expression Omnibus database. We identified the genes that are differentially expressed between SCRC and normal colon tissues, CCRC and healthy tissues, and between SCRC and CCRC using Student’s t-tests and Benjamini-Hochberg (BH) multiple testing corrections. The differentially expressed genes (DEGs) were then mapped to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and their enrichment for specific pathways was investigated using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool with a significance threshold of 0.1. Analysis of the potential interactions between the protein products of 220 DEGs (between CCRC and SCRC) was performed by constructing a PPI network using data from the high performance RDF database (P<0.1). The interaction between pathways was also analyzed in CCRC based on the PPI network. Our study identified thousands of genes differentially expressed in SCRC and CCRC compared to healthy tissues. The DEGs in SCRC and CCRC were enriched in cell cycle, DNA replication, and base excision repair pathways. The proteasome pathway was significantly enriched in SCRC but not in CCRC after BH adjustment. The PPI network showed that tumour necrosis factor receptor-associated factor 6 (TRAF6) and atrophin 1 (ATN1) were the most central genes in the network, with respective degrees of node predicted at 90 and 88. In conclusion, the preoteasome pathway was shown to be specifically enriched in SCRC. Furthermore, TRAF6 and ATN1 may be promising biomarkers for the distinction between serrated and conventional CRC.
Keywords: microarray, serrated colorectal carcinoma, conventional colorectal carcinoma, proteasome pathway, TRAF6, ATN1
Introduction
Colorectal carcinoma (CRC) is the third most commonly diagnosed cancer (1) and has been reported to occur through different pathways; approximately one third of CRC cases arise along the serrated pathway developing from sessile serrated adenoma (2). The term ‘serrated adenoma’ was first proposed in 1990 by Longacre and Fenoglio-Preiser (3). Morphological features can allow a preliminary distinction of forms of colorectal cancer. The morphological characteristics of serrated CRC (SCRC) include a more mature epithelium than that of conventional CRC (CCRC) (4). However, morphological features are not sufficient for diagnosis of this tumor type.
Serrated adenoma is related to genetic alterations, including DNA methylation, DNA microsatellite instability, K-ras mutation and loss of chromosome 1p (5). Sessile serrated adenomas can induce carcinomas with extensive CpG island promoter methylation, which can be either microsatellite-instable high or microsatellite stable (2). The gene mutated in colorectal cancer was shown to selectively repress β-catenin-dependent transcription, and was thus proposed to constitute a tumor suppressor gene in the SCRC pathway (6). The c-MYC oncogene was shown to be activated by the MAPK/ERK1/2 pathway via the genes K-ras and BRAF and by Wnt signalling in the serrated pathway (7). A mutation in BRAF was frequently observed in serrated adenoma tissues associated with DNA methylation (8). The genes encoding ephrin receptor B2, hypoxia-inducible factor 1-α, and patched were reported as important for the genesis of SCRC (4). The expression of genes p53, APC and CRAC was found altered in SCRC as well as in CCRC (9). However, the mechanism(s) underlying the pathogenesis of SCRC are still not well elucidated.
In this study, we identified differentially expressed genes (DEGs) between SCRC and CCRC and between each of these subtypes of CRC and healthy tissues. Then, DEGs were analyzed for their enrichment in molecular pathways, and a protein-protein interaction (PPI) network was constructed to identify the potential interactions between the genes’ products. Overall, this study provided information that may be useful for the clinical classification of CRC.
Materials and methods
Microarray data
The transcriptional profile data of SCRC samples, CCRC samples and healthy mucosa samples were downloaded from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). The data were derived from the accession numbers GSE4045 (4) and GSE8671 (10). GSE4045 comprises7 SCRC and 30 CCRC samples. CRC samples with serrated histology were from two population-based Finnish collections. The analysis of microarray data was performed on a GPL96 (HG-U133A) Affymetrix Human Genome U133A array. The GSE8671 dataset comprises 32 CCRC samples and 32 healthy mucosa samples, which were obtained from patients during colonoscopy. The analysis of microarray data was performed on a GPL570 (HG-U133-Plus_2) Affymetrix Human Genome U133 Plus 2.0 array. The raw data (CEL files) from both datasets were downloaded.
Normalization of microarray data and identification of DEGs
First, data from each dataset were separately normalized. The two sets of data were preprocessed using background correction and robust multi-array analysis (RMA) normalization (11) with the Affy package (12). From the chip expression profile data, we removed the probes that corresponded to multiple Entrez Gene IDs and retained the median of different probes representing the same Entrez Gene ID. We finally obtained expression profile data for 12,779 genes from 37 samples of the GSE4045 dataset and for 20,539 genes from 64 samples of the GSE8671 dataset. Second, the datasets derived from the different platforms were normalized between them. It has been reported that batch effects are critical in high-throughput data (13). Thus, cross-platform normalization (XPN) was used (14). We selected the 12,779 common genes between the two datasets and their expression data were normalized with the XPN method using the CONOR package (15). These data were used for further analyses. The DEGs were identified from the normalized final dataset using Student’s t-tests (P<0.05), followed by Benjamini-Hochberg (BH) multiple testing correction (16). A BH P-value <0.05 was set as the threshold.
Construction of a cancer global network
In order to explore expression patterns in the two CRC subtypes, we constructed a cancer global network. First, we downloaded the following 14 cancer-related pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (17): colorectal cancer, pancreatic cancer, glioma, thyroid cancer, acute myeloid leukemia, chronic myeloid leukemia, basal cell carcinoma, melanoma, renal cell carcinoma, bladder cancer, prostate cancer, endometrial cancer, small cell lung cancer and non-small cell lung cancer. Second, we screened the DEGs which were enriched in the 14 cancer pathways and then a network of these DEGs was constructed.
KEGG pathway enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) online enrichment tool (18,19) was adopted for pathway enrichment analysis, and the EASE score (a modified Fisher exact test) was used to identify significantly enriched KEGG pathways with a threshold value of 0.1. BH adjustment was performed on the raw P-values obtained from the pathway enrichment analysis.
PPI network construction
We constructed a PPI network for 149 DEGs selected from the GSE4045 dataset (P-value after BH adjustment <0.1) using the high performance RDF database (HPRD) (20). In the PPI network, a hub was defined as the node which has the most number of interactions with other nodes. PPI was visualized using the Cytoscape software (21).
Construction of an interactions network of pathways involved in SCRC and CCRC based on the PPI network
Significantly enriched pathways of DEGS were used to construct a pathway interaction network. We used the cumulative probability of the hypergeometric distribution model to identify whether any two pathways included a significant number of interactions (P<0.05). The cumulative probability formula was as follows:
where N represents the number of protein interactions in which all DEGs are involved; M represents the number of protein interactions which are related to genes in pathway 1 (there was at least one DEG in the interaction); n represents the number of protein interactions that are related to genes in pathway 2 (there was at least one DEG in the interaction); k represents the number of interactions between pathway 1 and 2 (there was at least one DEG in the interaction). If the cumulative probability was less than 0.1, two pathways were defined as interacting. We defined the degree as the number of interactions which one node had with other nodes.
Results
Identification of DEGs in SCRC and CCRC
In this study, we set two different significance thresholds to identify DEGs, listed in Table I. When P<0.05, the proportion of upregulated genes was higher in CCRC (62.6%, 1,713/2,736) compared to SCRC (36.9%, 1,012/2,736). Using a Fisher exact test, we found that there was a significant difference in the proportion of up- and downregulated genes between these two cancer subtypes (P=2.596e-08). Then, we adjusted the P-value using BH multiple testing correction.
Table I.
Identification of differentially expressed genes (DEGs), numbers obtained from application of different criteria.
| P<0.05 | |||
|---|---|---|---|
|
|
|||
| Total | Upregulated | Downregulated | |
| CCRC vs. SCRC | 1,819 | 602 | 1,299 |
| CCRC vs. control | 2,736 | 1,713 | 1,023 |
| SCRC vs. control | 2,123 | 1,012 | 1,121 |
CCRC, conventional colorectal carcinoma; SCRC, serrated colorectal carcinoma; control, healthy mucosa samples.
Global cancer network
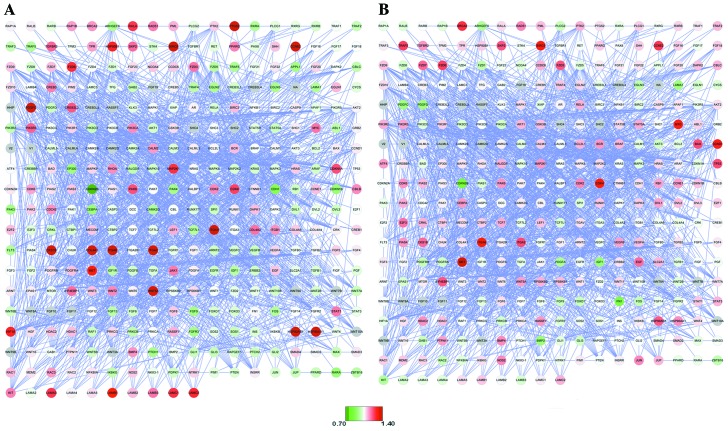
We constructed a network of 341 genes and 1,569 relation after identifying the DEGs which were enriched in the 14 cancer pathways (Fig. 1). Then, information on the expression profile of these genes was retrieved for DEGs in the two subtypes of CRC as shown in Fig. 1. In SCRC, there were 130 upregulated genes (48 genes of these appear downregulated in CCRC) and 184 downregulated genes (80 of these appear upregulated in CCRC). In CCRC, there were 162 upregulated and 152 downregulated genes. A total of 182 genes showed expression changes in the same direction in the two networks, comprising 100 downregulated and 82 upregulated genes.
Figure 1.
Global cancer network in (A) serrated colorectal carcinoma and (B) conventional colorectal carcinoma. Red circles represent upregulated genes and green circles downregulated genes.
After a Fisher exact test, there were 26 upregulated genes in SCRC and 25 upregulated genes in CCRC when P<0.05. In addition, there were 13 genes which were both upregulated between the two CRC subtypes with P<0.05. There were 12 genes with a fold-change (FC) >1.4 and one gene with FC<0.7 in SCRC, and 1 gene with FC>1.4 and no gene with FC<0.7 in CCRC.
Identification of biological pathways related to SCRC and CCRC
In this study, 2,736 DEGs from CCRC and 2,123 DEGs from SCRC (P<0.05; Table I) were selected for pathway enrichment analysis. Table II shows the top 9 most significantly enriched pathways in CCRC, in which are involved the genes with a BH-adjusted P-value <0.05. These significantly enriched pathways can be divided into three classes: pathways related to death (such as cell cycle and p53 signaling pathway); DNA replication and repair pathways (such as DNA replication, base and nucleotide excision repair, mismatch repair, and homologous recombination); and nucleotide metabolism-related pathways (such as purine and pyrimidine metabolism).
Table II.
Enriched pathways for differentially expressed genes (BH P<0.05) in conventional colorectal carcinoma. Source, Kyoto Encyclopedia of Genes and Genomes (KEGG), Homo sapiens (hsa) identification numbers.
| KEGG id. | Gene count | BH |
|---|---|---|
| 04110: Cell cycle | 61 | 2.53E-12 |
| 03030: DNA replication | 27 | 3.41E-10 |
| 00230: Purine metabolism | 55 | 5.07E-05 |
| 00240: Pyrimidine metabolism | 38 | 2.74E-04 |
| 03410: Base excision repair | 20 | 3.12E-04 |
| 03420: Nucleotide excision repair | 21 | 5.45E-03 |
| 03430: Mismatch repair | 14 | 8.23E-03 |
| 04115: p53 signaling pathway | 27 | 1.46E-02 |
| 03440: Homologous recombination | 15 | 2.47E-02 |
BH, P-value after Benjamin-Hochberg correction.
Table III shows the top 10 most significantly enriched pathways in SCRC, in which are involved the genes with a P<0.05. These pathways can also be divided in three classes: cell growth and death pathways (cell cycle); DNA replication and repair pathways (DNA replication and base excision repair); nucleotide metabolism-related pathways (such as purine and pyrimidine metabolism).
Table III.
Enriched pathways for differentially expressed genes (P<0.05) in serrated colorectal carcinoma. Source, Kyoto Encyclopedia of Genes and Genomes (KEGG), Homo sapiens (hsa) identification numbers.
| KEGG id. | Gene count | BH |
|---|---|---|
| 03050: Proteasome | 26 | 7.18E-08 |
| 04110: Cell cycle | 31 | 3.82E-01 |
| 04512: ECM-receptor interaction | 23 | 4.29E-01 |
| 05222: Small cell lung cancer | 22 | 7.11E-01 |
| 05200: Pathways in cancer | 64 | 7.68E-01 |
| 04914: Progesterone-mediated oocyte maturation | 22 | 8.08E-01 |
| 03030: DNA replication | 12 | 8.44E-01 |
| 00240: Pyrimidine metabolism | 23 | 9.28E-01 |
| 03410: Base excision repair | 11 | 9.84E-01 |
BH, P-value after Benjamin-Hochberg correction.
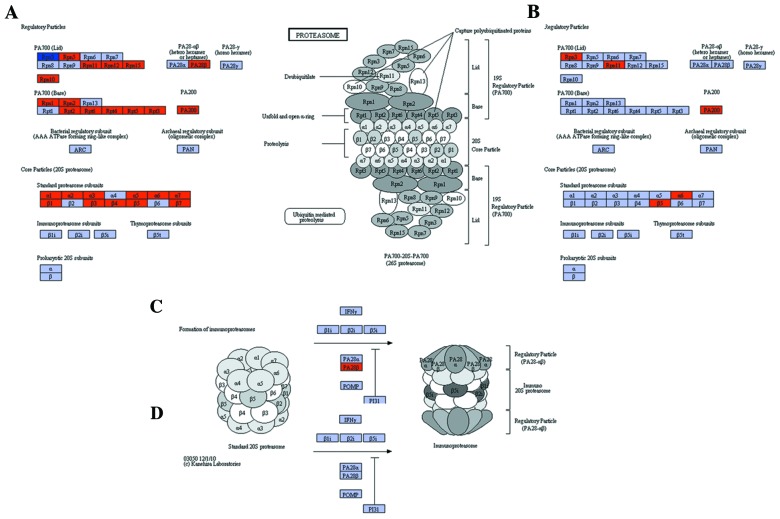
Following BH adjustment, the proteasome pathway was the only significantly enriched pathway in SCRC (P=7.18E-0.08). The proteasome pathway included 26 DEGs in SCRC, but was not enriched in CCRC. The proteasome pathway and the related DEGs are shown in Fig. 2. Nearly all DEGs in the proteasome pathway were upregulated genes, and the number of upregulated genes in SCRC (Fig. 2A and C) was higher compared to CCRC (Fig. 2B and D). The expression pattern of the gene Rpn3 was different between SCRC and CCRC: the gene was found to be downregulated in SCRC (Fig. 2B) and upregulated in CCRC (Fig. 2A).
Figure 2.
Differentially expressed genes in the proteasome pathway. Red boxes represent upregulated and dark blue boxes downregulated genes in cancer samples. Regulatory particles in (A) serrated and (B) conventional colorectal carcinoma and formation of immunoproteasomes in (C) serrated and (D) conventional colorectal carcinoma. Source, Kyoto Encyclopedia of Genes and Genomes (KEGG), Homo sapiens pathways.
Pathway interactions model based on PPI
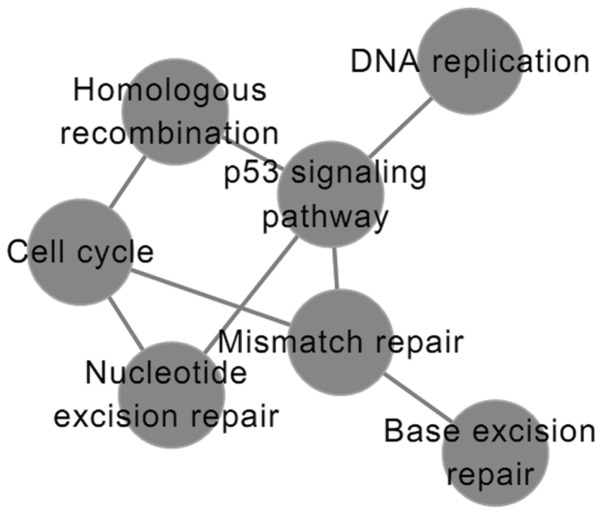
There was no interactions among pathways involved in SCRC (data not shown). However, there was some interaction between significantly enriched pathways involved in CCRC (Fig. 3). In the interaction network, p53 signaling, cell cycle and mismatch repair pathways showed the highest degree of nodes.
Figure 3.
Pathway network in conventional colorectal carcinoma. Nodes represent significantly enriched pathways. Edges represent the interaction between pathways.
PPI network
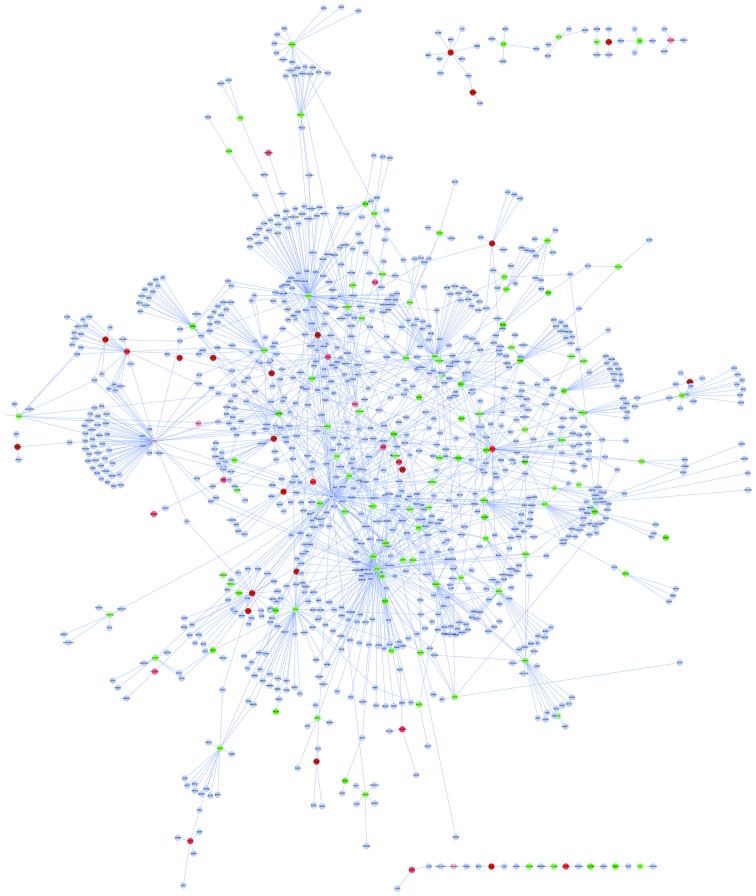
We constructed a PPI network for 149 DEGs involved in a total of 1,564 protein interactions. As shown in Fig. 4, we found 5 hub DEGs, i.e., DEGS that show the highest degree of nodes: tumour necrosis factor receptor-associated factor 6 (TRAF6, P=0.088453), atrophin 1 (ATN1, P=0.055616), integrin β1 (ITGB1, P=0.037011), fragile X-related gene 2 (FXR2, P=0.075519), kinase γ (IKBKG, P=0.094505) with degrees of node estimated at 90, 88, 62, 53 and 45, respectively (Table IV).
Figure 4.
Protein-protein interaction network of the 149 differential expressed genes (DEGs). Red nodes represent upregulated and green nodes downregulated gene products (proteins). Gray nodes represent non-differentially expressed proteins interacting with DEG gene products.
Table IV.
Information on the hub genes in the PPI network.
| Gene symbol | Degree | Dir | P-value_BH |
|---|---|---|---|
| TRAF6 | 90 | Downregulated | 0.088453 |
| ATN1 | 88 | Downregulated | 0.055616 |
| ITGB1 | 62 | Upregulated | 0.037011 |
| FXR2 | 53 | Downregulated | 0.075519 |
| IKBKG | 45 | Downregulated | 0.094505 |
PPI, protein-protein interaction; Dir, direction BH, Benjamini-Hochberg; TRAF6, tumour necrosis factor receptor-associated factor 6; ATN1, atrophin 1; ITGB1, integrin β1; FXR2, fragile X-related gene 2; IKBKG, inhibitor of nuclear factor κ-B kinase subunit γ.
Discussion
Serrated and conventional CRC are two subtypes of CRC. In this study, we attempted to identify the similarities and differences between these subtypes. DEGs were identified, and the enriched pathways in which these DEGs are involved were analyzed. We also constructed pathway interaction networks and PPI networks to further understand the molecular mechanisms underlying the pathogenesis of these two subtypes.
Based on KEGG pathway enrichment analysis, significantly enriched pathways in SCRC and CCRC included cell cycle, DNA replication, and base excision repair. These results suggest that a number of pathways are common between SCRC and CCRC.
The proteasome pathway was the only pathway that was enriched in SCRC but not in CCRC after BH adjustment. Proteasome is a subcellular organelle that is distributed in the cytosol, nucleus, endoplasmic reticulum and lysosome of eukaryotic cells (22). Its main function is the digestion of proteins, damaged or misfolded proteins, participation in the synthesis of peptides in the immune system, and regulation of the survival (half-life) of proteins that control the cell cycle. More than 80% of the proteins are processed by the proteasome in the cells (23). Proteasome dysfunction leads to disorders in cell-cycle regulation, cell hyperplasia, and imbalances in positive and negative signals (23). Inhibition of the proteasome function can lead to cell-cycle arrest, leading to cell death (24). Targeting the proteasome has been proposed as a novel approach in cancer therapy (25). Bortzomib is a proteasome inhibitor with antitumor activity against, for example, CRC, as shown in vitro (25), but it was revealed to be inactive in the clinic for patients with metastatic colorectal cancer (26). Our results showed that DEGs in CCRC were not significantly enriched for the proteasome pathway, which would provide an explanation for the clinical failure of bortzomib. It was also reported that the proteasome inhibitor MG-132 inhibits growth and stimulates cell apoptosis in CRC (27). In the present study, the proteasome pathway was enriched in SCRC, but not in CCRC. Therefore, the proteasome seems to be a pathway specifically involved in SCRC. The exact involvement of this pathway in SCRC merits further investigation.
In the CCRC pathway interaction network, we observed some interaction between pathways that was not observed in the SCRC pathway interaction network. The p53, cell cycle and mismatch repair pathways showed the highest degree of node. A mutation in p53 was previously reported to be detectable in the plasma and serum of patients with colorectal cancer or adenomas (28). Moreover, adenovirus-mediated transfer of genes of the p53 family induced cell-cycle arrest in colorectal cancer (29). It was also reported than mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating β-catenin or TCF signaling (30). Thus, our results suggest that the interaction between pathways may be important in the pathogenesis of conventional CRC.
In the PPI network, there were five hub genes, TRAF6, ATN1, ITGB1, FXR2 and IKBKG. TRAF6 had the highest degree of node. TRAF6 encodes a ubiquitin ligase. When activated, it can produce short protein chains. TRAF6 acts as a molecular switch, which allows activation of different signals. TGF with TRAF6 were reported to specifically activate the kinase TAK1 and other stress-activated kinases, leading to cell death (31). TRAF6 was shown to be an important oncogene for RAS-mediated oncogenesis in lung cancers (32). Ubiquitination plays a critical role in the activation of the nuclear factor-κB (NF-κB) signaling pathway, which has multiple functions in regulating cell proliferation, apoptosis and immune responses (33). There is no study to date on the involvement of TRAF6 in CRC. ATN1 encodes a nuclear corepressor expressed in nervous tissue. It interacts with tumor suppressors to control planar polarity (34). There are only a few reports on ATN1 and cancer (35,36). Therefore, TRAF6 and ATN1 may represent novel biomarkers, which are suitable for distinguishing SCRC and CCRC.
In conclusion, we found a number of significantly enriched pathways in SCRC and CCRC, which revealed certain aspects of the pathogenesis of these two subtypes of CRC. We also found that the proteasome pathway is significantly enriched only in SCRC. In addition, we identified the genes TRAF6 and ATN1, which showed the highest degree of node in the constructed PPI network. These results will be helpful for the understanding of the genesis and the specific therapy of SCRC and CCRC. However, further experiments are needed to confirm our results.
Acknowledgements
This study was supported by grants from the Shanghai Science and Technology Commission Foundation of Key Medical Research (no. 034119868 and 09411951600) and the Shanghai Health Research fund (no. 2007090, 20134034).
References
- 1.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol. 2011;42:1–10. doi: 10.1016/j.humpath.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Longacre TA, Fenoglio-Preiser CM. Mixed hyperplastic adenomatous polyps/serrated adenomas: A distinct form of colorectal neoplasia. Am J Surg Pathol. 1990;14:524–537. doi: 10.1097/00000478-199006000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Laiho P, Kokko A, Vanharanta S, et al. Serrated carcinomas form a subclass of colorectal cancer with distinct molecular basis. Oncogene. 2007;26:312–320. doi: 10.1038/sj.onc.1209778. [DOI] [PubMed] [Google Scholar]
- 5.Jass JR. Serrated route to colorectal cancer: back street or super highway? J Pathol. 2001;193:283–285. doi: 10.1002/1096-9896(200103)193:3<283::AID-PATH799>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Fukuyama R, Niculaita R, Ng KP, et al. Mutated in colorectal cancer, a putative tumor suppressor for serrated colorectal cancer, selectively represses beta-catenin-dependent transcription. Oncogene. 2008;27:6044–6055. doi: 10.1038/onc.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kriegl L, Vieth M, Kirchner T, Menssen A. Up-regulation of c-MYC and SIRT1 expression correlates with malignant transformation in the serrated route to colorectal cancer. Oncotarget. 2012;3:1182–1193. doi: 10.18632/oncotarget.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kambara T, Simms L, Whitehall V, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawyer E, Cerar A, Hanby A, et al. Molecular characteristics of serrated adenomas of the colorectum. Gut. 2002;51:200–206. doi: 10.1136/gut.51.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabates-Bellver J, Van der Flier LG, de Palo M, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 11.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 12.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 13.Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shabalin AA, Tjelmeland H, Fan C, Perou CM, Nobel AB. Merging two gene-expression studies via cross-platform normalization. Bioinformatics. 2008;24:1154–1160. doi: 10.1093/bioinformatics/btn083. [DOI] [PubMed] [Google Scholar]
- 15.Rudy J, Valafar F. Empirical comparison of cross-platform normalization methods for gene expression data. BMC Bioinformatics. 2011;12:467. doi: 10.1186/1471-2105-12-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thissen D, Steinberg L, Kuang D. Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J Educ Behav Stat. 2002;27:77–83. [Google Scholar]
- 17.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baolin L, Bo H. Network and Parallel Computing. Springer; Berlin: 2007. HPRD: a high performance RDF database; pp. 364–374. [Google Scholar]
- 21.Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters JM, Franke WW, Kleinschmidt JA. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994;269:7709–7718. [PubMed] [Google Scholar]
- 23.Adams J. Development of the proteasome inhibitor PS-341. Oncologist. 2002;7:9–16. doi: 10.1634/theoncologist.7-1-9. [DOI] [PubMed] [Google Scholar]
- 24.Zheng B, Georgakis GV, Li Y, et al. Induction of cell cycle arrest and apoptosis by the proteasome inhibitor PS-341 in Hodgkin disease cell lines is independent of inhibitor of nuclear factor-kappaB mutations or activation of the CD30, CD40, and RANK receptors. Clin Cancer Res. 2004;10:3207–3215. doi: 10.1158/1078-0432.ccr-03-0494. [DOI] [PubMed] [Google Scholar]
- 25.Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 26.O’Connor OA, Wright J, Moskowitz C, et al. Phase II clinical experience with the novel proteasome inhibitor bortezomib in patients with indolent non-Hodgkin’s lymphoma and mantle cell lymphoma. J Clin Oncol. 2005;23:676–684. doi: 10.1200/JCO.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 27.Hochwald SN, Lind DS, Malaty J, Copeland E, III, Moldawer LL, MacKay S. Antineoplastic therapy in colorectal cancer through proteasome inhibition. Am Surg. 2003;69:15–23. [PubMed] [Google Scholar]
- 28.Gocke CD, Benko FA, Kopreski MS, McGarrity TJ. p53 and APC mutations are detectable in the plasma and serum of patients with colorectal cancer (CRC) or adenomas. Ann NY Acad Sci. 2000;906:44–50. doi: 10.1111/j.1749-6632.2000.tb06589.x. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki Y, Morimoto I, Ishida S, Yamashita T, Imai K, Tokino T. Adenovirus-mediated transfer of the p53 family genes, p73 and p51/p63 induces cell cycle arrest and apoptosis in colorectal cancer cell lines: potential application to gene therapy of colorectal cancer. Gene Ther. 2001;8:1401–1408. doi: 10.1038/sj.gt.3301538. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Dong X, Mai M, et al. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat Genet. 2000;26:146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 31.Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS One. 2008;3:e4064. doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starczynowski DT, Lockwood WW, Deléhouzée S, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J Clin Investig. 2011;121:4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 34.Fanto M, Clayton L, Meredith J, et al. The tumor-suppressor and cell adhesion molecule Fat controls planar polarity via physical interactions with Atrophin, a transcriptional co-repressor. Development. 2003;130:763–774. doi: 10.1242/dev.00304. [DOI] [PubMed] [Google Scholar]
- 35.Mosakhani N, Lahti L, Borze I, et al. MicroRNA profiling predicts survival in anti-EGFR treated chemorefractory metastatic colorectal cancer patients with wild-type KRAS and BRAF. Cancer Genet. 2012;205:545–551. doi: 10.1016/j.cancergen.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Adélaïde J, Finetti P, Bekhouche I, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]