Abstract
Objective. Recently, many studies have shown that microRNAs (miRNA) exhibit altered expression in various cancers and may play an important role as prognostic biomarker of cancers. We performed a meta-analysis to evaluate the impact of miR-375 expression in solid tumors on patients' overall survival (OS). Methods. Studies were identified by searching PubMed, Embace, and Cochrane Library (last search update was in May 2014) and were assessed by further quality evaluation. The pooled hazard ratios (HRs) with 95% confidence intervals (CIs) for total and stratified analyses were calculated to investigate the association between miR-375 expression and cancer patients OS. Results. Our analysis results indicated that downregulation of miR-375 predicted poor OS (HR = 1.91, 95% CI 1.48–2.45, P < 0.001). Subgroup analyses showed that lower expression of miR-375 was significantly related with poor OS in patients with esophageal carcinoma (HR = 2.24, 95% CI 1.69–2.96, P < 0.001) and non-small-cell lung cancer (NSCLC) (HR = 1.71, 95% CI 1.31–2.24, P < 0.001). Conclusions. The findings from this meta-analysis suggest that miR-375 expression is associated with OS of patients with malignant tumors and could be a useful clinical prognostic biomarker.
1. Introduction
Cancer is one of the most common causes of death worldwide and has become a major public health issue [1]. Hence, newer cancer biomarkers with high sensitivity and specificity are essential for the proper detection, treatment, and prognosis of this fatal disease. MicroRNAs (miRNAs), small noncoding RNAs with a length of approximate 22 nucleotides, have been demonstrated to have high potential prognostic value in many cancers. MiRNAs biogenesis begins with transcription of primary transcripts (pri-miRNAs) in cell nucleus. Catalyzed by the ribonuclease Drosha and its essential cofactor DGCR8, pri-miRNAs release a 60–80-nucleotide precursor (pre-miRNA) which is exported to cell cytoplasm and cleaved by Dicer to generate a 22-nucleotide duplex. One strand of the duplex is incorporated into the RNA-induced silencing complex (RISC) by bounding with Argonaute, whereas the other is degraded. Base-pairing between the miRNAs and target mRNAs (usually in the 3′ untranslated region) guides RISC to complementary transcripts, leading to translation repression or the target mRNAs degradation [2]. It is estimated that miRNAs regulate 1/3 to 2/3 of human genes [3]. Thus, the dysregulation of the biogenesis and function of miRNAs is often associated with human diseases, especially malignancies [4]. Accumulating evidence has demonstrated that miRNAs act as oncogenes or tumor suppressors by targeting genes involved in cell differentiation, proliferation, survival, apoptosis, and metastasis (reviewed in [5]). The expression of numerous miRNAs is dysregulated in various cancers, which is often associated with diagnosis, staging, progression, prognosis, and response to clinical therapies (reviewed in [6]).
MicroRNA-375 (miR-375) was originally identified from murine pancreatic β-cell line MIN6 as a pancreatic islet-specific miRNA [7]. The miR-375 gene is located in an intergenic region between the beta-A2 crystallin (cryba2) and undescribed coiled-coil domain-containing protein 108 (ccdc108) genes in human chromosome 2q35 region [8], a genomic region conserving the synteny between humans and mice. Moreover, the sequences of pre-miR-375 in both species present a 100% homology, highlighting the high degree of conservation for this specific miRNA [8]. Further study revealed that miR-375 is a multifunctional miRNA participating in pancreatic islet development, glucose homeostasis, mucosal immunity, lung surfactant secretion, and more importantly tumorigenesis (reviewed in [9]). Recently, miR-375 has been found significantly dysregulated in many human cancers [10–15]. In addition, miR-375 has been found to have prognostic value in a variety of tumors. Multiple studies reported a significant association between low miR-375 and poor prognosis [12, 16–25]. However, insignificant or even opposite results have been found in some other studies [26–30]. According to the evidence we have gained so far, it is still not enough to come to a conclusion whether miR-375 could be used as a potent biomarker for prognosis. Therefore, a systematical and comprehensive meta-analysis was carried out to investigate the relationship between miR-375 expression and the survival of patients with cancer. To our knowledge, this meta-analysis is the first to evaluate the prognostic value of miR-375 in cancer patients.
2. Materials and Methods
This meta-analysis was carried out in accordance with the guidelines of meta-analysis of observational studies in epidemiology (MOOSE) [31].
2.1. Search Strategy
Studies were performed by searching PubMed, Embace, and Cochrane Library (updated by May 2014). The search strategy was “microRNA-375 OR miR-375” AND “tumor OR neoplasm OR cancer OR carcinoma.” No language limitation was applied. The reference lists of relative articles were also screened to further identify potential studies. The comprehensive database search was carried out independently by two authors (Y. Shao and Y. Geng). The disagreements were resolved by consensus.
2.2. Inclusion and Exclusion Criteria
Eligible studies included in this meta-analysis met the following criteria: (1) reporting explicit methods for the detection of miR-375 expression in tumor tissue or blood; (2) investigating the association between miR-375 expression and survival outcome, and the end-points being overall survival (OS); (3) reporting sufficient data to estimate the hazard ratio (HR) and 95% confidence intervals (CI) according to miR-375 expression. If a study reporting the same patient cohort was included in several publications, only the most recent or complete study was selected. Studies of case reports, letters, reviews, and animal trails were excluded. The titles and abstracts of the identified articles were evaluated independently by two reviewers and the irrelevant articles were excluded. The full text of the extracted articles was carefully examined for comprehensive evaluation. The disagreements were resolved by a discussion with the third reviewer (W. Gu).
2.3. Data Extraction and Qualitative Assessment
The data from all eligible studies were extracted by two investigators independently, which included first authors' surname, publication year, origin of population, sample number, tumor type, follow-up period, first-line therapies, source of miRNA, miR-375 assessment methods and the cut-off definition, and HR of miR-375 expression for OS as well as 95% corresponding interval (CI) and P value. If a study reported the results by both univariate and multivariate analyses, the latter was selected since it considered the confounding factors and was therefore more precise.
The quality of each study was assessed independently by two researchers according to the Newcastle-Ottawa Quality Assessment Scale (NOS) [32]. For quality, scores ranged from 0 (lowest) to 9 (highest), and studies with scores of 6 or more were rated as high quality.
2.4. Statistical Analysis
The high or low expression of miR-375 was defined according to the cut-off values provided by the authors. HRs and their 95% CIs were combined to evaluate the association between miR-375 expression and prognosis. If the statistical variables were described in the study, we pooled them directly. Otherwise, the statistical variables were calculated from available numerical data in the articles according to the methods described by Tierney et al. [33]. To reduce reading variability, the data from Kaplan-Meier survival curves was evaluated by three independent persons as described by Engauge Digitizer version 4.1. The additional information and original data needed for the meta-analysis were acquired by contacting with the corresponding authors of eligible articles. An observed HR greater than 1 indicated a worse prognosis in patients with miR-375 downregulation. Statistical heterogeneity was assessed by visual inspection of forest plots, by performing the Chi-square test (assessing the P value) and calculating the I 2 statistic [34, 35]. If the P value was less than 0.05 and/or I 2 exceeded 50%, indicating the presence of heterogeneity, a random-effects model (the DerSimonian-Laird method) was used. Otherwise, the fixed-effects model (the Mante-Haenszel method) was used. Subgroup analysis was further performed to explore the source of heterogeneity. Publication bias was estimated by visually assessing the asymmetry of an inverted funnel plot. Furthermore, Begg's test and Egger's test were performed to provide quantitative evidence of publication bias. If a publication bias was observed, it was adjusted by the use of the Duval and Tweedie trim-and-fill method [36]. All analyses were performed using STATA vision 12.0 (Stata Corporation, College Station, TX, USA). A P value less than 0.05 was considered to be statistically significant except where otherwise specified.
3. Results
3.1. Study Characteristics
According to the criteria mentioned in Materials and Methods, 504 abstracts were initially selected. However, 488 irrelevant abstracts were excluded. Sixteen full-text articles were reviewed for further evaluation. Of them, 3 were excluded because the data of HRs or OS were not available [28–30]. The remaining 13 articles contained 16 studies [12, 16–27], because one article included two independent cohort studies [16] and another article included three studies [12]. Thus, 16 studies were included in this meta-analysis, which were published between 2009 and 2014 (Figure 1). The total number of patients in all studies was 1,652, ranging from 37 to 249 patients. The category of cancers included esophageal carcinoma (8 studies), non-small-cell lung cancer (NSCLC, 3 studies), glioma, breast cancer, gastric cancer, head and neck squamous cell carcinoma (HNSCC), and pancreatic ductal adenocarcinoma (PDAC). Quantitative RT-PCR was used to detect miRNAs expression in all studies except one. The expression of miR-375 was detected in tumor tissues (11 studies) or blood samples (5 studies). The cut-off values of miR-375 varied in different studies. HRs were estimated in 8 studies and reported in the text of other studies. The major characteristics of the 16 eligible studies are listed in Table 1.
Figure 1.
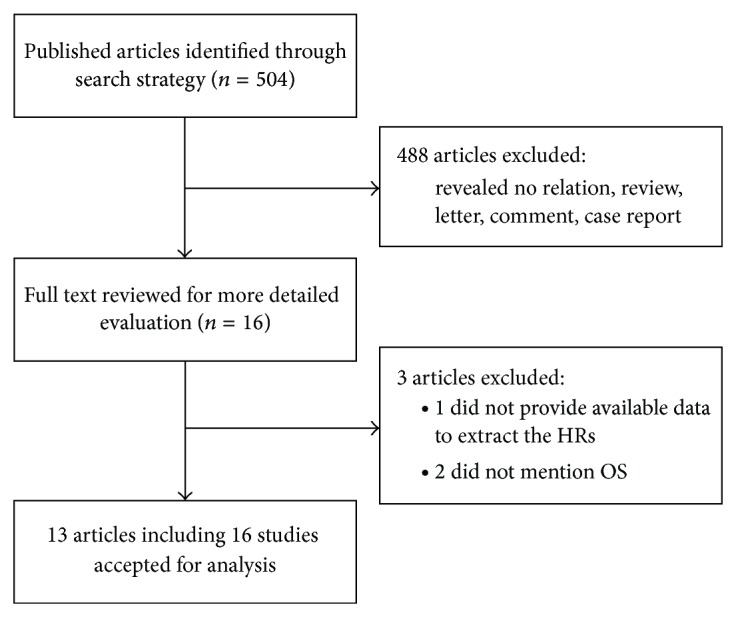
Flow chart depicting the selection of eligible studies.
Table 1.
Main characteristics of all studies included in the meta-analysis.
| Study | Origin of population |
Tumor type | Age (years, n) |
Gender (M/F) |
Sample number | Stage | Follow-up (months) |
Source of miRNA | miR-375 assay | Cut-off definition | Multivariate analysis | Survival analysis | HR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yu et al. 2014a [16] | China | NSCLC | <60, 96; ≥60, 68 |
86/78 | 164 | I–IV | Median 24 (rang: 0–56) |
Blood | qRT-PCR | Median | Yes | OS | Report |
| Yu et al. 2014b [16] | China | NSCLC | NR | NR | 53 | I–IV | Median 24 (rang: 0–56) |
Blood | qRT-PCR | Median | Yes | OS | Report |
| Wu et al. 2014 [17] | China | ESCC | Mean 61.4 (33–81) | 115/79 | 194 | I–IV | Median 31.3 | Blood | qRT-PCR | Mean | Yes | OS | Report |
| Li et al. 2012 [18] | China | NSCLC | Median 61 (37–75) | 70/26 | 96 | I–III | Median 30 (rang: 8–69) |
Tissue | qRT-PCR | Mean | Yes | OS | Report |
| Chang et al. 2012 [19] | China | Glioma | Median 42 (12–71) | 76/52 | 128 | I–IV | 60 | Tissue | qRT-PCR | Median | Yes | OS | Report |
| Madhavan et al. 2012 [20] | Germany | Breast | NR | 0/164 | 164 | IV | NR | Blood | qRT-PCR | Lower quartile | No | OS, PFS | DE |
| Zhang et al. 2011 [26] | China | Gastric | 30–83 | 43/22 | 65 | I–IV | Over 60 | Tissue | qRT-PCR | 2-fold | No | OS | DE |
| Harris et al. 2012 [27] | USA | HNSCC | 62.2 ± 11.9 | 85/38 | 123 | I–IV | NR | Tissue | qRT-PCR | Lower quartile | Yes | OS | Report |
| Ma et al. 2013 [21] | China | PDAC | NR | 44/34 | 78 | I–IV | NR | Tissue | qRT-PCR | Mean | Yes | OS | Report |
| Nguyen et al. 2010 [22] | USA, Canada | EADC | 32–80 | 35/23 | 58 | I–IV | Median 72 | Tissue | qRT-PCR | Median | Yes | OS | Report |
| Kong et al. 2012 [23] | China | ESCC | Mean 66 | 43/17 | 60 | I–IV | Over 60 | Tissue | qRT-PCR | Normal | No | OS, DFS | DE |
| Li et al. 2013 [24] | China | ESCC | ≤60, 105; >60, 144 |
136/113 | 249 | I–IV | Over 60 | Tissue | MISH | Normal | No | OS | DE |
| Komatsu et al. 2012 [25] | Japan | ESCC | Median 65 | 44/6 | 50 | 0–IV | 36 | Blood | qRT-PCR | Median | No | OS | DE |
| Mathe et al. 2009c [12] | USA, Canada | EADC | NR | NR | 63 | I–IV | Median 60 | Tissue | qRT-PCR | Median | No | OS | DE |
| Mathe et al. 2009d [12] | USA, Canada | EADC | NR | NR | 37 | I–IV | Median 60 | Tissue | qRT-PCR | Median | No | OS | DE |
| Mathe et al. 2009e [12] | USA, Japan | ESCC | <62, 28; ≥62, 42 |
52/18 | 70 | I–IV | Median 60 | Tissue | qRT-PCR | Median | No | OS | DE |
NSCLC: non-small cell lung cancer; ESCC: esophageal squamous cell carcinoma; HNSCC: head and neck squamous cell carcinoma; EADC: esophageal adenocarcinoma; qRT-PCR: quantitative real-time PCR; MISH: miRNA in situ hybridization; OS: overall survival; DFS: disease-free survival; PFS: progression-free survival; HR: hazard ratio; DE: data extrapolated; NR: not reported.
aStudy by Yu et al. [16] that evaluated the association between miR-375 expression and OS in a discovery cohort of 164 NSCLC patients.
bStudy by Yu et al. [16] that evaluated the association between miR-375 expression and OS in a validation cohort of 53 NSCLC patients.
cStudy by Mathe et al. [12] that evaluated the association between miR-375 expression and OS in a validation cohort of 63 EADC patients with Barrett's.
dStudy by Mathe et al. [12] that evaluated the association between miR-375 expression and OS in a validation cohort of 37 EADC patients without Barrett's.
eStudy by Mathe et al. [12] that evaluated the association between miR-375 expression and OS in a validation cohort of 70 ESCC patients.
3.2. Qualitative Assessment
Sixteen eligible studies included in our meta-analysis were assessed for quality according to the NOS. The quality of all included studies varied from 6 to 9, with a mean of 6.6. A higher value indicated a better methodology. Therefore, all studies were included in the subsequent analysis.
3.3. Meta-Analysis Results
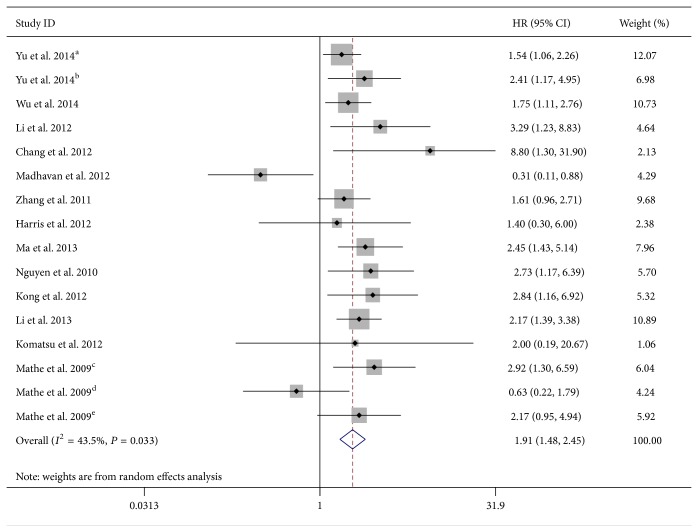
As the studies evaluating OS were of obvious statistical heterogeneity (I 2 = 43.2, P = 0.033), we used a random-effects model to pool the HRs. The result showed that downregulated miR-375 was significantly associated with poor OS outcome in various carcinomas, with the pooled HR of 1.91 (95% CI 1.48–2.45, P < 0.001) (Table 2; Figure 2).
Table 2.
The pooled associations between different situations of miR-375 expression and the prognosis of patients with solid tumors.
| Outcome subgroup | Number of patients | Number of studies | HR (95% CI) | P value | Heterogeneity | |
|---|---|---|---|---|---|---|
| I 2 | P | |||||
| Overall effect | 1652 | 16 | 1.91 (1.48–2.45)b | <0.001∗ | 43.2 | 0.033 |
| Tumor type | ||||||
| Esophageal carcinoma | 781 | 8 | 2.24 (1.69–2.96)a | <0.001∗ | 1.9 | 0.415 |
| NSCLC | 313 | 3 | 1.71 (1.31–2.24)a | <0.001∗ | 0.0 | 0.554 |
| Other cancers | 558 | 5 | 1.59 (0.71–3.58)b | 0.26 | 74.5 | 0.004 |
| miR-375 assay method | ||||||
| QRT-PCR | 1403 | 15 | 1.88 (1.42–2.48)b | <0.001∗ | 46.2 | 0.026 |
| MISH | 249 | 1 | 1.99 (1.28–3.10) | 0.002∗ | ||
| HR obtain method | ||||||
| Reported in text | 894 | 8 | 1.97 (1.57–2.47)a | <0.001∗ | 10.8 | 0.346 |
| Data extrapolated | 758 | 8 | 1.58 (0.99–2.51)b | 0.054 | 61.6 | 0.011 |
| Analysis type | ||||||
| Multivariate | 894 | 8 | 1.97 (1.57–2.47)a | <0.001∗ | 10.8 | 0.346 |
| Univariate | 758 | 8 | 1.58 (0.99–2.51)b | 0.054 | 61.6 | 0.011 |
| Source of miRNA | ||||||
| Tissue | 1027 | 11 | 2.16 (1.73–2.71)a | <0.001∗ | 15.7 | 0.295 |
| Blood | 625 | 5 | 1.40 (0.84–2.35)b | 0.197 | 63.5 | 0.027 |
| Patient origin | ||||||
| China | 1087 | 9 | 1.96 (1.62–2.37)a | <0.001∗ | 5.7 | 0.388 |
| USA | 351 | 5 | 1.97 (1.30–2.99)a | =0.001∗ | 36.2 | 0.180 |
| Other countries | 214 | 2 | 0.58 (0.10–3.24)b | 0.534 | 50.7 | 0.154 |
NSCLC: non-small-cell lung cancer; qRT-PCR: quantitative real-time PCR; MISH: miRNA in situ hybridization; HR: hazard ratio; CI: confidence intervals.
∗The difference was statistically significant.
aFixed-effects model.
bRandom-effects model.
Figure 2.
Forest plot of the relationship between lower miR-375 expression and overall survival (OS) in cancer patients with random-effects model. Different letters in superscript are represented in Table 1
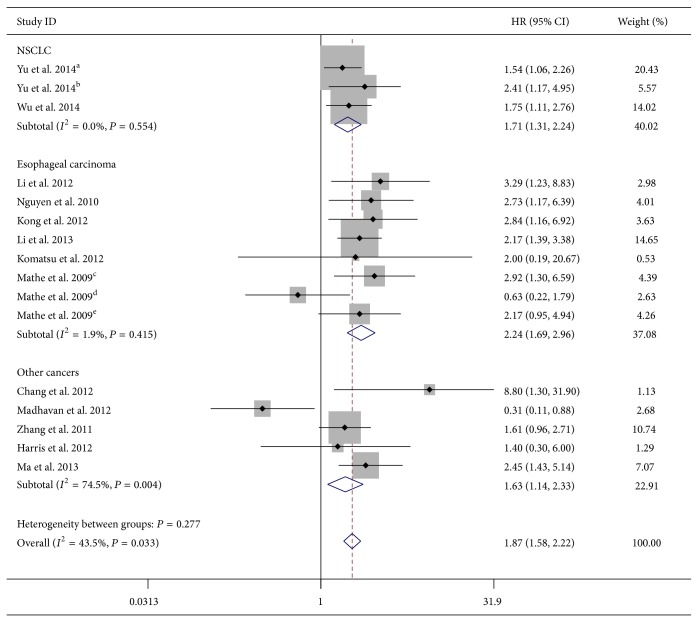
Considering the heterogeneity among these studies, the effect of miR-375 expression was further evaluated by subgroup analysis. The subgroups were classified according to the main characteristics such as tumor type, source of miRNA, miRNA assay method, type of method used to obtain the HR, patient origin, and analysis type. In the subgroup of tumor type, we found the downregulation of miR-375 was significantly associated with worse OS in esophageal carcinoma (HR = 2.24, 95% CI 1.69–2.96; P < 0.001; fixed-effects model) and NSCLC (HR = 1.71, 95% CI 1.31–2.24; fixed-effects model), without any heterogeneity in the data (I 2 = 1.9%, P = 0.415; I 2 = 0.0%,P = 0.554, resp.) (Table 2 and Figure 3). There was only one study that evaluated the association between lower miR-375 expression and OS in HNSCC, gastric cancer, breast cancer, PDAC, and glioma, respectively, and therefore, these tumors were defined as “other cancers.” Combined data from these five studies showed that decreased miR-375 expression was not correlated with poor OS (HR = 1.59, 95% CI 0.71–3.58; random-effects model) and with significant statistical heterogeneity (I 2 = 74.5%, P = 0.004) (Table 2). The association between lower miR-375 expression and worse OS outcome was statistically significant in other subgroups, including HR reported in text (HR = 1.97, 95% CI 1.57–2.47, fixed-effects model; P = 0.346 for heterogeneity test, I 2 = 10.8%), miR-375 assay by qRT-PCR (HR = 1.88, 95% CI 1.42–2.48, random-effects model; P = 0.026 for heterogeneity test, I 2 = 46.2%), tissue-source of miRNA (HR = 2.16, 95% CI 1.73–2.71, fixed-effects model; P = 0.295 for heterogeneity test, I 2 = 15.7%), multivariate analysis (HR = 1.97, 95% CI 1.57–2.47, fixed-effects model; P = 0.346 for heterogeneity test, I 2 = 10.8%), Chinese patients (HR = 1.96, 95% CI 1.62–2.37, fixed-effects model; P = 0.295 for heterogeneity test, I 2 = 15.7%), and American patients (HR = 1.97, 95% CI 1.30–2.99, fixed-effects model; P = 0.180 for heterogeneity test, I 2 = 36.2%) (Table 2). The expression of miR-375 did not show prognostic impact in subgroups of HRs by data extrapolated, univariate analysis, and blood-source of miRNA. The subgroup of miR-375 detected by MISH only included one study; therefore the result related entirely to the individual study.
Figure 3.
Forest plot of the relationship between lower miR-375 expression and overall survival (OS) in esophageal carcinoma and NSCLC with fixed-effects model.
3.4. Heterogeneity Analysis
Sensitivity analysis was performed by sequential omission of individual studies using the fixed-effects model, demonstrating that the study by Madhavan et al. [20] apparently influenced the overall results (Figure 4). When the Galbraith plot was analyzed, one study was identified as outliers of heterogeneity [20]. By excluding this study from the analysis, similar pooled HR and significance were obtained (HR = 1.96, 95% CI 1.65–2.33, P < 0.001) but heterogeneity was absent (P = 0.395, I 2 = 5.1%). We also conducted a meta-regression to explore the potential factors responsible for the heterogeneity. As a result, all these factors including tumor type, miR-375 assay method, follow-up time, publication year, patients origin, and cut-off values did not contribute to the heterogeneity obviously.
Figure 4.
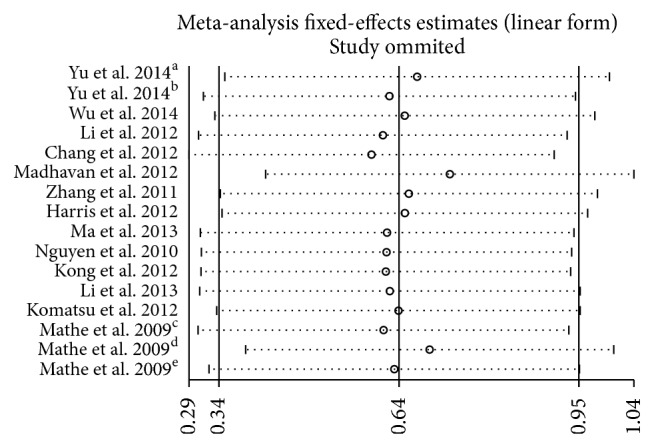
Sensitivity analysis for meta-analysis miR-375.
3.5. Publication Bias
Begg's funnel plot and Egger's test were used to evaluate the publication bias (Figure 5). The P values of Egger's and Begg's tests were all over 0.05 (P = 0.44 for Begg's test; P = 0.08 for Egger's test). Hence, there was no evidence for significant publication bias in the meta-analysis.
Figure 5.
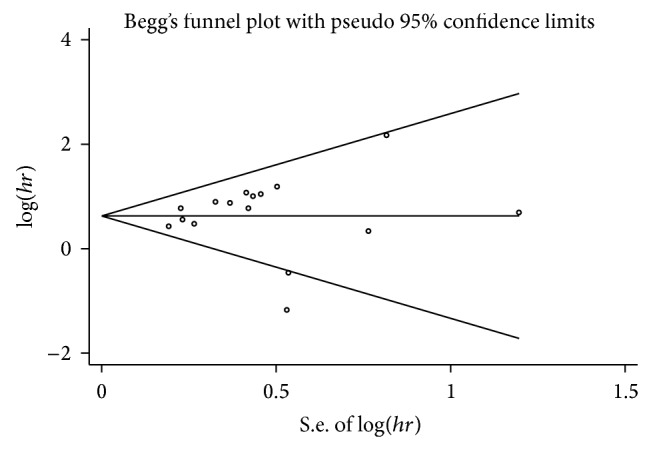
Funnel plot of lower miR-375 expression and overall survival in cancer patients.
4. Discussion
Recently, genome-wide miRNA expression profiling studies revealed that miR-375 is widely present in various tissues and organs, and its expression is significantly aberrant in malignant tumors, such as HNSCC, NSCLC, melanoma, glioma, hepatocellular, esophageal, gastric, breast, and prostate cancer [10–16, 19, 27]. It is indubitable that miR-375 is an important cancer-related miRNA. Increasing evidence has demonstrated that miR-375 is frequently downregulated in multiple types of cancer and acts as a tumor suppressor by repressing many critical oncogenes (Table 3). In hepatocellular carcinoma, the restoration of miR-375 in cancer cells decreased cell proliferation, clonogenicity, migration, and invasion and induced G1 arrest and apoptosis [10]. Moreover, MTDH was directly regulated by miR-375 in both hepatocellular carcinoma and HNSCC [10, 37, 38]. In gastric cancer, miR-375 was frequently downregulated and inhibited gastric cancer cell proliferation via targeting Janus kinase 2 [11]. Tsukamoto et al. also found a tumor suppressive role of miR-375 in gastric cancer. The ectopic expression of miR-375 reduced cell viability and induced apoptosis by targeting PDK1 and YWHAZ [39]. Similarly, in esophageal squamous cell carcinoma miR-375 was confirmed to inhibit cell proliferation, colony formation, and metastasis in vitro and in vivo [23]. Another study also verified the tumor-suppressive effect of miR-375 in esophageal cancer cell lines by targeting PDK1 [40]. However, there were still some contradictory views requiring adequate attention. It was reported that miR-375 was significantly upregulated in tumor tissues or serum of prostate carcinoma patients [14, 41, 42], but the role of miR-375 in prostate cancer was unclear. miR-375 was also upregulated in estrogen receptor alpha- (ERα-) positive breast cancer cell lines, which promoted cell proliferation and induced ERα upregulation via RASD1, a negative regulator of ERα [15]. In addition, higher expression of miR-375 was reported to result in progression of invasive lobular breast carcinoma [43]. Recently, miR-375 was found to participate in the process of epithelial-to-mesenchymal transition (EMT). It could partly reverse EMT-like properties in MCF-7 cells [44], which suggested its promoting role in tumor metastasis. Therefore, whether miR-375 acts as a tumor suppressor or an oncogene is unclear. Although plenty of studies focused on the potential application of miR-375 as a prognostic biomarker, the prognostic value of miR-375 expression in cancer patients is still controversial.
Table 3.
The dysregulated expression and target genes or pathways of miR-375 in cancer.
| Type of cancer | miR-375 expression | Validated target genes or pathways | Reference |
|---|---|---|---|
| HNSCC | Down | MTDH, LDHB, IGF1R | [37, 38, 45, 46] |
| ESCC | Down | PDK1, IGF1R | [23, 40] |
| Gastric cancer | Down | JAK2-STAT3, PDK1, YWHAZ, ERBB2 | [11, 39, 47–49] |
| HCC | Down | AEG-1, ATG7, YAP1 | [10, 50–52] |
| Lung cancer | Down | YAP1, CLDN1 | [53, 54] |
| Pancreatic cancer | Down | PDK1 | [55, 56] |
| Colorectal cancer | Down | PIK3CA | [57, 58] |
| Cervical cancer | Down | SP1 | [59, 60] |
| Breast cancer | Up | RASD1, IGF1R | [15, 61] |
| Prostate cancer | Up | Sec23A | [14] |
HNSCC: head and neck squamous cell carcinomas; ESCC: esophageal squamous cell carcinoma; HCC: hepatocellular carcinoma.
In the present study, we carried out a meta-analysis to evaluate the prognostic value of miR-375 by combining 16 studies with 1,652 patients. Our results demonstrated that lower miR-375 expression was associated with poor survival in patients with various carcinomas (HR = 1.91 95% CI 1.48–2.54, P < 0.001, random-effects model), indicating that miR-375 may serve as a positive prognostic marker for solid tumors. In the subgroup analyses, the association between lower miR-375 expression and worse OS was statistically significant in most subgroups, especially in esophageal carcinoma (HR = 2.24, 95% CI 1.69–2.96; P < 0.001; fixed-effects model) and NSCLC (HR = 1.71, 95% CI 1.31–2.24; fixed-effects model). Recently, there were only two studies exploring the relationship between miR-375 expression and the survival of cancer patients. Consistent with our results, they also indicated low miR-375 expression was significantly associated with poor OS of patients with esophageal carcinoma. However, the results of these two studies are not credible enough because they did not eliminate interference factors from their meta-analyses, including miR-375 expression in pericarcinoma tissues of the same patient cohort, which may possibly come to a conclusion with deviation.
In sensitivity analysis, the study of Madhavan et al. was identified as an outlier of heterogeneity. It indicated a positive correlation between upregulated miR-375 expression and poor OS in metastatic breast cancer (MBC) patients (HR = 0.31, 95% CI 0.11–0.88) [20], which may be attributed to mesenchymal-to-epithelial transition (MET), a process essential for successful colonization and establishment of metastasis induced by high miR-375 expression [44]. The significant prognostic effect of high miR-375 expression in this study with obvious heterogeneity possibly led to the negative statistical results in some subgroup analyses, including data extrapolated, univariate analysis, and blood-source of miRNA: in these subgroups, there was no association between lower expression of miR-375 and poor prognosis.
Although the predictive value of miR-375 was statistically proved by the meta-analysis in this study, it should be carefully comprehend for the following reasons. First, four studies which explored the association between miR-375 expression and prognosis were not included in the meta-analysis because they did not provide available data of HRs or OS, and three of them showed insignificant or even opposite results. These studies might influence the reliability of our results. Secondly, due to the lack of uniform cut-off value in miR-375 expression, the cut-off values applied by different researchers deviated from the actual value more or less, which may affect the validity of miR-375 as a predictive factor in cancer prognosis. Thirdly, several HRs were calculated based on the data extracted from the survival curve, bringing minor deviations. Fourthly, the statistical heterogeneity was not obvious in this meta-analysis, but the clinical and methodological heterogeneity existed in the baseline demographic characteristics, including population, tumor types and stages, the cut-off value of miR-375 expression, and duration of follow-up. Finally, although no significant publication bias was detected in this meta-analysis, the results still need to be verified by a large number of publications.
In conclusion, this meta-analysis summarized the global researches on the relationship of aberrant miR-375 expression and the prognosis of patients with cancer and clarified that downregulation of miR-375 is significantly associated with poor survival in patients with various types of carcinoma, especially in esophageal carcinoma and NSCLC. In view of the limitation of the current analysis, it should be cautious to appreciate the conclusion, and further prospective multicenter studies designed adequately with larger sample size are needed to verify the association between miR-375 and cancer prognosis as well as the efficiency of therapies.
Acknowledgments
The authors would like to acknowledge all the investigators, site staff, and their families involved in the study. They also thank Professor Jingting Jiang, M.D., for his editorial support.
Conflict of Interests
The authors declare that they have no conflict of interests.
Authors’ Contribution
Yingjie Shao and Yiting Geng contributed equally to this work.
Funding
This research project was supported by the National Natural Science Foundation of China (NSFC) (81171653) and the General Program of Jiangsu Province Department of Health (H201350).
References
- 1.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. A Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Kim V. N., Han J., Siomi M. C. Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 4.Friedman R. C., Farh K. K. H., Burge C. B., Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garzon R., Fabbri M., Cimmino A., Calin G. A., Croce C. M. MicroRNA expression and function in cancer. Trends in Molecular Medicine. 2006;12(12):580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Di Leva G., Croce C. M. MiRNA profiling of cancer. Current Opinion in Genetics and Development. 2013;23(1):3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., MacDonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 8.Baroukh N. N., van Obberghen E. Function of microRNA-375 and microRNA-124a in pancreas and brain. FEBS Journal. 2009;276(22):6509–6521. doi: 10.1111/j.1742-4658.2009.07353.x. [DOI] [PubMed] [Google Scholar]
- 9.Yan J.-W., Lin J.-S., He X.-X. The emerging role of miR-375 in cancer. International Journal of Cancer. 2014;135(5):1011–1018. doi: 10.1002/ijc.28563. [DOI] [PubMed] [Google Scholar]
- 10.He X.-X., Chang Y., Meng F.-Y., Wang M.-Y., Xie Q.-H., Tang F., Li P.-Y., Song Y.-H., Lin J.-S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31(28):3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 11.Ding L., Xu Y., Zhang W., Deng Y., Si M., Du Y., Yao H., Liu X., Ke Y., Si J., Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Research. 2010;20(7):784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 12.Mathe E. A., Nguyen G. H., Bowman E. D., et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clinical Cancer Research. 2009;15(19):6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avissar M., Christensen B. C., Kelsey K. T., Marsit C. J. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clinical Cancer Research. 2009;15(8):2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szczyrba J., Nolte E., Wach S., Kremmer E., Stöhr R., Hartmann A., Wieland W., Wullich B., Grässer F. A. Downregulation of Sec23A protein by miRNA-375 in prostate carcinoma. Molecular Cancer Research. 2011;9(6):791–800. doi: 10.1158/1541-7786.MCR-10-0573. [DOI] [PubMed] [Google Scholar]
- 15.Rocha Simonini P. D. S., Breiling A., Gupta N., Malekpour M., Youns M., Omranipour R., Malekpour F., Volinia S., Croce C. M., Najmabadi H., Diederichs S., Sahin Ö., Mayer D., Lyko F., Hoheisel J. D., Riazalhosseini Y. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor α in breast cancer cells. Cancer Research. 2010;70(22):9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 16.Yu H., Jiang L., Sun C., Guo L., Lin M., Huang J., Zhu L. Decreased circulating miR-375: a potential biomarker for patients with non-small-cell lung cancer. Gene. 2014;534(1):60–65. doi: 10.1016/j.gene.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Wu C., Li M., Hu C., Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Molecular Biology Reports. 2014;41(3):1257–1266. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Jiang Q., Xia N., Yang H., Hu C. Decreased expression of microRNA-375 in nonsmall cell lung cancer and its clinical significance. The Journal of International Medical Research. 2012;40(5):1662–1669. doi: 10.1177/030006051204000505. [DOI] [PubMed] [Google Scholar]
- 19.Chang C., Shi H., Wang C., Wang J., Geng N., Jiang X., Wang X. Correlation of microRNA-375 downregulation with unfavorable clinical outcome of patients with glioma. Neuroscience Letters. 2012;531(2):204–208. doi: 10.1016/j.neulet.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Madhavan D., Zucknick M., Wallwiener M., Cuk K., Modugno C., Scharpff M., Schott S., Heil J., Turchinovich A., Yang R., Benner A., Riethdorf S., Trumpp A., Sohn C., Pantel K., Schneeweiss A., Burwinkel B. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clinical Cancer Research. 2012;18(21):5972–5982. doi: 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 21.Ma M.-Z., Kong X., Weng M.-Z., Cheng K., Gong W., Quan Z.-W., Peng C.-H. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. Journal of Experimental and Clinical Cancer Research. 2013;32(1, article 71) doi: 10.1186/1756-9966-32-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen G. H., Schetter A. J., Chou D. B., Bowman E. D., Zhao R., Hawkes J. E., Mathé E. A., Kumamoto K., Zhao Y., Budhu A., Hagiwara N., Wang X. W., Miyashita M., Casson A. G., Harris C. C. Inflammatory and microRNA gene expression as prognostic classifier of Barrett's-associated esophageal adenocarcinoma. Clinical Cancer Research. 2010;16(23):5824–5834. doi: 10.1158/1078-0432.CCR-10-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong K. L., Kwong D. L. W., Chan T. H.-M., Law S. Y.-K., Chen L., Li Y., Qin Y.-R., Guan X.-Y. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61(1):33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 24.Li J., Li X., Li Y., Yang H., Wang L., Qin Y., Liu H., Fu L., Guan X.-Y. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0053582.e53582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komatsu S., Ichikawa D., Takeshita H., Konishi H., Nagata H., Hirajima S., Kawaguchi T., Arita T., Shiozaki A., Fujiwara H., Okamoto K., Otsuji E. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opinion on Biological Therapy. 2012;12(1):S53–S59. doi: 10.1517/14712598.2012.681373. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Yan Z., Zhang J., Gong L., Li W., Cui J., Liu Y., Gao Z., Li J., Shen L., Lu Y. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Annals of Oncology. 2011;22(10):2257–2266. doi: 10.1093/annonc/mdq758. [DOI] [PubMed] [Google Scholar]
- 27.Harris T., Jimenez L., Kawachi N., Fan J.-B., Chen J., Belbin T., Ramnauth A., Loudig O., Keller C. E., Smith R., Prystowsky M. B., Schlecht N. F., Segall J. E., Childs G. Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. American Journal of Pathology. 2012;180(3):917–928. doi: 10.1016/j.ajpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonsdottir K., Janssen S. R., Da Rosa F. C., Gudlaugsson E., Skaland I., Baak J. P. A., Janssen E. A. M. Validation of expression patterns for nine miRNAs in 204 lymph-node negative breast cancers. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0048692.e48692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlsen A. L., Joergensen M. T., Knudsen S., De Muckadell O. B. S., Heegaard N. H. H. Cell-free plasma microRNA in pancreatic ductal adenocarcinoma and disease controls. Pancreas. 2013;42(7):1107–1113. doi: 10.1097/MPA.0b013e318296bb34. [DOI] [PubMed] [Google Scholar]
- 30.Bryant R. J., Pawlowski T., Catto J. W. F., Marsden G., Vessella R. L., Rhees B., Kuslich C., Visakorpi T., Hamdy F. C. Changes in circulating microRNA levels associated with prostate cancer. British Journal of Cancer. 2012;106(4):768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stroup D. F., Berlin J. A., Morton S. C., Olkin I., Williamson G. D., Rennie D., Moher D., Becker B. J., Sipe T. A., Thacker S. B. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Journal of the American Medical Association. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 32.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 33.Tierney J. F., Stewart L. A., Ghersi D., Burdett S., Sydes M. R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8, article 16 doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins J. P. T., Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 35.Higgins J. P. T., Thompson S. G., Deeks J. J., Altman D. G. Measuring inconsistency in meta-analyses. British Medical Journal. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duval S., Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 37.Nohata N., Hanazawa T., Kikkawa N., Mutallip M., Sakurai D., Fujimura L., Kawakami K., Chiyomaru T., Yoshino H., Enokida H., Nakagawa M., Okamoto Y., Seki N. Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC) Journal of Human Genetics. 2011;56(8):595–601. doi: 10.1038/jhg.2011.66. [DOI] [PubMed] [Google Scholar]
- 38.Hui A. B. Y., Bruce J. P., Alajez N. M., Shi W., Yue S., Perez-Ordonez B., Xu W., O'Sullivan B., Waldron J., Cummings B., Gullane P., Siu L., Liu F.-F. Significance of dysregulated metadherin and microRNA-375 in head and neck cancer. Clinical Cancer Research. 2011;17(24):7539–7550. doi: 10.1158/1078-0432.CCR-11-2102. [DOI] [PubMed] [Google Scholar]
- 39.Tsukamoto Y., Nakada C., Noguchi T., Tanigawa M., Nguyen L. T., Uchida T., Hijiya N., Matsuura K., Fujioka T., Seto M., Moriyama M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3ζ . Cancer Research. 2010;70(6):2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 40.Li X., Lin R., Li J. Epigenetic silencing of MicroRNA-375 regulates PDK1 expression in esophageal cancer. Digestive Diseases and Sciences. 2011;56(10):2849–2856. doi: 10.1007/s10620-011-1711-1. [DOI] [PubMed] [Google Scholar]
- 41.Wach S., Nolte E., Szczyrba J., Stöhr R., Hartmann A., Ørntoft T., Dyrskjøt L., Eltze E., Wieland W., Keck B., Ekici A. B., Grässer F., Wullich B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. International Journal of Cancer. 2012;130(3):611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 42.Selth L. A., Townley S., Gillis J. L., Ochnik A. M., Murti K., Macfarlane R. J., Chi K. N., Marshall V. R., Tilley W. D., Butler L. M. Discovery of circulating microRNAs associated with human prostate cancer using a mouse model of disease. International Journal of Cancer. 2012;131(3):652–661. doi: 10.1002/ijc.26405. [DOI] [PubMed] [Google Scholar]
- 43.Giricz O., Reynolds P. A., Ramnauth A., Liu C., Wang T., Stead L., Childs G., Rohan T., Shapiro N., Fineberg S., Kenny P. A., Loudig O. Hsa-miR-375 is differentially expressed during breast lobular neoplasia and promotes loss of mammary acinar polarity. Journal of Pathology. 2012;226(1):108–119. doi: 10.1002/path.2978. [DOI] [PubMed] [Google Scholar]
- 44.Ward A., Balwierz A., Zhang J. D., Küblbeck M., Pawitan Y., Hielscher T., Wiemann S., Sahin Ö. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32(9):1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 45.Kinoshita T., Nohata N., Yoshino H., Hanazawa T., Kikawa N., Fujimura L., Chiyomaru T., Kawakami K., Enokida H., Nakagawa M., Okamoto Y., Seki N. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. International Journal of Oncology. 2012;40(1):185–193. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- 46.Luo J., Wu J., Li Z., et al. miR-375 suppresses IGF1R expression and contributes to inhibition of cell progression in laryngeal squamous cell carcinoma. BioMed Research International. 2014;2014 doi: 10.1155/2014/374598.374598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y., Jin J., Liu Y., et al. Snail-regulated MiR-375 inhibits migration and invasion of gastric cancer cells by targeting JAK2. PLoS ONE. 2014;9(7) doi: 10.1371/journal.pone.0099516.e99516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z.-Y., Zhang Z.-Z., Liu H., Zhao E.-H., Cao H. miR-375 inhibits the proliferation of gastric cancer cells by repressing ERBB2 expression. Experimental and Therapeutic Medicine. 2014;7(6):1757–1761. doi: 10.3892/etm.2014.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miao L., Liu K., Xie M., Xing Y., Xi T. miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis by blocking JAK2-STAT3 signaling. Cancer Immunology, Immunotherapy. 2014;63(7):699–711. doi: 10.1007/s00262-014-1550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang Y., Yan W., He X., Zhang L., Li C., Huang H., Nace G., Geller D. A., Lin J., Tsung A. MiR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143(1):177–187. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Liu A. M., Poon R. T. P., Luk J. M. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochemical and Biophysical Research Communications. 2010;394(3):623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 52.Furuta M., Kozaki K.-I., Tanaka S., Arii S., Imoto I., Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2009;31(5):766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 53.Nishikawa E., Osada H., Okazaki Y., Arima C., Tomida S., Tatematsu Y., Taguchi A., Shimada Y., Yanagisawa K., Yatabe Y., Toyokuni S., Sekido Y., Takahashi T. MiR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Research. 2011;71(19):6165–6173. doi: 10.1158/0008-5472.CAN-11-1020. [DOI] [PubMed] [Google Scholar]
- 54.Yoda S., Soejima K., Hamamoto J., et al. Claudin-1 is a novel target of miR-375 in non-small-cell lung cancer. Lung Cancer. 2014;85(3):366–372. doi: 10.1016/j.lungcan.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Zhou J., Song S., He S., Zhu X., Zhang Y. I., Yi B., Zhang B., Qin G., Li D. MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. International Journal of Molecular Medicine. 2014;33(4):950–956. doi: 10.3892/ijmm.2014.1638. [DOI] [PubMed] [Google Scholar]
- 56.Eun J. L., Gusev Y., Jiang J., Nuovo G. J., Lerner M. R., Frankel W. L., Morgan D. L., Postier R. G., Brackett D. J., Schmittgen T. D. Expression profiling identifies microRNA signature in pancreatic cancer. International Journal of Cancer. 2007;120(5):1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Tang Q., Li M., Jiang S., Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochemical and Biophysical Research Communications. 2014;444(2):199–204. doi: 10.1016/j.bbrc.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 58.Faltejskova P., Svoboda M., Srutova K., Mlcochova J., Besse A., Nekvindova J., Radova L., Fabian P., Slaba K., Kiss I., Vyzula R., Slaby O. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. Journal of Cellular and Molecular Medicine. 2012;16(11):2655–2666. doi: 10.1111/j.1582-4934.2012.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F., Li Y., Zhou J., Xu J., Peng C., Ye F., Shen Y., Lu W., Wan X., Xie X. MiR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. The American Journal of Pathology. 2011;179(5):2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilting S. M., Verlaat W., Jaspers A., Makazaji N. A., Agami R., Meijer C. J. L. M., Snijders P. J. F., Steenbergen R. D. M. Methylation-mediated transcriptional repression of microRNAs during cervical carcinogenesis. Epigenetics. 2013;8(2):220–228. doi: 10.4161/epi.23605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye X.-M., Zhu H.-Y., Bai W.-D., Wang T., Wang L., Chen Y., Yang A.-G., Jia L.-T. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer. 2014;14(1, article 134) doi: 10.1186/1471-2407-14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]