Abstract
The interaction between the transmembrane glycoprotein surface receptor CD40 expressed by skin epithelial cells (ECs) and its T-cell–expressed ligand CD154 was suggested to exacerbate inflammatory skin diseases. However, the full spectrum of CD40-mediated effects by ECs underlying this observation is unknown. Therefore, changes in gene expression after CD40 ligation of ECs were studied by microarrays. CD40-mediated activation for 2 hours stimulated the expression of a coordinated network of immune-involved genes strongly interconnected by IL8 and TNF, whereas after 24 hours anti-proliferative and anti-apoptotic genes were upregulated. CD40 ligation was associated with the production of chemokines and the attraction of lymphocytes and myeloid cells from peripheral blood mononuclear cells (PBMCs). Thus, CD40-mediated activation of ECs resulted in a highly coordinated response of genes required for the local development and sustainment of adaptive immune responses. The importance of this process was confirmed by a study on the effects of human papilloma virus (HPV) infection to the EC's response to CD40 ligation. HPV infection clearly attenuated the magnitude of the response to CD40 ligation and the EC's capacity to attract PBMCs. The fact that HPV attenuates CD40 signaling in ECs indicates the importance of the CD40-CD154 immune pathway in boosting cellular immunity within epithelia.
Introduction
CD40 is a 48-kDa transmembrane glycoprotein surface receptor also known as the tumor necrosis factor receptor superfamily member 5 (TNFRSF5). It is expressed at the cell surface of antigen-presenting cells of the hematopoietic lineage, including B cells, dendritic cells (DCs), Langerhans cells, and macrophages, and is also expressed by non-hematopoietic cells such as endothelial cells (Hollenbaugh et al., 1995), fibroblasts (Fries et al., 1995; Yellin et al., 1995), smooth muscle cells, and epithelial cells (ECs) (Galy and Spits, 1992). The ligand for CD40 is the type II membrane protein CD40L (CD154), which is primarily expressed on activated CD4+ T-helper cells. The CD40–CD154 interaction has a role in both cellular and humoral immune responses. Upon CD40 ligation, DCs mature and become activated to produce high levels of pro-inflammatory cytokines and chemokines, and upregulate major histocompatibility complex class II and co-stimulatory molecules such as CD80 and CD86. Together, these upregulated molecules facilitate effective priming of CD8+ T cells and stimulate activated CD8+ T cells to become cytotoxic effector cells (Ma and Clark, 2009). In B cells, CD40 ligation induces immunoglobulin isotype switching and differentiation as well as inhibits apoptosis by upregulating anti-apoptotic genes such as cIAPs, members of the BCL2 family, and MYC (Kehry, 1996; Laman et al., 1996). Deregulation of CD40–CD154 interaction can lead to various clinical conditions (Peters et al., 2009), such as autoimmune diseases, multiple sclerosis, allograft rejections, intraepithelial pre-malignancies, and inflammatory skin diseases such as psoriasis and subacute cutaneous lupus erythematosus (Caproni et al., 2007).
In the epidermis, CD40 is expressed at low levels by basal and parabasal layer ECs. ECs upregulate CD40 expression when stimulated with IFNγ (Denfeld et al., 1996; Gaspari et al., 1996; Peguet-Navarro et al., 1997), which is normally produced by effector cells of the innate immune system and by activated type 1 polarized (IFNγ-producing) CD40L-expressing CD4+ T-helper (Th1) cells that enter the skin (Swamy et al., 2010; van den Bogaard et al., 2013). Indeed, CD40 is highly expressed by ECs in T-cell–infiltrated psoriatic lesions (Denfeld et al., 1996). A limited number of in vitro studies on CD40 ligation of human primary IFNγ-stimulated ECs showed that these cells express ICAM-1 and secrete RANTES (CCL5), TNFα, IL-6, IL-8, and MCP-1 (CCL2) (Denfeld et al., 1996; Gaspari et al., 1996; Peguet-Navarro et al., 1997; Companjen et al., 2002; Pasch et al., 2004). In addition, there is evidence that CD40-activated ECs stop proliferating and start differentiating (Peguet-Navarro et al., 1997; Grousson et al., 2000; Concha et al., 2003; Villarroel Dorrego et al., 2006). However, the full spectrum of effects mediated by CD40 ligation on the response of ECs is still unknown.
The basal and parabasal layer ECs of squamous epithelia are a well-known target for different viruses (Andrei et al., 2010), including high-risk human papilloma virus (hrHPV). Chronic infections with hrHPV can last for many years, probably as a result of several sophisticated mechanisms employed by hrHPV to evade the hosts' innate immune response (Karim et al., 2011; Reiser et al., 2011; Karim et al., 2013). Interestingly, an in vivo model for EC-specific human-CD40 expression and activation showed that CD40 ligation on ECs enhanced DC migration and T-cell priming in a mouse model (Fuller et al., 2002), suggesting that ECs boost the activity of cells from the adaptive immune system. HPV-specific cellular immunity, however, develops quite late and slowly during persistent HPV infections (van der Burg and Melief, 2011), posing the question whether HPV may also impair pathways typically associated with activation of the adaptive immune response.
To obtain a better understanding of the outcome between the interaction of ECs and CD40 ligand–expressing CD4+ Th1 cells, we analyzed the genome-wide expression profiles of CD40-stimulated undifferentiated primary ECs. We observed that ECs react in a very coordinated manner to CD40 ligation with the induction of mainly immune-related genes and the attraction of immune cells. The parallel analysis of hrHPV-infected primary ECs revealed that hrHPV did not grossly change the gene expression pattern but attenuated the magnitude of the CD40-stimulated immune response, resulting in an impaired immune cell attraction. These data strengthen the notion that the CD40–CD154 pathway has an important role in protective epithelial immune responses.
Results
CD40 upregulation and functionality on ECs
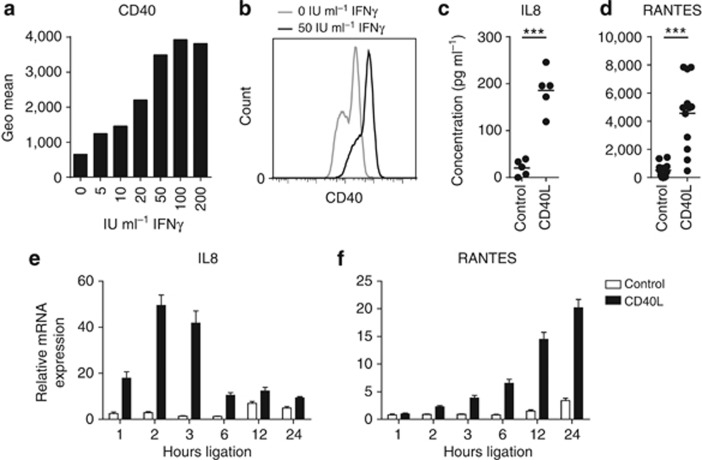
To study how ECs respond to CD40 ligation on a genome-wide scale, we mimicked the CD40–CD154 interaction between ECs and IFNγ-secreting CD4+ T cells. Basal CD40 levels on cultured ECs are too low for efficient in vitro ligation with CD154; however, ECs upregulate the expression of CD40 when stimulated with IFNγ (Denfeld et al., 1996; Gaspari et al., 1996; Peguet-Navarro et al., 1997). Therefore, we measured by flow cytometry the CD40 expression on primary undifferentiated ECs stimulated with increasing concentrations of IFNγ for 72 hours. In line with previous reports, CD40 expression was enhanced by IFNγ at all concentrations but became optimal at a concentration equal to or >50 IU ml−1 IFNγ for the primary ECs obtained from four different healthy donors (Figure 1a and b). Therefore, this dose was used in our subsequent studies.
Figure 1.
Epithelial cells (ECs) produce cytokines and chemokines upon CD40 ligation. (a) CD40 upregulation on vaginal ECs upon stimulation with 0, 5, 10, 20, 50, 100, or 200 IU ml−1 IFNγ for 3 days. The height of the bars represents the CD40 mean fluorescence intensity as determined by flow cytometry. (b) Histogram of CD40 expression on vaginal ECs stimulated 3 days with 0 and 50 IU ml−1 IFNγ. ELISA for IL8 (c) and RANTES (d) in cleared supernatants from IFNγ-pre-stimulated foreskin, vaginal, and cervical EC cultures (n=5–12) cocultured for 24 hours with control or CD40L-expressing L cells in the presence of IFNγ. *** Indicates P<0.0005. Reverse transcriptase-quantitative PCR of IL8 (e) and RANTES (f) expression by IFNγ-pre-stimulated vaginal ECs cocultured with L-control or L-CD40L cells in the presence of IFNγ for 0, 1, 2, 3, 6, 12, or 24 hours. Gene expression was normalized using GAPDH as the calibrator gene. Fold changes over 0 hours coculture were calculated and depicted. These data are representative for two to three independent experiments.
ECs were reported to secrete the pro-inflammatory chemokines IL8 (CXCL8) and RANTES (CCL5) upon CD40 ligation (Denfeld et al., 1996; Gaspari et al., 1996; Peguet-Navarro et al., 1997; Pasch et al., 2004). Indeed, this was also observed for CD40-expressing ECs stimulated with CD154-expressing L cells (CD40L) as compared with ECs cultured with control L cells (Figure 1c and d), showing that our ECs expressed functionally active CD40. To determine the optimal time points for measuring the response of CD40-ligated ECs on a genome-wide scale, ECs were stimulated for up to 24 hours with CD40L and the peak gene expression of IL8 and RANTES was determined. The highest expression of IL8 was detected after 2 hours (Figure 1e), whereas RANTES peaked after 24 hours of CD40 ligation (Figure 1f). We concluded that these two time points were most suited for studying early and late responses of ECs to CD40 ligation.
ECs upregulate genes involved in immune signaling and proliferation after CD40 ligation
The effects of CD40 ligation on four freshly isolated uninfected primary EC cultures from healthy donors of foreskin, vaginal, or cervical origin were studied by genome-wide expression profiling. These ECs are the natural target for hrHPV, which is most commonly transmitted by sexual contact. We verified that the cells were activated via CD40 by confirming the increased expression of IL8 (2 hours) and RANTES (24 hours) (Supplementary Figure S1 online), and subsequently subjected the samples to microarray analysis. Plots with microarray log2 intensities confirmed that IL8 and RANTES were upregulated after 2 and 24 hours, respectively (Supplementary Figure S1 online) and confirmed the results obtained by quantitative PCR.
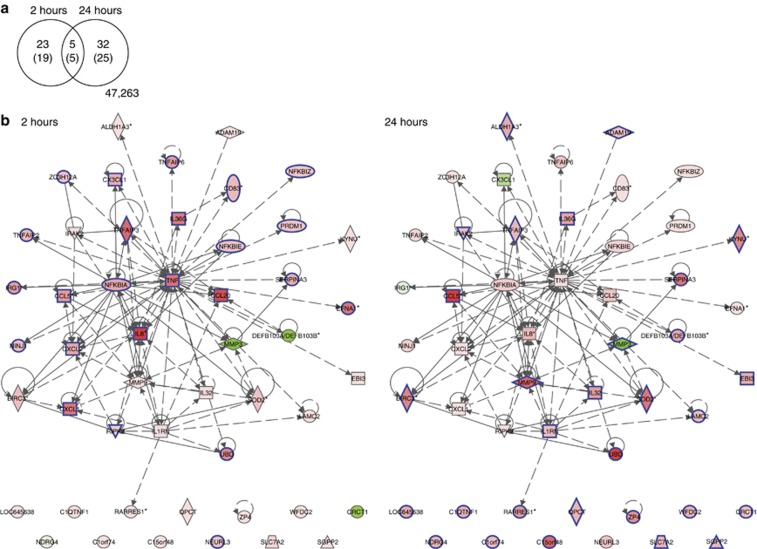
By using a false discovery rate ⩽0.05, the response to CD40 ligation in the four primary EC cultures was analyzed for genes that were at least twofold up- or downregulated (log2-fold change filter (LogFC) ⩾1) after 2 or 24 hours of stimulation. The response obtained in EC cultures with control cells was used to correct the results obtained with CD40-ligated ECs for both the time of coculture with L cells and total cell density. In total, 60 probes showed differential expression, representing 49 differentially expressed genes. Twenty-four genes were upregulated after 2 hours and 29 genes after 24 hours, whereas five genes were upregulated at both time points. One gene (MMP3) was significantly downregulated after 24 hours (Figure 2a; Supplementary Table S1 online).
Figure 2.
CD40 stimulation stimulates a highly coordinated immune response by epithelial cells (ECs). (a) Venn diagram depicting the overlap between 49 signature genes (60 microarray probes) differentially expressed at 2 and/or 24 hours; L-CD40L stimulation versus L-control stimulation with adjusted P-value ⩽0.05 and absolute log2-fold change ⩾1. Networks with expression changes at 2 (b) and 24 (c) hours were constructed of 49 connected CD40L signature genes using interaction data curated from literature and high-throughput screens by Ingenuity Pathway Analysis. The colors show the degree of upregulation (red) or downregulation (green) in the L-CD40L condition versus the L-control condition. The genes meeting the adjusted P-value ⩽0.05 and absolute log2-fold change ⩾1 thresholds, shown in the Venn diagram in a, are indicated by blue borders.
By Ingenuity Pathway Analysis (IPA), we explored whether these 49 differentially expressed genes were enriched for biological pathways and how they were connected. IPA enrichment analysis showed that the 24 genes differentially expressed after 2 hours of CD40 ligation were mainly involved in “cellular movement”, especially “leukocyte migration”, “cell-to-cell signaling and interaction”, and “cell death and survival”. The highest upregulated gene was IL8, followed by CCL20, TNFAIP3, TNF, CXCL1, EFNA1 (TNFAIP4), IL36G, and UBD, all having a LogFC⩾2. At 24 hours post stimulation the highest upregulated genes were CCL5 (RANTES), UBD, MMP9, C15orf48, SOD2, SerpinA3, and BIRC3 (cIAP2). The 30 genes differentially expressed at this time point are involved in “cellular movement”, “cell death and survival pathways”, “posttranslational modification”, and “protein degradation”.
According to the IPA knowledge database, 37 of these 49 differentially expressed genes formed a network (117 connections), including 23 out of the 24 genes differentially expressed after 2 hours and 19 out of the 30 genes differentially expressed after 24 hours (Figure 2b and c). The most interconnected genes within the center of the network were TNF and IL8, both upregulated only after 2 hours of CD40 ligation. These data indicated that CD40 stimulation of ECs results in a very coordinated reaction; first highly connected immune-involved genes that are able to recruit leukocytes or regulate cytokine expression are upregulated, and subsequently genes involved in the regulation of cell death and survival are upregulated.
CD40 ligation amplifies immune cell attraction to ECs
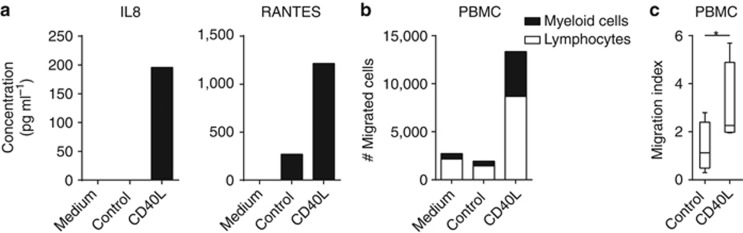
Many of the genes that were expressed by ECs after CD40 stimulation belonged to the “leukocyte migration” group, indicating that CD40–CD154 interactions between T cells and ECs may serve primarily to boost the attraction of immune cells. Therefore, as a second functional assay to study the impact of CD40 ligation, we assessed the capacity of ECs to induce immune cell migration after stimulation with CD40L or control cells. The culture supernatants were isolated and used in a trans-well system with peripheral blood mononuclear cells (PBMCs) seeded in the top wells. To confirm that CD40 ligation is associated with the production of chemokines belonging to the “leukocyte migration” group, the production of the representative cytokines IL8 and RANTES was measured. Their increased secretions are representative for the production of several chemoattractants following CD40 stimulation (Figure 3a). Indeed, higher numbers of PBMCs migrated toward the supernatants from CD40-ligated ECs when compared with supernatants of control ECs (Figure 3b and c). Analysis of the fraction of lymphocytes and myeloid cells in the migrated PBMCs suggested that the myeloid fraction in the total pool of migrated PBMCs was slightly more increased (Figure 3b). These data indicate that CD40 stimulation of ECs mainly results in the secretion of pro-inflammatory cytokines that aid ECs in the attraction of PBMCs.
Figure 3.
CD40 ligation induces immune cell migration toward epithelial cells (ECs). Example of a representative experiment of the (a) production of IL8 and RANTES in cleared supernatants of vaginal EC donors used for the migration assay by ELISA, and (b) peripheral blood mononuclear cell (PBMC) migration toward these cleared supernatants from vaginal EC donors prepared for the migration assay. PBMC numbers were determined by flow cytometry in the presence of FACS counting beads and subsequently gated on lymphocyte or myeloid cell fractions. Within the total cell numbers (total bar) the fractions of lymphocytes (white) and myeloid cells (black) are depicted. (c) Migration index of total PBMC toward indicated supernatants of ECs of foreskin or vaginal origin of four independent experiments. * Indicates P<0.05.
Persistent infection with hrHPV attenuates the intensity of the CD40-induced gene expression
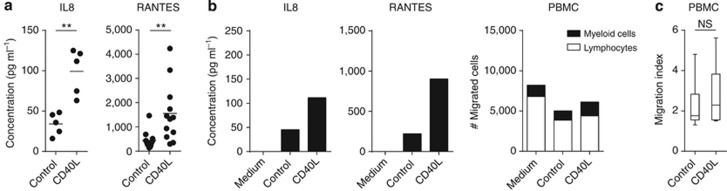
High-risk HPVs are known to deregulate the response of ECs to TNF (Termini et al., 2008). In view of the cellular mediators shared between the TNF and the CD40 pathway, we studied whether a persistent infection with hrHPV influences the gene expression pattern of CD40-stimulated ECs by genome-wide expression analysis. We confirmed the expression of CD40 after IFNγ stimulation at the cell surface of hrHPV-positive ECs as well as the expression of IL8 after 2 hours and RANTES after 24 hours of CD40 ligation (Supplementary Figure S1a–c online) and the secretion of these cytokines in the supernatant of hrHPV-infected ECs (Figure 5a). The gene expression profiles of four hrHPV-positive primary EC cultures, stably harboring HPV16 or HPV18 episomes, were compared with those of the four uninfected primary EC cultures. The expression of IL8 and RANTES of HPV-infected ECs after CD40 stimulation was verified by quantitative PCR (Supplementary Figure S1e online). The log2 intensity plots of these genes as measured by microarray (Supplementary Figure S1f online) showed that the results obtained by both methods were comparable.
Figure 5.
Human papilloma virus (HPV) infection hampers the enhanced attraction of immune cells by CD40-stimulated epithelial cells (ECs). (a) ELISA for IL8 and RANTES in cleared supernatants from IFNγ-pre-stimulated HPV-positive foreskin, vaginal, and cervical EC cultures (n=5–12) cocultured for 24 hours with control or CD40L-expressing L cells in the presence of IFNγ. ** Indicates P<0.005 using unpaired Welch-corrected t-test. (b) Example of a representative experiment of the production of IL8 and RANTES in cleared supernatants of vaginal EC donors used for the migration assay by ELISA, and peripheral blood mononuclear cell (PBMC) migration toward these cleared supernatants from vaginal EC donors prepared for the migration assay. (c) Migration index of total PBMC toward indicated HPV-positive foreskin, vaginal, and cervical EC supernatants of four independent experiments. NS indicates P is not significant.
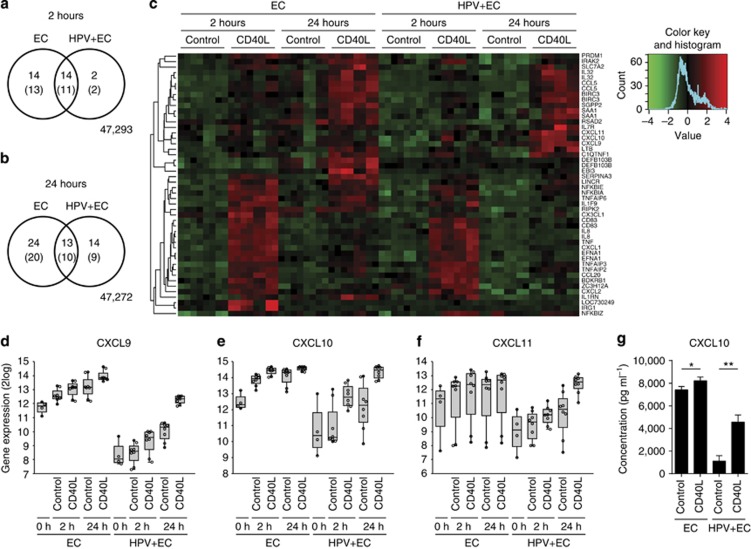
We studied differential gene expression in HPV-positive ECs after CD40 ligation. At 2 hours, HPV-positive ECs differentially expressed 13 genes, 11 of which overlapped with the 24 genes differentially expressed in uninfected ECs (Figure 4a). At 24 hours, HPV-positive ECs differentially expressed 19 genes, 10 of which overlapped with the 30 genes differentially expressed in uninfected ECs (Figure 4b). This was a first indication that HPV does not grossly alter the reaction to CD40. All differentially expressed genes, 65 in total, were analyzed by IPA and the resulting network (159 connections) was highly similar to the network of genes expressed by CD40-stimulated noninfected ECs (Supplementary Figure S3 online; Supplementary Table S1 online). There were no specific clusters of genes that were either up- or downregulated in HPV-positive ECs but not in uninfected ECs (Supplementary Figure S2 online); rather, the expression intensities of the differentially expressed genes were attenuated in HPV-positive ECs. Focusing on the immune-related genes (Figure 4c) revealed that the presence of hrHPV in ECs impaired the expression of 12 immune-related genes after 2 hours of CD40 stimulation, whereas one gene (BDKRB1) was enhanced. After 24 hours of stimulation, hrHPV impaired the expression of eight genes and upregulated seven immune-related genes in ECs. A closer look into the seven upregulated genes was carried out. Three genes, IL7R, LTB, and SAA1, showed similar upregulation in the uninfected ECs but did not reach our significance and fold change thresholds (Supplementary Figure S4 online). The remaining four genes, CXCL9, CXCL10, CXCL11, and RSAD2, were already strongly upregulated in uninfected ECs compared with HPV-positive ECs in response to the IFNγ pre-stimulation, and were not further increased by additional CD40 ligation (Supplementary Figure S4 online). In HPV-positive ECs, CD40 ligation resulted in the upregulation of these genes to levels similar to those in uninfected ECs (Supplementary Figure S4 online).
Figure 4.
HPV infection results in an attenuated response of epithelial cell (ECs) to CD40 ligation. Venn diagrams showing the overlapping genes between ECs and human papilloma virus (HPV)-positive ECs in their response to L-CD40L versus L-control stimulation for 2 (a) and 24 (b) hours; significance thresholds as in Figure 2a, numbers in brackets represent unique genes. (c) Heat map of differentially expressed immune-involved genes as determined by Ingenuity Pathway Analysis. Expression ratios for each condition compared with the 0 hour time point per cell line were mean-centered and scaled over all conditions. The genes were hierarchically clustered using cosine similarity and average linking. Microarray intensities for CXCL9 (d), 10 (e), and 11 (f) represented in a box plot. (g) CXCL10 concentration as measured by ELISA in supernatants of 24 hours IFNγ-stimulated and L-control or L-CD40L cocultured foreskin ECs and HPV-positive foreskin ECs (n=3). * Indicates P<0.05 and ** P<0.005.
hrHPV impairs CD40 ligation–mediated immune cell attraction to ECs
The T-cell–attracting chemokines CXCL9, 10, and 11 are known to be induced by IFNγ in various cell types, including ECs (Sauty et al., 1999; Kanda et al., 2007; Kanda and Watanabe, 2007; Ohta et al., 2008; Kawaguchi et al., 2009). Although CD40 stimulation salvaged the expression levels of CXCL9, CXCL10, and CXCL11 in HPV-positive ECs to similar levels found in noninfected ECs (Figure 4d–f), ELISA assays showed that hrHPV-positive ECs still secreted lower levels of CXCL9 and CXCL10 compared with noninfected ECs (Figure 4g and not shown). On average, the CD40-ligated HPV-positive ECs also produced lower amounts of IL8 and RANTES (Figure 5a), albeit that in some experiments the levels approached that of noninfected ECs. To obtain a broader view of the impact of HPV in CD40L-induced immune activation, their capacity to attract PBMCs was also tested. Notwithstanding the production of the earlier tested cytokines, no increased attraction of PBMCs to the supernatants of CD40L-stimulated HPV-positive ECs was observed (Figure 5b and c). This indicates that the production of other chemokines within the “leukocyte migration” group, those that are key in the attraction of PBMCs, must also have been impaired in HPV-positive ECs. In independent experiments, the absolute numbers of migrated PBMCs differed per primary EC culture and PBMC donor used; however, the increase in PBMC attraction following CD40 ligation was consistently and significantly higher in uninfected ECs (Figure 3c), but not in hrHPV+ ECs (Figure 5c). Together, these data show that hrHPV does not grossly alter, but rather attenuates, the intracellular response of ECs to CD40 ligation, resulting in a hampered ability of the HPV-positive ECs to attract immune cells.
Discussion
We studied the response of ECs to CD40 ligation, a major immune trigger of B- and T-cell immunity and a major cue for leukocyte migration toward the skin. Stimulation of ECs via CD40 resulted in a highly coordinated regulation of predominantly immune-related genes involved in the attraction, sustainment, and amplification of adaptive immune responses as well as resulted in the attraction of immune cells. Interestingly, hrHPV infection did not qualitatively alter the gene expression profile of CD40-stimulated ECs; instead, the extent of the response was attenuated. The fact that HPV attenuates CD40 signaling in ECs indicates the importance of the CD40–CD154 immune pathway in boosting immunity in epithelia.
Microarray expression studies showed that CD40 ligation of non-hematopoietic cells, such as endothelial cells (Pluvinet et al., 2008), pancreatic cells (Klein et al., 2008), renal proximal tubule ECs (Li and Nord, 2005), smooth muscle cells (Stojakovic et al., 2007), microglia (Ait-Ghezala et al., 2005), and ECs (this report), generally results in the upregulation of genes involved in immunity and inflammatory responses, cell fate, and cell adhesion. The response of ECs to CD40 stimulation is alike that of muscle cells and pancreatic cells. Endothelial cells seem to have a broader response as they also upregulate genes involved in the viral immune surveillance system, e.g., the 2′-5′-oligoadenylate/RNase L system and guanylate-binding proteins (GBP1–4), potentially to keep the vasculature from harmful consequences and prevent the spread of systemic viral infection in the host (Pluvinet et al., 2008). ECs are well equipped with viral sensors, which can launch an antiviral response upon infection (Karim et al., 2011), and the CD40 pathway may help to establish efficient adaptive B- and T-cell immunity to expand the precision of protection after the initial innate immune cell response.
Interestingly, we found that late CD40-mediated responses in ECs involved the upregulation of the anti-apoptosis genes cIAP2 and BCL3 as well as the negative regulator of proliferation RARRES1. These observations may explain earlier findings that ECs do not go into apoptosis but rather stop proliferating after CD40 ligation (Peguet-Navarro et al., 1997). We are currently exploring this further. The response of ECs to CD40 stimulation is paralleled by B cells, which respond to CD40 ligation by preventing apoptosis through the upregulation of several anti-apoptotic genes, including cIAPs, MYC, and BCL2 members (Kehry, 1996; Laman et al., 1996).
CD40 stimulation of DCs has been thoroughly studied as it has a key role in the activation, maturation, and T-cell priming capacity of DCs. Upon CD40 stimulation, DCs produce pro-inflammatory cytokines and chemokines, upregulate human leukocyte antigen class I and II as well as the co-stimulatory molecules CD86 and CD80 (Ma and Clark, 2009). This allows DCs to convey the appropriate signals to T cells required for them to become effector cells. Candidate gene studies showed that ECs can express CD40, human leukocyte antigen class I and II, CD86 but not CD80 (Black et al., 2007; Ortiz-Sanchez et al., 2007; Romero-Tlalolini et al., 2013), as well as the co-stimulatory molecules CD83 and ICAM-1 and a number of cytokines after being exposed to IFNγ and CD40 activation (Denfeld et al., 1996; Gaspari et al., 1996; Peguet-Navarro et al., 1997; Companjen et al., 2002; Pasch et al., 2004). This may allow CD40-stimulated ECs to process and present antigen to effector/memory CD4+ and CD8+ T cells (Black et al., 2007) as well as to amplify immune responses. However, it is not likely that such activated ECs function as professional antigen-processing cells as it was shown that CD40L-activated ECs fail to prime allogeneic T-cell reactions, underlining the difference of CD40 ligation on professional and nonprofessional antigen-processing cells (Grousson et al., 2000).
The pathogenesis of skin diseases such as psoriasis is based on an influx of immune cells into psoriatic lesions where cytokine levels are elevated. Our results sustain the notion that tissue-infiltrating T cells may exacerbate the disease via the production of IFNγ and the interaction with CD40 on ECs. The resulting cytokines may amplify the immune response via the attraction of more immune cells, thereby forming a loop in EC stimulation and cytokine production. The involvement of ECs in the exacerbation of disease has been questioned, as CD40 expression on ECs in vivo can be weak (Ohta and Hamada, 2004). However, we and others have shown that CD40 expression is rapidly upregulated (at least temporarily) under the influence of physiological doses of IFNγ, and thus weak steady-state expression does not preclude robust action under conditions of immune activation.
HPV attenuates the extent of the ECs' response to CD40 ligation, suggesting that HPV interferes with CD40 ligation–induced signal transduction and subsequent canonical and noncanonical NFκB activation (Ma and Clark, 2009; Gommerman and Summers deLuca, 2011; Hostager and Bishop, 2013). Several research groups have reported that hrHPV deregulates NFκB activation following the activation of pattern recognition receptors (PRRs) (Karim et al., 2011; Reiser et al., 2011) or the TNF receptor (Termini et al., 2008). We and others have previously shown that hrHPV attenuates the pattern recognition receptor-induced (Karim et al., 2013) and TNFR-induced (Takami et al., 2007) NFκB pathway activation by upregulating UCHL1, a cellular deubiquitinase/E3 ligase. Therefore, the expression of UCHL1, or other non-identified modulators, may explain how HPV mediates the attenuation of CD40 ligation–induced gene expression.
Surprisingly, PBMCs were more attracted to supernatants of non-CD40-ligated HPV-positive ECs than to uninfected ECs, implying that supernatants of HPV-positive ECs contain higher cytokine levels compared with supernatants of uninfected ECs. However, not only in this study but also in previous studies (Karim et al., 2011; Karim et al., 2013) we observed that hrHPV generally downregulates the basal expression and secretion of many pro-inflammatory cytokines. Recent literature has shown that metabolism intermediates can act as inflammatory signals (Tannahill et al., 2013), implying that a simple difference in cell density can affect basal immune cell attraction. Although both the HPV-positive and uninfected ECs have been treated exactly the same throughout the experiments, HPV-positive ECs proliferate faster than uninfected ECs, and as such the supernatants may contain higher metabolite levels to mediate CD40-independent PBMC attraction toward HPV-positive cells. In HPV-positive ECs, despite the higher basal numbers of attracted PBMCs, CD40 stimulation does not result in an increased number of PBMCs attracted, whereas in uninfected ECs this is the case.
In conclusion, epithelial cells show a coordinated response to CD40 ligation, mainly inducing the expression of genes involved in leukocyte migration, cell-to-cell signaling and interaction, as well as cell death and survival. HPV attenuates the extent of CD40 signaling, resulting in lower amounts of chemoattractants produced and a failure to enhance immune cell migration. These data suggest that progression of inflammatory skin diseases may be driven by highly programmed immune activation scenarios in ECs, which have their evolutionary basis in the ECs' response to infections.
Materials and Methods
Ethics statement
The use of discarded human foreskin, cervical, and vaginal keratinocyte tissues to develop cell lines for these studies was approved by the Institutional Review Board at the Pennsylvania State University College of Medicine and by the Institutional Review Board at Pinnacle Health Hospitals. The Medical Ethical Committee of the Leiden University Medical Center approved the human tissue sections (healthy foreskin, healthy cervix, HPV16- or 18-positive cervical neoplasias) used for staining. All sections and cell lines were derived from discarded tissues and de-identified; therefore, no informed consent was necessary.
Cell culture
Primary cultures of human ECs were established from foreskin, vaginal, and cervical tissues as previously described (Karim et al., 2011) and grown in keratinocyte serum-free medium (K-SFM; Medium 154 supplemented with HKGS kit, Invitrogen, Breda, The Netherlands). The cells morphologically and biochemically resembled ECs in both monolayer and organotypic raft cultures, as indicated by keratin expression, hemidesmosome, and desmosome structures, and in their ability to differentiate into full-thickness epithelium (Meyers et al., 1997; McLaughlin-Drubin et al., 2004). By using the microarray data, the cells were verified to express high levels of keratin (KRT) 10, 14, 17, and 19, and low levels of KRT18 (Supplementary Figure S5 online), a signature specific for keratinocytes (Moll et al., 2008; Bononi et al., 2012). EC lines stably maintaining the full episomal HPV genome following electroporation (HPV-positive ECs) were grown in a monolayer culture using E medium in the presence of mitomycin C–treated J2 3T3 feeder cells (Meyers et al., 1997; McLaughlin-Drubin et al., 2004) for two passages and were then adapted to K-SFM for one passage before experimentation. Because primary ECs have a limited life span and do not survive long enough to undergo a mock electroporation procedure similar to that used to obtain HPV-positive ECs, normal undifferentiated primary ECs were used as control. J2 3T3 mouse fibroblasts and L cells were cultured in Dulbecco's modified Eagle's medium supplemented with 8% fetal bovine serum, 2 mM l-glutamine, and 1% penicillin–streptomycin (complete Dulbecco's modified Eagle's medium) (Gibco-BRL, Invitrogen).
CD40 ligation on ECs
Uninfected ECs or HPV-positive ECs were seeded at 1.5 × 105 cells per well in six-well plates in K-SFM and allowed to attach for 24 hours, after which the cells received fresh K-SFM containing 50 IU ml−1 IFNγ (Immunotools, Friesoythe, Germany) for 72 hours. Control or CD40L-expressing L cells were harvested, irradiated (4,800–5,200 rad), and resuspended in K-SFM containing 50 IU ml−1 IFNγ. L cells were cocultured with ECs in a 1:1 ratio for indicated time points, after which the supernatant was collected, the L cells were removed, and the RNA of the ECs was harvested. CD40L expression and functionality of the L cells were validated, as was the percentage of residual L cells after coculture (<1% data not shown).
RNA expression analyses and ELISA
Total RNA was isolated using the NucleoSpin RNA II kit (Machery-Nagel, Leiden, The Netherlands) according to the manufacturer's instructions. Total RNA (0.5–1.0 μg) was reverse transcribed using the SuperScript III First Strand synthesis system from Invitrogen. TaqMan PCR was performed using TaqMan Universal PCR Master Mix and pre-designed, pre-optimized primers and probe mix for RANTES (CCL5), IL8, and GAPDH (Applied Biosystems, Foster City). Threshold cycle numbers (Ct) were determined using the CFX PCR System (Bio-Rad, Veenendaal, The Netherlands), and the relative quantities of complementary DNA per sample were calculated using the ΔΔCt method using GAPDH as the calibrator gene. ELISA's for CCL2, RANTES, IL8, and CXCL10 were performed according to the manufacturer's instructions (PeproTech, London, UK). Statistical differences in cytokine production were evaluated using a Welch-corrected t-test, correcting for possible unequal variances between the groups.
Gene expression profiling
Four primary EC cultures were used, HVK (vaginal), HCK (cervical), HFK_1, and HFK_2 (both foreskin), as well as four EC cell lines stably maintaining episomal HPV16 or 18, HVK16 (vaginal), HVK18 (vaginal), HCK18 (cervical), and HPV16 (foreskin). Cells were harvested at five conditions: 0, 2 and, 24 hours of 50 IU ml−1 IFNγ in combination with either L-control or L-CD40L cells. Stimulated 2- and 24-hour samples were generated in duplo. Total RNA for these 72 samples was isolated as stated above. The microarray experiment was performed by ServiceXS according to their protocols (ServiceXS, Leiden, The Netherlands). Briefly, total RNA was analyzed by Lab-on-a-Chip. All RNA showed a RNA integrity number score of >9.5. Total RNA was reverse-transcribed, amplified, and biotin labeled. cRNA was hybridized to Illumina (San Diego, CA) Human HT-12 v4 BeadChips in a randomized manner and scanned with the Illumina iScan. Samples passed quality control as assessed by Illumina GenomeStudio software. Values for missing bead types on the HumanHT-12 BeadChip were estimated using the k-Nearest Neighbor (k-NN) algorithm (Troyanskaya et al., 2001) in Illumina's BeadStudio Gene Expression Module (v3.3+).
Microarray data preprocessing
The expression array data were analyzed using R2.14.1 and Bioconductor (R Development Core Team, 2008). The data were normalized using the Bioconductor package lumi version 2.6.0 (Du et al., 2008; Lin et al., 2008), resulting in log2-transformed normalized intensities. Quality control plots were generated using limma version 3.10.2 (Smyth, 2005) and mpm version 1.0–22 (Wouters et al., 2003; Wouters, 2011). Uninfected and HPV-positive ECs correlated in separate blocks, and within these blocks the next level similarity was at the cell line level, and within cell line at the exposure level, indicating that the data behaved as expected (data not shown). All microarray data are MIAME compliant and the raw data have been deposited in the MIAME compliant database Gene Expression Omnibus with accession number GSE54181, as detailed on the MGED Society website http://www.mged.org/Workgroups/MIAME/miame.html.
Analysis of differentially gene expression
Differentially expressed genes were identified using maanova version 1.24.0 (Wu; Wu et al., 2003). We modeled the cell line effect as a random effect and indicated the technical replicates in the model. We calculated test statistics for testing the null hypotheses of no difference in expression between L-CD40L-stimulated and L-control-stimulated cells at 2 and 24 hours for uninfected ECs as well as HPV-positive ECs for each gene. We applied the Fs statistic, which uses a shrinkage estimator for gene-specific variance components based on the James–Stein estimator. To correct for multiple testing, false discovery rates were calculated using the q-value method (Dabney; Storey, 2002). The ranking and selection of the genes are based on these adjusted P-values.
Functional genomics analyses
The networks were constructed using Ingenuity Pathway Analysis (IPA version 17199142; Ingenuity systems, www.ingenuity.com). The list of differentially expressed genes was used to generate the network. All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathway Knowledge Base.
Box plot representations
Boxplots are drawn as a box, containing the 1st quartile up to the 3rd quartile of the data values. The median is represented as a line within the box. Whiskers represent the values of the outer two quartiles. These whiskers are, however, maximized at 1.5 times the size of the box (also known as inter-quartile distance). If one or more values outside of the whiskers are present, then this is indicated with a single mark “o” next to the implicated whisker. Plots were generated using the webtool R2: microarray analysis and visualization platform (http://r2.amc.nl).
Migration assays
IFNγ pre-stimulated (HPV-positive) ECs were cocultured with L cells for 3 hours, after which the L cells were removed. The ECs were cultured for a subsequent 24 hours with fresh K-SFM. Cleared (HPV-positive) EC supernatants were added to the lower compartment of a trans-well plate (Corning). The upper compartment was filled with PBMCs, which were allowed to migrate for 16 hours, after which the cells in the lower compartment were counted by flow cytometry in the presence of counting beads (Invitrogen) according to the manufacturer's protocol. Myeloid cells and lymphocytes were differentiated by their respective size in the forward scatter/side scatter plot (data not shown). To normalize for biological differences between PBMC donors and EC cultures, a migration index was calculated of the total number of PBMCs migrated toward the indicated stimulation over the medium control. The statistical significance of differences in migration toward supernatants of EC cultures stimulated with CD40L or control L cells was assessed using a paired t-test.
Flow cytometry
Expression of CD40 on ECs was analyzed by flow cytometry using FITC-coupled mouse anti-human CD40 antibodies (BD Biosciences, Breda, The Netherlands). A total of 50,000 cells per live gate were recorded using the BD FACS Calibur with Cellquest software (BD Bioscience) and data were analyzed using Flowjo (Treestar, Olten, Switzerland).
Acknowledgments
This work was supported by the Netherlands Organization for Health Research (NOW/ZonMw) TOP grant 91209012. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
- CD40
cluster of differentiation 40
- CD40L
cluster of differentiation 40 ligand
- DC
dendritic cell
- EC
epithelial cell
- hrHPV
high-risk human papilloma virus
- IPA
Ingenuity Pathway Analysis
- KC
keratinocyte
- K-SFM
keratinocyte serum-free medium
- PBMC
peripheral blood mononuclear cell
CM has received speaker honoraria from Merck, Quest Diagnostics, GSK, and Bristol-Myers. CM has performed research funded by Merck, The Phillip Morris External Research Program, NexMed, GSK, OriGenix, and Interferon Sciences Inc. The remaining authors state no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
References
- Ait-Ghezala G, Mathura VS, Laporte V, et al. Genomic regulation after CD40 stimulation in microglia: relevance to Alzheimer's disease. Brain Res Mol Brain Res. 2005;140:73–85. doi: 10.1016/j.molbrainres.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Andrei G, Duraffour S, Van den Oord J, et al. Epithelial raft cultures for investigations of virus growth, pathogenesis and efficacy of antiviral agents. Antiviral Res. 2010;85:431–449. doi: 10.1016/j.antiviral.2009.10.019. [DOI] [PubMed] [Google Scholar]
- Black AP, Ardern-Jones MR, Kasprowicz V, et al. Human keratinocyte induction of rapid effector function in antigen-specific memory CD4+ and CD8+ T cells. Eur J Immunol. 2007;37:1485–1493. doi: 10.1002/eji.200636915. [DOI] [PubMed] [Google Scholar]
- Bononi I, Bosi S, Bonaccorsi G, et al. Establishment of keratinocyte colonies from small-sized cervical intraepithelial neoplasia specimens. J Cell Physiol. 2012;227:3787–3795. doi: 10.1002/jcp.24088. [DOI] [PubMed] [Google Scholar]
- Caproni M, Torchia D, Antiga E, et al. The CD40/CD40 ligand system in the skin of patients with subacute cutaneous lupus erythematosus. J Rheumatol. 2007;34:2412–2416. [PubMed] [Google Scholar]
- Companjen AR, van der Wel LI, Boon L, et al. CD40 ligation-induced cytokine production in human skin explants is partly mediated via IL-1. Int Immunol. 2002;14:669–676. doi: 10.1093/intimm/dxf033. [DOI] [PubMed] [Google Scholar]
- Concha M, Vidal MA, Moreno I, et al. Evidence for modulation of human epidermal differentiation and remodelling by CD40. Br J Dermatol. 2003;148:1105–1114. doi: 10.1046/j.1365-2133.2003.05300.x. [DOI] [PubMed] [Google Scholar]
- Dabney A.qvalue: Q-value estimation for false discovery rate control. . http://bioc.ism.ac.jp/2.9/bioc/html/qvalue.html ( R package version 1.28.0).
- Denfeld RW, Hollenbaugh D, Fehrenbach A, et al. CD40 is functionally expressed on human keratinocytes. Eur J Immunol. 1996;26:2329–2334. doi: 10.1002/eji.1830261009. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Fries KM, Sempowski GD, Gaspari AA, et al. CD40 expression by human fibroblasts. Clin Immunol Immunopathol. 1995;77:42–51. doi: 10.1016/0090-1229(95)90135-3. [DOI] [PubMed] [Google Scholar]
- Fuller BW, Nishimura T, Noelle RJ. The selective triggering of CD40 on keratinocytes in vivo enhances cell-mediated immunity. Eur J Immunol. 2002;32:895–902. doi: 10.1002/1521-4141(200203)32:3<895::AID-IMMU895>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Galy AH, Spits H. CD40 is functionally expressed on human thymic epithelial cells. J Immunol. 1992;149:775–782. [PubMed] [Google Scholar]
- Gaspari AA, Sempowski GD, Chess P, et al. Human epidermal keratinocytes are induced to secrete interleukin-6 and co-stimulate T lymphocyte proliferation by a CD40-dependent mechanism. Eur J Immunol. 1996;26:1371–1377. doi: 10.1002/eji.1830260629. [DOI] [PubMed] [Google Scholar]
- Gommerman JL, Summers deLuca L. LTbetaR and CD40: working together in dendritic cells to optimize immune responses. Immunol Rev. 2011;244:85–98. doi: 10.1111/j.1600-065X.2011.01056.x. [DOI] [PubMed] [Google Scholar]
- Grousson J, Ffrench M, Concha M, et al. CD40 ligation alters the cell cycle of differentiating keratinocytes. J Invest Dermatol. 2000;114:581–586. doi: 10.1046/j.1523-1747.2000.00905.x. [DOI] [PubMed] [Google Scholar]
- Hollenbaugh D, Mischel-Petty N, Edwards CP, et al. Expression of functional CD40 by vascular endothelial cells. J Exp Med. 1995;182:33–40. doi: 10.1084/jem.182.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostager BS, Bishop GA. CD40-Mediated Activation of the NF-kappaB2 Pathway. Front Immunol. 2013;4:376. doi: 10.3389/fimmu.2013.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda N, Shimizu T, Tada Y, et al. IL-18 enhances IFN-gamma-induced production of CXCL9, CXCL10, and CXCL11 in human keratinocytes. Eur J Immunol. 2007;37:338–350. doi: 10.1002/eji.200636420. [DOI] [PubMed] [Google Scholar]
- Kanda N, Watanabe S. Prolactin enhances interferon-gamma-induced production of CXC ligand 9 (CXCL9), CXCL10, and CXCL11 in human keratinocytes. Endocrinology. 2007;148:2317–2325. doi: 10.1210/en.2006-1639. [DOI] [PubMed] [Google Scholar]
- Karim R, Meyers C, Backendorf C, et al. Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PloS One. 2011;6:e17848. doi: 10.1371/journal.pone.0017848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim R, Tummers B, Meyers C, et al. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response. PLoS Pathog. 2013;9:e1003384. doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S, Ishiguro Y, Imaizumi T, et al. Retinoic acid-inducible gene-I is constitutively expressed and involved in IFN-gamma-stimulated CXCL9-11 production in intestinal epithelial cells. Immunol Lett. 2009;123:9–13. doi: 10.1016/j.imlet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Kehry MR. CD40-mediated signaling in B cells. Balancing cell survival, growth, and death. J Immunol. 1996;156:2345–2348. [PubMed] [Google Scholar]
- Klein D, Timoneri F, Ichii H, et al. CD40 activation in human pancreatic islets and ductal cells. Diabetologia. 2008;51:1853–1861. doi: 10.1007/s00125-008-1092-y. [DOI] [PubMed] [Google Scholar]
- Laman JD, Claassen E, Noelle RJ. Functions of CD40 and its ligand, gp39 (CD40L) Crit Rev Immunol. 1996;16:59–108. doi: 10.1615/critrevimmunol.v16.i1.40. [DOI] [PubMed] [Google Scholar]
- Li H, Nord EP. CD40/CD154 ligation induces mononuclear cell adhesion to human renal proximal tubule cells via increased ICAM-1 expression. Am J Physiol Renal Physiol. 2005;289:F145–F153. doi: 10.1152/ajprenal.00317.2004. [DOI] [PubMed] [Google Scholar]
- Lin SM, Du P, Huber W, et al. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DY, Clark EA. The role of CD40 and CD154/CD40L in dendritic cells. Semin Immunol. 2009;21:265–272. doi: 10.1016/j.smim.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Christensen ND, Meyers C. Propagation, infection, and neutralization of authentic HPV16 virus. Virology. 2004;322:213–219. doi: 10.1016/j.virol.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Meyers C, Mayer TJ, Ozbun MA. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K, Shigeishi H, Taki M, et al. Regulation of CXCL9/10/11 in oral keratinocytes and fibroblasts. J Dent Res. 2008;87:1160–1165. doi: 10.1177/154405910808701211. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Hamada Y. In situ Expression of CD40 and CD40 ligand in psoriasis. Dermatology. 2004;209:21–28. doi: 10.1159/000078582. [DOI] [PubMed] [Google Scholar]
- Ortiz-Sanchez E, Chavez-Olmos P, Pina-Sanchez P, et al. Expression of the costimulatory molecule CD86, but not CD80, in keratinocytes of normal cervical epithelium and human papillomavirus-16 positive low squamous intraepithelial lesions. Int J Gynecol Cancer. 2007;17:571–580. doi: 10.1111/j.1525-1438.2007.00904.x. [DOI] [PubMed] [Google Scholar]
- Pasch MC, Timar KK, van Meurs M, et al. In situ demonstration of CD40- and CD154-positive cells in psoriatic lesions and keratinocyte production of chemokines by CD40 ligation in vitro. J Pathol. 2004;203:839–848. doi: 10.1002/path.1581. [DOI] [PubMed] [Google Scholar]
- Peguet-Navarro J, Dalbiez-Gauthier C, Moulon C, et al. CD40 ligation of human keratinocytes inhibits their proliferation and induces their differentiation. J Immunol. 1997;158:144–152. [PubMed] [Google Scholar]
- Peters AL, Stunz LL, Bishop GA. CD40 and autoimmunity: the dark side of a great activator. Semin Immunol. 2009;21:293–300. doi: 10.1016/j.smim.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluvinet R, Olivar R, Krupinski J, et al. CD40: an upstream master switch for endothelial cell activation uncovered by RNAi-coupled transcriptional profiling. Blood. 2008;112:3624–3637. doi: 10.1182/blood-2008-03-143305. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2008. R: a language and enviroment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0 http://www.R-project.org
- Reiser J, Hurst J, Voges M, et al. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J Virol. 2011;85:11372–11380. doi: 10.1128/JVI.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Tlalolini MA, Chavez Olmos P, Garrido E. Differential DNA methylation patterns in the CD86 gene controls its constitutive expression in keratinocytes. Biochem Biophys Res Commun. 2013;438:54–60. doi: 10.1016/j.bbrc.2013.07.023. [DOI] [PubMed] [Google Scholar]
- Sauty A, Dziejman M, Taha RA, et al. The T cell-specific CXC chemokines IP-10, Mig, and I-TAC are expressed by activated human bronchial epithelial cells. J Immunol. 1999;162:3549–3558. [PubMed] [Google Scholar]
- Smyth GK.2005Limma: linear models for microarray dataIn: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds)Bioinformatics and Computational Biology Solutions using R and Bioconductor Springer: New York, NY, USA [Google Scholar]
- Stojakovic M, Krzesz R, Wagner AH, et al. CD154-stimulated GM-CSF release by vascular smooth muscle cells elicits monocyte activation—role in atherogenesis. J Mol Med. 2007;85:1229–1238. doi: 10.1007/s00109-007-0225-y. [DOI] [PubMed] [Google Scholar]
- Storey J. A direct approach to false discovery rates. J R Stat Soc Ser B. 2002. pp. 479–498.
- Swamy M, Jamora C, Havran W, et al. Epithelial decision makers: in search of the “epimmunome”. Nat Immunol. 2010;11:656–665. doi: 10.1038/ni.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami Y, Nakagami H, Morishita R, et al. Ubiquitin carboxyl-terminal hydrolase L1, a novel deubiquitinating enzyme in the vasculature, attenuates NF-kappaB activation. Arterioscler Thromb Vasc Biol. 2007;27:2184–2190. doi: 10.1161/ATVBAHA.107.142505. [DOI] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termini L, Boccardo E, Esteves GH, et al. Characterization of global transcription profile of normal and HPV-immortalized keratinocytes and their response to TNF treatment. BMC Med Genomics. 2008;1:29. doi: 10.1186/1755-8794-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- van den Bogaard EH, Tjabringa GS, Joosten I, et al. Crosstalk between keratinocytes and T Cells in a 3D microenvironment: a model to study inflammatory skin diseases. J Invest Dermatol. 2013;134 (3:719–727. doi: 10.1038/jid.2013.417. [DOI] [PubMed] [Google Scholar]
- van der Burg SH, Melief CJ. Therapeutic vaccination against human papilloma virus induced malignancies. Curr Opin Immunol. 2011;23:252–257. doi: 10.1016/j.coi.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Villarroel Dorrego M, Whawell SA, Speight PM, et al. Transfection and ligation of CD40 in human oral keratinocytes affect proliferation, adhesion and migration but not apoptosis in vitro. Clin Exp Dermatol. 2006;31:266–271. doi: 10.1111/j.1365-2230.2005.02018.x. [DOI] [PubMed] [Google Scholar]
- Wouters L.2011. Exploratory graphical analysis of multivariate data, specifically gene expression data with different projection methods: principal component analysis, correspondence analysis, spectral map analysis. http://CRAN.R-project.org/package=mpm
- Wouters L, Gohlmann HW, Bijnens L, et al. Graphical exploration of gene expression data: a comparative study of three multivariate methods. Biometrics. 2003;59:1131–1139. doi: 10.1111/j.0006-341x.2003.00130.x. [DOI] [PubMed] [Google Scholar]
- Wu H.maanova: Tools for analyzing Micro Array experiments. http://research.jax.org/faculty/churchill
- Wu H, Kerr MK, Cui XQ, et al. 2003MAANOVA: a software package for the analysis of spotted cDNA microarray experimentsIn: Parmigiani G, Garret ES, Irizarry RA, Zeger SL (eds)The Analysis of Gene Expression Data: An Overview of Methods and Software Springer: New York, NY, USA; 313–341. [Google Scholar]
- Yellin MJ, Winikoff S, Fortune SM, et al. Ligation of CD40 on fibroblasts induces CD54 (ICAM-1) and CD106 (VCAM-1) up-regulation and IL-6 production and proliferation. J Leukocyte Biol. 1995;58:209–216. doi: 10.1002/jlb.58.2.209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.