Abstract

Colon cancer is a major cause of cancer-related deaths worldwide. Adjuvant chemotherapy significantly reduces mortality in stage III colon cancer; however, it is only marginally effective in stage II patients. There is also increasing evidence that right-side colon cancer is different from left-side colon cancer. We have observed that the genes altered in expression between the poor and good prognosis tumors vary significantly depending on whether the malignancy originates on the right or left side of the colon. We have identified NADPH oxidase 4 (NOX4) to be highly predictive of relapse in stage II left-side colon cancer, whereas integrin alpha 3 beta 1 (ITGA3) is predictive of relapse in stage II right-side colon cancer. To investigate the underlying molecular mechanisms, we are analyzing the effect of ITGA3 and NOX4 silencing via RNA interference and pharmacological inhibition on global protein expression patterns via iTRAQ labeling and mass spectrometry in colon cancer cells. On the basis of bioinformatic analysis, the functions of these genes were assessed through phenotypic assays, revealing roles in cell migration and reactive oxygen species generation. These biomarkers for relapse risk are of clinical interest and lead to insight into how a tumor progresses to metastasis.
Keywords: ITGA3, NOX4, iTRAQ, Q-Exactive, colon cancer
Introduction
There is accumulating evidence that cancer of the ascending and transverse (right side) colon is different from cancer of the descending and sigmoid (left side) colon. These two segments of the colon have differences in anatomy, embryology, physiology, epidemiology, and gene expression.1−3 Right- and left-side colon cancers also follow different molecular pathways to carcinogenesis and relapse.2,4 This growing evidence indicates that consideration of the location of the primary tumor could have significant implications for the evaluation of data. The identification of distinct molecular subtypes of colon cancer has potentially widespread implications for improved diagnosis, prognosis, treatment selection, and therapeutic evaluation.
Using microarray data (GSE14333) from 102 right-side and 95 left-side colon carcinomas, we have shown that different single gene prognostic biomarkers are found separately for right-sided and left-sided colon cancer.4 The expression levels of these genes in the primary tumors separate patients into a poor prognosis group that is likely to experience tumor recurrence or metastasis and a good prognosis group, with higher survival probability. Higher integrin alpha 3 (ITGA3) expression levels were found to be strongly associated with relapse in right-side colon cancer, whereas elevated NADPH oxidase 4 (NOX4) expression was identified as a significant predictor of relapse in left-side colon cancer. Both ITGA3 and NOX4 are much less prognostic for tumors arising in the opposite side of the colon. These biomarkers for relapse risk are of clinical interest and may lead to insight into how a tumor progresses to metastasis.
ITGA3 belongs to the integrin family of cell surface receptors. Integrin-ligand binding on the extracellular matrix activates diverse signaling pathways that have been implicated in migration, proliferation, cell survival, and gene expression.5−7 Although we are the first to associate elevated ITGA3 expression with metastatic potential in colon cancer, ITGA3 expression has previously been correlated with increased invasiveness in gastric carcinomas8 and has been identified as a biomarker for cervical lymph node metastasis for tongue squamous cell carcinoma.9 However, other integrin family members have been associated with colon cancer clinical outcome or disease progression. Integrin alpha 5 beta 6 and alpha 5 beta 3 expression levels are elevated in liver metastatic tissue compared to that in primary colon cancer tissue.10,11 Increased integrin alpha 5 beta 3 expression is also correlated with reduced relapse-free intervals and overall survival.11 The protein product, ITGB3, has been shown to be upregulated with reactive oxygen species (ROS) production, and the silencing of ITGB3 reduces migratory and invasive potential.12
NOX4 is the catalytic, transmembrane subunit in the multiprotein complex, NADPH oxidase. The NADPH oxidase enzyme family’s primary function is the production of ROS, which act as secondary messengers within cells.13 ROS levels have been implicated in inflammation and carcinogenesis due to their involvement in many critical processes within cells including angiogenesis, proliferation, and DNA damage responses.14,15 NOX4-mediated ROS production has been shown to increase epithelial motility in breast cancer cells.16−18 ROS generated by NOX4 also contributes to urothelial carcinogenesis and melanoma tumorigenesis through regulation of cell cycle progression.19,20 NOX4 is the most widely distributed isoform with constitutive activity; therefore, alterations in its expression may have major consequences, including the phenotypic effects previously mentioned.13
In this study, the roles of ITGA3 and NOX4 in colon cancer are investigated through gene silencing via RNA interference (RNAi) and isobaric tag for relative and absolute quantification (iTRAQ) protein profiling. The proteomic data was analyzed by pathway analysis, and the functional implications of ITGA3 and NOX4 were further explored by phenotypic assays and mRNA expression levels. These results indicate that aberrant expression of ITGA3 and NOX4 contribute to cell migration and ROS production, possibly through the action of microtubule regulating proteins and mitochondrial associated proteins, contributing to cellular capabilities that enable more aggressive primary and metastatic lesions. The results of this study have implications for understanding tumor progression in colon cancer, as determining the molecular species associated with relapse-associated ITGA3 and NOX4 will contribute to our knowledge of colon cancer recurrence and patient relapse.
Experimental Details
Reagents
Cell culture reagents, transfection reagents Oligofectamine and Lipofectamine RNAiMAX, and phosphate buffered saline (PBS) were obtained from Invitrogen (Gaithersburg, MD). All siRNAs were obtained from Qiagen Inc. (Germantown, MD). GoTaq RT-PCR detection reagent was acquired from Promega (Madison, WI). Protease inhibitor cocktail was purchased from Roche Diagnostics (Indianapolis, IN). Mass spectrometry solvents including, acetonitrile (ACN), and water with 0.1% formic acid (FA), were purchased from Burdick and Jackson (Muskegon, MI). iTRAQ reagent 8 plex multiplex kit and buffer kit were procured from Sciex (Framingham, MA). All reagents not specified were acquired from Sigma-Aldrich (St. Louis, MO).
Cell Culture and siRNA Transfections
Colorectal cancer cell lines (SW480, SW620, DLD-1, SW837, HCT 116, and HT29) were purchased from the American Type Culture Collection (ATCC, Manassas, VA). These cell lines were used within 3 months of resuscitation of frozen aliquots thawed from liquid nitrogen. The provider assured the authentication of these cell lines by cytogenetic analysis. The cell lines SW1116, LS513, and SW1463, provided by Dr. Marion Grade, were confirmed to be absent of mycoplasma contamination using the MycoAlert mycoplasma dectection kit (Lonza, Cologne, Germany), and cell line cross-contamination was excluded using short tandem repeat profiling.21 Cell lines were maintained in RPMI 1640 for SW480, SW620, DLD1, SW1116, LS513, SW837, and SW1463 or McCoy’s 5A for HCT116 and HT29, all supplemented with 10% fetal bovine serum (FBS) and 2 mmol/L l-glutamine and grown in 5% CO2 at 37 °C. SW620 cells were transfected (5000 cells/0.35 μL Oligofectamine) with one of two siRNA oligonucleotides targeting the human NOX4 transcript (40 nmol/L siNOX4.1 or siNOX4.2). HCT116 cells were transfected (5000 cells/0.25 μL Lipofectamine RNAiMAX) with one of two siRNA oligonucleotides targeting the human ITGA3 transcript (20 nmol/L siITGA3.1 or siITGA3.7). AllStar negative control siRNA (20 nmol/L) was used as a negative control, and a siRNA targeting Polo-like kinase 1 (PLK1) (20 nmol/L siPLK1.7) was used as a positive control. All transfections were conducted in triplicate. Transfections were evaluated 48 h post-transfection by quantitative RT-PCR. Total RNA was extracted using the RNeasy kit (Qiagen, Germantown, MD) following the manufacturer’s instructions. cDNA was generated using the high-capacity reverse transcriptase cDNA kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Normal human colon RNA isolated post-mortem from a donor was used for comparison with RNA from CRC cell lines (Applied Biosystems). Quantitative RT-PCR was performed in triplicate with a real-time PCR system, StepOnePlus (Applied Biosystems, Foster City, CA), using the following conditions: 95 °C for 2 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s. The primer sequences used are included in Supporting Information Table S1. Relative expression was quantified using the comparative cycle threshold (CT) (2–ΔΔCT) method using YWAHZ as the endogenous control. Transfection efficiency was also evaluated on the basis of PLK1 expression in the positive control sample (Supporting Information Figure S1A).
VAS2870 NOX4 Inhibition and Measurement of Superoxide Production
SW620 cells were treated with a range of VAS2870 concentrations (0–100 μmol/L) and assayed for superoxide production 24 h post-treatment. After media removal, cells were loaded with 20 μmol/L 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA) for 1 h at 37 °C. Cells were then rinsed with PBS and lysed. End point readings (485EX/528EM) were acquired with a Spectramax M5 96-well plate reader (Molecular Devices, Sunnyvale, CA). Response values were fitted with the five parameter logistic model to determine the IC50. The VAS2870 IC50 for SW620 cells was determined to be 18 μmol/L based on inhibitor values of superoxide production compared to that of untreated cells. SW620 cells were treated with the NOX inhibitor VAS2870 (18 μmol/L) in triplicate for 72 h prior to proteomic evaluation.
Preparation of Mass Spectrometric Samples
Proteins were extracted 72 h post-transfection on ice and sonicated twice for 60 s in the presence of 500 mM TEAB, 0.01% SDS, 1 mmol/L NaF, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, and a protease inhibitor cocktail. The total protein concentration was determined using a BCA protein assay kit (Thermo Scientific, Waltham, MA) and bovine serum albumin standards (Thermo Scientific). Fifty micrograms of each sample was reduced with 5 mmol/L Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) at 60 °C for 1 h and alkylated with 10 mmol/L iodoacetic acid (IAA) at RT for 10 min in the dark. The proteins were digested with TPCK treated trypsin (protein/trypsin, 20:1) overnight at 37 °C and then reduced to 20 μL. iTRAQ labels were thawed and resuspended in 60 μL of isopropanol. Peptides were labeled at RT for 2 h. To avoid label biases, labels were varied between samples in different experiments. The labeled peptides were quenched with 100 μL of HPLC-grade water, dried, and pooled. Excess labeling reagents were removed from the pooled peptide samples with a SCX SpinTip sample preparation kit (Protea Biosciences, Morgantown, WV) following the manufacturer’s instructions. High-pH RP fractionation was performed using 50 mg C18 SepPak cartridges (Waters, Milford, MA) and 10 mmol/L ammonium bicarbonate buffers at pH 10. The C18 SepPak cartridge was prepared by washing with 3 mL of 80% ACN and then equilibrated with 3 mL of 1% ACN. Samples were resuspended in 100 μL of 0.1% formic acid (FA), loaded onto the cartridge, and then washed with 3 mL of 1% ACN. The peptide samples were fractionated using elution buffers with concentrations of 5, 10, 15, 20, 23, 27, 30, and 80% ACN.
Mass Spectrometry Analysis
Liquid chromatography electrospray ionization tandem mass spectrometry (LC–ESI–MS/MS) was performed on a nanoAcquity ultra performance LC system (100 μm × 100 mm C18 BEH column) (Waters, Milford, MA) coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptides were eluted using a binary solvent system with 0.1% formic acid (A) and 0.1% formic acid in acetonitrile (B). The following linear gradient was used for ITGA3 samples: 95–70% A in 60 min, 70–50% A in 15 min, washed at 15% A for 5 min, and equilibrated with 95% A for 10 min at a 1000 nL/min flow rate. The following linear gradient was used for NOX4 samples: 95–70% A in 140 min, 70–50% A in 20 min, washed at 15% A for 10 min, and equilibrated with 95% A for 10 min at a 1000 nL/min flow rate. The mass spectrometer was operated in a Top 12 data-dependent mode with automatic switching between MS and MS/MS. Source ionization parameters were as follows: spray voltage, 1.8 kV; capillary temperature, 280 °C; and s-lens level, 50.0. Full-scan MS mode (400–1800 m/z) was operated at a resolution of 70 000 with automatic gain control (AGC) target of 1 × 106 ions and a maximum ion transfer (IT) of 20 ms. Ions selected for MS/MS (fixed first mass 50 m/z) were subjected to the following parameters: resolution, 17 500; AGC, 1 × 105 ions; maximum IT, 80 ms; 2.0 m/z isolation window; normalized collision energy, 32.0; underfill ratio, 0.05%; and dynamic exclusion, 40.0 s. All samples were run with duplicate injections.
All raw files were analyzed using the Thermo Proteome Discoverer (1.3.0.339) software platform. The files were searched using the Mascot (2.2.04) search engine against the UniProt human (72 390 entries) and decoy human databases with a strict FDR of 0.01. The following Mascot parameters were used: trypsin was selected as the enzyme; two missed cleavages were allowed; precursor mass tolerance was set to 20 ppm; fragment mass tolerance was set to 0.05 Da; modifications of protein N-terminal glutamine deamidation, methionine oxidation, carbamidomethylation of cysteine; and iTRAQ 8plex labels at the N-termini and at lysine and tyrosine side chains were allowed. Strict maximum parsimony principle was applied considering only peptide spectrum matches (PSMs) with medium confidence or higher and delta Cn better than 0.15. An integration tolerance window of 20 ppm and most confident centroid integration method were applied, with only unique peptides considered for quantification.
iTRAQ Data Analysis
Control sample (siNeg) aliquots were individually labeled with iTRAQ reagents, combined, and analyzed by LC–MS. The variation measured in resulting protein ratios was used to set a fold-change threshold that encompasses experimental variation and variation associated with the biological system. The fold-change threshold was set at 1.3, which is three times the standard deviation measured in the protein profiling of the control samples. A p value (p > 0.05) criterion (between untreated/control ratios and sample/control ratios) was also used to determine differentially expressed proteins.
Cell Migration Assays
HCT116 cells (150 000) were plated in 12-well plates and transfected with 3.1 μL of transfection reagents and 20 nmol/L AllStar negative control siRNA, siITGA3.1, or siITGA3.7. Forty eight hours post-transfection, a p200 pipet tip was used to create a scratch of the cell monolayer, and the media was changed. Images of the wound region were acquired every 4 h post-wound. The analysis of open wound area in the images was conducted using Tscratch software.22 Student’s t tests were performed to determine statistical significance between the control transfected and siRNA silenced cells (p value < 0.05). Changes in open wound areas between control and gene-silenced cells were used as a measure of cell mobility.
Cell migration was assessed using the QCM Chemotaxis 8 μm cell migration assay system (Millipore, Bedford, MA) according to the manufacturer’s instructions. Briefly, 50 000 SW620 or HCT116 cells transfected with control siRNA or gene-specific siRNA were seeded into the migration chamber in serum-free medium. For experiments with an inhibitor, cells were seeded in the presence of 20 μmol/L VAS2870. Medium containing 10% FBS was placed in the feeder tray. After allowing cell migration for 24 h, cells that had migrated through the membrane were lysed, stained, and quantified using fluorescence readings at 480EX/520EM. The experiments were performed in triplicate and repeated on two independent cell transfections/treatments.
Caspase Activity and Cell Viability Assays
Following siRNA transfection or VAS2870 treatment, cells were subjected to the ApoLiveGlo assay kit (Promega, Madison, WI) following the manufacturer’s instructions at 48 h post-transfection. The viability of cells was assessed with fluorescence readings (560EX/590EM). The caspase-3/7 activity, as a measure of apoptosis, was quantified with the luminescence of each sample measured with 500 ms integration. The experiments were performed in triplicate and repeated on two independent cell transfections/treatments.
Statistical Analysis
Data was analyzed by Student’s t test and considered statistically significant if p < 0.05. All data are mean ± SD of three replicate measurements unless otherwise indicated.
Results and Discussion
ITGA3 and NOX4 Modulation
In this study, we examined the global quantitative differences in protein expression following manipulation of the expression level or activity of two genes implicated in the progression of colorectal cancer. ITGA3 and NOX4 were silenced in HCT116 and SW620 colon cancer cells, respectively. These cell lines were specifically selected on the basis of ITGA3 and NOX4 gene expression analysis in a panel of colorectal cancer cell lines. Expression analysis revealed elevated ITGA3 expression in HCT116 cells and elevated NOX4 expression in SW620 cells (Supporting Information Figure S2). Furthermore, while previous manuscripts have cited the origin of SW62023 and HCT11624 as left and ride side, respectively, we also determined that their gene expression patterns are highly consistent with these designations. As an example, the gene expression pattern of the gene PRAC, which is highly expressed on the left but nearly undetectable on the right, confirms these designations.4 HCT116 and SW620 cell lines also have molecular and genetic characteristics consistent with primary tumors found in the right and left colon. HCT116 is microsatellite unstable (MSI) and has CpG island methylation phenotype (CIMP) high status, which are more frequently seen in right-sided colon cancer.25,26 SW620 cells have molecular phenotypes more common in left-sided colon cancer, including chromosomal instability (CIN) and mutation of the p53 gene.26,27
The NADPH oxidase inhibitor VAS2870 was used to treat SW620 cells at the determined IC50 (18 μmol/L) (Supporting Information Figure S1B).13,28 At this concentration, VAS2870 had no effect on cell viability (Supporting Information Figure S1C). Although VAS2870 is not a NOX4 isoform specific inhibitor, expression profiling of the Nox family in a panel of colorectal cell lines revealed NOX4 expression to be the highest of any Nox homologue in the SW620 cell line (Supporting Information Figure S3). The proteomic alterations associated with VAS2870 inhibition of NOX-mediated ROS were evaluated in addition to the effects of siRNA-mediated NOX4 gene silencing.
Quantitative Protein Profiling
The workflow for the combined use of RNA interference and iTRAQ protein quantification is summarized in Figure 1. Differential expression was determined using a combined criterion involving both expression fold changes and a p-value cutoff. Combined criterion based on expression fold changes and a p-value threshold will address the reduction in quantitative accuracy from precursor interference associated with iTRAQ labeling. Our analysis considered only proteins found in two or more biological replicates. The fold-change cutoff was determined on the basis of the global standard deviation associated with experimental variation inherent to the iTRAQ labels and biological variation of the replicate control samples (Supporting Information Figure S4). This strategy combats the dynamic range compression inherent in the chemical isobaric labeling used for quantitative analysis. A statistical threshold (t test p value) was also employed such that the expression changes associated with gene silencing were different (p value < 0.05) from changes associated with the untreated and control cells to account for artifacts accompanying the transfection conditions.
Figure 1.
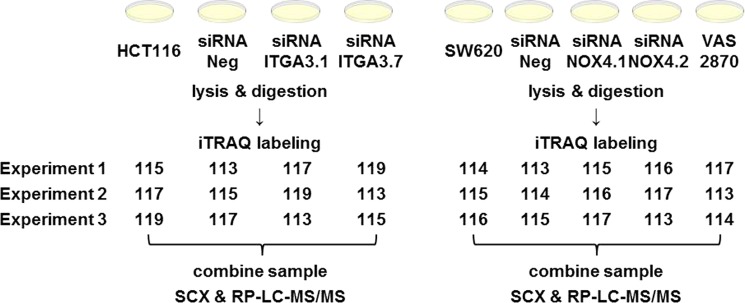
Experimental design workflow. HCT116 cell were silenced with siITGA3 siRNAs, whereas NOX4 was inhibited in SW620 cells with siRNAs or VAS2870. Proteins samples were digested, labeled with iTRAQ reagents, fractionated, and analyzed with a nanoAcquity UPLC coupled to a Q-Exactive mass spectrometer.
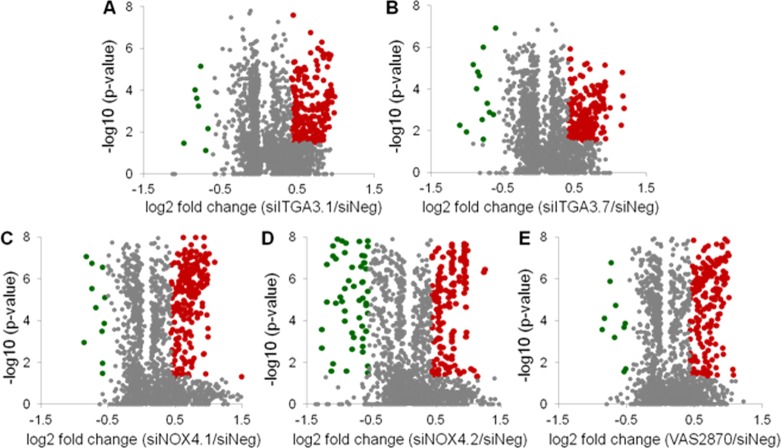
On the basis of the aforementioned criterion, 238 and 216 proteins were found to be significantly changed in expression following the silencing of ITGA3 by one of two different siRNAs. Following the silencing of NOX4, 244 and 258 proteins were found to be altered in abundance, whereas 236 proteins were found to have differential expression following NOX4 inhibition with VAS2870. The overall proteomic profiling results are displayed by Volcano plots with up- and downregulated proteins highlighted in red and green, respectively (Figure 2). To account for siRNA off-target effects, the number of differential proteins was further condensed by requiring significant regulation for both gene-specific siRNAs (Supporting Information Figure S5). The subsequent lists of differential proteins were used to conduct pathway analysis (Supporting Information Information).
Figure 2.
Volcano plots displaying the statistical p-value test with the magnitude of the abundance change to identify protein expression changes that are statistically significant. (A, B) Proteins changed in expression with ITGA3 silencing. (C–E) Protein altered in expression with NOX4 gene silencing or chemical inhibition. Green data points represent downregulated proteins, and red data points represent upregulated proteins.
Pathway Analysis
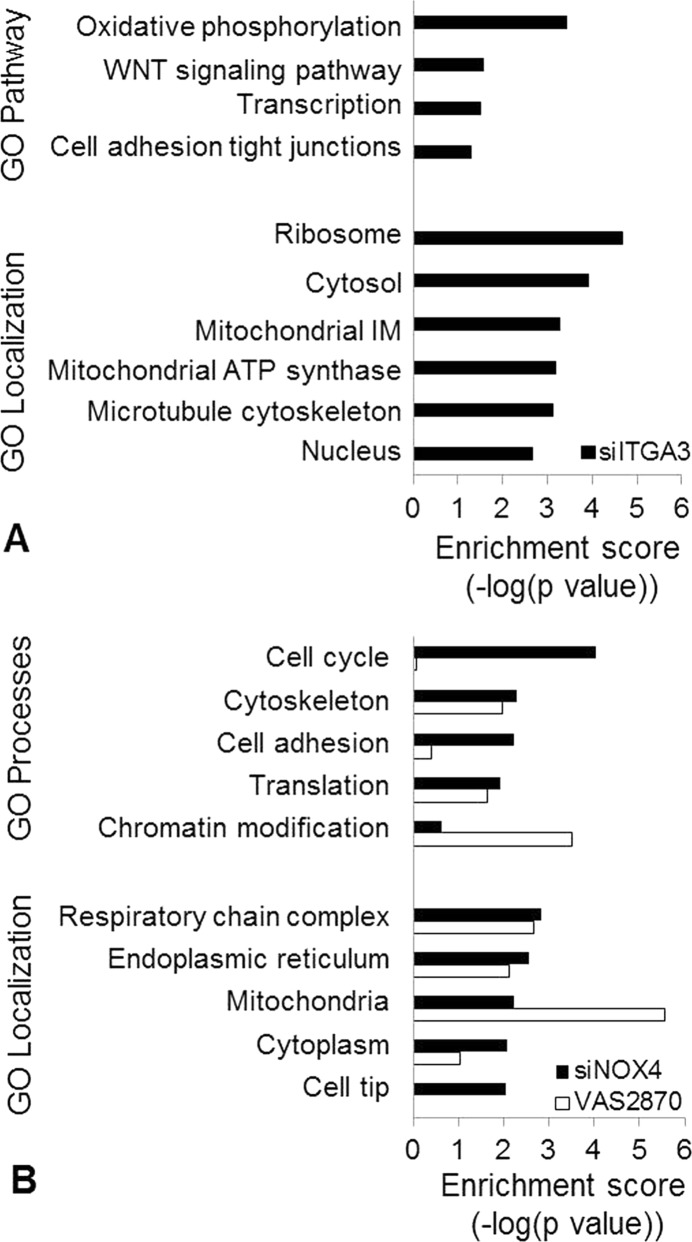
Gene ontology analysis was conducted using GeneGo MetaCore pathway map and process network enrichment to identify functional themes over-represented in the regulated protein species. Pathway and biological process analysis indicate that genes involved in oxidative phosphorylation (p value = 3.7 × 10–4) and cell adhesion (p value = 4.9 × 10–2) were enriched in the differentially expressed proteins following ITGA3 silencing. Furthermore, altered proteins were also significantly localized to the microtubule cytoskeleton (p value = 7.8 × 10–4) (Figure 3A).
Figure 3.
Pathway analysis using GeneGo Metacore enrichment analysis to identify common functional themes over-represented in the differentially expressed proteins. Metacore functional analysis was evaluated on the basis of the prioritization, and statistical significance (p value) was assigned to the pathways, processes, and localizations. The biological themes and cellular locations with the highest enrichment score for (A) siITGA3 and for (B) siNOX4 (black) and VAS2870 (white) responsive proteins are shown.
Pathway analysis was conducted separately for proteins with altered expression following NOX4 gene silencing and NOX4 small molecule inhibition. Proteins associated with NOX4 gene silencing versus protein activity inhibition were found to have differentially prioritized biological processes and cellular locations. Following NOX4 gene silencing, proteins involved in cell cycle (p value = 9.1 × 10–5), cytoskeleton (p value = 5.1 × 10–3), and cell adhesion (p value = 5.7 × 10–3) were found to be changed in expression, whereas proteins altered in abundance as a result of NOX4 inhibition with the small molecule VAS2870 were most significantly involved in chromatin modification (p value = 3.1 × 10–4). NOX4 gene silencing and activity inhibition resulted in altered expression of proteins from similar cellular locations overall. However, siRNA introduction resulted in abundance changes in proteins over-represented in the cell tip (p value = 9.1 × 10–3), whereas proteins changed in response to VAS2870 treatment showed a greater relevance in the mitochondria (p value = 2.7 × 10–6) (Figure 3B).
Phenotypic and Functional Investigation
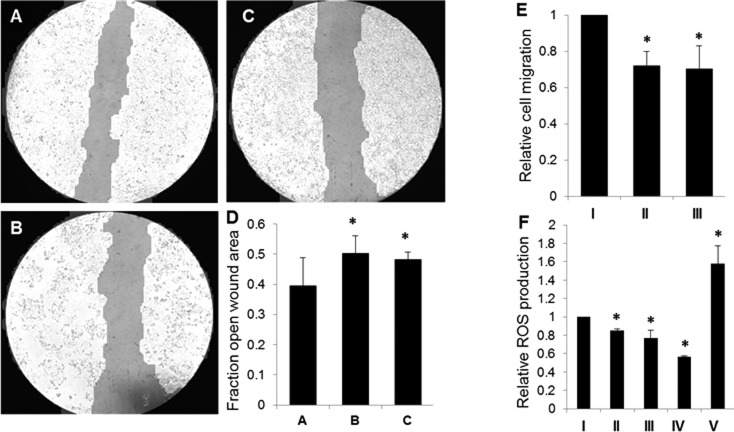
On the basis of the pathway analysis for proteins found to have differential expression following ITGA3 gene silencing, corresponding cellular phenotypes were investigated. Wound healing assays reveal that colon cancer cells with reduced expression levels of ITGA3 have diminished migratory capability as compared to that of control siNeg cells (Figure 4A–D). Migration assays further support these results, indicating that ITGA3 (Figure 4E) plays a role in providing cells with enhanced ability for movement. The redox status of colon cancer cells also changed in response to ITGA3 silencing. Measurement of ROS production showed decreased ROS in siITGA3 transfected cells relative to that in siNeg cells (Figure 4F). These results suggest a role for ITGA3-mediated ROS production and cell migration in advanced colon cancer.
Figure 4.
ITGA3 phenotypic investigation based on results of gene ontology analysis. (A–D) Wound closure assay to assess cell mobility following gene expression manipulation. Wound closure for (A) negative control siNeg cells, (B) siITGA3.1 cells, and (C) siITGA3.7 cells was monitored for 24 h post-transfection. Representative images for each condition are shown. (D) Relative quantitative analysis of wound closure was conducted using Tscratch software. (E) QCM migration assay shows a decrease in cell migration for gene silenced (II, III) cells relative to that for control colls (I). (F) H2DCF-DA assay to assess relative ROS production shows decreased ROS in ITGA3 siRNA cells (II, III) relative to that in siNeg cells (I). HCT116 cells treated with VAS2870 as a negative control (IV) show decreased ROS, whereas cells treated with H2O2 as a positive control (V) have increased levels of ROS. All data is shown as mean ± SD; * p < 0.05. These results suggest a role for ITGA3-mediated ROS production and cell migration in advanced colon cancer.
Pathway analysis of the differential proteins associated with NOX4 gene silencing and activity inhibition lead to the investigation of the effect of NOX4 expression levels on ROS production and cell migration. Our previously published study revealed a decrease in ROS production with siRNA-mediated NOX4 gene silencing.4 We further investigated the link between NOX4 and ROS by correlating NOX4 expression levels with ROS production in the patient-matched primary (SW480) and metastatic (SW620) CRC cell line model. The data shows a direct correlation between NOX4 expression and ROS production, with both being reduced in primary SW480 cells compared to that in metastatic SW620 cells (Figure 5A). We also assessed the effect of NOX4 expression levels on cell migration. NOX4 gene silencing and small molecule inhibition both lead to a decrease in cell migration (Figure 5B). This data further supports a role for ROS generation and cell mobility in advanced colon cancer and also indicates an association with NOX4 expression.
Figure 5.
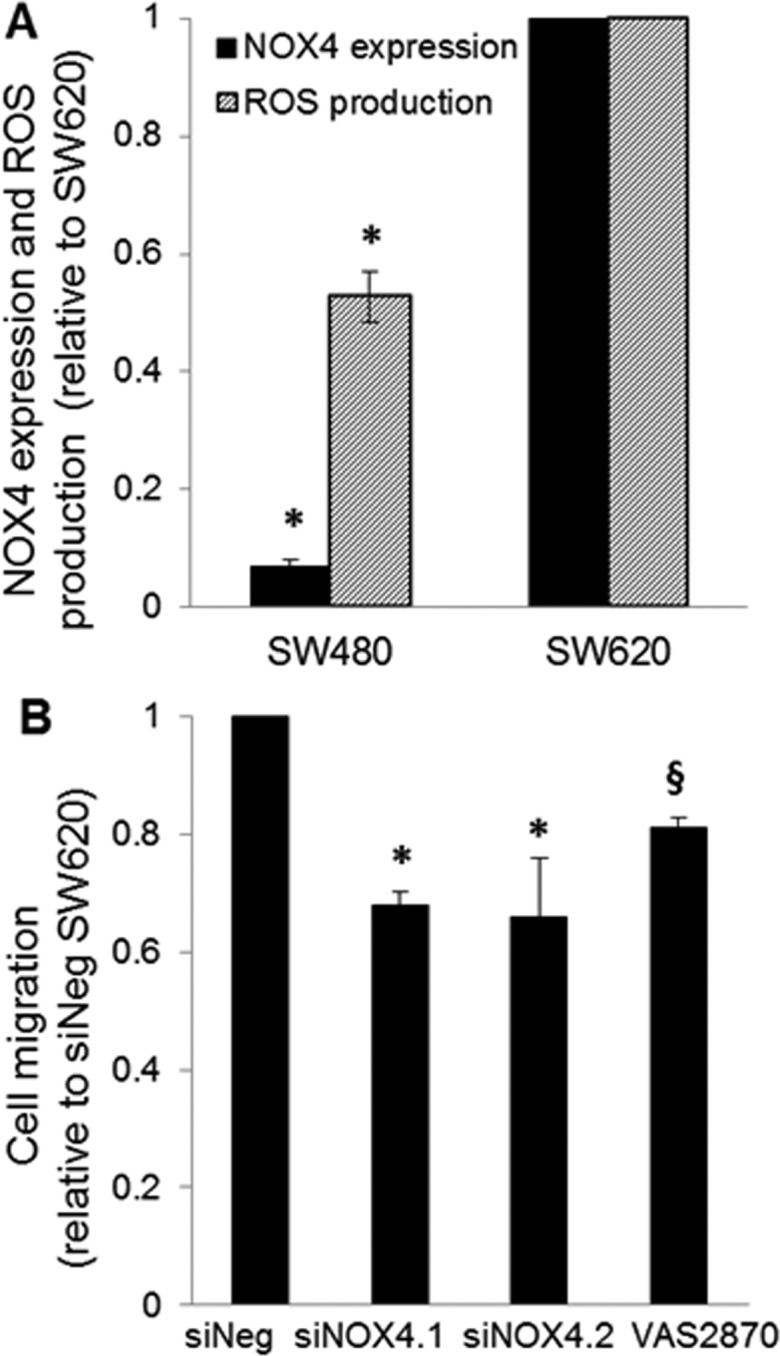
NOX4 phenotypic investigation based on results of gene ontology analysis. (A) NOX4 expression and ROS production in patient-matched SW480 and SW620 cells. There is higher ROS production and NOX4 expression in the metastatic lymph node derived cell line (SW620) as compared to that in the primary adenocarcinoma cell line (SW480). NOX4 expression is based on qRT-PCR, and ROS production was quantified using a H2DCF-DA assay. (B) QCM migration assay shows a decrease in cell migration with NOX4 gene silencing and chemical inhibition. All data is shown as mean ± SD; * p < 0.05, § p < 0.1.
Relevance between Differential Protein Expression and Tumor Stage in Primary Tissues
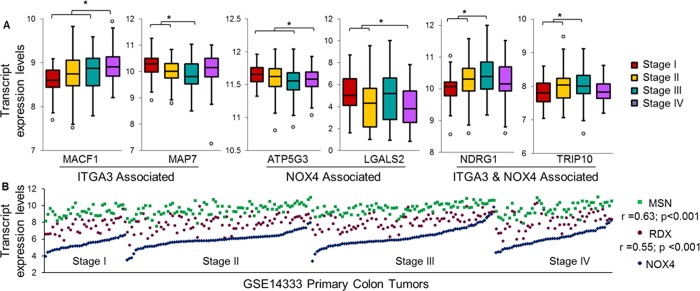
Selected findings were further investigated for clinical significance. The expression of transcripts corresponding to proteins altered in expression with ITGA3 or NOX4 inhibition was examined in relation to the pathological stage of colon carcinoma progression. Microarray data (GSE14333) from 250 primary colon cancer tumors was used to explore the correlation between the molecular species and Duke’s tumor stage (a summary of the patient characteristics can be found in Supporting Information Table S2). Our analysis revealed abundance trends throughout colon tumorigenesis for several genes of interest that agree with the changes in abundance directionality observed in the global proteomic profiling. For example, silencing of ITGA3 and NOX4 resulted in increased relative abundance of ANP32B and LGALS2 at the protein level, respectively. ANP32B and LGALS2 show a significant reduction in expression as the primary tumors progress in tumorigenesis. The same relationships in gene expression are seen in the global proteomic profiling and primary tissue samples. Representative trends for altered proteins correlating with tumor stage are shown in Figure 6A. We also used the microarray data to explore the transcript correlation between NOX4 and individual genes with differential expression in the protein profiling. MSN and RDX were found to have significant transcript expression correlations with NOX4 (Figure 6B). These combined results show that many of the altered gene products correlate with stage progression in a manner consistent with their expression relationship to ITGA3 and NOX4 in the proteomic profiling.
Figure 6.
Relevance between altered protein expression and tumor stage. (A) Several proteins found to be altered following ITGA3, NOX4, or ITGA3 and NOX4 silencing also show altered transcript expression according to tumor stage among 250 primary colon tumors (t test; * p < 0.05). (B) Two genes were found to have significant correlation with NOX4 expression across the primary tumors. Data shown as transcript expression levels based on microarray results using Affymetrix Human Genome U133Plus 2.0 arrays and quantile normalization from the GEO data set GSE14333.
Discussion
This study links the expression levels of protein mediators to their functional effects in the cancer cell through the combined use of RNA interference and protein quantification. The protein abundance changes induced by siRNA-mediated gene silencing and chemical inhibition were analyzed with iTRAQ protein quantification and pathway analysis.
Oxidative phosphorylation is the metabolic pathway by which mitochondria synthesize ATP from the oxidation of nutrients and produce reactive oxygen species (ROS) as byproducts. Mitochondrial function may play a key role in controlling cancer cells because it acts as an energy supplier and ROS regulator. Phenotypic analysis of ITGA3-silenced cells revealed altered expression of oxidative phosphorylation proteins, altered control of apoptosis and proliferation, and redox imbalance, which could suggest mitochondrial dysfunction. This data also supports a role in tumorigenesis, as ITGA3 expression correlates with deregulated cellular metabolism, a common cancer phenotype.29 These results are further supported by previous reports of altered ITGA3 and ITGB3 expression in association with oxidative stress.12,30 We have also shown ITGA3 to have a role in cell mobility and migration. This is not surprising given that ITGA3 is an integrin protein and integrin-mediated cell migration and adhesion pathways have been well-defined in the literature.31
NOX family enzymes have wide tissue distributions, but the individual members’ expression levels vary from tissue to tissue. NOX4 is found to be highly expressed in renal, breast, and ovarian cells, and NOX4 expression increases with the development of melanoma, hepatic, and ovarian cancers.32,33 NOX4 was found to attenuate migration in these contexts. Contrary to our expectations, we have shown, for the first time, that the NOX4 isoform is also be abundant in advanced colon cancer and correlates with metastatic potential.4 This study revealed an association between NOX4 and motility in colon cancer, consistent with tissue types previously found to have elevated NOX4 expression.
A decrease in cell migration was observed following NOX4 gene silencing and functional inhibition in colon cancer cells. These results suggest that ROS produced by NOX4 regulate cell mobility. ROS generation by Nox family NADPHs play a key role in regulatory and signaling events including migration, cytoskeletal organization, and gene transcription. Nox-mediated ROS production is required for mobility in several cell types; however, the mechanism(s) by which ROS contributes to cell migration is not well-defined. The signaling pathways most often associated with ROS include MAPK, P13K, Rho-GTPase, and NF-κB, whereas TGF-β and SMAD3 have specifically been linked to NOX4-mediated ROS in breast epithelial cells.17,34
Both ITGA3 and NOX4 silencing resulted in decreased cell migration and ROS generation. However, the data suggests that these phenotypic capabilities may be mediated by diverse genes and pathways. Proteins with cell motility modulating potential that were globally altered in expression included NGRG1 and TGRIP10. NDRG1 and TRIP10 have been proposed to have many diverse cellular mechanisms of regulation and to participate in multiple cellular processes.35,36 This diverse involvement may explain the expression alterations of these two proteins in both the ITGA3 and NOX4 data sets. However, cell motility-related MACF1 and MAP7 were unique to ITGA3 silencing, whereas NOX4 activity inhibition resulted in changes to CTTN, GNB2L1, and RDX. MACF1 is known to interact with microtubules and coordinate microtubule dynamics,37 whereas CTTN and RDX act as an actin scaffolding protein and bind to actin filaments, respectively, both regulating the actin network.38,39Unlike NOX4, which produces ROS as a primary product, ITGA3 seems to generate ROS as a byproduct as a result of integrin engagement, where the subsequent change in redox signaling may attenuate or even cause cellular migration. Although altered cellular mobility and redox status are linked to both right- and left-side colon cancer through the expression levels of ITGA3 and NOX4, these findings support the hypothesis that right- and left-side colon cancer follow different pathways to relapse.
Conclusions
Overall, this study links the expression levels of protein mediators to their functional effects in the cancer cell through the combined use of RNA interference and protein quantification to capture biologically significant changes that define human colon tumorigenesis. This approach proved to be a powerful method to elucidate protein function after accounting for iTRAQ labeling dynamic range compression and possible siRNA off-target effects. Furthermore, many of the proteins found to change in expression following ITGA3 and NOX4 inhibition were discovered to have clinical relevance by correlating expression with tumor stage in colon cancer.
Acknowledgments
The authors thank the Notre Dame Mass Spectrometry and Proteomics Facility for their assistance and Dr. Marian Grade for the generous donation of the LS513, SW1116, and SW1463 CRC cell lines. K.M.B. was supported by the Notre Dame Chemistry Biochemistry Biology Interface (CBBI) program and NIH training grant T32GM075762 and by the Indiana Clinical and Translational Sciences Institute (CTSI) Program and NIH grant TL1 TR000162. T.N.W. was supported by the Notre Dame College of Science REU program.
Supporting Information Available
Table S1: qRT-PCR primer sequences. Table S2: Patient characteristics from GSE14333. Figure S1: (A) Confirmation of siRNA gene silencing. (B) VAS2870 IC50 determination. (C) Confirmation that VAS2870 treatment at the IC50 does not affect SW620 cell viability. Figure S2: ITGA3 and NOX4 expression levels in colorectal cancer cell line panel. Figure S3: NOX family expression levels in colon cancer cell lines. Figure S4: (A) Descriptive statistics of global proteomics profiling of control samples. (B) Threshold for biological significance. (C) Normal distribution of global proteomic data. Figure S5. Venn diagrams displaying overlap of differentially expressed proteins for each treatment in the ITGA3 and NOX4 data sets. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Li F. Y.; Lai M. D. Colorectal cancer, one entity or three. J. Zhejiang Univ., Sci., B 2009, 10, 219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopetta B. Are there two sides to colorectal cancer?. Int. J. Cancer 2002, 101, 403–8. [DOI] [PubMed] [Google Scholar]
- Bufill J. A. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann. Int. Med. 1990, 113, 779–88. [DOI] [PubMed] [Google Scholar]
- Bauer K. M.; Hummon A. B.; Buechler S. Right-side and left-side colon cancer follow different pathways to relapse. Mol. Carcinog. 2012, 51, 411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M.; Haan K.; Baker S. E.; Fitchmun M.; Todorov I.; Weitzman S.; Jones J. C. A cell signal pathway involving laminin-5, α3β1 integrin, and mitogen-activated protein kinase can regulate epithelial cell proliferation. Mol. Biol. Cell 1999, 10, 259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar A.; Shome S. G.; Lamar J.; Stirling L.; Iyer V.; Pumiglia K.; DiPersio C. M. α3β1 integrin promotes keratinocyte cell survival through activation of a MEK/ERK signaling pathway. J. Cell Sci. 2004, 117, 4043–54. [DOI] [PubMed] [Google Scholar]
- Wen T.; Zhang Z.; Yu Y.; Qu H.; Koch M.; Aumailley M. Integrin α3 subunit regulates events linked to epithelial repair, including keratinocyte migration and protein expression. Wound Repair Regener. 2010, 18, 325–34. [DOI] [PubMed] [Google Scholar]
- Ura H.; Denno R.; Hirata K.; Yamaguchi K.; Yasoshima T. Separate functions of alpha2beta1 and alpha3beta1 integrins in the metastatic process of human gastric carcinoma. Surg. Today 1998, 28, 1001–6. [DOI] [PubMed] [Google Scholar]
- Kurokawa A.; Nagata M.; Kitamura N.; Noman A. A.; Ohnishi M.; Ohyama T.; Kobayashi T.; Shingaki S.; Takagi R. Diagnostic value of integrin α3, β4, and β5 gene expression levels for the clinical outcome of tongue squamous cell carcinoma. Cancer 2008, 112, 1272–81. [DOI] [PubMed] [Google Scholar]
- Yang G. Y.; Xu K. S.; Pan Z. Q.; Zhang Z. Y.; Mi Y. T.; Wang J. S.; Chen R.; Niu J. Integrin αvβ6 mediates the potential for colon cancer cells to colonize in and metastasize to the liver. Cancer Sci. 2008, 99, 879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonlaufen A.; Wiedle G.; Borisch B.; Birrer S.; Luder P.; Imhof B. A. Integrin α(v)β(3) expression in colon carcinoma correlates with survival. Mod. Pathol. 2001, 14, 1126–32. [DOI] [PubMed] [Google Scholar]
- Lei Y. H.; Kai; Gao C.; Lau Q. C.; Pan H.; Xie K.; Li L.; Liu R.; Zhang T.; Xie N.; Nai H. S.; Wu H.; Gong Q.; Zhao X.; Nice E.; Huang C.; Wei Y. Proteomics identification of ITGB3 as a key regulator in reactive oxygen species-induced migration and invasion of colorectal cancer cells. Mol. Cell. Proteomics 2011, 10, M110.005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhöfer S.; Kleikers P. W.; Radermacher K. A.; Scheurer P.; Rob Hermans J. J.; Schiffers P.; Ho H.; Wingler K.; Schmidt H. H. The NOX toolbox: validating the role of NADPH oxidases in physiology and disease. Cell. Mol. Life Sci. 2012, 69, 2327–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner M. Y.; Arbiser J. L. Targeting NADPH oxidases for the treatment of cancer and inflammation. Cell. Mol. Life Sci. 2012, 69, 2435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyemi U.; Dupuy C. The emerging role of ROS-generating NADPH oxidase NOX4 in DNA-damage responses. Mutat. Res. 2012, 751, 77–81. [DOI] [PubMed] [Google Scholar]
- Tobar N.; Guerrero J.; Smith P. C.; Martinez J. NOX4-dependent ROS production by stromal mammary cells modulates epithelial MCF-7 cell migration. Br. J. Cancer 2010, 103, 1040–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau H. E.; Casterline B. W.; Rada B.; Korzeniowska A.; Leto T. L. Nox4 involvement in TGF-β and SMAD3-driven induction of the epithelial-to-mesenchymal transition and migration of breast epithelial cells. Free Radical Biol. Med. 2012, 53, 1489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Liu Z.; Hu X. Inhibiting cancer metastasis via targeting NAPDH oxidase 4. Biochem. Pharmacol. 2013, 86, 253–66. [DOI] [PubMed] [Google Scholar]
- Shimada K.; Fujii T.; Anai S.; Fujimoto K.; Konishi N. ROS generation via NOX4 and its utility in the cytological diagnosis of urothelial carcinoma of the urinary bladder. BMC Urol. 2011, 11, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaura M.; Mitsushita J.; Furuta S.; Kiniwa Y.; Ashida A.; Goto Y.; Shang W. H.; Kubodera M.; Kato M.; Takata M.; Saida T.; Kamata T. NADPH oxidase 4 contributes to transformation phenotype of melanoma cells by regulating G2–M cell cycle progression. Cancer Res. 2009, 69, 2647–54. [DOI] [PubMed] [Google Scholar]
- Masters J. R.; Thomson J. A.; Daly-Burns B.; Reid Y. A.; Dirks W. G.; Packer P.; Toji L. H.; Ohno T.; Tanabe H.; Arlett C. F.; Kelland L. R.; Harrison M.; Virmani A.; Ward T. H.; Ayres K. L.; Debenham P. G. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 8012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geback T.; Schulz M. M.; Koumoutsakos P.; Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. BioTechniques 2009, 46, 265–74. [DOI] [PubMed] [Google Scholar]
- Flatmark K.; Maelandsmo G. M.; Martinsen M.; Rasmussen H.; Fodstad O. Twelve colorectal cancer cell lines exhibit highly variable growth and metastatic capacities in an orthotopic model in nude mice. Eur. J. Cancer 2004, 40, 1593–8. [DOI] [PubMed] [Google Scholar]
- Eshleman J. R.; L E.; Bowerfind G. K.; Parsons R.; Vogelstein B.; Willson J. K.; Veigl M. L.; Sedwick W. D.; Markowitz S. D. Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer. Oncogene 1995, 10, 33–37. [PubMed] [Google Scholar]
- Sugai T.; Habano W.; Jiao Y. F.; Tsukahara M.; Takeda Y.; Otsuka K.; Nakamura S. Analysis of molecular alterations in left- and right-sided colorectal carcinomas reveals distinct pathways of carcinogenesis: proposal for new molecular profile of colorectal carcinomas. J. Mol. Diagn. 2006, 8, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed D.; Eide P. W.; Eilertsen I. A.; Danielsen S. A.; Eknaes M.; Hektoen M.; Lind G. E.; Lothe R. A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman S.; Adlard J. Left and right sided large bowel cancer. BMJ. 2002, 324, 931–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler K.; Altenhoefer S. A.; Kleikers P. W.; Radermacher K. A.; Kleinschnitz C.; Schmidt H. H. VAS2870 is a pan-NADPH oxidase inhibitor. Cell. Mol. Life Sci. 2012, 69, 3159–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaini G.; Sgarbi G.; Baracca A. Oxidative phosphorylation in cancer cells. Biochim. Biophys. Acta 2011, 1807, 534–42. [DOI] [PubMed] [Google Scholar]
- Weigel A. L.; Handa J. T.; Hjelmeland L. M. Microarray analysis of H2O2-, HNE-, or tBH-treated ARPE-19 cells. Free Radical Biol. Med. 2002, 33, 1419–32. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A.; Horwitz A. R. Integrins in cell migration. Cold Spring Harbor Perspect. Biol. 2011, 3, a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K. A.; Kulawiec M.; Owens K. M.; Li X.; Desouki M. M.; Chandra D.; Singh K. K. NADPH oxidase 4 is an oncoprotein localized to mitochondria. Cancer Biol. Ther. 2010, 10, 223–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz A.; Ge Y.; Markel S.; Chiu A.; Matsumoto L.; van Balgooy J.; Roy K.; Doroshow J. H. Expression of NADPH oxidase homologues and accessory genes in human cancer cell lines, tumours and adjacent normal tissues. Free Radical Res. 2009, 43, 523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Liu Z.; Hu X. Inhibiting cancer metastasis via targeting NAPDH oxidase 4. Biochem. Pharmacol. 2013, 86, 253–66. [DOI] [PubMed] [Google Scholar]
- Ellen T. P.; Ke Q.; Zhang P.; Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis 2008, 29, 2–8. [DOI] [PubMed] [Google Scholar]
- Hsu C. C.; Leu Y. W.; Tseng M. J.; Lee K. D.; Kuo T. Y.; Yen J. Y.; Lai Y. L.; Hung Y. C.; Sun W. S.; Chen C. M.; Chu P. Y.; Yeh K. T.; Yan P. S.; Chang Y. S.; Huang T. H.; Hsiao S. H. Functional characterization of Trip10 in cancer cell growth and survival. J. Biomed. Sci. 2011, 18, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. J.; Lin C. M.; Lin C. S.; Perez-Olle R.; Leung C. L.; Liem R. K. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 2006, 20, 1933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M. L.; Wang M. R. CTTN (EMS1): an oncogene contributing to the metastasis of esophageal squamous cell carcinoma. Cell Res. 2007, 17, 298–300. [DOI] [PubMed] [Google Scholar]
- Arpin M.; Chirivino D.; Naba A.; Zwaenepoel I. Emerging role for ERM proteins in cell adhesion and migration. Cell Adhes. Migr. 2011, 5, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.