SUMMARY
DNA methylation at the fifth position of cytosine (5mC) is an important epigenetic modification that affects chromatin structure and gene expression. Recent studies have established a critical function of the Ten-eleven translocation (Tet) family of proteins in regulating DNA methylation dynamics. Three Tet genes have been identified in mammals, and they all encode for proteins capable of oxidizing 5mC as part of the DNA demethylation process. While regulation of Tet expression at the transcriptional level is well documented, how TET proteins are regulated at post-translational level is poorly understood. In this study, we report that all three TET proteins are direct substrates of calpains, a family of calcium-dependent proteases. Specifically, calpain1 mediates TET1 and TET2 turnover in mouse ES cells, and calpain2 regulates TET3 level during differentiation. This study provides the first evidence that TET proteins are subject to calpain-mediated degradation.
INTRODUCTION
The ten-eleven translocation (Tet) family of proteins was initially described when the gene encoding the founding member TET1 was identified as a fusion partner of the mixed lineage leukemia (MLL) gene in acute myeloid leukemia (Ono et al., 2002). However, TET proteins were not at a central stage till they were found to oxidize 5mC to 5-hydroymethylcytosine (5hmC) as part of the DNA demethylation process (Ito et al., 2010; Tahiliani et al., 2009). Subsequent studies demonstrated that TET proteins further oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which are removed through base excision repair, thus completing the demethylation process (He et al., 2011; Ito et al., 2011). Expressions of TET proteins are tightly regulated at the transcriptional level. For example, in mouse embryonic stem cells (mESC) both Tet1 and Tet2 are positively regulated by Oct4, and their mRNA levels decrease dramatically upon mESC differentiation. In contrast, Tet3 is significantly up-regulated during differentiation (Koh et al., 2011). In addition to transcription, two recent studies reported that microRNA (miR-22) regulates Tet mRNA in leukemia and breast cancers (Song et al., 2013a; Song et al., 2013b). However, regulation of TET proteins at the post-translational level is less understood. One recent study suggests that IDAX and CXXC5 interact with TET2 and regulate its stability through caspase-dependent degradation (Ko et al., 2013). It is not clear whether TET1 and TET3 are subjected to a similar regulation.
Four major proteolytic systems mediate protein turnover: proteasome, lysosome, caspase and calpain. Proteasomes are best known for degrading proteins that are modified by polyubiquitylation (Glickman and Ciechanover, 2002); Lysosomes mediate the bulk breakdown of proteins or organelles (Pan et al., 2008); Caspases are a family of cysteine proteases involved in proteins cleavage during programmed cell death (Cohen, 1997). Finally, calpains are a family of calcium-dependent cysteine proteases with 14 members identified in human (Storr et al., 2011). So far calpain1 and calpain2 (µ- and m-calpains, respectively) are the best characterized members. Known substrates for calpain include structural proteins, signaling molecules and transcriptional factors (Suzuki et al., 2004). Dysregulation of calpains have been linked to a number of human diseases such as muscular dystrophy, diabetes and Alzheimer's disease (Zatz and Starling, 2005). Moreover, calpains have been implicated in stem cell maintenance and differentiation (Santos et al., 2012; Yajima and Kawashima, 2002). Due to the ubiquitous expression pattern and large number of family members, novel calpain substrates and biological functions of calpain-mediated protein cleavage await to be identified.
In this study, we took advantage of the various chemical inhibitors for different protein turnover pathways and identified calpains as major players that mediate TET protein turnover. We then use a well-established protocol to differentiate mESC towards neural progenitor cells (NPCs) to demonstrate that calpain1 and calpain2 are responsible for TET protein turnover in ESCs and NPCs, respectively.
RESULTS
Post-Translational Regulation of TET Proteins
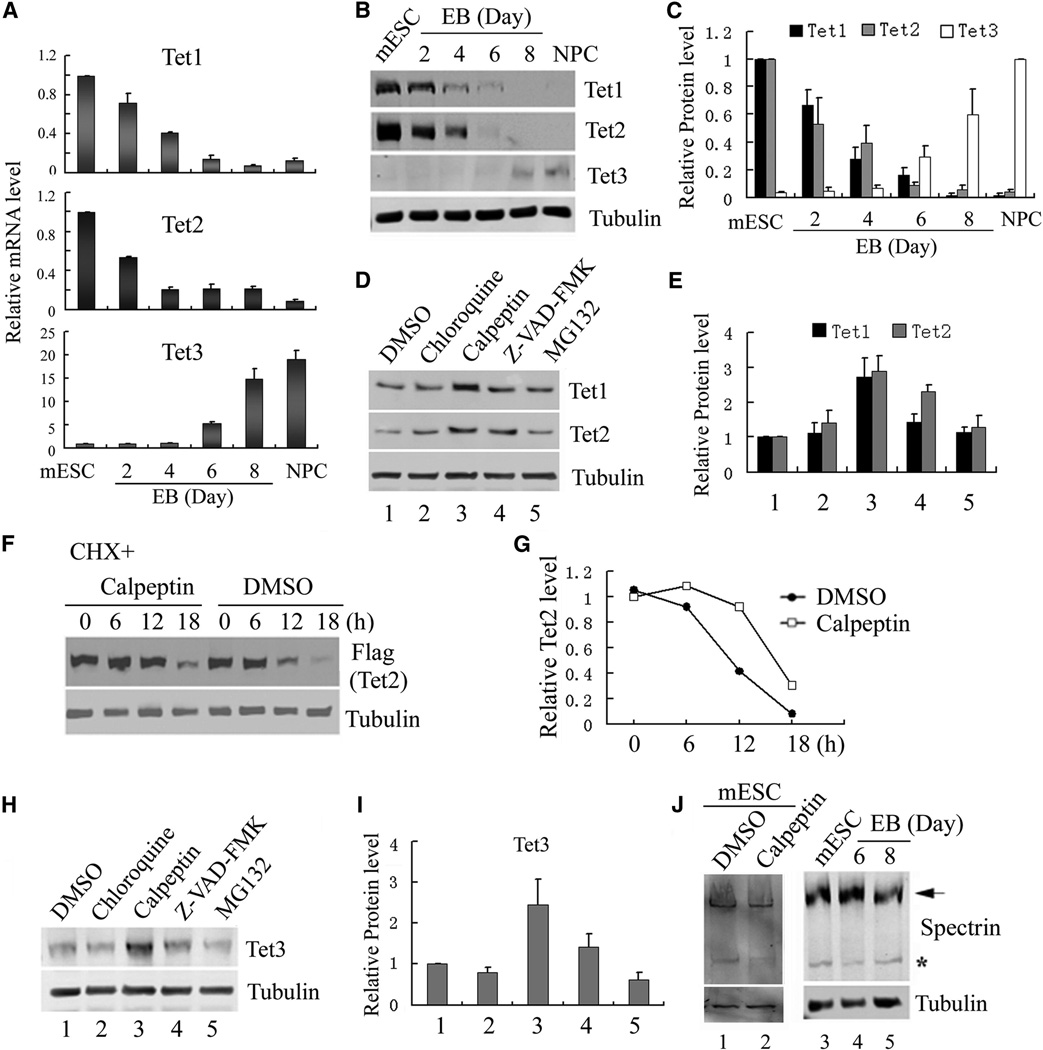
The three Tet genes have distinct expression profiles, while Tet1 and Tet2 are down-regulated during ESC differentiation, Tet3 is up-regulated in the same process (Koh et al., 2011). To systematically examine the relationship between Tet mRNA and protein levels, we utilized an embryonic body (EB)-based protocol to differentiate mESC into NPCs (Fig. S1A) (Bibel et al., 2007). Successful differentiation was verified by significant up-regulation of the neural marker Nestin (Fig. S1B). We then examined TET expression change during differentiation by RT-qPCR and Western blot. We found that while both Tet1 and Tet2 are down-regulated during mESC differentiation, Tet3 is up-regulated (Fig. 1A). Western blot analysis revealed that TET protein levels correlate with mRNA levels (Fig. 1B, C), suggesting TET expression is largely controlled at the transcription level. Nevertheless, the rapid protein turnover of TET1 and TET2 between EB days 2 and 6 suggests a possible post-translational regulation. To explore this possibility, we analyzed the effect of various proteolytic pathways on TET protein turnover by focusing on ESCs for TET1 and TET2, and EB day8 for TET3. We treated cells with inhibitors of the four major proteolytic pathways: proteasome (MG132), lysosome (chloroquine), calpain (calpeptin), and caspase (Z-VAD-FMK) and found that calpeptin treatment induced the most significant accumulation of TET1 and TET2 proteins, and a less prominent effect was observed by inhibiting caspase. However, no significant effect was observed when treated with lysosome or proteasome inhibitors (Fig. 1D, E). We confirmed the effectiveness of MG132 as well as chloroquine (Fig. S1C, D). Thus, lysosome and proteasome are not essential for TET protein turnover.
Figure 1. Regulation of TET protein levels by transcription and protein stability.
(A) RT-qPCR analysis of Tet mRNA levels during mESC to NPC differentiation. While Tet1 and Tet2 levels decrease during differentiation, Tet3 level is significantly up-regulated. Data represent the mean of three independent experiments, and Tet levels in mESCs are set as 1.
(B, C) Representative Western blot (B) and quantification of three repeats (C) demonstrate that TET protein levels generally follow mRNA levels during NPC differentiation.
(D, E) Representative Western blot analysis of TET1 and TET2 levels in mESCs treated with chloroquine, calpeptin, Z-VAD-FAM and MG132 for 24h. Quantification of three independent experiments was shown in panel E.
(F, G) Calpeptin increases the half-life of Flag-TET2 protein. Western blot (F) and quantification (G) of the Flag-TET2 levels in the presence or absence of calpeptin upon inhibition of protein translation by cycloheximide.
(H, I) Representative Western blot analysis of TET3 in day 7 embryoid body (EB) treated with chloroquine, calpeptin, Z-VAD-FAM and MG132 for 24h, and the results were quantified in (I).
(J) Calpain activity is detectable in mESCs and during their differentiation. Western blot analysis of mESC lysate with a spectrin antibody identified both full length (arrow) and cleaved spectrin (*), a marker for calpain activity. Spectrin cleavage is detectable during mESC differentiation (lanes 4, 5), and was prevented by calpeptin treatment (compare lane 1 and 2).
To further evaluate the role of calpeptin in stabilizing TET proteins, we attempted to determine the half-life of TET by cycloheximide treatment which blocks protein synthesis. Because mESCs are sensitive to cycloheximide, we expressed Tet2 in 293T cells and then treated the cells with cycloheximide. We found that calpeptin extended TET2 half-life from 10 hours to 16 hours (Fig. 1F, G), supporting a role of calpains in TET2 degradation. In addition to mESCs, we also analyzed the effect of the various proteolysis pathways on TET3 stability in EBs and observed a similar effect by calpeptin treatment (Fig. 1H, I).
The above results suggest that calpains are likely responsible for TET turnover. Next we examined calpain activity in mESC and EBs by monitoring the cleavage of spectrin, a well characterized calpain substrate (Czogalla and Sikorski, 2005). Western blot analysis of EB day6 and day8 lysates clearly showed a lower band matching cleaved spectrin, which disappeared following calpeptin treatment (Fig. 1J), suggesting calpain activity is present in both self-renewing and differentiated mESC. Collectively, the above results suggest that calpains-mediated proteolysis play a role in regulating TET protein stability and caspases may also contribute to this process. Since the role of caspases has been recently reported (Ko et al., 2013), we focus our study on calpain-mediated regulation of TET proteins.
Tet Proteins are Direct Substrates of Calpains
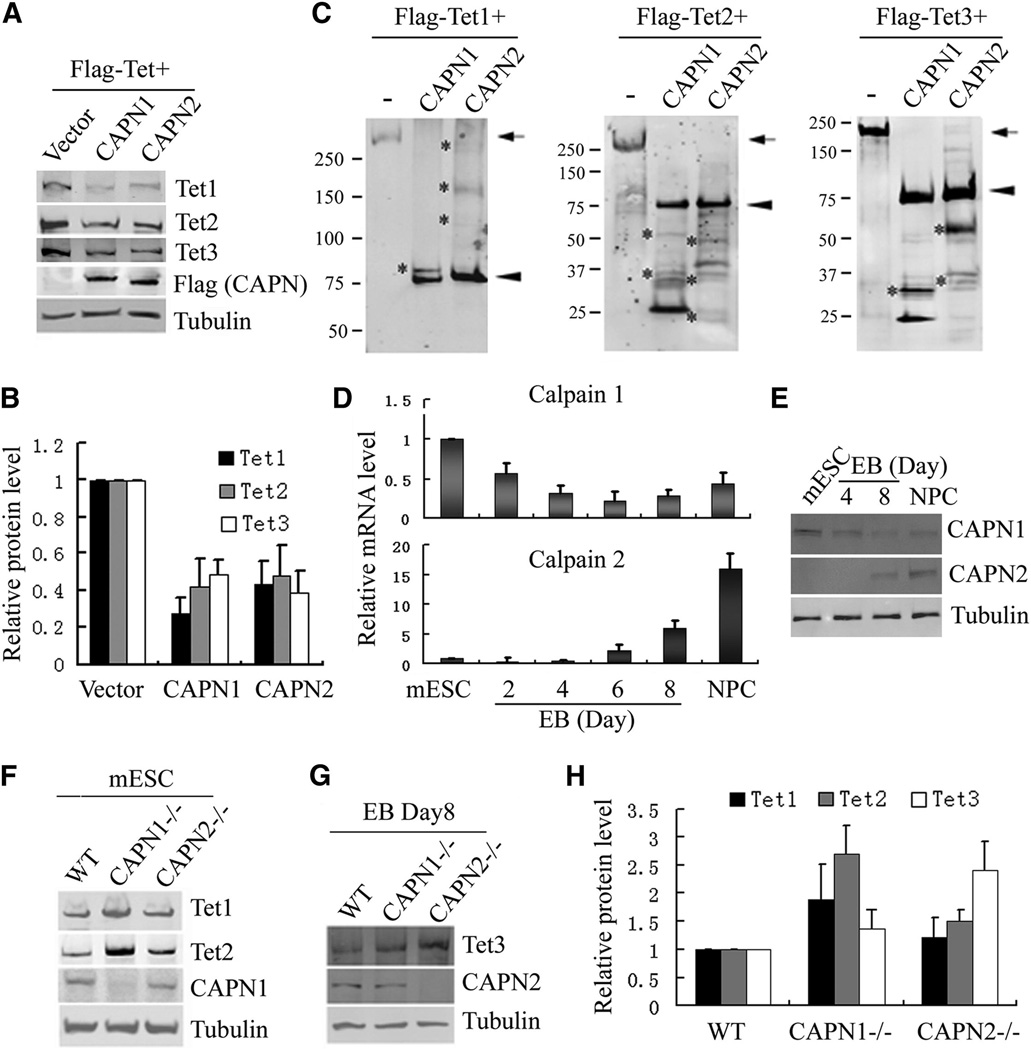
To directly address the role of calpains in regulating TET stability, we asked whether exogenously expressed TET2 can be down-regulated by co-expression of calpain1 or calpain2, two of the best characterized calpains. As shown in Fig. 2A and 2B, TET levels are significantly decreased by co-expression of either calpain 1 or calpain 2. To examine if calpains directly cleave TET proteins, we performed calpain cleavage assays in vitro using purified calpain1, calpain2 and three TET proteins. Results shown in Fig. 2C demonstrate that all three TET proteins are efficiently cleaved by both calpain1 and calpain2. The variable sizes of the cleavage products (Fig. 2C and S2A) suggest multiple cleavages sites. The proteolytic activity of calpain1 and calpain2 is not due to contaminating proteases as neither calpain1 nor calpain2 cleaved RNF4 under the same conditions (Fig. S2C).
Figure 2. Tet proteins are direct substrates of calpain1 and calpain2.
(A, B) Representative Western blot analysis (A) and quantification of three independent repeats (B) demonstrate that exogenously expressed TET protein levels can be reduced by co-expression of calpain1 or calpain2 in 293T cells.
(C) Western blot analysis demonstrates that both calpain 1 and calpain 2 can cleave all three Tet proteins in vitro. Purified Flag-Tet1, Tet2 and Tet3 were incubated with buffer alone, or purified Flag-calpain1 or calpain2 at room temperature for 30min before Western blot analysis using Flag antibody. Cleaved products are indicated by (*), full length TET (arrow), calpains (arrow head).
(D, E) RT-qPCR (D) and Western blot (E) analysis demonstrate that calpain1 and calpain2 are reversely expressed in mESCs and NPCs. Data represent the mean of three independent experiments, and value from mESC is normalized as 1.
(F) Western blot analysis demonstrates that both TET1 and TET2 levels are increased in calpain1 knockout mESCs, while calpain2 knockout have little effect. Calpain1 and calpain2 knockout mESC were generated by CRISPR.
(G) Western blot analysis of the TET3 levels in day 8 EB demonstrates calpain2 knockout increases TET3 levels, while the effect of calpain1 knockout is modest.
(H) Quantification of three independent experiments (F and G), value in WT cells is set as 1.
To test if calpain1 and calpain2 regulate TET protein stability in vivo, we analyzed the expression profiles of calpain1 and calpain2 during mESC differentiation. RT-qPCR analysis indicated that calpain1 level is relatively high in mESCs, while calpain2 is mainly expressed in NPCs (Fig. 2D, E). Considering Tet expression profiles (Fig. 1A), we hypothesize that calpain1 mainly regulates TET1 and TET2 stability in mESCs, while calpain2 regulates TET3 during differentiation. To test this possibility, we utilized the CRISPR-based genome editing technology (Cong et al., 2013; Mali et al., 2013) and generated calpain1 and calpain2 knockout mESCs (Fig. S2D). Targeting sequences were designed against exons of the N-terminal part of the transcript (Fig. S2D), and no off-target was identified based on the established criteria (Hsu et al., 2013). The genotypes were determined by DNA sequencing. A clone with frame shifts on both alleles is chosen and further confirmed by Western blot analysis (Fig. 2F, G). As expected, both TET1 and TET2 levels are increased in calpain1 knockout mESC compared to control (Fig. 2F). Due to a low calpain2 level in mESCs, the effect of calpain2 knockout is less apparent (Fig. 2F). However, when the knockout mESCs are induced to differentiate, significant increase in TET3 levels is observed in calpain2−/− EBs, which is less apparent in calpain1−/− cells (Fig. 2G). The observed effect is likely mediated at the protein level as Tet mRNA level is not significantly altered by calpain knockout (Fig. S2F, G). These results strongly suggest that calpains regulate TET protein levels in vivo and the regulation exhibits isoform and cell differentiation state specificity.
Calpains Regulate TET Functions in mESC Maintenance and Differentiation
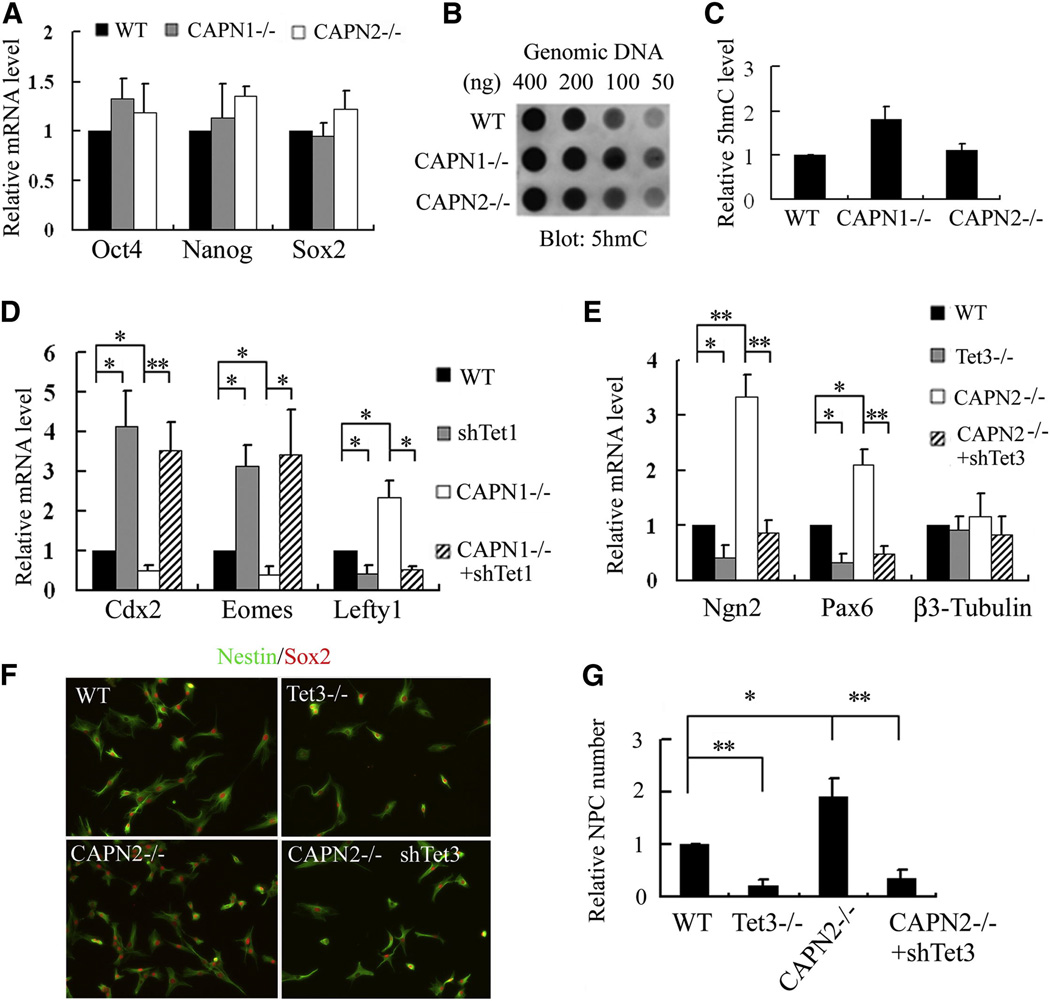
TET proteins play complicated roles in mESCs (Wu and Zhang, 2011). While Tet1 and Tet2 double knockout results in a depletion of 5hmC and dysregulation of hundreds of genes, yet the mESCs remain pluripotent (Dawlaty et al., 2013). To understand the role of calpain-mediated TET cleavage in mESCs, we focused on some known functions of TET proteins. Because calpains functionally antagonize TET proteins, we anticipate that depletion of calpains and TET proteins result in opposite phenotypes. Similar to Tet1/2 double knockout, calpain1−/− or calpain2−/− mESCs exhibit typical mESC morphology (Fig. S2E) and no obvious defect in self-renewal was observed. Consistently, the levels of the key pluripotency factors, including Oct4, Sox2 and Nanog, are not significantly altered by calpain knockout (Fig. 3A). Consistent with the report that 5hmC generation depends on TET1 and TET2 (Dawlaty et al., 2013), dot blot analysis revealed a two-fold increase in 5hmC levels in calpain1−/− mESCs, while calpain2 −/− had little effect (Fig. 3B, C). This result is consistent with the fact that calpain1, but not calpain2, is expressed in mESCs and regulates TET1/2 protein levels (Fig. 2F). Although not affecting pluripotency, knockdown of Tet in mESCs does affect the expression of lineage-specific transcription factors. For example, trophectoderm marker Cdx2 and Eomes are significantly up-regulated in Tet1 knockdown cells, while expression of other markers such as Lefty1 is decreased (Ito et al., 2010; Koh et al., 2011). We confirmed this observation and importantly obtained an opposite effect in calpain1−/− mESCs presumably due to the stabilization of TET1 proteins (Fig. 3D). To rule out the possibility that the gene expression change in calpain1−/− cells is caused by other calpain1 substrates, we knocked down Tet1 in calpain1−/− mESCs, and the expression profiles of these genes were reversed (Fig. 3D and Fig. S3D). These data suggest that although calpain1 knockout does not affect mESC maintenance, it affects 5hmC generation and lineage-specific gene expression in a way opposite to Tet1 knockdown, consistent with a role of calpain1 in regulating TET1 and TET2 stability.
Figure 3. Effects of calpain-mediated TET cleavage on gene expression and NPC differentiation.
(A) RT-qPCR analysis demonstrates that calpain1 or calpain2 knockout in mESCs does not affect pluripotent gene expression. Data represent the mean of three independent experiments, and value in WT mESC is set as 1.
(B, C) Dot blot analysis (B) and densitometry quantification of three repeats (C) demonstrate that calpain1 knockout, but not calpain2, increased 5hmC levels in mESCs.
(D) RT-qPCR analysis demonstrates that Tet1 knockdown in mESC enhances trophectoderm lineage genes (Cdx2 and Eomes) expression, and inhibits Lefty1. Knockout of calpain1 opposes this tendency, which is rescued by Tet1 knockdown. Data represent the mean of three independent experiments, and value in WT mESC is set as 1.
(E) RT-qPCR analysis demonstrates that during differentiation to NPC (EB day 8), Tet3 knockout significantly reduces the expression of neuronal markers Ngn2 and Pax6, while calpain2 knockout enhanced their expression, which is reversed by Tet3 knockdown. In contrast, β3-tubulin expression is not affected by either Tet3 or calpain2. Value in WT EB is set as 1.
(F) Immunostaining demonstrates generation of Nestin and Sox2 double positive NPCs. After EB disassociation and a 48h adherent culture, Nestin and Sox2 positive NPC were successfully generated from all WT and knockout cells.
(G) TET3 and calpain2 have opposite effect on mESC differentiation to NPC. While Tet3−/− significantly reduced NPC generation, CAPN2−/− enhanced the differentiation efficiency, which is abolished by Tet3 knockdown. Number from WT cells is normalized to 1. P<0.05 (*), P<0.01 (**).
Since calpain2 regulates TET3 levels in EB differentiation (Fig. 2G), we next analyzed the biological relevance of this enzyme-substrate pair during mESC differentiation. TET3 plays an important role in regulating expression of some neural transcription factors such as Pax6 and Ngn2 during neurogenesis in Xenopus (Xu et al., 2012). To test if this mechanism is conserved in mammals, we generated Tet3−/− mESC with a published CRISPR guiding sequence (Fig. S3A) (Wang et al., 2013). Clones carrying frame shifts on both alleles were selected. Consistent with previous report (Wang et al., 2013), Tet3 knockout does not affect mESC morphology or self-renewal (Fig. S3B). EB-based differentiation followed by RT-qPCR analysis demonstrated that the expression levels of Pax6 and Ngn2 were significantly reduced in Tet3 knockout mESCs (Fig. 3E), suggesting a functional conservation of Tet3 between Xenopus and mammals. Importantly, both Pax6 and Ngn2 are up-regulated in calpain2−/− EBs, and shRNA-mediated Tet3 knockdown in calpain2−/− cells abolished this up-regulation (Fig. 3E and Fig. S3E). However, manipulation of calpain2 or Tet3 does not affect the expression of other neuronal marker genes, such as β3-tubulin (Fig. 3E). This suggests that although calpain2 and TET3 affect the expression of certain neural genes, they are not master regulators that drive differentiation from mESCs to NPCs.
EBs are composed of a mixed cell population that includes non-neural lineage cells. To study the effect of calpain2 and Tet3 on differentiation efficiency from mESCs to NPCs, we disassociated EBs and switched to monolayer culture in chemically defined N2 medium, which enrich NPCs by eliminating none NPCs and intermediates. The surviving cells showed typical bipolar NPC morphology, and were positive for Nestin and Sox2 (Fig. 3F). Although NPCs were successfully generated from all four groups of cells, the yield differs significantly (Fig. 3G). The increased NPC differentiation efficiency in calpain2−/− mESCs is likely due to the increase in TET3 levels as Tet3 knockdown in calpain2−/− cells suppressed NPC generation (Fig. 3G). This result suggests that calpain2-mediated degradation of TET3 modulate neuronal gene expression program and the efficiency of in vitro neural differentiation. Up-regulation of calpain2 during NPC differentiation may be part of a negative feedback mechanism that prevents hyper-activation of Tet3.
DISCUSSION
In this study, we provide evidence that TET proteins are direct substrates of calpains. Specifically, calpain1 modulates TET1 and TET2 levels in mESCs, while calpain2 promotes TET3 turnover during neural differentiation. Calpain-mediated regulation of TET proteins is physiologically relevant, as it affects global 5hmC level and expression of certain lineage-specific genes in mESCs, as well as mESC differentiation.
Cell differentiation is a highly orchestrated process with dynamic proteomic changes as unwanted proteins are degraded. The importance of major proteolytic systems including proteasome, caspase, calpain and lysosome has been implicated in cell differentiation (Buckley et al., 2012; Fujita et al., 2008; Guan et al., 2013). Utilizing inhibitors against these proteolytic systems, we identified calpains as important regulators of TET protein turnover (Fig. 1D–I). We also observed a modest effect by inhibiting caspase (Fig. 1D, E), which is consistent with a recent report (Ko et al., 2013). In fact, calpain and caspase are proteases that share many properties and substrates (Wang, 2000). While we focused on calpains in this study, the relative contribution of calpain and caspase in regulating TET protein turnover remains to be determined. It worth noting that while we observed an effect of calpains and caspases on TET turnover, no obvious effect was detected by inhibiting proteasomes, indicating that the uniquitylation pathway does not play a major role in regulating TET protein turnover.
It is well-known that calpain-mediated cleavage can either result in protein turnover or generate functional truncated proteins. Calpain-mediated TET cleavage likely results in turnover because TET degradation products observed in vitro (Fig. S2A) were undetectable in mESCs or 293T cells co-transfected with TET2 and calpains, suggesting that the cleaved TET fragments are unstable and are quickly turned over in vivo. Moreover, the wide spectrum of TET degradation products suggests many cleavage sites, making it difficult to generate mutant TET proteins that are resistant to calpains, which would otherwise be useful tools in functional studies. However, the fact that knocking down Tet in calpain−/− cells can rescue the calpain−/− phenotypes strongly supports the biological relevance of this enzyme-substrate pair (Fig. 3D, E, G). In this study, we have tested only two of the bested characterized calpains, and the role of the other 12 calpains in regulating TET stability remains unknown.
Tet protein levels are consistent with their mRNA levels, suggesting a dominant regulation at the transcriptional level (Fig. 1A, B), yet post-translational mechanism may be required to fine-tune TET protein level and function. Considering the large numbers of calpain substrates, and the difficulty in generating calpain-resistant TET mutants, we choose to study the function of calpain-mediated TET degradation by focusing on some known TET functions, such as 5hmC generation and expression of some lineage-specific genes. The opposite effects from depletion of Tet and calpain, and the observation that Tet knockdown reverses the phenotypes of calpain knockout (Fig. 3 B–G) strongly support a role of calpain-mediated TET protein degradation. Given that calpains are calcium-dependent proteases, studying calpain-Tet in physiological contexts such as neuron activation is of great relevance. In addition, cancer cells may prove to be another useful model in understanding calpain-mediated TET degradation as calpain levels are frequently elevated while TET are down-regulated in cancer cells (Storr et al., 2011; Yang et al., 2012). Our findings provide mechanistic basis for these future studies.
EXPERIMENTAL PROCEDURES
Differentiation of Neural Progenitors
Experiment was performed as described (Bibel et al., 2007). 4 × 106 ESCs were plated into non-adherant dish in differentiation medium (ES medium with 10% FBS and no LIF) to form embryoid body. On day 4, 5µM retinoic acid was applied. On day8, embryoid bodies were disassociated and cultured in N2 medium in PORN/laminin-coated plates.
Knockout Calpains by CRISPR
Design of targeting constructs was described in (Hsu et al., 2013). To knockout calpains, CRISPR constructs were co-transfected with a puromycin resistant vector. After puromycin selection, single clones were picked, and the genotypes were determined by sequencing. Clones with frame shifts on both alleles were selected for further analysis.
In vitro calpain assay
Proteins were exogenously expressed and purified from 293T cells. TET proteins were incubated with calpain1, calpain2 or elution buffer as control. 1mM CaCl2 was added and the reaction was performed at room temperature for 30 minutes before being stopped by adding laemmli buffer.
More details are available at supplemental experiment procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Feng Zhang for the px330 vector, Li Shen and Hao Wu for Tet1 and Tet3 shRNA lentiviral vectors; Hao Wu and Damian Sendler for critical reading of the manuscript. This study is supported by GM68804 and U01DK089565 from NIH. YZ is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interests.
REFERENCES
- Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nature protocols. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell stem cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. The Biochemical journal. 1997;326(1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czogalla A, Sikorski AF. Spectrin and calpain: a 'target' and a 'sniper' in the pathology of neuronal cells. Cell Mol Life Sci. 2005;62:1913–1924. doi: 10.1007/s00018-005-5097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Developmental cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita J, Crane AM, Souza MK, Dejosez M, Kyba M, Flavell RA, Thomson JA, Zwaka TP. Caspase activity mediates the differentiation of embryonic stem cells. Cell stem cell. 2008;2:595–601. doi: 10.1016/j.stem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiological reviews. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Guan JL, Simon AK, Prescott M, Menendez JA, Liu F, Wang F, Wang C, Wolvetang E, Vazquez-Martin A, Zhang J. Autophagy in stem cells. Autophagy. 2013;9:830–849. doi: 10.4161/auto.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013 doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science (New York, NY. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- Pan T, Kondo S, Le W, Jankovic J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson's disease. Brain. 2008;131:1969–1978. doi: 10.1093/brain/awm318. [DOI] [PubMed] [Google Scholar]
- Santos DM, Xavier JM, Morgado AL, Sola S, Rodrigues CM. Distinct regulatory functions of calpain 1 and 2 during neural stem cell self-renewal and differentiation. PloS one. 2012;7:e33468. doi: 10.1371/journal.pone.0033468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, et al. The Oncogenic MicroRNA miR-22 Targets the TET2 Tumor Suppressor to Promote Hematopoietic Stem Cell Self-Renewal and Transformation. Cell stem cell. 2013a;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al. MicroRNA-Antagonism Regulates Breast Cancer Stemness and Metastasis via TET-Family-Dependent Chromatin Remodeling. Cell. 2013b;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nature reviews. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53(Suppl 1):S12–S18. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KK. Calpain and caspase: can you tell the difference? Trends in neurosciences. 2000;23:20–26. doi: 10.1016/s0166-2236(99)01536-2. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima Y, Kawashima S. Calpain function in the differentiation of mesenchymal stem cells. Biological chemistry. 2002;383:757–764. doi: 10.1515/BC.2002.079. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2012;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatz M, Starling A. Calpains and disease. The New England journal of medicine. 2005;352:2413–2423. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.