Summary
The abundant, liver-specific, microRNA miR-122 forms extensive base-pairing interactions with the 5’ noncoding region of the hepatitis C virus (HCV) RNA genome, protecting the viral RNA from degradation. We discovered that the 5’-3’ exoribonuclease Xrn2, which plays a crucial role in the transcription termination of RNA polymerase II, modulates HCV RNA abundance in the cytoplasm but is counteracted by miR-122-mediated protection. Specifically, Xrn2 depletion results in increased accumulation of viral RNA, while Xrn2 overexpression diminishes viral RNA abundance. Depletion of Xrn2 did not alter translation or replication rates of HCV RNA, but affected viral RNA stability. Importantly, during sequestration of miR-122, Xrn2 depletion restored HCV RNA abundance, arguing that Xrn2 depletion eliminates the miR-122 requirement for viral RNA stability. Thus, Xrn2 has a cytoplasmic, antiviral function against HCV that is counteracted by HCV subversion of miR-122 to form a protective oligomeric complex at the 5’ end of the viral genome.
Keywords: HCV, miR-122, Xrn2, antiviral
Introduction
Hepatitis C Virus (HCV) is a hepatotropic, positive-sense, enveloped RNA virus that belongs to the Flaviviridae. HCV can establish persistent infections that lead to chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (Hoofnagle, 2002). The HCV 9.6 kb RNA genome contains conserved 5′ and 3′ noncoding regions (NCRs) that are important for viral replication and translation. An internal ribosome entry site (IRES) directs the synthesis of a large, 3010-amino acid viral polyprotein, which is co- and post-translationally cleaved by host and viral proteases to yield mature structural and nonstructural proteins (Bartenschlager et al., 2011).
HCV infects an estimated 170 million people worldwide (Lavanchy, 2011), making it a serious global health problem. Advances in anti-HCV pharmacology include the recently FDA-approved NS5B inhibitor Sofosbuvir (Gilead Sciences, Foster City, CA) and NS3 protease inhibitors Simeprevir (Janssen Pharm., Titusville, NJ), Telaprevir (Vertex Pharm., Cambridge, MA) and Boceprevir (Merck, Whitehouse Station, NJ). Recently, in phase II clinical trials by Santaris Pharma (Copenhagen, Denmark), treatment with Miravirsen, a modified oligonucleotide to sequester liver-specific microRNA miR-122, resulted in 1000-fold lower HCV titers in HCV patients (Janssen et al., 2013). Thus, miR-122 is an exciting, host cell target to combat HCV. Mechanistically, two miR-122 molecules form an oligomeric complex at the 5’ proximal end of the HCV RNA genome, thereby enhancing viral RNA abundance by stabilizing the viral genome (Jopling et al., 2008; Jopling et al., 2005; Li et al., 2013; Machlin et al., 2011; Shimakami et al., 2012). Here, we show that miR-122 protects the HCV RNA genome from degradation by the 5’-3’ exoribonuclease Xrn2, whose role was thought to be limited to the transcription termination of RNA polymerase II (Pol II) in the nucleus (Kim et al., 2004; West et al., 2004). This finding argues for an unprecedented cytoplasmic antiviral function for Xrn2.
Results
Subcellular localization and steady-state abundance of Xrn2 during HCV infection
The 5’-3’ exoribonuclease Xrn2 executes known nuclear functions in RNA metabolism, including Pol II transcription termination (Kim et al., 2004; West et al., 2004). However, it was recently reported that Xrn2 degrades mature miRNAs in C. elegans (Chatterjee and Grosshans, 2009). To examine whether Xrn2 affects the turnover of cytoplasmic, mature miR-122 in human cells, we first monitored the subcellular localization of Xrn2 in uninfected and HCV genotype 2a (JFH1)-infected Huh7 cells. Nuclear and cytoplasmic extracts were prepared and the distribution of Xrn2, nuclear lamin A/C and cytoplasmic GAPDH were determined by Western blot analysis. Figure S1A shows that Xrn2 is found abundantly in the nucleus, but a fraction of this protein also resides in the cytoplasm. No changes in the subcellular distribution of Xrn2 during HCV infection were observed. To examine more thoroughly the steady-state abundance of Xrn2 during HCV infection, Xrn2 abundance was monitored at different times after infection. Western blot analysis showed that Xrn2 slightly increased during infection, concomitantly with the expression of the HCV NS5A protein (Fig. S1B). This finding suggested the possibility that Xrn2 could either be facilitating or restricting the HCV replicative cycle.
Effects of Xrn2 depletion on HCV protein and RNA abundances
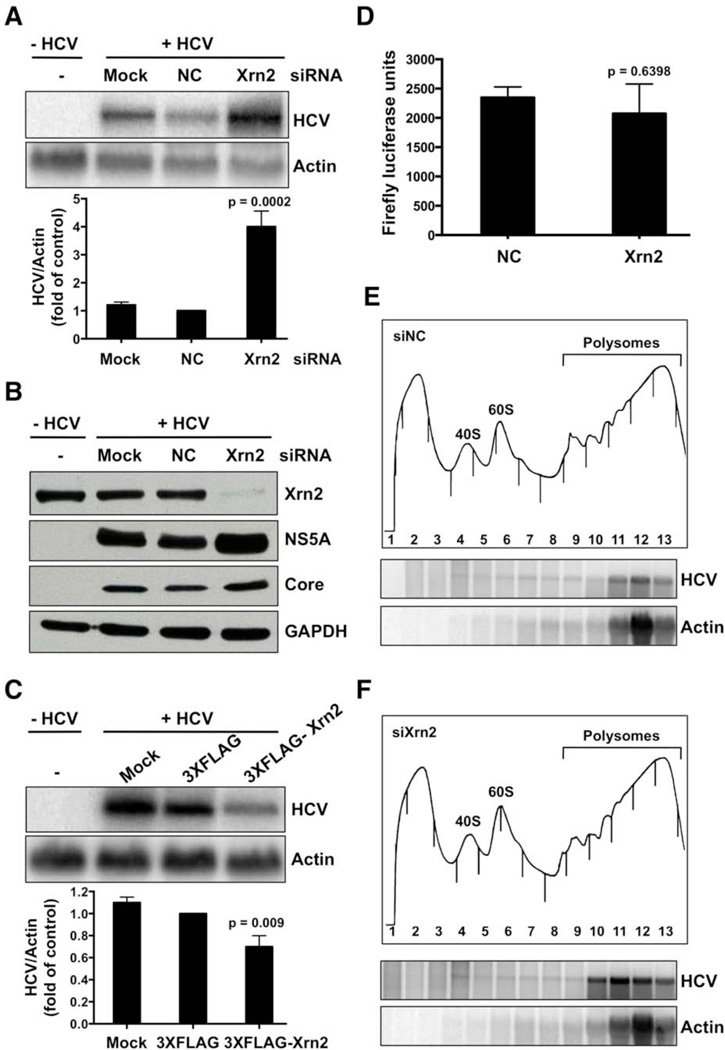
To determine whether Xrn2 plays a role in the HCV life cycle, Xrn2 was depleted by siRNA-mediated gene knockdown. Depletion of Xrn2 increased HCV RNA, and HCV NS5A and core protein abundances (Figures 1A,B). Similar effects on viral RNA and protein abundances were also observed in HCV genotype 1b (5B) replicon cells when Xrn2 was depleted (Figures S1C and D). These findings suggest that enhancement of HCV RNA and protein abundances during Xrn2 depletion is not confined to the HCV genotype 2. To exclude off -target effects of the Xrn2 siRNA, additional siRNAs that target different regions of the Xrn2 ORF were tested. These siRNAs also enhanced the abundance of HCV RNA and proteins (Figures S1E and F). In contrast, overexpression of flagged Xrn2 resulted in a 40% decrease in HCV RNA abundance (Figure 1C), arguing that Xrn2 is antiviral.
Figure 1. Effects of Xrn2 depletion on HCV RNA and protein abundance.
(A) Effects on RNA abundance. Cells were mock-transfected (Mock), or transfected with control (NC) or Xrn2 siRNAs. One day later, cells were infected with HCV as indicated. RNA abundances were measured by Northern blot analysis three days after infection. The Northern blot is representative of at least three independent replicates (top). Quantitation of HCV RNA abundance (bottom). Error bars represent standard error of the mean. P value was determined by Student’s t-test. (B) siRNA-mediated depletion of Xrn2. Abundances of HCV NS5A and core proteins, and GAPDH were examined. Immunoblot shown is representative of at least three independent replicates. (C) Overexpression of Xrn2. 3XFLAG-Xrn2 or 3XFLAG plasmids were transfected one day prior to infection with HCV. HCV and actin RNA abundances were measured by Northern blot analysis three days after infection. The Northern blot shown represents at least three independent replicates (top). Quantitation of HCV RNA abundance (bottom). Error bars represent standard error of the mean. P value was determined by Student’s t-test. (D) Effects of Xrn2 depletion on HCV IRES-mediated translation, measured by luciferase assay as described in the text. The data shown represent at least three independent replicates. Error bars represent standard error of the mean. P-value was determined by Student’s t-test. (E and F) Polysomal profile from Xrn2-depleted cells (top). siRNAs were transfected one day prior to infection. Three days after infection, cell lysates from HCV-infected, siXrn2-treated or siNC-treated samples were separated in 10–60% sucrose gradients. 40S and 60S ribosomal subunits and polysomes are indicated in the A260nm profile. Northern blot analyses of the sucrose gradient fractions (bottom). The Northern blots shown represent at least three independent replicates.
We next assessed whether Xrn2 depletion affected the expression of Xrn1, the other member of the 5’-3’ exoribonuclease family. Western blot analysis of Xrn1 abundance showed that Xrn2 depletion had no effect on Xrn1 protein abundance (Fig. S1G). Importantly, Figures S1H and I show that depletion and overexpression of Xrn2 have no effect on cell viability or apoptosis. These results identify a role for cytoplasmic Xrn2 in the regulation of HCV RNA and protein abundances.
Effects of Xrn2 depletion on HCV IRES-mediated translation
To identify effects of Xrn2 on HCV IRES-mediated translation, translation of dicistronic mRNAs, containing the HCV IRES in the intergenic region, was measured. Specifically, cells were depleted of Xrn2 by siRNAs on day one, transfected with dicistronic plasmids the next day, and first cistron-encoding Renilla luciferase and second cistron-encoding firefly luciferase activities were measured 24 hours later. Figure 1D shows that HCV IRES-mediated translation was not affected during Xrn2 knockdown, indicating that Xrn2 does not modulate HCV IRES activity.
To examine further the apparent lack of effect of Xrn2 on HCV IRES-mediated translation, polysomal mRNAs were analyzed from cell lysates obtained from Xrn2-depleted or control siRNA HCV-infected cells. Sucrose gradient analysis showed that Xrn2 depletion had little effect on polysomal profile traces (Fig. 1E and F), arguing that ribosome density on cellular mRNAs was not affected during Xrn2 depletion. However, an increase in abundance of HCV RNA that was associated with polysomes was observed in Xrn2-depleted extracts (Fig. 1F). To confirm that RNAs associated with polysomal fractions were indeed translated, lysates were treated with 2mM puromycin, an elongation inhibitor that releases ribosomes from mRNAs after high salt treatment (Figure S1J). Northern blot analysis of sucrose gradient fractions showed that actin and viral RNAs from both control siRNA and Xrn2-depleted samples had decreased sedimentation upon puromycin treatments (Fig. S1J). Therefore, we conclude that Xrn2 depletion caused the accumulation of translatable HCV RNA, thus increasing viral protein accumulation. These results point to a role for Xrn2 in decreasing HCV RNA replication or stability.
Effects of Xrn2 depletion on rates of HCV RNA replication
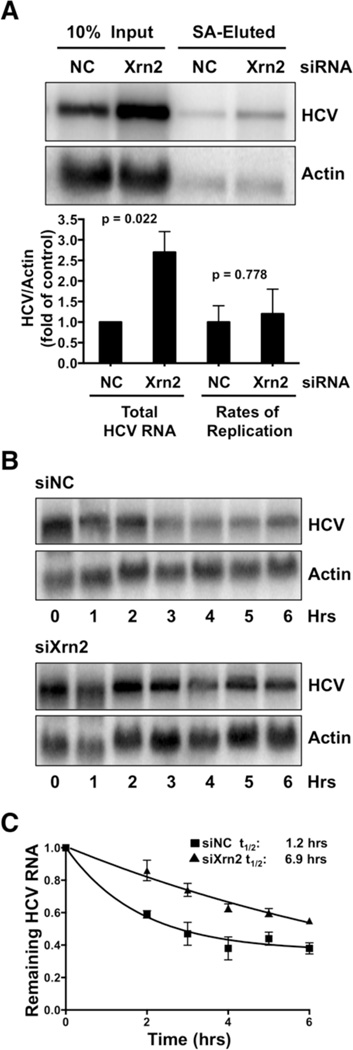
To examine whether Xrn2 modulated the rate of HCV RNA synthesis, rates of viral and host cell RNA replication were measured in the presence or during depletion of Xrn2, using a modified protocol initially described by Cleary and colleagues (Cleary et al., 2005). Briefly, control or siRNAs against Xrn2 were transfected into cells one day prior to HCV infection. Infected cells were subsequently incubated with 4-thiouridine (4SU) three days after infection. The modified nucleoside is converted by host enzymes to 4SU-triphosphate, which is incorporated into newly synthesized RNA. After extraction, 4SU-labeled RNA was biotinylated and isolated using streptavidin beads (Norman and Sarnow, 2010). Total and streptavidin-eluted RNAs were then visualized in Northern blots. Figure 2A reveals, as expected, a significant increase in total HCV RNA in Xrn2-depleted samples. Inspection of streptavidin-eluted RNAs also showed a significant increase in levels of newly synthesized HCV RNA in Xrn2-depleted samples. Next, the amount of newly synthesized RNA was divided by the amount of total RNA. No significant change in viral RNA replication rates (Fig. 2A) during efficient depletion of Xrn2 was observed. This finding argues that Xrn2 depletion enhances the abundance of HCV RNA without altering the rates of replication.
Figure 2. Effects of Xrn2 Depletion on HCV RNA synthesis and decay.
(A) Effects of Xrn2 depletion on rates of HCV RNA synthesis. siRNAs were transfected one day prior to infection. Abundances of total and newly synthesized HCV and actin RNAs were examined by Northern blot analysis three days after infection (top) (Norman and Sarnow, 2010). The Northern blot shown is representative of at least three independent replicates. Replication rates of HCV RNA correspond to the ratio of newly synthesized HCV RNA to input RNA. Error bars represent standard error of the mean. P-values were determined by Student’s t-test. (B) Effects of Xrn2 depletion on HCV RNA decay. Control (siNC) or Xrn2 (siXrn2) siRNAs were transfected one day prior to infection. Three days after infection cells were treated with 25 µM of the MK-0608 nucleoside analog of HCV NS5B, and RNAs were extracted at indicated times. HCV RNA levels were measured by Northern blot analysis. Northern blots shown represent at least three independent experiments. (C) One-phase decay graph of HCV RNA. Data from cells harvested at time 0 were set to 1. Data shown represent remaining HCV RNA following addition of MK-0608. Error bars represent standard error of the mean. Estimated half-life (t1/2) of HCV RNA under these conditions is indicated; ± 95% confidence intervals (CI) are 0.54–2.24 and 5.82–8.42 in hours for NC and Xrn2 respectively (see Fig. S7). The data shown represent the means of at least three independent replicates.
Effects of Xrn2 depletion on HCV RNA stability
To determine whether Xrn2 affects the decay rate of HCV RNA, cells were depleted of Xrn2 by siRNAs and subsequently infected with HCV. Three days after infection, the nucleoside analog MK-0608 (Migliaccio et al., 2003) was added to block further viral RNA synthesis. RNA was extracted at different times after MK-0608 addition and HCV RNA abundance was monitored by Northern blot analysis. Viral RNAs from Xrn2-depleted samples were degraded much more slowly than those from control samples (Figure 2B). Specifically, HCV RNA was five times more stable in the absence of Xrn2, displaying a half-life of 6.9 hours as compared to 1.2 hours for siRNA control samples (Fig. 2C; Fig. S2). These observations indicate that Xrn2 decreases the stability of the viral RNA genome, likely by its exoribonuclease activity.
HCV RNA – Xrn2 interaction and HCV RNA protection by miR-122
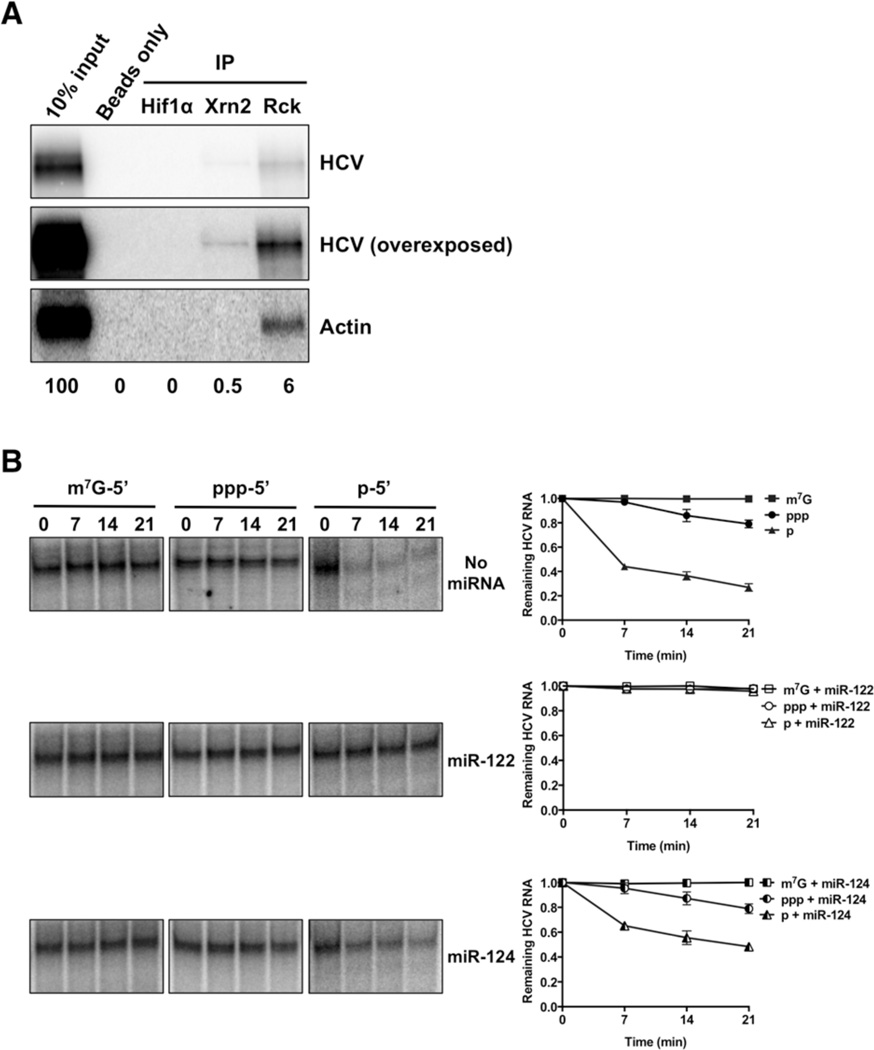
Next, we examined whether Xrn2 associates with the viral RNA genome in JFH1-infected Huh7 cells. RNA co-immunoprecipitation experiments were performed using antibodies that recognize the C-terminus of Xrn2; Hif1α and RCK antibodies were used as negative and positive controls, respectively. Northern blot analysis of immunoprecipitation reactions showed that Xrn2 associates reproducibly with a small fraction of viral RNA (Fig. 3A).
Figure 3. HCV RNA – Xrn2 interaction and protection by miR-122.
A) Xrn2 association with HCV RNA. Representative Northern blot of co-immunoprecipitation reactions performed with antibodies against Hif1-α, Xrn2, and Rck. Lane 1 corresponds to 10% of input RNA. (B) Protection of viral RNA by miR-122 from Xrn2-mediated degradation. 3’-32P-labelled capped- (m7G-5’), monophosphorylated- (p-5’), and triphosphorylated- (ppp-5’) HCV reporter RNAs were incubated with recombinant human Xrn2 in the absence (top) or presence of miR-122 (middle) or miR-124 (bottom) for the indicated periods of time, and subsequently analyzed by denaturing gel electrophoresis. Quantitation of radiolabeled RNA is on the right. Results from at least three independent replicates are shown. Error bars represent standard error of the mean. Values were normalized to the 0-minute no-Xrn2 control.
To determine whether HCV RNA is susceptible to the exoribonuclease activity of Xrn2, we tested the activity of recombinant Xrn2 on various substrate mRNAs containing different 5’ end moieties. The known targets for Xrn2 contain 5’ monophosphates ((Henry et al., 1994; Kim et al., 2004; Morlando et al., 2004; Qu et al., 1999; Stevens and Poole, 1995; West et al., 2004). Thus, the 5’ triphosphate moiety of the 5’ end of HCV (Li et al., 2013) should be a poor substrate for Xrn2. However, the original characterization of Xrn2 by Stevens reported that the protein degrades 5’ triphosphate-containing RNA templates as well (Stevens and Maupin, 1987). When the nuclease activity of recombinant Xrn2 on 5’ monophosphorylated-, 5’ triphosphorylated-, and 5’ capped-HCV domain I-containing RNAs was tested in in-vitro assays (Figure 3B, top), a preference for 5’ monophosphorylated RNA was observed. No nuclease activity was observed for capped RNAs, and modest degradation of 5’ triphosphorylated RNA (Fig. 3B, top). This finding argues that full-length HCV RNA may be a direct target for Xrn2.
To assess whether miR-122 functions in protecting the 5’-end of viral RNA from Xrn2 degradation, the susceptibility of modified HCV domain I-containing RNAs to Xrn2 attack was tested in the presence of miR-122 or miR-124. When HCV RNAs were incubated with miR-122, 5’ monophosphorylated-, 5’ triphosphorylated-, and capped-HCV RNAs were protected from degradation by Xrn2 (Fig. 3B middle). This finding shows that the recombinant Xrn2 protein does not contain endonuclease activity. In contrast, the unrelated miR-124 failed to provide protection of 5’ monophosphorylated or 5’ triphosphorylated HCV RNA domain I RNAs from Xrn2-mediated degradation (Fig. 3B bottom). These results provide strong evidence that Xrn2 associates with HCV RNA and that miR-122 protects the 5’-end of the viral genome from degradation by Xrn2.
Enhancement of HCV RNA abundance during sequestration of miR-122 and concomitant depletion of Xrn2
MicroRNA-122, which has a 5’ monophosphate, is known to stabilize HCV RNA. To determine whether Xrn2 degrades miR-122, Xrn2 was depleted by siRNA-mediated gene knockdown. Figure S3A shows that Xrn2 depletion had no effect on miR-122 abundance in both uninfected and HCV-infected cells. These results show that, unlike mature microRNAs in C. elegans (Chatterjee et al., 2009), human miR-122 is not degraded by Xrn2.
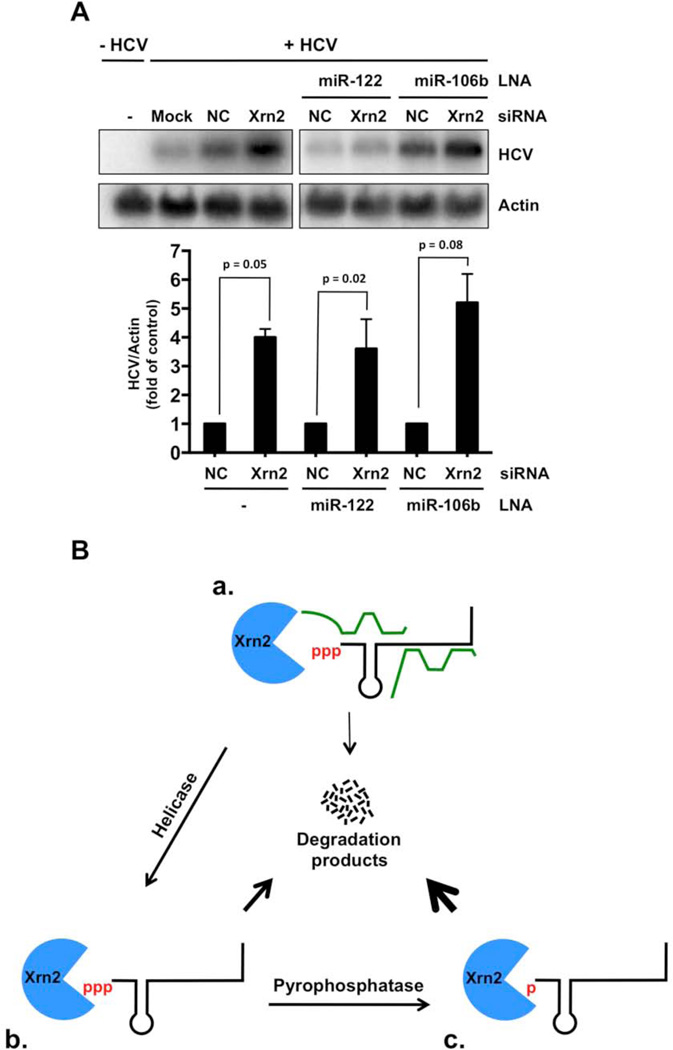
Next, we investigated whether Xrn2 depletion eliminates the miR-122 requirement for HCV RNA abundance. Briefly, control or Xrn2 siRNAs were transfected into Huh7 cells, which were infected with HCV one day later. Modified locked nucleic acids (LNAs) with base complementarity to miR-122 or control miR-106b (Elmen et al., 2008; Machlin et al., 2012) were transfected one day after infection with HCV. RNA samples were prepared and analyzed in Northern blots (Figure 4A). As expected, samples from cells that did not receive miR-122 LNAs, or that received control miR-106 LNAs, showed a four-fold increase in HCV RNA abundance when Xrn2 was depleted (Fig. 4A). Sequestration of miR-122 by LNAs resulted in a loss of HCV RNA. However, siRNA-mediated Xrn2 depletion restored HCV RNA abundance by four-fold even when miR-122 was sequestered (Fig. 4A; Fig. S3B and C). One would expect that HCV RNA abundance in siXRN2/miR-122LNA and siNC/106LNA treated cells should be the same if miR-122 and Xrn2 act together. The siXRN2/miR-122 LNA ratio is approximately 0.75× in Figs. 4A and S3B, but is 1× in Fig. S3C. This variability could be explained if not every LNA-treated, infected cell received the Xrn2 siRNA, or if Xrn2 affects HCV RNA abundance that is independent of miR-122. Finally, to confirm that the anti-miR-122 LNAs were functional, the effects of miR-122-specific and control LNAs were tested on luciferase reporter mRNAs that contain four miR-122 seed match sites in their 3’-noncoding regions, which are expected to destabilize the reporter mRNA in the presence of miR-122. Treatment with miR-122 LNAs resulted in a considerable increase in luciferase activity, while addition of miR-106 LNAs did not affect luciferase activity (Figure S3D). These findings further argue that miR-122 protects the 5’-end of the HCV RNA genome from direct Xrn2-mediated degradation.
Figure 4. HCV RNA abundance during sequestration of miR-122 and depletion of Xrn2.
(A) Indicated siRNAs were transfected one day prior to infection. 0.5 nM miR-122 or 0.5 nM miR-106b LNA were transfected one day post-infection, and total RNA was harvested three days after infection. HCV and γ-actin RNA abundance was measured by Northern blot analysis (top). The Northern blot shown represents three independent experiments. Quantitation of HCV RNA abundance (bottom). HCV RNA abundances were normalized to γ-actin levels. Data from cells transfected with control siRNAs were set to 1. Error bars represent standard error of the mean. P-values determined by Student’s t-test. (B) Model of protection of the 5’-end of the HCV genome by miR-122 from Xrn2-mediated 5’-3’ degradation. (a) In the presence of miR-122, Xrn2 produces few viral degradation products (thin arrow). (b) Loss of miR-122-mediated protection renders the 5’-triphosphate of viral genome susceptible to Xrn2 degradation. Helicases could remove miR-122 from the viral genome, resulting in larger amounts of degradation products (thick arrow). (c) Removal of the 5’ triphosphate moiety of HCV RNA by a pyrophosphohydrolase could generate 5’-monophosphates that would be degraded by Xrn2 (very thick arrow).
Discussion
Our recent model on the stabilization of the HCV RNA genome by miR-122 (Machlin et al., 2011) led us to investigate nuclease activities from which HCV RNA is being protected. Through a series of siRNA-mediated Xrn2 depletion experiments we found that miR-122 protected the 5’-end of HCV RNA from degradation by Xrn2. Depletion of Xrn2 caused an upregulation in HCV RNA and protein abundance, without altering viral RNA replication rates, but rather by increasing HCV RNA stability. This finding presented a conundrum, because a recent report showed that the 5’ end of HCV RNA contains a triphosphate moiety (Li et al., 2013), while Xrn2 is known to preferentially degrade 5’ monophosphorylated RNAs. However, cell-free stability assays demonstrated that RNAs containing 5’ triphosphate-bearing 5’ noncoding regions from HCV were susceptible to degradation by Xrn2. Importantly, addition of miR-122, but not miR-124, provided protection to 5’ monophosphorylated- and 5’ triphosphorylated HCV RNAs against Xrn2-mediated degradation. Ultimately, Xrn2 depletion was able to rescue HCV RNA abundance during miR-122 sequestration in infected cells, arguing that depletion of Xrn2 eliminates the miR-122 requirement for viral RNA stability.
The 5’-3’ exoribonuclease Xrn2 has important functions in nuclear RNA metabolism. Especially, it has crucial functions in Pol I (El Hage et al., 2008) and Pol II transcription termination (Kim et al., 2004; West et al., 2004). The known cellular functions of Xrn2 are primarily linked to its exoribonuclease activity in the nucleus, and a cytoplasmic function for this protein had not been previously described. There is precedent that nuclear proteins can have distinct functions in the cytoplasm. For example, the nuclear poly(A) binding protein PABP2 modulates poly(A) polymerase to polyadenylate mRNAs in the nucleus but modulates poly(A) deadenylase to shorten 3’ poly(A) sequences in the cytoplasm (Benoit et al., 2005). Thus, the different functions of PABP2 are modulated by different interacting proteins in these compartments (Benoit et al., 2005), as may be the case for nuclear and cytoplasmic Xrn2.
In our current model, extensive base-pair interactions between miR-122 and the 5’-end of HCV RNA protect the integrity of the viral genome from Xrn2-mediated degradation (Fig. 4Ba). HCV RNA stability could be further regulated by RNA helicases that could remove miR-122 and unwind secondary structures to induce viral mRNA decay (Fig. 4Bb). Such RNA helicases could be virus-encoded proteases, such as NS3, or host factors. For example, a recent study predicted an interaction between Xrn2 and the RIG-I-like receptor, double-stranded RNA helicase LGP2 (Li et al., 2011), suggesting that LPG2 could be a co-factor of Xrn2. Because 5’-monophosphates are the preferred substrate for Xrn2, pyrophosphohydrolases may also play an important role in converting the 5’-triphosphate of HCV RNA into a 5’-monophopsphate for more efficient nucleolytic degradation by Xrn2 (Fig. 4Bc). For example, the pyrophosphohydrolase Rai1 stabilizes and enhances the exoribonuclease activity of yeast Xrn2, Rat1 (Xiang et al., 2009). However, the human ortholog of Rai1, Dom3Z, is highly divergent and does not appear to interact with Xrn2 (Xiang et al., 2009). Thus, exploring the presence and action of other pyrophosphohydralases in HCV infection would be important.
A previous study suggested that transfected type 1a HCV RNA is degraded by both Xrn1 and the exosome complex, whereas replicating RNA in infected cells is degraded primarily by Xrn1 (Li et al., 2013). However, miR-122 sequestration did not restore viral RNA abundance during Xrn1 depletion (Li et al., 2013). In contrast, Scheller et al. (Scheller et al., 2009) and Pager et al. (Pager et al., 2013) did not find that depletion of Xrn1 significantly affected HCV RNA abundance of type 1b and type 2 HCV genotype, respectively. It is thus possible that differences in cell type or viral genotype could account for these different observations.
Xrn2 mRNA abundance increased in cells infected with the negative-stranded respiratory syncytial virus (Ternette et al., 2011). We noted that treatment of cells with interferon α did not induce Xrn2 mRNA abundance (Fig. S4), indicating that Xrn2 is not an interferon-stimulated gene. Thus, the observed enhancement of Xrn2 steady-state protein abundance during HCV infection may reflect stabilization of Xrn2 protein. Evidence for this scenario comes from studies in C. elegans where Xrn2 protein is stabilized and activated by PAXT-1 (Miki et al., 2014). Curiously, the Xrn2-interacting domain in PAXT-1 also resides in human NKRF/NRF protein, which is involved in the transcriptional repression of certain NF-κB responsive genes and in the constitutive silencing of the interferon-β promoter (Miki et al., 2014). Finally, Xrn2 has been shown to be part of the type 1 interferon network (Li et al., 2011), which is consistent with Xrn2 being an antiviral effector. This antiviral effector is apparently so potent that HCV has evolved a mechanism to evade it by subversion of miR-122.
Experimental Procedures
Cell culture and plasmid transfections
Huh7 cells were maintained in DMEM and supplemented with 10% FBS, 1% nonessential amino acids and 200 µM L-glutamine. Huh7 cells were seeded in 60 mm tissue culture plates. The following day, Huh7 cells were transfected with 1 µg of 3XFLAG or 3XFLAG-Xrn2 plasmid using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocol. LNA oligonucleotides were delivered to cells at concentrations of 0.5, 2.5, and 25 nM using Dharmafect I reagent (Dharmacon) according to the manufacturer's protocol.
Small interfering RNA (siRNA) transfections
The siRNA sequence for Xrn2 #1 is as follows: siXrn2, 5’-GGAUGCAGCUGAUGAGAAAdTdT-3’. Oligonucleotides were resuspended in RNase-free water to a 50 µM final concentration. Sense and antisense strands were combined in annealing buffer (150 mM HEPES (pH 7.4), 500 mM potassium acetate, 10 mM magnesium acetate) to a final concentration of 20 µM, denatured for 1 min at 95 °C, and annealed for 1 h at 37 °C. Negative-control (NC) siRNA #3 had the following sequence: 5’-AUGUAUUGGCCUGUAUUAGUU-3’. Huh7 cells were seeded in 60 mm tissue culture plates. The following day, Huh7 cells were transfected with 2.5 nM of siRNA duplexes and Dharmafect I reagent (Dharmacon). Twenty-four hours post-transfection, cells were infected with JFH-1 virus as described, and harvested 3 days after infection. The efficiency of siRNA depletion was examined by Western blot analysis.
JFH-1 infections and lysate preparations
Huh7 cells were infected at 37°C with the JFH-1 infectious HCV virus (Wakita et al., 2005) at a multiplicity of infection (moi) of 0.01. Three days after infection cells were harvested. For Western blot analysis, cells were washed with phosphate-buffered saline (PBS) and pelleted at room temperature and lysed in RIPA buffer (100 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% sodium deoxycholic acid, 1% Triton X-100, and 0.1% SDS) containing Complete EDTA-free protease inhibitors (Roche) for 15 min on ice and pelleted for 15 min at 16,000 × g at 4 °C.
Subcellular protein fractionation and Western blot analysis
Cytoplasmic and nuclear fractions were obtained by NE-PER kit (Thermo Scientific) according to the manufacturer’s protocol. For Western blot analysis, 30 µg cell lysate was mixed with 5× protein sample buffer (200 mM Tris–HCl (pH 6.8), 50% glycerol, 25% SDS and β-mercaptoethanol), denatured for 5 min at 95 °C, and loaded onto a 8% polyacrylamide-SDS gel. After electrophoresis, proteins were transferred to a PVDF membrane (Millipore) for 1 h at 100 V at 4 °C. Membranes were blocked at room temperature for 1 h with 5% nonfat dry milk in TBS (50 mM Tris–HCl (pH 7.4), and 150 mM NaCl) with 0.1% Tween 20 (TBS/T), and then incubated overnight at 4 °C with primary antibodies. Membranes were washed 3× 10 min with TBS/T prior incubating with horseradish peroxidase-conjugated secondary antibodies diluted in 5% milk/TBS/T for 1 h at room temperature. After washing 3× 10 min with TBS/T, bound antibodies were labeled with Pierce ECL Western Blotting Substrate (Thermo Scientific) and detected after exposure to Biomax Light Film (Kodak).
The following primary antibodies were used for western blot analysis: anti-Lamin A/C (N-18) (1:1000; Santa Cruz Biotechnologies sc-6215), anti-Actin (1:1,000, Sigma A2066), anti-GAPDH (1:100,000, EMD Chemicals CB1001), anti-HCV core (C7–50) (1:1000; Abcam ab2740), anti-HCV NS5A (1:10,000), and anti-Xrn2 (1:10,000, Bethyl Laboratories A301-101A). Donkey anti-mouse-HRP (sc-2314) and donkey anti-rabbit-HRP (sc-2313) were purchased from Santa Cruz Biotechnologies and used at 1:10,000.
Northern blot analysis
To visualize specific mRNAs, 10 µg TRIzol-extracted RNA was resuspended in loading buffer (1% MOPS-EDTA-sodium acetate (MESA), 67% formaldehyde, and 50% formamide), denatured for 15 min at 65 °C and separated in 1% agarose/6.7% formaldehyde gels. Samples were separated in gels in MESA buffer, containing 18% formaldehyde, at 100 V. RNA was transferred and UV cross-linked to a Zeta probe membrane (BioRad). Detection of HCV and actin RNA (HCV (nucleotides 84–374) and γ-actin (nucleotides 685–1171) was performed using the ExpressHyb hybridization buffer (Clontech) and α-32P-dATP-RadPrime DNA labeled (Invitrogen) probes. Autoradiographs were quantitated using ImageQuant (GE Healthcare).
Sucrose gradient analysis
Cells were incubated with fresh medium 4 h prior to harvest. Cells were then treated with 100µg/ml cycloheximide for 3 min at 37 °C, followed by 2× washes with cold PBS containing 100 µg/ml cycloheximide. Lysates were prepared, and gradient separation and fractionation were performed as previously described (Wehner et al., 2010). Fractions were incubated with 0.5 mg/ml proteinase K (New England Biolaboratories) in the presence of 1% SDS, extracted and precipitated at −20 °C overnight. RNA was pelleted at 14,000 rpm for 15 min at 4 °C and resuspended in water.
RNA co-immunoprecipitation
Cells were lysed in polysome lysis buffer (300 mM NaCl, 15 mM Tris–HCl (pH 7.5), 15 mM MgCl, and 1% TritonX-100) with Ribonucleoside Vanadyl Complex (VRC NEB S1402S) and complete protease inhibitors. Lysates were incubated with antibodies for 1 hr at 4 °C with rotation. Protein A Dynabeads were added to lysate/antibody solution and incubated at 4 °C for 30 min. Beads were collected by using a magnetic rack (Dynal) and washed 3× with 3 volumes polysome lysis buffer. Beads were collected, RNAs were extracted by adding 1 ml of TRIzol to each reaction and analyzed by Northern blotting as previously described.
Xrn2 nuclease activity in-vitro assays
To generate 5’ triphosphate-containing RNAs, RNA substrates were prepared by labeling T7 IVT RNA oligonucleotides corresponding to 1–47 nucleotides of HCV genotype 1b (HCV domain I) with 32P-pCp and T4 RNA ligase (NEB) (20 µL of reaction; 1× T4 RNA ligase buffer (NEB), 1 mM ATP, 0.1 µg BSA, 10% (v/v) DMSO; 100 pmol RNA, 1 µM 32P-pCp (PerkinElmer) and 10 U T4 RNA ligase) overnight at 4 °C. Excess ATP and 32P-pCp was removed using a spin column (BioRad 732–6221, MicroBio Spin-6). To generate 5’ monophosphorylated RNAs, 3’-end labeled RNAs were incubated with the 5’ pyrophosphohydrolase enzyme (NEB RppH) for 1 hr at 37 °C according to manufacturer’s protocol. 3’-end labeled RNA was capped using the ScriptCap m7G Capping System (CELLSCRIPT C-SCCE0610) according to the manufacturer’s protocol. RNA was extracted with phenol/chloroform, re-extracted with chloroform, and precipitated in 100% ethanol with 1× volume of 5M ammonium acetate and 1 µl of 20 mg/ml glycogen (Roche). RNA was resuspended in water. In-vitro assays were carried out in Xrn2 buffer (20 mM HEPES (pH 7.5), 100 mM KCl, 4 mM MgCl2, 5% (v/v) glycerol, 1 mM DTT) with 0.766 µg of human Xrn2 (OriGene TP323980) and ~2 pmol RNA substrate in a volume of 10 µl at 30 °C for 20 min to 60 min. Reactions were quenched by addition of 3 µl RNA-loading dye (8M urea and 50mM EDTA pH 8.0), samples were heated to 95°C for 5 min. miR-122 or miR-124 (1 µM final) were incubated with labeled RNAs for 10min at 65°C, then placed on ice for 10min. Reactions were then incubated at 30°C for 20min before addition rXrn2. Reaction products were then resolved on a 10% polyacrylamide/urea gel. Autoradiographs were quantitated using ImageQuant (GE Healthcare).
Supplementary Material
Highlights.
Cytoplasmic function for nuclear Xrn2 in the hepatitis C virus (HCV) infectious cycle
HCV RNA abundance increases upon Xrn2 depletion
Xrn2 destabilizes the viral RNA genome
MicroRNA miR-122 protects the 5’ end of HCV RNA from degradation by Xrn2
Acknowledgements
We thank Gabriele Fuchs for many discussions and the gift of the domain I-HCV plasmid, and Tzu-Chun Chen for her critical input. We greatly appreciate Karla Kirkegaard’s many helpful comments. Thanks to James Manley for the 3XFLAG-Xrn2 expression plasmid and to Tim Tellinghuisen for the NS5A antibodies. This study was supported by grants from the National Institutes of Health (AI47365, AI069000). C.D.S. acknowledges support from the Stanford Genome Training Program (SGTP; NIH/NHGRI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartenschlager R, Penin F, Lohmann V, Andre P. Assembly of infectious hepatitis C virus particles. Trends in microbiology. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Developmental cell. 2005;9:511–522. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5' exonuclease Rat1 in yeast. Genes & development. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5' end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. The EMBO journal. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. The New England journal of medicine. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell host & microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Lavanchy D. Evolving epidemiology of hepatitis C virus. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. Competing and noncompeting activities of miR-122 and the 5' exonuclease Xrn1 in regulation of hepatitis C virus replication. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. Combating hepatitis C virus by targeting microRNA-122 using locked nucleic acids. Current gene therapy. 2012;12:301–306. doi: 10.2174/156652312802083558. [DOI] [PubMed] [Google Scholar]
- Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, et al. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. The Journal of biological chemistry. 2003;278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- Miki TS, Grosshans H. The multifunctional RNase XRN2. Biochemical Society transactions. 2013;41:825–830. doi: 10.1042/BST20130001. [DOI] [PubMed] [Google Scholar]
- Miki TS, Richter H, Ruegger S, Grosshans H. PAXT-1 promotes XRN2 activity by stabilizing it through a conserved domain. Molecular cell. 2014;53:351–360. doi: 10.1016/j.molcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B, Bozzoni I. Coupling between snoRNP assembly and 3' processing controls box C/D snoRNA biosynthesis in yeast. The EMBO journal. 2004;23:2392–2401. doi: 10.1038/sj.emboj.7600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager CT, Schutz S, Abraham TM, Luo G, Sarnow P. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology. 2013;435:472–484. doi: 10.1016/j.virol.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LH, Henras A, Lu YJ, Zhou H, Zhou WX, Zhu YQ, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie JP. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Molecular and cellular biology. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Diez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A, Maupin MK. A 5'----3' exoribonuclease of human placental nuclei: purification and substrate specificity. Nucleic acids research. 1987;15:695–708. doi: 10.1093/nar/15.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A, Poole TL. 5'-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5'-exonuclease-1. The Journal of biological chemistry. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- Ternette N, Wright C, Kramer HB, Altun M, Kessler BM. Label-free quantitative proteomics reveals regulation of interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) and 5'-3'-exoribonuclease 2 (XRN2) during respiratory syncytial virus infection. Virology journal. 2011;8:442. doi: 10.1186/1743-422X-8-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature medicine. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner KA, Schutz S, Sarnow P. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Molecular and cellular biology. 2010;30:2006–2016. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ. Human 5' --> 3' exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.