Abstract
Synthetic linear antimicrobial peptides with cationic α-helical structures, such as BP100, have potent and specific activities against economically important plant pathogenic bacteria. They are also recognized as valuable therapeutics and preservatives. However, highly active BP100 derivatives are often phytotoxic when expressed at high levels as recombinant peptides in plants. Here we demonstrate that production of recombinant phytotoxic peptides in transgenic plants is possible by strictly limiting transgene expression to certain tissues and conditions, and specifically that minimization of this expression during transformation and regeneration of transgenic plants is essential to obtain viable plant biofactories. On the basis of whole-genome transcriptomic data available online, we identified the Os.hsp82 promoter that fulfilled this requirement and was highly induced in response to heat shock. Using this strategy, we generated transgenic rice lines producing moderate yields of severely phytotoxic BP100 derivatives on exposure to high temperature. In addition, a threshold for gene expression in selected tissues and stages was experimentally established, below which the corresponding promoters should be suitable for driving the expression of recombinant phytotoxic proteins in genetically modified plants. In view of the growing transcriptomics data available, this approach is of interest to assist promoter selection for specific purposes.
Introduction
Antimicrobial peptides (AMPs) are key components of innate immunity in plants and animals, and are also produced by microbes in antibiosis processes. A significant proportion are strongly cationic and have linear structures that adopt an amphipathic α-helical conformation that binds to the phospholipid membranes of target microbes before the hydrophobic face is inserted into the membrane bilayer. This unique mode of action explains the lack of resistance in target pathogens, and makes AMPs valuable novel therapeutic agents against bacteria, fungi, viruses, parasites and tumor cells. Improved or synthetic AMPs have been designed with increased potency against selected pathogens [1]–[5]. As an example, the synthetic undecapeptide BP100 (KKLFKKILKYL-NH2) is effective against Xanthomonas vesicatoria in pepper, Erwinia amylovora in apple and Pseudomonas syringae in pear [3] with the same efficacy as standard antibiotics, while being biocompatible, as determined by acute oral toxicity tests in mice [6]. Plant expression of recombinant cationic α-helical peptides such as BP100 is preferable for industrial and phytosanitary applications. We have recently showed that active BP100-derived peptides can be expressed as recombinant peptides in plants [7], [8], demonstrated by the increased resistance of GM plants to some rice pathogens [7] and by in vitro growth inhibition assays [8]. These recombinant BP100-derived peptides include endoplasmic reticulum (ER) retention motifs to minimize toxicity to the host plant. This did not affect the antimicrobial activity of the products in vitro, demonstrated using bacterial growth inhibition tests. Following transient expression in Nicotiana benthamiana and stable expression in Arabidopsis thaliana seedlings, the peptides accumulated in large ER-derived vesicles, along with typical ER luminal proteins [8]. However, the authors found that the recombinant peptides were often toxic to the host during later developmental stages. Similarly, most transgenic Oryza sativa plants constitutively expressing recombinant ER-targeted BP100-derived peptides failed to achieve maturity, with only a few peptides coupling potent antimicrobial and low hemolytic activity accumulating in transgenic rice lines, with a yield of up to 0.5% total soluble protein (TSP) [8]. Not many cationic α-helical peptides can be expressed in transgenic plants following this strategy, although they have potent activities against other types of pathogenic cells making them valuable as novel therapeutics and preservatives.
High temperature stress is one of the most common abiotic stresses among many world crops. Plants have evolved various physiological and molecular mechanisms to resist heat stress. Based on the expression data from different plant species, it has been estimated that high temperatures affect approximately 2% of the plant genome (review in [9]). Exhaustive identification of heat stress-responsive genes has been carried out by means of transcriptomics [10]–[15]. Two groups of genes have been found, signaling components (e.g. protein kinases and transcription factors) and functional genes such as heat shock proteins (Hsps) [16], although many heat shock genes still have unknown functions. Hsps are functionally linked to molecular chaperones that are essential for maintenance and restoration of protein homeostasis. Protein denaturation occurring during stress triggers high transcription of hsp genes by the binding of active heat shock factors (Hsfs) to heat shock elements. Alterations in expression of a high number of genes in response to stress occur in complex systems. Cross-talk seems to exist between the regulatory pathways in response to different abiotic stresses such as temperature, and osmotic or oxidative, and biotic stresses [17]. On fusing several heat-shock promoters to reporter genes, they have been found to be regulated during various stress situations, have organ and developmental stage-specific basal expression levels and are induced by stress [18].
Heat-shock induced genes can be highly up-regulated at the transcriptional level by exposure to temperatures above those of normal plant growth. Therefore, induction can be easily carried out, is inexpensive and does not require the use of hormones or chemicals. Here we explored the use of heat-shock inducible promoters with minimal basal activity during normal plant development, to allow expression of recombinant phytotoxic peptides in transgenic rice. Rice has emerged as a powerful platform for large-scale production of recombinant proteins; it has easy cropping conditions and is a self-pollinating crop [19], [20]. It is a most suitable crop for genetic manipulation due to its small genome size and the well-established gene transfer technology. Moreover, the availability of the rice complete genome sequence, the exponential growth of profiling studies reporting gene expression data for rice, and the development of databases and bioinformatics tools, provide a possible way to identify genes related to heat stress responses, and with specific expression patterns.
Materials and Methods
In silico identification of candidate heat shock promoters
Miamexpress-EMBL-EBI (http://www.ebi.ac.uk/miamexpress/) and GEO-NCBI (http://nlmcatalog.nlm.nih.gov/geo/) were used to identify published microarray experiments questioning the late response of rice seedlings to heat shock stress. In the experiment with accession number GSE14275, the authors [10] state that rice Affymetrix microarrays were hybridized with RNA extracted from 14-day-old rice seedlings grown in a growth chamber with a daily photoperiodic cycle of 14 h light and 10 h dark, at between 28–30°C, with or without being subjected to 42 °C for 3 h. The data from this experiment was extracted using the RMA software [21], which includes background adjustment, quantile normalization and summarization, and used to identify probes for subsequent analysis.
Rice gene expression data in different organ and developmental stages, and in response to various stress conditions, was obtained from the CREP database (Collection of Rice Expression Profiles; http://crep.ncpgr.cn/crep-cgi/home.pl) and the microarray hybridization results available at the Miamexpress-EMBL-EBI and GEO-NCBI websites.
The NetAffx-analysis center (www.affymetrix.com) allowed identification of the gene associated to every probe, and the sequence was retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank). Likely promoter sequences were identified using the PlantProm DB, an annotated and non-redundant database of proximal promoter sequences [22], [23].
Primer design and amplification of rice promoter sequences
Primer blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to design primer pairs specifically targeting the identified promoter sequences, blocking the reverse primer at the −1 position (immediately upstream of the ATG translation start codon). Two primer pairs (Table S1) were designed to amplify 553 and 1016 bp fragments of the pHsp18 and pHsp82 promoters. They each included an enzyme restriction site (KpnI and SpeI at the distal and proximal promoter end, respectively) to facilitate the subsequent cloning steps.
Oryza sativa L. ssp. japonica, v. Senia was grown under controlled conditions at 28±1°C and with a 16 h light / 8 h dark photoperiod with fluorescent Sylvania Cool White lamps. Genomic DNA was extracted from 1 g of leaf samples, obtained from young plants, using the commercial NucleoSpin Plant II kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions.
PCR was in a final volume of 50 µl 1x reaction buffer with the appropriate concentrations of MgCl2 and primers (Fisher Scientific SL, Madrid, Spain) (Table S1), 200 µM dNTPs and 1 unit Expand High Fidelity DNA polymerase (Roche Diagnostics GmbH, Mannheim, Germany). The reaction conditions were as follows: 3 min at 94°C; 10 cycles of 15 s at 94°C, 30 s at the appropriate annealing temperature (Table S1) and 1 min at 72°C; 20 cycles of 15 s at 94°C, 30 s at the same annealing temperature and 1 min, plus an additional 5 s for each successive cycle, at 72°C; and a final extension of 10 min at 72°C.
Construction of plant transformation vectors
The pHsp18 and pHsp82 promoters were separately subcloned into the KpnI and SpeI sites of a pBluescriptIIKS + derived vector having a polylinker fragment with the KpnI, SpeI and BamHI restriction sites, followed by the A. tumefaciens nopaline synthase nos terminator sequence. The sequences encoding BP100.2 [7], BP100-DsRed-tag54 and DsRed-tag54 [8] were subcloned into these plasmids using the SpeI and BamHI restriction sites.
For the BP100.2 plasmids, the sequence encoding the N. tabacum pathogenesis related protein PR1a signal peptide [24] fused to BP100.2 [7] was PCR amplified using pAHC17-bp100.2 [7] as template and the primers PR1a_Spe and BP100KDEL_Bam, with the SpeI and BamHI restriction sequences respectively. For DsRed-tag54 control plasmids, the sequence encoding the Petrosilinum hortense chalcone synthase 5′ untranslated region plus the codon-optimized leader peptide derived from the heavy chain of the murine mAb24 monoclonal antibody [25] fused to DsRed [26], the epitope tag54 [27] and the KDEL ER retention motif, were PCR amplified using pCdsred-tag54 [8] as template and the primers CHS_Spe and TagKDEL_Bam, that included the required restriction sites. As an additional control, a BP100-DsRed-tag54 plasmid was generated similarly to DsRed-tag54, and including the BP100 antimicrobial sequence placed N-terminal to DsRed-tag54. The coding sequence was obtained by amplification of the pCbp100-dsred-tag54 DNA [8] with the same CHS_Spe and TagKDEL_Bam primers. PCR was in a final volume of 50 µl 1x reaction buffer with the appropriate concentrations of MgCl2 and primers (Fisher Scientific SL, Madrid) (Table S1), 200 µM dNTPs and 1 unit Expand High Fidelity DNA polymerase (Roche Diagnostics Corporation). Reaction conditions were as follows: 3 min at 94°C; 10 cycles of 15 s at 94°C, 30 s at 49°C and 1 min at 72°C; 20 cycles of 15 s at 94°C, 30 s at 58°C and 1 min, plus additional 5 s each successive cycle, at 72°C; and a final extension of 10 min at 72°C.
On sequence verification (Macrogen, Seoul, Korea) using the CLCbio software (Aarhus, Denmark), the complete constructs, bp100.2, bp100-dsred-tag54 and dsred-tag54 plus promoter and terminator elements, were directionally inserted into the KpnI and SbfI sites of pCAMBIA1300. The resulting binary vectors were transferred into A. tumefaciens strain EHA105 by cold shock [28].
Production of transgenic rice plants
Commercial japonica rice (Oryza sativa L.) var. Senia was transformed by A. tumefaciens to obtain transgenic rice lines expressing the chimeric proteins mentioned above, using hygromycin resistance as the selection trait. The control plasmid, pCambia1300 (transferring only the hptII selection gene), was transformed in parallel. Embryonic calluses derived from mature embryos were transformed as previously described [29]. Hygromycin resistant T0 plants were grown to maturity under standard greenhouse conditions to obtain the T1 generation. Leaf samples of individual T0 plants at the 5-leaf vegetative stage were used to extract genomic DNA and assess the presence and copy number of the transgene by real-time PCR (qPCR), targeting the coding region of every transgene as previously described [8]. The actin endogenous gene was used to normalize the Ct values. The number of fertile genetically modified (GM) plants containing every plasmid was recorded to calculate the transformation efficiency compared to that of the control pCambia 1300 plasmid (number of fertile GM plants obtained per initial callus. This value was then normalized with that obtained for the control plasmid).
Analysis of gene expression: plant material, RNA isolation and RT-PCR
Plant material: heat shock treatment
Wild type rice seeds (var. Senia) were surface sterilized and germinated in vitro in sterile MS medium [30], including vitamins (2.2 g/L MS medium, 8 g/L agar and 30 g/L sucrose), under controlled conditions (28±1°C temperature and a 16 h light / 8 h dark photoperiod with fluorescent Sylvania Cool White lamps). After one week (two-leaf vegetative stage, V2), groups of 15 plants were treated at 42°C for 0, 1, 2, 4, 8 and 16 h. For each treatment, three groups of five plants were collected as biological replicates, immediately frozen in liquid nitrogen and stored at −80°C.
For each GM event, 15 T1 seeds were surface sterilized and in vitro cultured, as mentioned above, up to the V2 stage. They were treated at 42°C for 0 and 2 h and their leaves individually frozen in liquid nitrogen. The presence of the transgene was individually assessed in every plant using the Phire Plant Direct PCR Kit (Thermo Scientific, Lithuania) combined with the DsRed_for and Nos.te_rev (constructs with dsred-tag54 and bp100-dsred-tag54) or the P82_for and Nos.te_rev (constructs with bp100.2) primer pairs (Table S1), according to the manufacturer's instructions. Samples giving a positive PCR signal (i.e. harboring the transgene) were mixed for subsequent analyses.
Plant material: monitoring of the transformation process
The transformation to obtain transgenic rice was monitored using A. tumefaciens strain EHA105, with or without the basic pCambia1300 vector (with hptII). Embryonic calluses derived from rice mature embryos were transformed and, when hptII was transformed, selected with hygromycin [29]. Samples of ten independent calluses or events were taken at eight different stages for each transformation.
For embryo extraction, 120 hygromycin-resistant seeds (harboring hptII) and 80 non-transformed seeds were surface-sterilized and their embryos were immediately extracted and frozen, or germinated in 500 µL water or 500 µL water with 45 mg/L hygromycin B (40 seeds per treatment and embryo type). Germination was in a culture chamber (28±1°C with a 16 h light/8 h dark photoperiod, under fluorescent Sylvania Cool White lamps) for two and five days prior to embryo extraction and immediate freezing in liquid nitrogen. Untransformed seeds were not treated with hygromycin B.
RNA isolation and reverse transcription (RT) coupled to qPCR
Samples were homogenized in liquid nitrogen and 100 mg was used to extract RNA with the Trizol reagent (Invitrogen, Karlsruhe, Germany) based protocol. The DNase I digestion (Ambion, Grand Island, NY) was carried out according to the manufacturer's protocol, and RNA concentration and quality was assessed by UV absorption at 260 and 280 nm using a NanoDrop ND1000 spectrophotometer (Nanodrop technologies, Wilmington, DE). RT-qPCR was carried out as previously described [7]. For each sample, cDNA was synthesized with random primers in duplicate and qPCR reactions targeting bp100.2 and dsred-tag54 transgenes, and Os.hsp18 and Os.hsp82 rice genes were carried out in triplicate. The qPCRs were in a final volume of 20 µl containing 1X SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), the appropriate concentrations of primers (Fisher Scientific SL, Madrid, Spain) (Table S1) and 1 µl cDNA. The reaction conditions were as follows: 10 min at 95°C for initial denaturation; 50 cycles of 15 s at 95°C and 1 min at 60°C; and a final melting curve program of 60 to 95°C with a heating rate of 0.5°C/s. Melting curve analyses produced single peaks, with no primer-dimer peaks or artifacts, indicating the reactions were specific. All reactions had a linearity coefficient exceeding 0.995 and efficiency values above 0.95. The ef1α gene was used for normalization, its suitability having been confirmed using the geNORM v3.4 statistical algorithm ([31]; M values below 0.5 in our samples). Triplicate biological samples, each containing at least five plants, were analyzed in each case.
Protein extraction and western blot analyses
For each GM event, 15 T1 seeds were surface sterilized and in vitro cultured as mentioned above, up to the V2 stage. Rice seedlings were treated at 42°C for 2 h, then transferred to 28±1°C with a 16 h light/8 h dark photoperiod under fluorescent Sylvania Cool White lamps for two days, to allow recombinant protein accumulation and DsRed maturation, and then immediately frozen in liquid nitrogen. The Phire Plant Direct PCR Kit (Thermo Scientific, Lithuania) was used in combination with the SYDsRed_for and SYDsRed_rev primers (Table S1), according to the manufacturer's instructions, to individually identify transgenic T1 plants. Transgenic seedlings were homogenized in liquid nitrogen and TSP extracted as previously described [8]. Insoluble proteins in the samples expressing bp100-dsred-tag54 were re-extracted by boiling for 10 min in the same buffer supplemented with 8 M urea and 1% SDS. Protein concentration was determined using the Sigma Bradford Reagent and bovine serum albumin as standard. Twenty µg TSP were separated by PAGE, using 18% (w/v) SDS polyacrylamide gels, and electrotransferred to nitrocellulose membranes. Twenty-five to 1 pmol chemically synthesized controltag54 (GQNIRDGIIKAGPAVAVVGQATQIAKAGPAKDWEHLKDEL), mixed with 20 µg TSP extracted from untransformed rice seedlings, were included to allow quantification of the recombinant peptides. They were hybridized overnight with the mAb54k monoclonal antibody [27] (1∶1,500 dilution) at 4°C, and with horseradish peroxidase-conjugated anti-mouse IgG as the secondary antibody (GE Healthcare Life sciences) (1∶10,000 dilution) for 1 h at room temperature. The hybridization signal was detected by ECL chemiluminescence (Luminata Forte HRP Chemiluminescence Detection Reagents, Millipore – Darmstadt, Germany).
Phenotype evaluation
T1 seeds carrying pHsp82::bp100.2, pHsp82::dsred-tag54 (three independent events per construct) or hptII, and conventional Senia seeds, were surface sterilized and in vitro cultured as mentioned above, up to the V2 stage (height, 6±1 cm). They were all treated at 42°C for 2 h and allowed to recover for three days under the same conditions. Plant growth was monitored daily by measuring the height of the aerial part. Transgenic plants were identified using the Phire Plant Direct PCR Kit (Thermo Scientific, Lithuania) as mentioned above, and only those harboring the transgene (at least five plants per event) were included in statistical analyses. As an additional control, five untransformed Senia plants were grown in parallel and not subjected to heat shock.
Confocal microscopy
Transgenic rice seedlings, obtained as mentioned above, were treated at 42°C for 2 h and transferred to the standard conditions for two days to allow accumulation of the recombinant protein and DsRed maturation. Radicles were observed under an FV1000 Olympus confocal microscope and red fluorescent images collected with 543 nm excitation using a 550–600 nm emission window. The ImageJ software (http://rsb.info.nih.gov/ij/) was used to calculate the number and size of fluorescent spots and quantify the DsRed fluorescence.
Results
Rationale of the approach and selection of suitable promoters
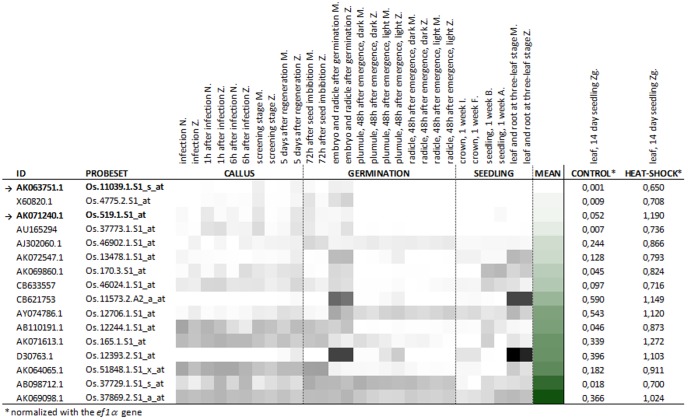
With the increasingly abundant transcriptomics analytical tools and datasets available, we used an in silico approach to initially select a small number of candidate heat shock promoters. The EMBL-EBI Array Express website was searched for datasets on transcription profiling of the Oryza sativa in response to heat shock. A single microarray experiment was found (E-GEOD-14275, [10]) that questioned 14-day old seedlings, grown at 28–30°C, and with or without heat stress at 42°C. Expression data was available for a total of 22,905 sequences. Those with the highest expression levels had fluorescence intensities up to 13,800, and 16,300 units in microarrays hybridized with RNA from control and heat-shock treated seedlings, respectively. Only 15 and 17 sequences had values above 10,000 fluorescence units. Up to 19 sequences, corresponding to 16 genes, had fluorescence intensities more than 5,500 units higher in heat-shock treated than in control seedlings (Table S2). This included three sequences (AK071613.1, AK071240.1 and CB621753) with very high expression values under induction conditions (above 10,000 fluorescence units) and sequences with the greatest changes in expression in response to heat stress treatment (up to 615-fold, AK063751.1). These sequences were initially selected to drive the expression of phytotoxic peptides in rice.
The transcription patterns of these 16 genes were subsequently compared with the Collection of Rice Expression Profiles (CREP) database (http://crep.ncpgr.cn/crep-cgi/home.pl), a compilation of quantitative transcriptomes of 39 tissues of various rice genotypes, from the indica and japonica subspecies, obtained using the commercial Affymetrix Rice GeneChip microarray. We specifically focused on the tissues and developmental stages with special relevance in transformation and regeneration to obtain GM plants. They included callus at different steps of agrotransformation and hygromycin based selection, and imbibed seeds and seedlings at various developmental stages. Multiple gene expression profiles were retrieved using the Multi-genes Chronologer tool (Figure 1). The Os.519.1.S1_at sequence (AK071240.1, encoding a putative 18 kDa Class II Heat Shock Protein) combined low expression levels in all 14 analyzed tissues considered relevant for transformation and very high expression in heat-shock treated seedlings (0.073 and 1.190 normalized fluorescence units). Although it had some expression in control seedlings (0.052 normalized fluorescence units), this promoter was chosen to drive the expression of phytotoxic peptides in plants. A second selected sequence, Os.11039.1.S1_s_at (AK063751.1, encoding a putative Heat Shock Protein 82), had extremely low expression values in the 14 tissues and control seedlings but only moderate expression in response to high temperature treatment (0.033, 0.001 and 0.650 normalized fluorescence units, respectively). There was no induction of Os.519.1.S1_at or Os.11039.1.S1_s_at upon cold, salt or drought stresses, as assessed in silico using profiling datasets with GSE6901 as the reference, reporting transcriptome analyses of 7-day old seedlings of Indica rice variety IR64 subjected to 4±1°C, 200 mM NaCl or insufficient moisture in their roots for 3 h (Figure S1).
Figure 1. A summary of the expression profiles of a selection of sequences induced by high temperature, in callus and seedlings of eight rice genotypes [Bala (B), FL478 (F), IR29 (I), Minghui 63 (M), Zhenshan 97 (Z), indica subspecies, and Azucena (A), Nipponbare (N) and ZhongHua 11 (Zg), japonica subspecies], from published in silico data.
Cluster analysis of 16 genes with the highest expression or induction in response to heat-shock, in a total of 14 tissues considered relevant to the process of obtaining transgenic plants (grey scale). The overall mRNA levels in these tissues and developmental stages were estimated using the mean expression values (green scale). Dark to light color scale represents high to low expression levels, the darkest color corresponding to 90162 (grey scale) and 19788 (green scale) normalized fluorescence units. Arrows indicate the two sequences whose promoters were selected to produce transgenic plants.
Expression patterns of the two selected genes in response to heat shock and during the process of Agrobacterium tumefaciens mediated stable transformation of rice using the hptII selection gene
Specific real-time PCR (qPCR) assays were developed to target the coding sequences of Os.hsp18 and Os.hsp82 and, coupled to reverse transcription, used to experimentally assess the expression patterns of the selected genes in a range of developmental stages and conditions. Treatment of rice plants with high temperature resulted in increased expression of the two genes (Figure 2). Os.hsp18 and Os.hsp82 were highly induced by exposure to 42°C for 2 to 4 h, reaching expression levels of 294 and 516-fold that of ef1α in leaves and 326 and 796-fold in roots. There was a decrease in gene expression after these time points.
Figure 2. Transcriptional response to high temperature of Os.hsp18 and Os.hsp82 in rice var. Senia.
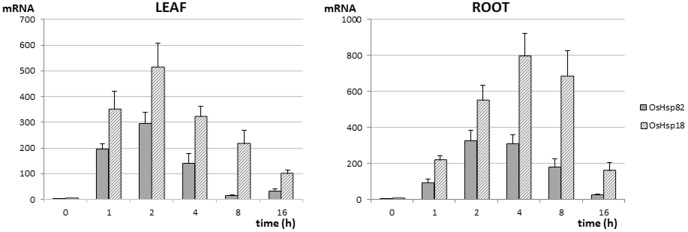
Plants at the two leaf vegetative stage were exposed to 42°C for 0, 1, 2, 4, 8 and 16 h, then the mRNA levels of the selected sequences, from leaf and root samples, were analyzed by RT-qPCR. Three biological replicates, each of five plants, with two experimental replicates, were analyzed per treatment. The ef1α reference gene (gNORM M value <0.5 in these samples) was used for normalization and the given mRNA values correspond to fold expression vs. the reference gene.
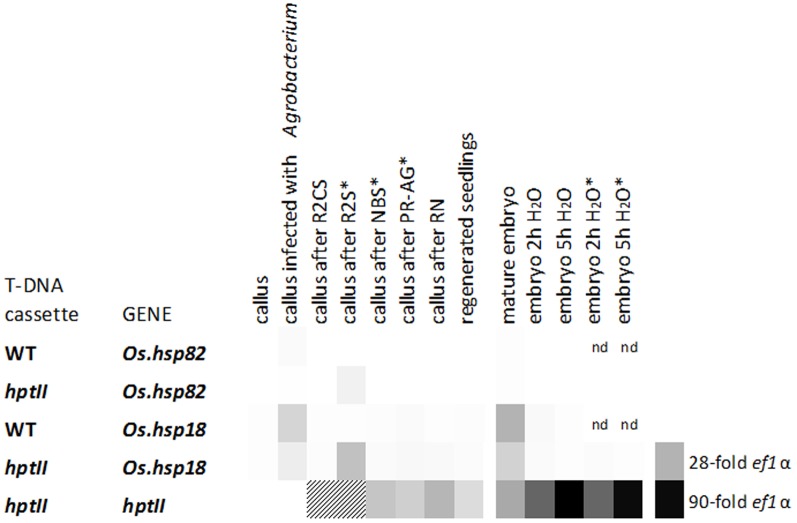
Additionally, their expression levels were determined during A. tumefaciens-mediated transformation and selection using the hygromycin resistance phenotype (Figure 3). Senia wild type seeds were induced to generate primary callus and the standard transformation procedure was followed, using A. tumefaciens carrying pCAMBIA1300 (with the hptII selection gene) to produce transgenic plants. A. tumefaciens not transformed with the binary plasmid was used as control, without hygromycin as a selection agent. Mature and imbibed embryos from untransformed and transgenic S-hptII plants were also analyzed. Os.hsp18 was clearly expressed at higher levels than Os.hsp82 in all tested tissues and stages. Expression was up to 28-fold that of the reference gene in transformed callus incubated for two weeks in hygromycin selection medium and that of mature embryos, and up to 15-fold that of ef1α upon A. tumefaciens infection. Seed imbibition rapidly resulted in mRNA decline. Os.hsp82 had a similar expression pattern, although the level was less than that of ef1α in virtually all tested samples. As a control, we measured the mRNA levels of the hptII transgene, regulated by the CaMV 35S strong constitutive promoter. They were substantially higher than those of Os.hsp18, and especially Os.hsp82, at all stages in which there were uniquely transgenic cells (i.e. after hygromycin selection).
Figure 3. mRNA expression profiles of Os.hsp18, Os.hsp82 and the hptII transgene, driven by the CaMV 35S promoter, in rice var. Senia.
Different stages during transformation with A. tumefaciens, either carrying pCAMBIA1300 (hptII) or no T-DNA (WT), and expression in mature and imbibed embryos, are shown. The following stages of callus are shown: 6 weeks after induction of embryogenic callus; immediately after 15 min infection with A. tumefaciens; after a 3-day incubation in R2CS medium; after a 2-week incubation in R2S selection medium; 3 weeks after transfer to NBS selection medium; after an 8 to 10-day incubation in PR-AG selection medium; 3–4 weeks after induction of regeneration in RN medium; and two weeks after seedling culture in P medium. Embryos extracted from mature seeds, either before or after imbibition in water for two or five hours, were also analyzed. When the hptII DNA cassette was transformed, transgenic cells were selected using hygromycin (asterisk). Three biological replicates, each of five calluses, with two experimental replicates, were analyzed at each stage. The ef1α reference gene (M values <0.5 in these samples) was used for normalization. Dark to light color scale represents high to low expression level. All values were in the 90 to 0.01-fold range. nd: not determined. Striped: underestimated values due to callus samples containing a mixture of transgenic and non-transgenic cells.
Os.hsp18 and Os.hsp82 promoter fragments regulate the expression of transgenes in response to heat treatment
The sequences corresponding to the promoters of Os.hsp18 and Os.hsp82 were retrieved from the Plant Promoter Database, and 553 and 1,016 bp promoter fragments of Os.hsp18 and Os.hsp82, respectively, were PCR amplified from rice var. Senia genomic DNA. Their 3′ end was at position −1 relative to the ATG translation start codon. Os.hsp18 and Os.hsp82 promoter fragments, pHsp18 and pHsp82, were cloned and sequence verified. The activity of these promoter fragments was experimentally assessed using the Discosoma spp. red fluorescent reporter protein, DsRed [26], and the epitope tag54 sequence to detect recombinant proteins through the specific mAb54 antibody [27]. The sequence encoding the DsRed-tag54-KDEL reporter (hereafter, DsRed-tag54) was placed in-frame with the sequence encoding the signal peptide of the murine monoclonal antibody mAb24 under the control of the Os.hsp18, Os.hsp82 or Zm.ubi [32] promoters and nos terminator. All constructs were sequence verified, introduced in A. tumefaciens and used to transform rice plants, along with the hptII marker for hygromycin selection. All constructs yielded hygromycin-resistant plants and those derived from different calluses were considered independent events.
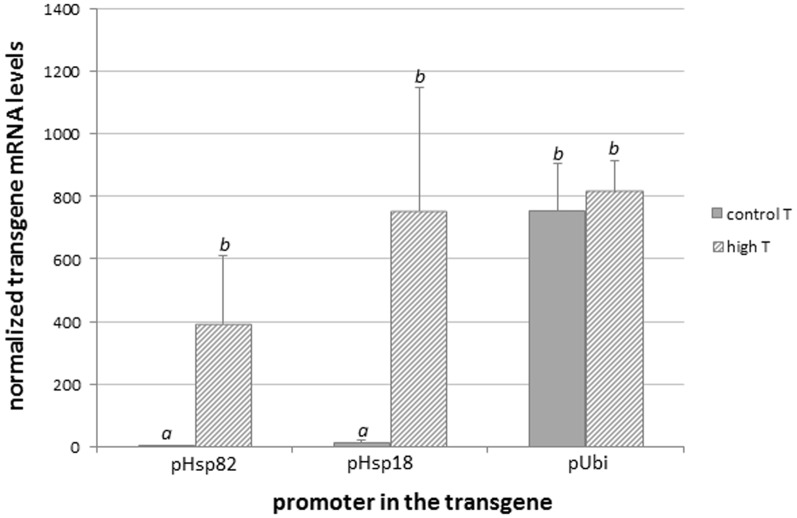
The transgene copy number and expression profile were assessed in three independent events representing each promoter. The ratio of dsred-tag54 to the actin reference gene sequence was close to 0.5 (mean and SD, 0.63±0.27), as determined by qPCR using leaf genomic DNA from T0 plants, suggesting single-copy insertions. Transgene expression was quantified by RT-qPCR in leaf samples of transgenic T1 plants at the two-leaf vegetative stage (V2), with or without exposure to 42°C for 2 h (Figure 4). Significant amounts of dsred-tag54 mRNA were only observed when plants were kept under control temperature conditions, where the transgene was driven by the constitutive pUbi promoter. The expression values, normalized with the ef1α constitutive gene, were 0.47 to 2.5, 0.73 to 18.84 and 603 to 904 for pHsp82, pHsp18 and pUbi, respectively. One-way ANOVA, P = 0.000, gave two groups in Tukey's b posttest with α<0.05. Upon heat treatment, the pHsp18 promoter reached transgene mRNA levels from 214 to 1,117-fold that of ef1α, similar to those reached with the pUbi promoter. The levels of expression of the transgene was slightly lower with the pHsp82 promoter, 208 to 636-fold that of ef1α, but statistically similar (one-way ANOVA P = 0.279). As expected, no mRNA encoding DsRed-tag54 was detected in untransformed rice plants. This proved that, after exposure to heat stress, the pHsp82, and especially the pHsp18 promoter fragments, had the capacity to drive the expression of a reporter gene to levels similar to those achieved by a frequently used strong constitutive promoter.
Figure 4. Transgene mRNA expression levels in leaves of GM rice plants in which the reporter dsred-tag54 was regulated by three different promoters, pHsp82, pHsp18 and pUbi.
Three independent events were analyzed carrying every construct. For each event, T1 seeds were germinated under control conditions and sets of eight plants at the V2 stage were either subjected to 42°C for 2 h, or not, and their leaves were individually sampled. Sets of five plants harboring the transgene (i.e. giving positive transgene signal using the Phire Plant kit) were collectively analyzed by RT-qPCR targeting the sequence encoding DsRed-tag54, and normalized against ef1α. Mean and SD values corresponding to each promoter are shown. Letters indicate statistically different transgene mRNA values (One-way ANOVA, Tukey's b posttest α<0.05).
Use of Os.hsp18 and Os.hsp82 promoters to drive the expression of phytotoxic BP100 derivatives in rice
The capacity of the pHsp18 and pHsp82 promoter fragments to drive the expression of phytotoxic cationic α-helical antimicrobial peptides in stably transgenic plants was evaluated using BP100.2. The antimicrobial peptide has high phytotoxic and hemolytic activities and its constitutive expression in transgenic rice is incompatible with the survival of GM plantlets [7]. The sequence encoding BP100.2 was placed in-frame with the sequence encoding the signal peptide of the N. tabacum pathogenesis related protein PR1a, under the control of the pHsp18 and pHsp82 promoters. After sequence verification, the constructs were introduced in A. tumefaciens and used to transform rice plants along with the hptII marker. The empty vector, with only the hptII selection gene, was transformed in parallel. Joint analysis of the transformation efficiencies achieved for constructs encoding DsRed-tag54 and BP100.2, under the control of the pHsp18, pHsp82 and pUbi promoters, showed that four out of five constructs yielded hygromycin-resistant plants with efficiencies similar to that of the empty vector (mean efficiency and SD, 97±18%, Table S3). As expected, this included all vectors encoding the non-phytotoxic DsRed-tag54 reporter, irrespective of the specific promoter regulating its expression (constitutive pUbi or heat-shock inducible pHsp18 and pHsp82).
Remarkably, the vector encoding BP100.2 under the control of the high temperature inducible promoter pHsp82 produced fertile transgenic rice plants with the same efficiency. The presence of the bp100.2 or dsred-tag54 transgene was confirmed in all T0 events using qPCR. Conversely, no GM plants were obtained with the construct encoding the highly phytotoxic BP100.2 antimicrobial peptide under the control of the pHsp18 promoter (transformation efficiency <3%). The efficiency of this transformation was at least 30-fold below that of the control plasmid, which can be considered not workable. This is in agreement with the higher activity of the pHsp18 promoter, compared to pHsp82, during transformation.
Transgene expression profiles were assessed in three independent events carrying sequences encoding BP100.2 under the control of pHsp82 (pHsp82::bp100.2), using transgenic plants encoding DsRed-tag54 regulated by the same promoter (pHsp82::dsred-tag54) as the control. Hygromycin-resistant plants derived from different calluses were considered independent events. The ratio of bp100.2 to the reference gene sequence was close to 0.5 in these events (mean and SD, 0.53±0.29) as determined by qPCR using leaf genomic DNA from T0 plants, suggesting single-copy insertions. Transcript levels of the bp100.2 transgene were, prior to heat shock, 2.18 to 5.70-fold the level of ef1α (Figure 5), i.e. similar to those of dsred-tag54 (One-way ANOVA P = 0.161). Upon heat shock, pHsp82 drove transcription to similar levels for the phytotoxic bp100.2 (275 to 357-fold the level of ef1α) and the control dsred-tag54 (One-way ANOVA P = 0.592). Thus, the pHsp82 promoter is a suitable tool to achieve transcription of transgenes encoding phytotoxic recombinant peptides in stably transformed GM plants.
Figure 5. Transgene mRNA expression levels in leaves of GM rice plants harboring pHsp82::bp100.2 or pHsp82::dsred-tag54 constructs.
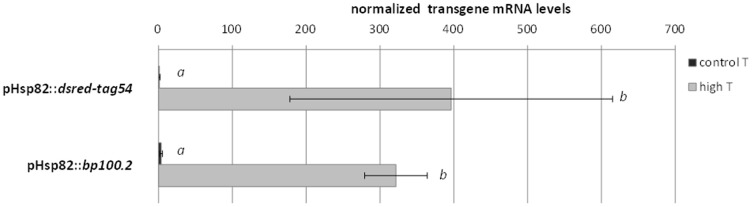
Three independent events of each construct were analyzed. For each event, T1 seeds were germinated under control conditions and sets of eight plants at the V2 stage were either subjected to 42°C for 2 h (black bars) or not (grey bars), and their leaves individually sampled. Sets of five plants harboring the transgene (i.e. giving positive transgene signal using the Phire Plant kit) were collectively analyzed by RT-qPCR targeting the sequence encoding DsRed-tag54 or BP100.2, and normalized against ef1α. Mean and SD values corresponding to every transgene are shown. Different letters indicate statistically different transgene mRNA values (One-way ANOVA, Tukey's b posttest α<0.05).
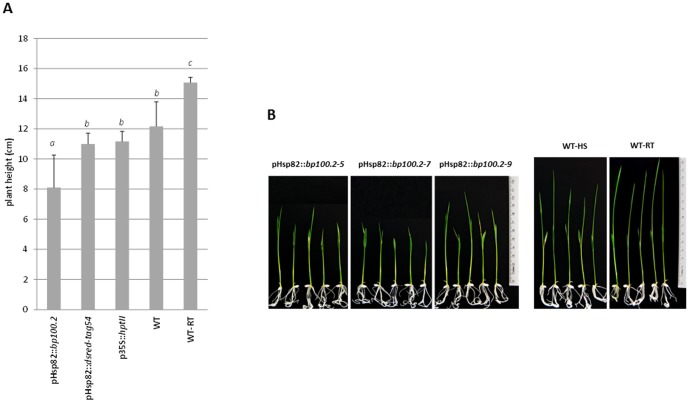
Recombinant BP100.2 is difficult to detect due to its low extinction coefficient (low content of aromatic amino acids) and lack of immunogenicity [8]. We used its phytotoxic character to indirectly confirm the synthesis of BP100.2 in GM plants in response to heat treatment. We predicted that, in the presence of the phytotoxic recombinant BP100.2, transgenic plants would show altered phenotype after heat shock as compared to control plants. As shown in Figure 6, untransformed plants not subjected to heat treatment were taller than any temperature-stressed groups. Twenty-four hours after heat shock, plants expressing bp100.2 were shorter than those either untransformed or expressing dsred-tag54 or hptII (One-way ANOVA P = 0.000, three groups after Tukey b posttest with α<0.05) and had more serious symptoms of leaf wilting. This strongly suggested that phytotoxic recombinant BP100.2 was produced in these plants.
Figure 6. Growth of transgenic rice plants carrying pHsp82::bp100.2 after heat shock.
T1 seeds of rice plants carrying pHsp82::bp100.2, pHsp82::dsred-tag54 (three independent events per construct) and p35S::hptII were germinated for 1 day in the presence of hygromycin, and five plants per event were then in vitro cultured, without the selection agent, up to the V2 stage (height, 6±1 cm). Five untransformed Senia plants were grown in parallel (WT). They were all treated at 42°C for 2 h and the length of their aerial parts was measured after 24 h incubation under standard conditions. Five untransformed plants were not subjected to heat stress, as an additional control (WT-RT). (A) Means and SD of the height of the different groups are shown. Letters indicate statistically significant differences (One-way ANOVA, Tukey b posttest with α value <0.05). (B) The phenotypic effects of heat treatment on transgenic plants carrying pHsp82::bp100.2 (three independent events) and untransformed plants (WT-HS) are shown. WT-RT is shown as control.
Production of recombinant reporter proteins in GM rice plants using high temperature inducible promoters
Phytotoxic recombinant BP100 derivatives in transgenic plants were quantified by fusion of BP100 to a reporter moiety encompassing the fluorescent DsRed and the tag54 epitope. A construct was obtained in which the pHsp82 promoter drove the expression of BP100-DsRed-tag54-KDEL (hereafter, BP100-DsRed-tag54), with the same signal peptide and regulatory elements as the pHsp82::bp100.2 construct. It was sequence verified and used to transform rice plants using the same Agrobacterium-based strategy, achieving similar transformation efficiency as the control (109%). Synthesis of bp100-dsred-tag54 mRNA was assessed by RT-qPCR in three independent events with single copy insertions (as determined by qPCR of T0 leaves using ef1α as reference, 0.52±0.15). Residual mRNA levels in leaves of V2 plants grown under control temperature were in the range of 0.07 to 0.3-fold actin mRNA levels, but increased to 142 to 275-fold after a 2 h exposure to heat shock. These values were not statistically distinguishable from those of pHsp82-regulated transgenes expressing bp100.2 and dsred-tag54 in the same conditions (One-way ANOVA, P = 0.524 and P = 0.205 for control and heat treated plants, respectively).
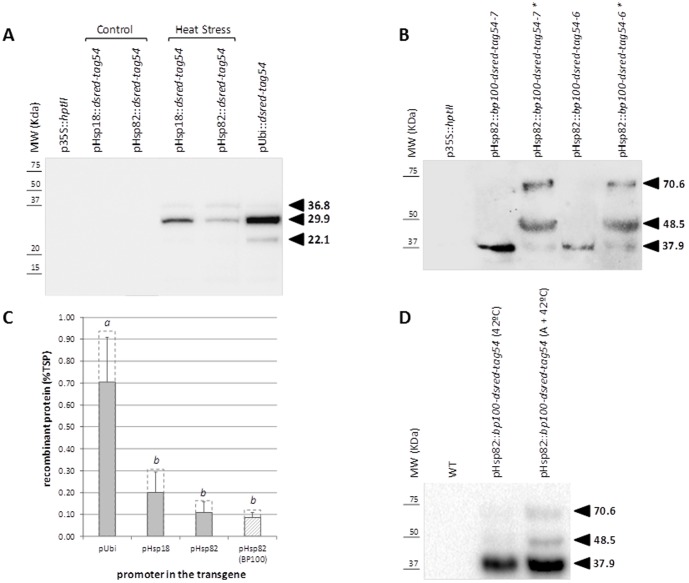
Transgenic plants from three independent events carrying pHsp82::bp100-dsred-tag54 were grown to the V2 stage under controlled conditions, heat-induced for 2 h at 42°C and allowed to accumulate recombinant BP100-DsRed-tag54 for an additional two days. Three events of transgenic plants expressing dsred-tag54 under the regulation of the pHsp82 and pHsp18 promoters were used as control. Plants with the constitutive pUbi promoter driving the expression of the same recombinant protein were included in the assay. As shown in Figure 7A, TSP extracts from all transgenic plants expressing dsred-tag54 produced a band of 29.9 kDa in western blot assays, which is similar to the DsRed-tag54 anticipated size, and a band of about 36.8 kDa. There was an additional secondary band of 22.1 kDa that probably corresponds to a cleavage product originated by boiling TSP before SDS-PAGE [33]. TSP extracted from all three samples expressing bp100-dsred-tag54 under the control of pHsp82 produced a band of an apparent MW of 37.9 kDa (Figure 7B), which is in agreement with the 36.8 kDa band detectable in extracts of plants producing recombinant DsRed-tag54. When higher amounts of protein extracts were analyzed, a faint band of the expected size (31.5 kDa) was also visible (Figure S2). The BP100 portion of the recombinant molecule seemed to strongly enhance the proportion of the slower band, which was probably the result of incomplete denaturation or interaction of different molecules. Subsequent analysis of the insoluble protein fraction showed that a significant proportion of immune-reactive proteins, with apparent MW of 37.9, 48.5 and 70.6 kDa, could only be extracted in an SDS and urea-based, strongly denaturing buffer (Figure 7B). As expected, no immune-reactive bands were observed upon analysis of TSP from untransformed plants, those uniquely expressing the hygromycin resistance selection marker, or those transformed with pHsp82::dsred-tag54 and not subjected to high temperature (Figure 7A). Thus, phytotoxic recombinant BP100 derivatives were produced and accumulated after heat induction in transgenic rice using the pHsp82 heat inducible promoter. Quantification of the immune-reactive bands (Figure 7C) showed that the pUbi promoter led to approximately 0.9% TSP of DsRed-tag54, while promoters derived from Os.hsp18 and Os.hsp82 led to recombinant DsRed-tag54 levels up to 0.3 and 0.15% TSP, respectively, after heat shock. Finally, use of the same pHsp82 promoter to drive the expression of the phytotoxic BP100 fusion protein led to similar accumulation levels, up to 0.12% TSP in these conditions.
Figure 7. Recombinant DsRed-tag54 and BP100-DsRed-tag54 accumulation in transgenic rice seedlings.
Western blot analysis of proteins from GM plants carrying: (A) p35S::hptII, pUbi::dsred-tag54, pHsp18::dsred-tag54 and pHsp82::dsred-tag54, either treated at 42°C (heat shock) or not (control); (B) pHsp82::bp100-dsred-tag54 (two independent events are shown) treated at 42°C. Recombinant protein in TSP was extracted from rice seedlings (five plants per event) and 20 µg of TSP per lane was boiled for five minutes and separated by SDS-PAGE before transfer to nitrocellulose filters. In lanes marked with an asterisk, insoluble pHsp82::bp100-dsred-tag54 pellets were re-extracted in 8 M urea and 1% SDS buffer. Recombinant proteins were detected using the mAb54k antibody (diluted 1∶1,500) and the horseradish peroxidase-labeled anti-mouse IgG secondary antibody (diluted 1∶10,000) followed by ECL chemiluminescent detection. Five to 20 pmol of chemically synthesized controltag54 mixed with 20 µg TSP from wild type rice was used as standards for quantification. GM rice carrying p35S::hptII was used as control. (C) Recombinant protein accumulation values. Means and SD of three independent events per construct are shown in filled boxes; letters indicate statistically different values; values corresponding to the highest producer event are shown in dashed boxes. (D) Western blot analysis of proteins from GM plants of a single event carrying pHsp82::dsred-tag54, either directly subjected to 42°C or with a 6 h initial step of gradual temperature increase up to 42°C (A+42°C) (five plants per condition). Recombinant proteins were extracted in 8 M urea and 1% SDS buffer. Senia (WT) was used as control.
One week after germination, pHsp82::bp100-dsred-tag54 and control pHsp82::dsred-tag54 transgenic plantlets from three different events per construct were induced by heat shock, and the recombinant chimeric protein allowed to accumulate and mature for three days prior to confocal microscopy of the plantlet radicles. All analyzed transgenic plants displayed red fluorescence, further confirming the accumulation of the recombinant proteins. Fluorescence intensities were measured in four fields per radicle. The p82::bp100-dsred-tag54 and control p82::dsred-tag54 plants expressed similar levels of recombinant proteins, with mean and SD values of 1258±420 and 981±286 fluorescence units per field, respectively (One-way ANOVA P = 0.135). This demonstrated that recombinant phytotoxic BP100-derived peptides could be accumulated to levels similar to those of the DsRed-tag54 reporter in stable GM plants constructed with the pHsp82 temperature inducible promoter. The DsRed-tag54 fluorescence showed a typical ER reticular pattern accompanied by a few spherical structures, attributable to the known effects of high temperatures on the internal organization of the cell, such as fragmentation of the Golgi system and the ER [34]. In contrast, fluorescence in pHsp82::bp100-dsred-tag54 radicles was essentially in numerous and widely distributed vesicles, obscuring the ER network (Figure S3), in agreement with BP100 inducing ER-derived protein bodies [8].
Discussion
Biotechnological production of cationic α-helical antimicrobial peptides that have potent activities against pathogenic microorganisms is of great interest [35], but their constitutive expression is not compatible with the viability and fertility of host plants [7]. This problem has been previously approached by expression of peptides (i) with suboptimal antimicrobial activity linked to lower phytotoxicity [36], and (ii) with rational sequence modification to achieve a targeted reduction in phytotoxicity without affecting antimicrobial potency [8]. Modifications often alter the properties of the peptides not only in terms of antibacterial activity but also the range of target bacterial species [3], [5], [37]. This makes the design of a peptide with optimal properties difficult [38]. A tool to produce highly active peptides of interest in plants, irrespective of their phytotoxic character, is desirable. Inducible promoters could potentially reduce the negative impact of toxic recombinant peptides on the transgenic plant by forcefully restraining the presence of the peptide during plant development, while allowing high expression upon induction. The probable harmful effects of accumulation of recombinant phytotoxic compounds after induction, to the plant biofactory, are irrelevant since the main objective is recovery of the recombinant product. We predicted that it would only be possible to successfully generate transgenic plants carrying transgenes that encoded phytotoxic peptides by using promoters extremely inactive throughout transformation and regeneration, due to the vulnerability of the plant material during this process. Due to the practicality and low cost of using high temperature as an inducing agent in plants, and the extensive literature on the plant response to heat shock [39]–[44], we focused on heat shock promoters to assess the feasibility of obtaining recombinant phytotoxic antimicrobial peptides in rice, using as model, highly active and phytotoxic peptides derived from BP100.
Profiling technologies such as microarray hybridization have been widely used to systematically investigate transcriptomic changes during plant development, in different tissues and in response to diverse environmental conditions, including heat shock [45]–[47]. We selected candidate promoters in silico on the unique basis of the massive transcriptome information on publically available web sites (MIAMExpress, CREP), specifically focusing on studies on prolonged heat shock transcriptional response [10]. A similar strategy has recently been used to identify abiotic stress-inducible promoters intended to drive the overexpression of genes encoding nontoxic, stress resistance proteins in transgenic plants, with the aim of reducing the stunted growth and decrease in yield that may be caused by constitutive overexpression of these transgenes [48]. In contrast, here we selected candidate promoters for the expression of phytotoxic recombinant peptides, in plants, to drive high gene expression after heat treatment with minimal gene expression under any other condition, especially focusing on callus during agrotransformation and different tissues in the germination and seedling developmental stages.
The four sequences which best fulfilled these requirements [AK063751.1 (Os.hsp82), X60820.1, AU165294 and AK071240 (Os.hsp18), Figure 1] encoded heat shock proteins belonging to the Hsp90 (Os.hsp82) and the small Hsp (sHsp) families (X60820.1, AU165294 and Os.hsp18, UniProt database). Heat shock proteins are found in every organism [49], and play important roles in cell protection against deleterious effects of stress e.g. acting as molecular chaperones [49]–[55] and in cellular functions related to growth and development [49], [56]. Small Hsps are the most abundant, heterogeneous and stress-responsive group of Hsp in higher plants [57]–[60]. The genes for more than 20 sHsps have been identified in rice, with specific spatial and temporal regulation under stress and in developmental stages [42], [61]. Specifically, the Os.hsp18 gene, encoding the cytosolic class II Hsp18.0, had very high expression upon induction, although our microarray analysis showed that it retained some activity in developmental stages and conditions that are fundamental to the production of GM plants. Hsp90 are, in general, constitutively expressed and among the most abundant proteins in cells, although their expression increases in response to stress [39]. Os.hsp82 had extremely low expression in control tissues and reached high levels of expression in response to heat, but Os.hsp18 and especially Os.hsp82 were not significantly induced by other stress conditions, such as cold, NaCl and drought, in agreement with previous publications [10], [62], [63]. Their moderate mRNA levels in seeds, as determined using the CREP platform, would most probably allow fertility of the plants. Experimental assessment of the expression patterns of these two genes in the rice genotype of interest, Senia, allowed ratification of the Os.hsp18 and Os.hsp82 candidate sequences [10], [62], [64]. In a thorough analysis of A. tumefaciens-mediated transformation of Senia rice and selection using the hygromycin resistance trait, Os.hsp18 and Os.hsp82 mRNA levels were consistently 20 and 60-fold lower, respectively, than in leaves under induction conditions (the highest values were a transient response to Agrobacterium infection and hygromycin treatment) and about 10 and 300-fold lower, respectively, than those of the hptII transgene regulated by the p35S constitutive promoter. We therefore considered that Os.hsp82, and perhaps Os.hsp18, promoters would be able to drive the expression of transgenes encoding phytotoxic peptides in rice, possibly the latter with higher yields, and allow survival of GM plants.
Five hundred to 1,000 base pair promoter fragments of Os.hsp18 and Os.hsp82 regulated the expression of a reporter sequence in leaves of transgenic rice plants, in a similar manner as the endogenous genes. The low transgene mRNA levels measured prior to heat shock in GM plants carrying pHsp18::dsred-tag54 and pHsp82::dsred-tag54 (about 10 and 1-fold those of the reference gene, respectively) increased to approximately 700 and 400-fold that of the reference gene 2 hours after heat treatment. Even though different GM events had different transgene mRNA levels, globally the expression of endogenous Os.hsp18 and Os.hsp82 genes was similar to, respectively, that of the dsred-tag54 transgene driven by the pHsp18 and pHsp82 promoters (One-way ANOVA P = 0.465 and P = 0.463, respectively). In addition, western blot analyses and confocal microscopy showed accumulation of recombinant DsRed-tag54 specifically upon heat shock in leaves and roots carrying pHsp18::dsred-tag54 (data not shown) and pHsp82::dsred-tag54. The selected pHsp18 and pHsp82 promoter fragments retained the essential elements to drive heat shock induction in these tissues, and they performed as expected in GM plants. This is in agreement with previous publications where regulatory elements including the TATA box were in silico predicted in the two 5′ proximal regions [65], [66].
Use of the pHsp18 promoter to drive the expression of the phytotoxic peptide BP100.2 did not allow survival of fertile transgenic plants. Remarkably, transgenic plants producing BP100.2 and BP100-DsRed-tag54, also phytotoxic [8], were obtained with pHsp82, with the same efficiency as those expressing a non-toxic recombinant protein such as the reporter DsRed-tag54. After heat treatment, leaves of plants carrying pHsp82::bp100.2 and pHsp82::bp100-dsred-tag54 produced levels of transgene mRNA similar to those harboring the dsred-tag54 reporter under the control of the same pHsp82 promoter, indicating that these transgenes were fully functional. As BP100 is difficult to detect due to lack of antigenicity and a low extinction coefficient [8], a phenotypic approach was indirectly used to prove the synthesis of BP100.2 in these cells. High temperatures are known to cause wilting and a significant decline in relative growth rate [67]. Growth of pHsp82::bp100.2 plants after a high temperature shock was slower than that of untransformed or transgenic control plants and there was considerably more wilting of their leaves, which supported the production of the phytotoxic recombinant BP100.2 peptide in these cells.
In view of the somewhat higher expression of Os.hsp18 than Os.hsp82 during transformation and regeneration (about 5–10 fold), the finding that pHsp82, but not pHsp18, was able to drive the expression of phytotoxic recombinant peptides in plants supports our initial hypothesis and experimental approach. It is essential that promoter sequences to regulate the expression of phytotoxic transgenes in plants are chosen on the basis of their extremely low basal activity in the tissues, the developmental stages and under the conditions for production of GM plants and germination. No viable transgenic plants were obtained that expressed BP100.2 under the control of 1000-bp promoter fragments of the Os.37773.1.s1_at (AU165294) and Os.165.1.s1_at genes, with mRNA levels above those of Os.hsp18 during the transformation process (Figure 1). These results place the threshold of basal expression of a phytotoxic transgene during production of the GM plant at very low levels, similar to those of the Os.hsp82 gene. We then re-evaluated the initially selected heat stress-responsive genes on the basis of this threshold. Five additional sequences (AB110191.1, AU165294, AK069860.1, X60820.1 and AB098712.1, Table S2) had higher expression than Os.hsp82 after heat shock, and mRNA levels below Os.hsp18 in control conditions. Four of them were expressed at levels above the Os.hsp18 threshold during transformation (Figure 1), so their promoters would not be suitable for producing GM plants by driving the expression of phytotoxic components. As X60820.1 had expression patterns similar to Os.hsp82, its promoter most probably would be suitable to drive this expression. However, it is unlikely that yields would be substantially above those of pHsp82, as deduced from its moderate transcriptional response to heat shock.
Recombinant BP100 derivatives were produced in transgenic plants in response to heat treatment through regulation of the pHsp82 promoter. Three days after induction, BP100-DsRed-tag54 represented 0.12% of seedling proteins solubilized in urea and SDS containing buffer, determined by western blot using the tag54 epitope. Using the same promoter to drive the expression of DsRed-tag54 yielded up to 0.15% TSP, harsher denaturing conditions not significantly increasing the yield (data not shown). Although an anti-tag54 reactive band of the anticipated size was visible in TSP from heat-treated pHsp82::bp100-dsred-tag54 leaves, the major bands, especially those only extracted under strongly denaturing conditions, had slower mobility, which is compatible with remaining BP100-driven molecular interactions. This could be explained by the unusual physicochemical properties of BP100 (highly cationic, pI = 11.5), known to result in interactions with many components of the cell and reduce the efficiency of isolation from complex matrices [8]. In agreement, after the same temperature treatment, BP100-DsRed-tag54 was accumulated at statistically similar levels to the reporter DsRed-tag54, as determined by quantification of the fluorescence signal in confocal micrographs (i.e. when no extraction is needed). We have previously described [8] how the BP100 sequence induces the formation of ER-derived vesicles or protein bodies (PB), functioning similarly to the tandemly repeated VPGXG elastin-like motif [68], hydrophobins [69] and the γ-zein derived Zera polypeptide [70]. Recombinant BP100-DsRed-tag54 accumulates in these vesicles, together with other luminal ER proteins, in transiently transformed N. benthamiana leaves [8]. Here we observed that recombinant BP100-DsRed-tag54 had a similar vesicular pattern in radicles of transgenic rice after high temperature induction, further establishing that recombinant BP100 derivatives accumulate in newly formed ER-derived vesicles in different host plant species.
It is therefore clear that phytotoxic compounds such as BP100 can be expressed in stably transformed plants using a strategy based on inducible promoters. Constitutive expression of this type of transgene has only previously been achieved in transient systems, the harmful effects of the recombinant proteins strongly interfering with normal growth and development of the transgenic plants. With the pHsp82 promoter, we estimate a yield of about 0.1% TSP in our initial induction conditions. Larkindale and Vierling [71] have showed that different acclimation schemes resulted in different yields of Hsp in Arabidopsis. Notably, a gradual temperature increase led to higher transcript levels than heat shock without acclimation. Here, use of an acclimation treatment (6 h gradual increase prior to 2 h at 42°C) increased the recombinant protein yield 3-fold (Figure 7D). Thus, this approach allowed obtaining up to about 0.3% total extracted phytotoxic recombinant proteins. This is about half the yield of the BP100.gtag peptide, specifically designed to display extremely low toxicity, achieved using the constitutive maize pUbi promoter in the same rice system [8]. Recombinant Cry1B has been produced in rice at similar yields (0.2% TSP) under the control of the maize proteinase inhibitor (Mpi) wound-inducible promoter [72]. Higher yields of recombinant Acidothermus cellulolyticus endoglucanase E1, 1.3% TSP, have been achieved in transgenic tobacco using the tomato Rubisco small subunit (RbcS-3C) light-inducible promoter [73]. Note that this type of highly inducible promoter does not allow production of phytotoxic compounds.
In a prospective exercise, our experimental approach was used to identify alternative promoter sequences with increased performance to drive the expression of phytotoxic peptides in plants. Transcriptome-wide datasets are available for rice seedlings under control and a range of abiotic stress conditions, such as salinity, desiccation, suboptimal temperature (4°C), and various minerals, elicitor and hormone treatments, and were analyzed according to the above mentioned criteria. A total of 23 sequences were identified with high expression under induction conditions (above that of Os.hsp82) and minimal expression levels in control conditions (below that of Os.hsp18) (Figure S4). On assessment of their expression patterns in callus and during the transformation process and production of transgenic plants (taken as the mean of the microarray normalized data in these tissues), there were only four sequences with mRNA levels below Os.hsp82, strongly suggesting that their promoters would serve to drive the expression of phytotoxic proteins in transgenic plants. They were induced by drought, NaCl, phosphorus and the heavy metals chromium (VI) and arsenic. A single sequence, Os.49245.1.S1_at, reached very high levels after drought and NaCl induction (similar to Os.hsp18 after heat shock), making its promoter the best candidate. Five additional sequences had mRNA levels below Os.hsp18 but above Os.hsp82 in the analyzed tissues and developmental stages. Their promoters might also be suitable to drive the expression of toxic compounds in GM plants, but their expression levels after induction were below that of Os.hsp18. The best use of our promoter-selection strategy would most likely be achieved by using longer-term expression data to select sequences with the longest span of transcriptional response to the stimulus, which we could speculate would result in higher accumulation of the recombinant proteins.
In conclusion, we produced phytotoxic α-helical antimicrobial peptides derived from BP100 in plants on the basis of strict regulation of transgene transcription. A requirement was that transcription was below a rigorous threshold in specific plant tissues and developmental stages, particularly during transformation and regeneration of GM plants. The heat shock induced promoter, pHsp82, fulfilled this condition and was capable of driving production of phytotoxic recombinant peptides in plants, although the yield is rather low for most commercial applications. In addition, we demonstrate that, thanks to the increasing information available, in silico analysis of transcriptome profiles is a suitable and inexpensive approach to select promoters with specific activity patterns.
Supporting Information
A summary of the expression profiles of 16 genes selected as having the highest expression or induction in response to heat-shock in 7-day IR64 ( indica ) seedlings subjected to cold, salt and drought stress for 3 h. Data on 14-day ZhongHua 11 (japonica) seedlings subjected to heat shock for 3 h are also shown. Dark to light color scale represents high to low expression levels, black corresponding to 11,309 normalized fluorescence units. Arrows indicate the two sequences whose promoters were selected to produce transgenic plants.
(TIF)
Recombinant BP100-DsRed-tag54 accumulation in transgenic rice seedlings. Western blot analysis of proteins from GM plants carrying pHsp82::dsred-tag54 and pHsp82::bp100-dsred-tag54 (two independent events taken as example) treated at 42°C. Recombinant protein in TSP was extracted from rice seedlings (five plants per event) and 30 µg of TSP per lane was boiled for five minutes and separated by SDS-PAGE before transfer to nitrocellulose filters. Recombinant proteins were detected using the mAb54k antibody (diluted 1∶1,500) and the horseradish peroxidase-labeled anti-mouse IgG secondary antibody (diluted 1∶10,000) followed by ECL chemiluminescent detection.
(TIF)
Confocal micrographs of rice radicles from transgenic plantlets carrying pHsp82:: bp100-dsred-tag54 (A and C) and control pHsp82:: dsred-tag54 (B and D), at the V2 developmental stage, subjected to 42°C for 2 hours and further incubated under control growth conditions for three days in a culture chamber. (A and B), DsRed fluorescence; (C and D), bright field. Scale bars: 0.5 µm.
(TIF)
A summary of the expression profiles of a selection of sequences induced by temperature, drought and NaCl stress, and hormone treatment, in callus and seedlings of rice, based on the following in silico published data [74]–[79] . Genes with mRNA levels below that of Os.hsp18 in control seedlings (control) and above that of Os.hsp82 after induction (induced) are shown. Normalized expression levels in 28 tissues, relevant to the process of obtaining transgenic plants, of three rice genotypes: Minghui 63 (M, indica), Zhenshan 97 (Z, indica) and Nipponbare (N, japonica) (grey scale). The mRNA levels in these tissues and developmental stages were estimated using the mean expression values (green scale). Dark to light color scale represents high to low expression levels, the darkest corresponding to 37,405 (grey scale) and 12,480 (green scale) normalized fluorescence units.
(TIF)
Primer sequences and PCR conditions used. Additional restriction sites are underlined.
(DOCX)
Sequences with the greatest expression changes in response to heat stress in rice, extracted from publically available microarray hybridization data. Details on the sequence (representative public ID, Affymetrix code and description) and the mRNA expression in response to treatment at 42°C for 3 h [normalized fluorescence units in rice seedlings under control (control) and heat-shock (heat-shock) conditions; fold change (fold) and difference (HS-C) of normalized fluorescence intensities in the two conditions are also indicated]. Note that the sequences with the same Affymetrix number correspond to the same gene. Dark to light scale of shading represents high to low expression level. The promoters of the two sequences indicated in bold were selected to produce transgenic plants.
(DOCX)
Transformation efficiencies of the different constructs encoding the DsRed-tag54 reporter or the phytotoxic BP100.2 AMP, under the control of a rice heat-shock (pHsp18 and pHsp82) or a maize constitutive (pUbi) promoter.
(DOCX)
Acknowledgments
We thank J. Messeguer, E. Melé (IRTA, Cabrils) and E. Montesinos (UdG) for valuable suggestions; M. Amenós (CRAG, Barcelona) for technical assistance in confocal microscopy; and the scientific writer S. Burgess for revision of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the Spanish Ministerio de Economía y Competitividad (reference AGL2010-17181/AGR). The research group belongs to 2014SGR697, recognized by the Catalonian Government. N.C. and C.R. received fellowships from Generalitat de Catalunya. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Monroc S, Badosa E, Besalú E, Planas M, Bardají E, et al. (2006) Improvement of cyclic decapeptides against plant pathogenic bacteria using a combinatorial chemistry approach. Peptides 27: 2575–2584. [DOI] [PubMed] [Google Scholar]
- 2. Cavallarín L, Andreu D, San Segundo B (1998) Cecropin A-derived peptides are potent inhibitors of fungal plant pathogens. Mol Plant Microbe Interact 11: 218–227. [DOI] [PubMed] [Google Scholar]
- 3. Badosa E, Ferré R, Planas M, Feliu L, Besalú E, et al. (2007) A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 28: 2276–2285. [DOI] [PubMed] [Google Scholar]
- 4. Marcos JF, Muñoz A, Pérez-Paya E, Misra S, López-García B (2008) Identification and rational design of novel antimicrobial peptides for plant protection. Annu Rev Phytopathol 46: 273–301. [DOI] [PubMed] [Google Scholar]
- 5. López-García B, Pérez-Paya E, Marcos JF (2002) Identification of novel hexapeptides bioactive against phytopathogenic fungi through screening of a synthetic peptide combinatorial library. Appl Environ Microbiol 68: 2453–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montesinos E, Bardaji E (2008) Synthetic antimicrobial peptides as agricultural pesticides for plant-disease control. Chem Biodivers 5: 1225–1237. [DOI] [PubMed] [Google Scholar]
- 7. Nadal A, Montero M, Company N, Badosa E, Messeguer J, et al. (2012) Constitutive expression of transgenes encoding derivatives of the synthetic antimicrobial peptide BP100: impact on rice host plant fitness. BMC Plant Biol 12: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Company N, Nadal A, La Paz J-L, Martínez S, Rasche S, et al. (2014) The production of recombinant cationic α-helical antimicrobial peptides in plant cells induces the formation of protein bodies derived from the endoplasmic reticulum. Plant Biotechnol J 12: 81–92. [DOI] [PubMed] [Google Scholar]
- 9. Qu A-L, Ding Y-F, Jiang Q, Zhu C (2013) Molecular mechanisms of the plant heat stress response. Biochem Biophys Res Commun 432: 203–207. [DOI] [PubMed] [Google Scholar]
- 10. Hu W, Hu G, Han B (2009) Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci 176: 583–590 10.1016/j.plantsci.2009.01.016 [DOI] [PubMed] [Google Scholar]
- 11. Qin D, Wu H, Peng H, Yao Y, Ni Z, et al. (2008) Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genomics 9: 432 10.1186/1471-2164-9-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ginzberg I, Barel G, Ophir R, Tzin E, Tanami Z, et al. (2009) Transcriptomic profiling of heat-stress response in potato periderm. J Exp Bot 60: 4411–4421 10.1093/jxb/erp281 [DOI] [PubMed] [Google Scholar]
- 13. Mittal D, Madhyastha DA, Grover A (2012) Genome-wide transcriptional profiles during temperature and oxidative stress reveal coordinated expression patterns and overlapping regulons in rice. PLoS One 7 10.1371/journal.pone.0040899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y-F, Wang Y, Tang Y, Kakani VG, Mahalingam R (2013) Transcriptome analysis of heat stress response in switchgrass (Panicum virgatum L.). BMC Plant Biol 13: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang X, Rerksiri W, Liu A, Zhou X, Xiong H, et al. (2013) Transcriptome profile reveals heat response mechanism at molecular and metabolic levels in rice flag leaf. Gene 530: 185–192 10.1016/j.gene.2013.08.048 [DOI] [PubMed] [Google Scholar]
- 16. Todaka D, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K (2012) Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice 5: 6 10.1186/1939-8433-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hadiarto T, Tran L-SP (2011) Progress studies of drought-responsive genes in rice. Plant Cell Rep 30: 297–310 10.1007/s00299-010-0956-z [DOI] [PubMed] [Google Scholar]
- 18. Khurana N, Chauhan H, Khurana P (2013) Wheat Chloroplast Targeted sHSP26 Promoter Confers Heat and Abiotic Stress Inducible Expression in Transgenic Arabidopsis Plants. PLoS One 8: e54418 10.1371/journal.pone.0054418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramessar K, Capell T, Christou P (2008) Molecular pharming in cereal crops. Phytochem Rev 7: 579–592 10.1007/s11101-008-9087-3 [DOI] [Google Scholar]
- 20. Wakasa Y, Takaiwa F (2013) The use of rice seeds to produce human pharmaceuticals for oral therapy. Biotechnol J 8: 1133–1143 10.1002/biot.201300065 [DOI] [PubMed] [Google Scholar]
- 21. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 22. Yamamoto YY, Obokata J (2008) ppdb: a plant promoter database. Nucleic Acids Res 36: D977–D981 10.1093/nar/gkm785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hieno A, Naznin HA, Hyakumachi M, Sakurai T, Tokizawa M, et al.. (2013) Ppdb: Plant Promoter Database Version 3.0. Nucleic Acids Res: 1–5. [DOI] [PMC free article] [PubMed]
- 24. Cornelissen BJ, Horowitz J, van Kan JA, Goldberg RB, Bol JF (1987) Structure of tobacco genes encoding pathogenesis-related proteins from the PR-1 group. Nucleic Acids Res 15: 6799–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaquero C, Sack M, Chandler J, Drossard J, Schuster F, et al. (1999) Transient expression of a tumor-specific single-chain fragment and a chimeric antibody in tobacco leaves. Proc Natl Acad Sci USA 96: 11128–11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, et al. (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17: 969–973. [DOI] [PubMed] [Google Scholar]
- 27. Rasche S, Martin A, Holzem A, Fischer R, Schinkel H, et al. (2011) One-step protein purification: Use of a novel epitope tag for highly efficient detection and purification of recombinant proteins. Open Biotechnol J 5: 1–6. [Google Scholar]
- 28.Sambrook J, Russell D (2001) Molecular Cloning: a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 29. Sallaud C, Meynard D, van Boxtel J, Gay C, Bes M, et al. (2003) Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106: 1396–1408 10.1007/s00122-002-1184-x [DOI] [PubMed] [Google Scholar]
- 30. Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497. [Google Scholar]
- 31. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218. [DOI] [PubMed] [Google Scholar]
- 33. Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY (2000) The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA 97: 11990–11995 10.1073/pnas.97.22.11990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richter K, Haslbeck M, Buchner J (2010) The Heat Shock Response: Life on the Verge of Death. Mol Cell 40: 253–266 10.1016/j.molcel.2010.10.006 [DOI] [PubMed] [Google Scholar]
- 35.Rajasekaran K, Cary J, Jaynes J, Montesinos E (2012) Small wonders: peptides for disease control. Washington, DC: Oxford University Press: American Chemical Society: Oxford University Press. [Google Scholar]
- 36. Osusky M, Zhou G, Osuska L, Hancock RE, Kay WW, et al. (2000) Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat Biotechnol 18: 1162–1166. [DOI] [PubMed] [Google Scholar]
- 37. Monroc S, Badosa E, Feliu L, Planas M, Montesinos E, et al. (2006) De novo designed cyclic cationic peptides as inhibitors of plant pathogenic bacteria. Peptides 27: 2567–2574. [DOI] [PubMed] [Google Scholar]
- 38. Badosa E, Moiset G, Montesinos L, Talleda M, Bardají E, et al. (2013) Derivatives of the Antimicrobial Peptide BP100 for Expression in Plant Systems. PLoS One 8: e85515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Efeoğlu B (2009) Heat Shock Proteins and Heat Shock Response in Plants. J Sci 22: 67–75. [Google Scholar]
- 40. Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, et al. (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 10.1016/j.pbi.2007.04.011 [DOI] [PubMed] [Google Scholar]
- 41. Sarkar NK, Thapar U, Kundnani P, Panwar P, Grover A (2013) Functional relevance of J-protein family of rice (Oryza sativa). Cell Stress Chaperones 18: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sarkar NK, Kim Y-K, Grover A (2009) Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10: 393 10.1186/1471-2164-10-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarkar NK, Kundnani P, Grover A (2013) Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 18: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh A, Singh U, Mittal D, Grover A (2010) Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes. BMC Genomics 11: 95 10.1186/1471-2164-11-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sarkar NK, Kim Y-K, Grover A (2014) Coexpression network analysis associated with call of rice seedlings for encountering heat stress. Plant Mol Biol 84: 125–143. [DOI] [PubMed] [Google Scholar]
- 46. Jung Y-J, Lee S-Y, Moon Y-S, Kang K-K (2012) Enhanced resistance to bacterial and fungal pathogens by overexpression of a human cathelicidin antimicrobial peptide (hCAP18/LL-37) in Chinese cabbage. Plant Biotechnol Rep 6: 39–46 10.1007/s11816-011-0193-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamakawa H, Hakata M (2010) Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol 51: 795–809 10.1093/pcp/pcq034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rerksiri W, Zhang X, Xiong H, Chen X (2013) Expression and promoter analysis of six heat stress-inducible genes in rice. ScientificWorldJournal 2013: 397401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lindquist S, Craig E (1988) The heat-shock proteins. Annu Rev Genet 22: 631–677. [DOI] [PubMed] [Google Scholar]
- 50. Lee GJ, Roseman AM, Saibil HR, Vierling E (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16: 659–671 10.1093/emboj/16.3.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blumenthal C, Bekes F, Wrigley C, Barlow E (1990) The Acquisition and Maintenance of Thermotolerance in Australian Wheats. Aust J Plant Physiol 17: 37 10.1071/PP9900037 [DOI] [Google Scholar]
- 52. Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61: 243–282 10.1146/annurev.physiol.61.1.243 [DOI] [PubMed] [Google Scholar]
- 53. Iba K (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53: 225–245 10.1146/annurev.arplant.53.100201.160729 [DOI] [PubMed] [Google Scholar]
- 54. Sorensen JG, Kristensen TN, Loeschcke V (2003) The evolutionary and ecological role of heat shock proteins. Ecol Lett 6: 1025–1037. [Google Scholar]
- 55. Young R, Elliott T (1989) Stress proteins, infection, and immune surveillance. Cell 59: 5–8. [DOI] [PubMed] [Google Scholar]
- 56. Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr Opin Biotechnol 16: 123–132 10.1016/j.copbio.2005.02.001 [DOI] [PubMed] [Google Scholar]
- 57. Vierling E (1991) The Roles of Heat Shock Proteins in Plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620. [Google Scholar]
- 58. Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf K-D (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13: 183–197 10.1007/s12192-008-0032-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waters ER, Lee GJ, Vierling E (1996) Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot 47: 325–338. [Google Scholar]
- 60. Sun W, Van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1–9. [DOI] [PubMed] [Google Scholar]
- 61. Ouyang Y, Chen J, Xie W, Wang L, Zhang Q (2009) Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol Biol 70: 341–357 10.1007/s11103-009-9477-y [DOI] [PubMed] [Google Scholar]
- 62. Sarkar NK, Kim YK, Grover A (2009) Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ham D-J, Moon J-C, Hwang S-G, Jang CS (2013) Molecular characterization of two small heat shock protein genes in rice: their expression patterns, localizations, networks, and heterogeneous overexpressions. Mol Biol Rep: 1–12. [DOI] [PubMed]
- 64. Chang PFL, Jinn TL, Huang WK, Chen Y, Chang HM, et al. (2007) Induction of a cDNA clone from rice encoding a class II small heat shock protein by heat stress, mechanical injury, and salicylic acid. Plant Sci 172: 64–75 10.1016/j.plantsci.2006.07.017 [DOI] [Google Scholar]
- 65. Civán P, Svec M (2009) Genome-wide analysis of rice (Oryza sativa L. subsp. japonica) TATA box and Y Patch promoter elements. Genome 52: 294–297 10.1139/G09-001 [DOI] [PubMed] [Google Scholar]
- 66. Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, et al. (2007) Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics 8: 8–67 10.1186/1471-2164-8-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: An overview. Environ Exp Bot 61: 199–223 10.1016/j.envexpbot.2007.05.011 [DOI] [Google Scholar]
- 68. Conley AJ, Joensuu JJ, Menassa R, Brandle JE (2009) Induction of protein body formation in plant leaves by elastin-like polypeptide fusions. BMC Biol 7: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Joensuu JJ, Conley AJ, Lienemann M, Brandle JE, Linder MB, et al. (2010) Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol 152: 622–633 10.1104/pp.109.149021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Torrent M, Llompart B, Lasserre-Ramassamy S, Llop-Tous I, Bastida M, et al. (2009) Eukaryotic protein production in designed storage organelles. BMC Biol 7: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Larkindale J, Vierling E (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 10.1104/pp.107.112060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Breitler JC, Vassal JM, del Mar Catala M, Meynard D, Marfà V, et al. (2004) Bt rice harbouring cry genes controlled by a constitutive or wound-inducible promoter: protection and transgene expression under Mediterranean field conditions. Plant Biotechnol J 2: 417–430 10.1111/j.1467-7652.2004.00086.x [DOI] [PubMed] [Google Scholar]
- 73. Dai Z, Hooker BS, Anderson DB, Thomas SR (2000) Expression of Acidothermus cellulolyticus endoglucanase E1 in transgenic tobacco: biochemical characteristics and physiological effects. Transgenic Res 9: 43–54 10.1023/A:1008922404834 [DOI] [PubMed] [Google Scholar]
- 74. Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J 276: 3148–3162 10.1111/j.1742-4658.2009.07033.x [DOI] [PubMed] [Google Scholar]
- 75. Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, et al. (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143: 1467–1483 10.1104/pp.106.091900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sharma R, Mohan Singh RK, Malik G, Deveshwar P, Tyagi AK, et al. (2009) Rice cytosine DNA methyltransferases - gene expression profiling during reproductive development and abiotic stress. FEBS J 276: 6301–6311 10.1111/j.1742-4658.2009.07338.x [DOI] [PubMed] [Google Scholar]
- 77. Zheng L, Huang F, Narsai R, Wu J, Giraud E, et al. (2009) Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol 151: 262–274 10.1104/pp.109.141051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dubey S, Misra P, Dwivedi S, Chatterjee S, Bag SK, et al. (2010) Transcriptomic and metabolomic shifts in rice roots in response to Cr (VI) stress. BMC Genomics 11: 648 10.1186/1471-2164-11-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kang K, Park S, Natsagdorj U, Kim YS, Back K (2011) Methanol is an endogenous elicitor molecule for the synthesis of tryptophan and tryptophan-derived secondary metabolites upon senescence of detached rice leaves. Plant J 66: 247–257 10.1111/j.1365-313X.2011.04486.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A summary of the expression profiles of 16 genes selected as having the highest expression or induction in response to heat-shock in 7-day IR64 ( indica ) seedlings subjected to cold, salt and drought stress for 3 h. Data on 14-day ZhongHua 11 (japonica) seedlings subjected to heat shock for 3 h are also shown. Dark to light color scale represents high to low expression levels, black corresponding to 11,309 normalized fluorescence units. Arrows indicate the two sequences whose promoters were selected to produce transgenic plants.
(TIF)
Recombinant BP100-DsRed-tag54 accumulation in transgenic rice seedlings. Western blot analysis of proteins from GM plants carrying pHsp82::dsred-tag54 and pHsp82::bp100-dsred-tag54 (two independent events taken as example) treated at 42°C. Recombinant protein in TSP was extracted from rice seedlings (five plants per event) and 30 µg of TSP per lane was boiled for five minutes and separated by SDS-PAGE before transfer to nitrocellulose filters. Recombinant proteins were detected using the mAb54k antibody (diluted 1∶1,500) and the horseradish peroxidase-labeled anti-mouse IgG secondary antibody (diluted 1∶10,000) followed by ECL chemiluminescent detection.
(TIF)
Confocal micrographs of rice radicles from transgenic plantlets carrying pHsp82:: bp100-dsred-tag54 (A and C) and control pHsp82:: dsred-tag54 (B and D), at the V2 developmental stage, subjected to 42°C for 2 hours and further incubated under control growth conditions for three days in a culture chamber. (A and B), DsRed fluorescence; (C and D), bright field. Scale bars: 0.5 µm.
(TIF)
A summary of the expression profiles of a selection of sequences induced by temperature, drought and NaCl stress, and hormone treatment, in callus and seedlings of rice, based on the following in silico published data [74]–[79] . Genes with mRNA levels below that of Os.hsp18 in control seedlings (control) and above that of Os.hsp82 after induction (induced) are shown. Normalized expression levels in 28 tissues, relevant to the process of obtaining transgenic plants, of three rice genotypes: Minghui 63 (M, indica), Zhenshan 97 (Z, indica) and Nipponbare (N, japonica) (grey scale). The mRNA levels in these tissues and developmental stages were estimated using the mean expression values (green scale). Dark to light color scale represents high to low expression levels, the darkest corresponding to 37,405 (grey scale) and 12,480 (green scale) normalized fluorescence units.
(TIF)
Primer sequences and PCR conditions used. Additional restriction sites are underlined.
(DOCX)
Sequences with the greatest expression changes in response to heat stress in rice, extracted from publically available microarray hybridization data. Details on the sequence (representative public ID, Affymetrix code and description) and the mRNA expression in response to treatment at 42°C for 3 h [normalized fluorescence units in rice seedlings under control (control) and heat-shock (heat-shock) conditions; fold change (fold) and difference (HS-C) of normalized fluorescence intensities in the two conditions are also indicated]. Note that the sequences with the same Affymetrix number correspond to the same gene. Dark to light scale of shading represents high to low expression level. The promoters of the two sequences indicated in bold were selected to produce transgenic plants.
(DOCX)
Transformation efficiencies of the different constructs encoding the DsRed-tag54 reporter or the phytotoxic BP100.2 AMP, under the control of a rice heat-shock (pHsp18 and pHsp82) or a maize constitutive (pUbi) promoter.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.