Abstract
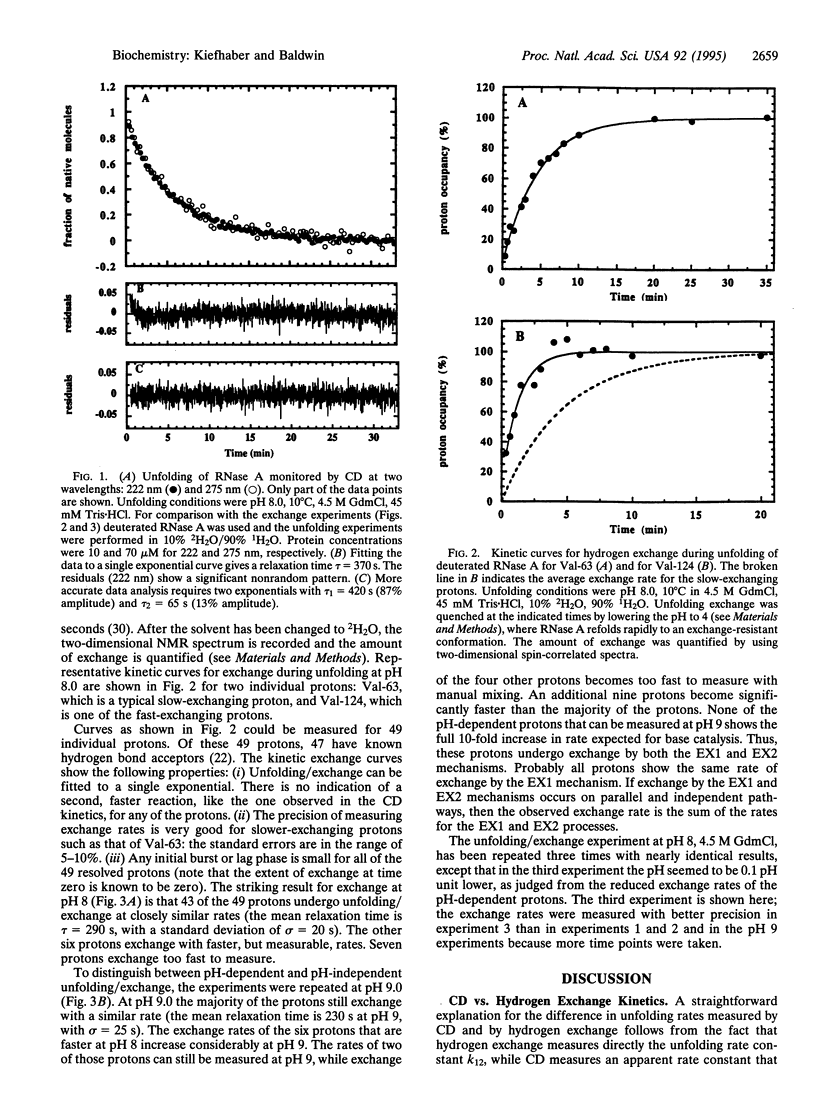
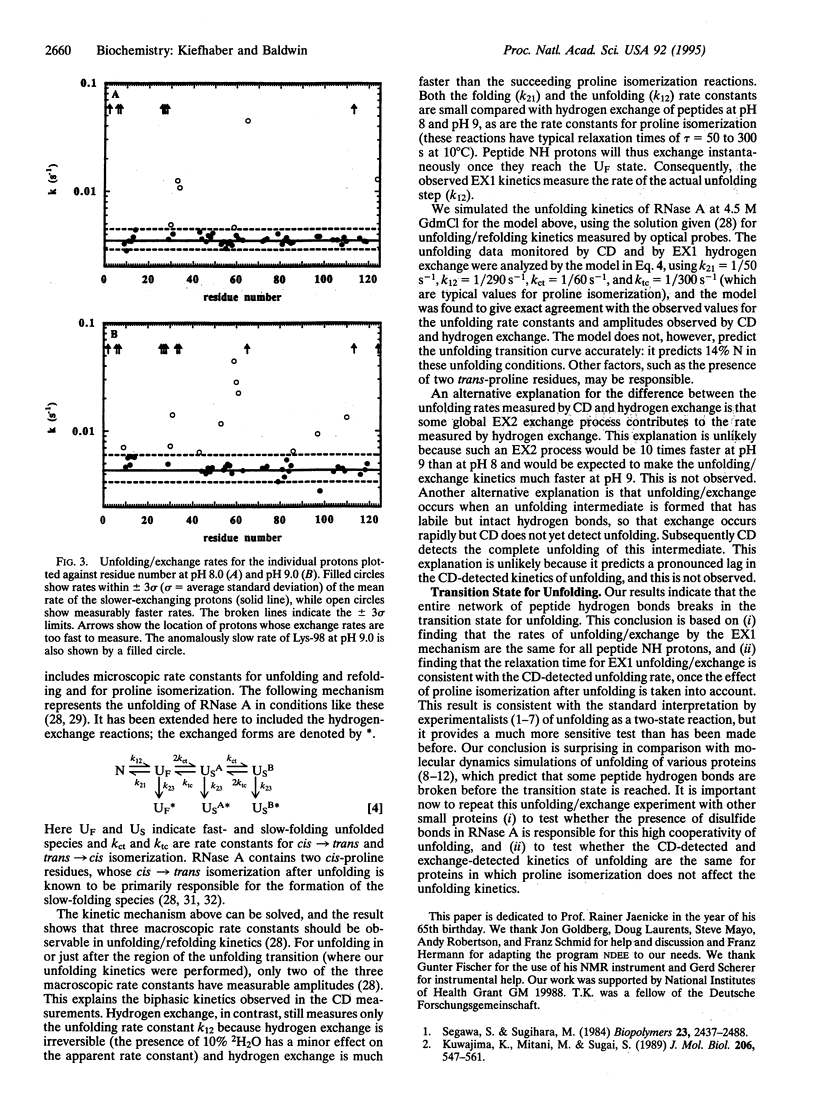
A sensitive test for kinetic unfolding intermediates in ribonuclease A (EC 3.1.27.5) is performed under conditions where the enzyme unfolds slowly (10 degrees C, pH 8.0, 4.5 M guanidinium chloride). Exchange of peptide NH protons (2H-1H) is used to monitor structural opening of individual hydrogen bonds during unfolding, and kinetic models are developed for hydrogen exchange during the process of protein unfolding. The analysis indicates that the kinetic process of unfolding can be monitored by EX1 exchange (limited by the rate of opening) for ribonuclease A in these conditions. Of the 49 protons whose unfolding/exchange kinetics was measured, 47 have known hydrogen bond acceptor groups. To test whether exchange during unfolding follows the EX2 (base-catalyzed) or the EX1 (uncatalyzed) mechanism, unfolding/exchange was measured both at pH 8.0 and at pH 9.0. A few faster-exchanging protons were found that undergo exchange by both EX1 and EX2 processes, but the 43 slower-exchanging protons at pH 8 undergo exchange only by the EX1 mechanism, and they have closely similar rates. Thus, it is likely that all 49 protons undergo EX1 exchange at the same rate. The results indicate that a single rate-limiting step in unfolding breaks the entire network of peptide hydrogen bonds and causes the overall unfolding of ribonuclease A. The additional exchange observed for some protons that follows the EX2 mechanism probably results from equilibrium unfolding intermediates and will be discussed elsewhere.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin R. L. Protein folding. Matching speed and stability. Nature. 1994 May 19;369(6477):183–184. doi: 10.1038/369183a0. [DOI] [PubMed] [Google Scholar]
- Caflisch A., Karplus M. Molecular dynamics simulation of protein denaturation: solvation of the hydrophobic cores and secondary structure of barnase. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1746–1750. doi: 10.1073/pnas.91.5.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. L., Baase W. A., Schellman J. A. Low-temperature unfolding of a mutant of phage T4 lysozyme. 2. Kinetic investigations. Biochemistry. 1989 Jan 24;28(2):691–699. doi: 10.1021/bi00428a042. [DOI] [PubMed] [Google Scholar]
- Chen X., Matthews C. R. Thermodynamic properties of the transition state for the rate-limiting step in the folding of the alpha subunit of tryptophan synthase. Biochemistry. 1994 May 24;33(20):6356–6362. doi: 10.1021/bi00186a040. [DOI] [PubMed] [Google Scholar]
- Daggett V., Levitt M. A model of the molten globule state from molecular dynamics simulations. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5142–5146. doi: 10.1073/pnas.89.11.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Fajardo J. E., Shatkin A. J. Translation of bicistronic viral mRNA in transfected cells: regulation at the level of elongation. Proc Natl Acad Sci U S A. 1990 Jan;87(1):328–332. doi: 10.1073/pnas.87.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Honeycutt J. D., Thirumalai D. The nature of folded states of globular proteins. Biopolymers. 1992 Jun;32(6):695–709. doi: 10.1002/bip.360320610. [DOI] [PubMed] [Google Scholar]
- Houry W. A., Rothwarf D. M., Scheraga H. A. A very fast phase in the refolding of disulfide-intact ribonuclease A: implications for the refolding and unfolding pathways. Biochemistry. 1994 Mar 8;33(9):2516–2530. doi: 10.1021/bi00175a022. [DOI] [PubMed] [Google Scholar]
- Itzhaki L. S., Evans P. A., Dobson C. M., Radford S. E. Tertiary interactions in the folding pathway of hen lysozyme: kinetic studies using fluorescent probes. Biochemistry. 1994 May 3;33(17):5212–5220. doi: 10.1021/bi00183a026. [DOI] [PubMed] [Google Scholar]
- Jackson S. E., Fersht A. R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991 Oct 29;30(43):10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Kohler H. H., Schmid F. X. Kinetic coupling between protein folding and prolyl isomerization. I. Theoretical models. J Mol Biol. 1992 Mar 5;224(1):217–229. doi: 10.1016/0022-2836(92)90585-8. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Schmid F. X. Kinetic coupling between protein folding and prolyl isomerization. II. Folding of ribonuclease A and ribonuclease T1. J Mol Biol. 1992 Mar 5;224(1):231–240. doi: 10.1016/0022-2836(92)90586-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. A consideration of alternative models for the initiation of translation in eukaryotes. Crit Rev Biochem Mol Biol. 1992;27(4-5):385–402. doi: 10.3109/10409239209082567. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989 Nov;9(11):5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie. 1994;76(9):815–821. doi: 10.1016/0300-9084(94)90182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Features in the 5' non-coding sequences of rabbit alpha and beta-globin mRNAs that affect translational efficiency. J Mol Biol. 1994 Jan 7;235(1):95–110. doi: 10.1016/s0022-2836(05)80019-1. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M., Shatkin A. J. Sequences and properties of two ribosome binding sites from the small size class of reovirus messenger RNA. J Biol Chem. 1977 Oct 10;252(19):6895–6908. [PubMed] [Google Scholar]
- Kuwajima K., Mitani M., Sugai S. Characterization of the critical state in protein folding. Effects of guanidine hydrochloride and specific Ca2+ binding on the folding kinetics of alpha-lactalbumin. J Mol Biol. 1989 Apr 5;206(3):547–561. doi: 10.1016/0022-2836(89)90500-7. [DOI] [PubMed] [Google Scholar]
- Li A., Daggett V. Characterization of the transition state of protein unfolding by use of molecular dynamics: chymotrypsin inhibitor 2. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10430–10434. doi: 10.1073/pnas.91.22.10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. T., MacDougald O. A., Diehl A. M., Lane M. D. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark A. E., van Gunsteren W. F. Simulation of the thermal denaturation of hen egg white lysozyme: trapping the molten globule state. Biochemistry. 1992 Sep 1;31(34):7745–7748. doi: 10.1021/bi00149a001. [DOI] [PubMed] [Google Scholar]
- Peabody D. S., Subramani S., Berg P. Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol Cell Biol. 1986 Jul;6(7):2704–2711. doi: 10.1128/mcb.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico M., Bruix M., Santoro J., Gonzalez C., Neira J. L., Nieto J. L., Herranz J. Sequential 1H-NMR assignment and solution structure of bovine pancreatic ribonuclease A. Eur J Biochem. 1989 Aug 15;183(3):623–638. doi: 10.1111/j.1432-1033.1989.tb21092.x. [DOI] [PubMed] [Google Scholar]
- Robertson A. D., Purisima E. O., Eastman M. A., Scheraga H. A. Proton NMR assignments and regular backbone structure of bovine pancreatic ribonuclease A in aqueous solution. Biochemistry. 1989 Jul 11;28(14):5930–5938. doi: 10.1021/bi00440a033. [DOI] [PubMed] [Google Scholar]
- Sali D., Bycroft M., Fersht A. R. Surface electrostatic interactions contribute little of stability of barnase. J Mol Biol. 1991 Aug 5;220(3):779–788. doi: 10.1016/0022-2836(91)90117-o. [DOI] [PubMed] [Google Scholar]
- Schultz D. A., Schmid F. X., Baldwin R. L. Cis proline mutants of ribonuclease A. II. Elimination of the slow-folding forms by mutation. Protein Sci. 1992 Jul;1(7):917–924. doi: 10.1002/pro.5560010710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa S., Sugihara M. Characterization of the transition state of lysozyme unfolding. I. Effect of protein-solvent interactions on the transition state. Biopolymers. 1984 Nov;23(11 Pt 2):2473–2488. doi: 10.1002/bip.360231122. [DOI] [PubMed] [Google Scholar]
- Serrano L., Matouschek A., Fersht A. R. The folding of an enzyme. III. Structure of the transition state for unfolding of barnase analysed by a protein engineering procedure. J Mol Biol. 1992 Apr 5;224(3):805–818. doi: 10.1016/0022-2836(92)90563-y. [DOI] [PubMed] [Google Scholar]
- Shaw M. W., Choppin P. W., Lamb R. A. A previously unrecognized influenza B virus glycoprotein from a bicistronic mRNA that also encodes the viral neuraminidase. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4879–4883. doi: 10.1073/pnas.80.16.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusher L. B., Gillman E. C., Martin N. C., Hopper A. K. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature. 1986 Nov 6;324(6092):34–38. doi: 10.1038/324034a0. [DOI] [PubMed] [Google Scholar]
- Tirado-Rives J., Jorgensen W. L. Molecular dynamics simulations of the unfolding of apomyoglobin in water. Biochemistry. 1993 Apr 27;32(16):4175–4184. doi: 10.1021/bi00067a004. [DOI] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. Early folding intermediate of ribonuclease A. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8197–8201. doi: 10.1073/pnas.87.21.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. A., Lamb R. A. Effect of mutations and deletions in a bicistronic mRNA on the synthesis of influenza B virus NB and NA glycoproteins. J Virol. 1989 Jan;63(1):28–35. doi: 10.1128/jvi.63.1.28-35.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C., Simon I., Tüchsen E. Hydrogen exchange and the dynamic structure of proteins. Mol Cell Biochem. 1982 Oct 29;48(3):135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]



