Abstract
Eukaryotic elongation factor eEF1A transits between the GTP- and GDP-bound conformations during the ribosomal polypeptide chain elongation. eEF1A*GTP establishes a complex with the aminoacyl-tRNA in the A site of the 80S ribosome. Correct codon–anticodon recognition triggers GTP hydrolysis, with subsequent dissociation of eEF1A*GDP from the ribosome. The structures of both the ‘GTP’- and ‘GDP’-bound conformations of eEF1A are unknown. Thus, the eEF1A-related ribosomal mechanisms were anticipated only by analogy with the bacterial homolog EF-Tu. Here, we report the first crystal structure of the mammalian eEF1A2*GDP complex which indicates major differences in the organization of the nucleotide-binding domain and intramolecular movements of eEF1A compared to EF-Tu. Our results explain the nucleotide exchange mechanism in the mammalian eEF1A and suggest that the first step of eEF1A*GDP dissociation from the 80S ribosome is the rotation of the nucleotide-binding domain observed after GTP hydrolysis.
INTRODUCTION
A number of eukaryotic translation factors demonstrate RNA-binding properties; however, quite a few of them directly interact with both tRNA and ribosomes. eEF1A initiates the ribosomal peptide elongation process by the formation of the eEF1A*GTP*aminoacyl-tRNA complex and the timely arrival of the aminoacyl-tRNA to the A site facilitates the selection of the correct anticodon by the mRNA-programmed ribosome. GTP hydrolysis on eEF1A leads to the steady positioning of the aminoacyl-tRNA in the A site, which subsequently triggers the transpeptidation reaction. After adopting the GDP conformation, eEF1A is thought to leave the ribosome for the eEF1Bαβγ complex, which catalyzes the exchange of GDP for GTP. This sequence of events has only been established for the bacterial elongation factor EF-Tu (1), although this is also commonly believed to be valid for the eukaryotic elongation factors (2,3). However, peculiarities of the eukaryotic eEF1A function during translation have been described (4–9).
High-resolution crystal structures of the eubacterial homolog of eEF1A, EF-Tu, were obtained for the GTP- (or, more precisely, GDPNP), GDP- and (GDPNP+aminoacyl-tRNA)-bound forms (10–12). Archaeal elongation factors 1A (aEF1A) were crystallized in both the GDP- (13) and GTP-form complexed with Pelota (14) or termination factor RF1 (15). The only studies to date on the crystal structure of eukaryotic eEF1A have described the co-crystallization of the yeast elongation factor 1A (eEF1Ay) with the truncated guanine exchange factor (GEF) (16,17). Subsequently, this structure served as a universal model to explain molecular features of any eukaryotic homolog in any nucleotide-bound conformation. Numerous attempts to crystallize the higher eukaryotic eEF1A in a GTP or GDP form were unsuccessful, and a precise understanding of the eEF1A function was not possible.
Here, we report the X-ray crystal structure of a natively folded and a post-translationally modified rabbit isoform 2 of eEF1A (eEF1A2) in a complex with GDP. The results suggest the dissociation of eEF1A*GDP from 80S ribosome is a multistage process, the first step of which is a GTP hydrolysis-induced rotation of the nucleotide-binding domain from the ribosome. Notably, the data obtained are inconsistent with the currently accepted concept of the Mg2+-dependent nucleotide exchange in eEF1A (16–18). The absence of Mg2+ contribution to the binding and dissociation of GDP explains the similar eEF1A affinity for GDP and GTP, contrary to the bacterial homolog EF-Tu. The current model refines a mechanism of the guanine exchange process in eEF1A, which is important for understanding the ribosomal polypeptide elongation in mammalian cells and contributes to the concept of mechanics of G proteins functioning.
MATERIALS AND METHODS
Purification and crystallization of the rabbit eEF1A isoform 2 was performed as described (19). Recombinant eEF1Bα (Homo Sapiens) was produced essentially as described previously (20). Briefly, eEF1Bα was cloned into the pGEX-6P-1 vector (GE Healthcare, Buckinghamshire, UK). The expression of GST-fusion protein was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3 h at 37°C in BL21(DE3)pLysE bacteria strain (Stratagene, La Jolla, CA, USA). Cells were harvested and disrupted by sonication followed by centrifugation. GST-eEF1Bα protein was purified from clear lysate on glutathione-agarose beads (Sigma-Aldrich, St. Louis, MO, USA). The beads were extensively washed and GST-eEF1Bα was eluted stepwise using a glutathione containing buffer solution. The GST moiety was removed by incubation with a Prescission protease according to the manufacturer's instructions (GE Healthcare). eEF1Bα was further purified using a Q-sepharose column and a 150–400 mM NaCl linear gradient. Pure eEF1Bα, as judged by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, was dialyzed against 50% glycerol solution containing 30 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM DTT and stored at –20°C.
Crystals of the eEF1A2*GDP complex were obtained as previously described (19). For structure determination, crystals of the eEF1A2*GDP complex were soaked in the mother liquor solution containing 0.5 mM GdCl2 before freezing in liquid nitrogen. The diffraction data were collected on ID14–4 (ESRF) and processed using the XDS package (21). The phasing procedure was performed using SHARP (22). The model was built and refined using CCP4i suite program for crystallography (REFMAC, COOT) (23). The final model was checked with MolProbity (24). The structure files and coordinates of the eEF1A2*GDP complex were deposited in the Protein Data Bank. All the figures were drawn using PyMOL (the PyMOL MolecularGraphics System, Version 1.5.0.4 Schrödinger, LLC) or Chimera (25).
The guanine nucleotide exchange rate of eEF1A2 was determined by a filter binding assay mainly as in (6). For kinetic measurements eEF1A2*[3H]GDP was prepared by incubation of 8 μM eEF1A2 with 8 μM [3H]GDP (GE Healthcare, 1500 Ci/mol) in 135 μl of 45 mM Tris-HCl, pH 7.5, containing 0.5 mM DTT, 10 mM magnesium chloride, 100 mM NH4Cl, 1 mg/ml bovine serum albumin (BSA) and 25% glycerol for 10 min at 37°C. The reaction mixture was placed at 25°C and was diluted into 1110 μl of exchange buffer (20 mM Tris-HCl, pH 7.5, 10 mM magnesium chloride, 50 mM NH4Cl and 10% glycerol). The reaction mixture was then divided into two parts of 622 μl each. The exchange reaction was performed at 25°C and was initiated by the addition of 155.5 μl of exchange buffer containing 750 μM GDP in the presence or absence of eEF1Bα. Aliquots of 100 μl were withdrawn at different times and immediately filtered through nitrocellulose filters (Millipore, Billerica, MA, USA, pore size 0.45 μm). The filters were washed three times with 1 ml of ice-cold washing buffer (20 mM Tris-HCl, pH 7.5, 10 mM magnesium chloride, 100 mM NH4Cl and 0.1 mg/ml BSA), dried and then were counted in a liquid scintillator. To evaluate the effect of ethylenediaminetetraacetic acid (EDTA) on the exchange reaction, 10 mM magnesium chloride in all buffer solutions was substituted with 10 mM EDTA. The time courses depicted in the figure were obtained by averaging four independent kinetics experiments; the error bars represent standard deviations. The data were evaluated by fitting to a single exponential function (y = A1*exp(-x/t1) + y0) using OriginPro 8 software (OriginLab, Northampton, MA, USA).
RESULTS
Overview of the structure
The crystal structure of the eEF1A2*GDP complex has been solved and refined at 2.7 Å resolution (19) (Table 1). The asymmetric unit contains two copies (molecules A and B) arranged in a ‘head to tail’ dimer configuration (Figure 1A). The physiological relevance of the dimer is uncertain although evidence for the presence of eEF1A dimers in a cellular context has recently been obtained which suggests a role in actin bundling (26) and control of eEF1A via phosphorylation (27).
Table 1. Data collection and refinement statistics.
| eEF1A2 | |
|---|---|
| Data collection | |
| X-ray source | ESRF ID14-EH4 |
| Wavelength (Å) | 0.979 |
| Space group | P6122 |
| Cell dimensions | |
| a (Å) | 135.4 |
| b (Å) | 135.4 |
| c (Å) | 304.6 |
| aResolution range (Å) | 25–2.7 (2.82–2.7) |
| aCompleteness (%) | 98.6 (95.7) |
| a,bRsym I (%) | 8.8 (86.6) |
| I/σI | 18.5 (2.8) |
| aTotal reflections | 341,059 (34,459) |
| aUnique reflections | 45,528 (4,452) |
| aMultiplicity | 7.5 (7.7) |
| Phasing statistics | |
| cFOM (centric/acentric) | 0.065/0.378 |
| Phasing power (iso/ano) | -/1.68 |
| Refinement statistics | |
| R-factor (%) | 20.2 |
| Rfree (%) | 25.5 |
| Bond length (Å) | 0.015 |
| Bond angle (°) | 1.51 |
| Mean B-factor | |
| Protein | 62.4 |
| Ligand | 44.1 |
| Ion | 51.5 |
| Water | 46.5 |
| No. atoms | |
| Residues | 6 884 |
| Ligand | 56 |
| Ion | 2 |
| water | 9 |
| Ramachandran plot | |
| Allowed regions (%) | 97.4 |
| Disallowed regions (%) | 2.6 |
aValues in parentheses are for the highest resolution shell.
bRsym (I) = [∑hkl∑i|<Ihkl> - Ihkl,i|]/[∑hkl∑i|Ihkl|], where i is the number of reflection hkl.
cFigure Of Merit = 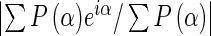 , where P(α) is the phase probability distribution and α is the phase.
, where P(α) is the phase probability distribution and α is the phase.
Figure 1.
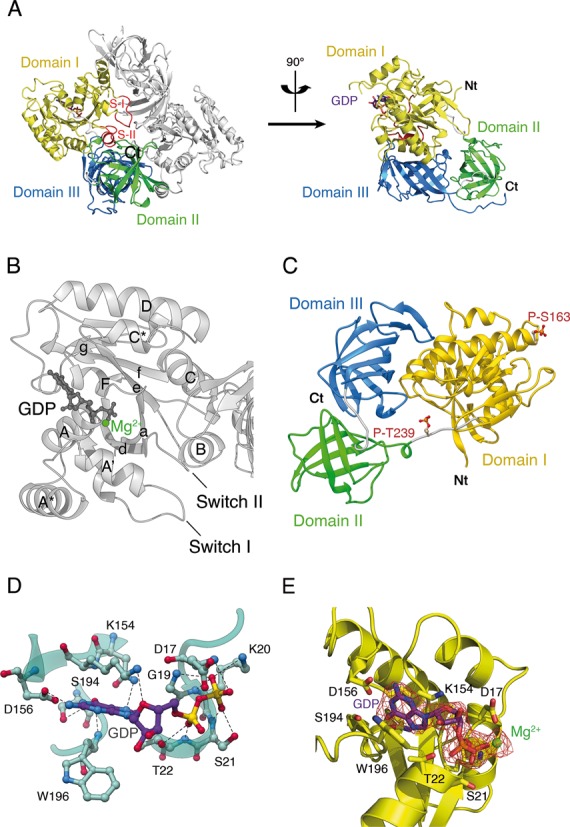
Overall structure of Oryctolagus cuniculus eEF1A2*GDP. (A) eEF1A2 is crystallized as a dimer. Three domains of eEF1A2 are colored as follows: domain I in yellow, domain II in green and domain III in blue. The Switch I and II regions are designated as S-I and S-II. The GDP is shown as a ball-and-stick representation. The N-terminus and the C-terminus are marked as Nt and Ct, respectively. (B) Presentation of the helices and beta-folds in domain I of eEF1A2*GDP. The α-helices are labeled by upper case letters, and β-strands are labeled by lower case letters. (C) Location of phosphorylated Thr239 and Ser163 in eEF1A2. (D) Network of interactions in the nucleotide-binding pocket of the GDP-bound eEF1A2. Magnesium ion is not shown for sake of clarity. (E) Electron density map corresponding to the molecule of GDP bound to eEF1A. Magnesium ion is colored in green.
Molecule A, in comparison with molecule B, displays a weaker electron density around Gly50 and contains 10 additional residues in the C-terminal region. The weak electron density around Gly50 of molecule A is consistent with the significantly increased mobility of helix A* in the eEF1A2 structure as determined from the high B-factor value and subsequent molecular dynamics simulation studies (data not shown). The local conformations of some sections of the molecules A and B fluctuate (root-mean-square (rms) deviation 0.068 Å for all Сα atoms), and results in a minor (<1 Å) shift of the GDP position. The synchronous fluctuations of Mg2+ and GDP are observed to favor the contribution of GDP to the stabilization of Mg2+ in the eEF1A2*GDP complex. Three structural domains are present in eEF1A2, domain I (4–234), domain II (241–328) and domain III (337–445) connected by linker sequences (Figure 1A). The nomenclature and location of α-helices and β-strands in the tertiary structure is depicted in Figure 1B. Studies on the native eEF1A2 structure allowed for the detection of phosphorylated Thr239 and Ser163 residues (Figure 1C). To the best of our knowledge, Thr239 phosphorylation in eEF1A2 has never been reported and has not been described by numerous phosphorylation prediction programs. The phosphorylation of Ser163 in eEF1A2 has been predicted by several programs, but has not been shown experimentally.
The main contacts between GDP and eEF1A2 are depicted in Figure 1D. Two oxygen atoms of the Asp156 side chain form H-bonds with N1 and N2 atoms of the guanine ring, respectively. Nitrogen from a peptide bond of Trp196 and the Ser194 side chain form H-bond with O6 atom of the guanine base. Asn153 forms H-bonds with N7 atom of GDP. Lys154 is linked to the ribose ring. Gly19 and Lys20 interact with O1 atom of the β-phosphate moiety. Peptide groups of Asp17 and Ser21 form H-bond with O3 and O2 atoms of the β-phosphate, correspondingly. Thr22, a part of the Walker motif, binds O2 atom of the α-phosphate. Electron density map corresponding to the molecule of GDP bound to eEF1A is shown in Figure 1E. In the mammalian eEF1A2 the GDP binding site is similar to that of the yeast and archaeal homologs.
There are no contacts between domains I and II of eEF1A2. Domain III has an eight stranded β-barrel-type structure, which interacts with domain I through a large interface including helices B and C. Interestingly, the unstructured C-terminal tail observed in molecule A is not free but linked to the ‘Domain II–linker-Domain III’ surface.
A, A* and A’ helices of eEF1A2 in nucleotide exchange
A comparative analysis of eEF1A2*GDP with the aEF1A*GTP (3AGJ), aEF1A*GDP (1SKQ) and eEF1Ay*eEF1Bα (1IJF) structures reveals a unique orientation for the A′ and A* helices in eEF1A2*GDP. While the N-terminal end of the helix A* is situated in a similar position in all molecules, the C-terminus is rotated approximately 50° in eEF1A2*GDP compared to aEF1A*GDP or eEF1Ay, and is only rotated ∼23° relative to aEF1A*GTP (Figure 2A and B). Helix A′ of eEF1A2*GDP is shifted right compared to both aEF1A*GDP and eEF1Ay.
Figure 2.
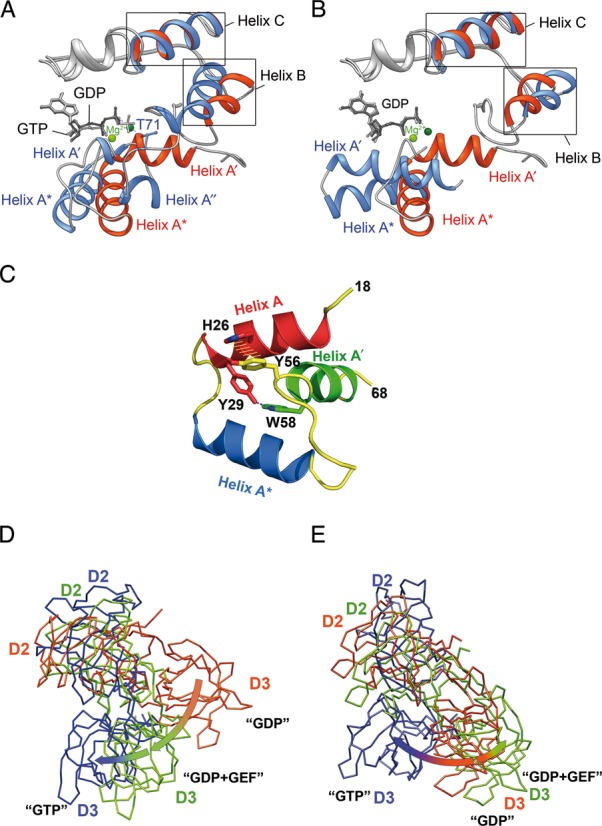
Main conformational rearrangements upon GTP hydrolysis. (A and B) A′ and A* helical arrangement in the mammalian and archaeal elongation factors. eEF1A2*GDP is superimposed with aEF1A*GTP (A) and aEF1A*GDP (B). The structure of eEF1A2 is colored in red with GDP in dark gray, aFF1A*GDP and aEF1A*GTP are colored blue with GDP or GTP in light gray. Mg2+ ions are colored light green in eEF1A2*GDP and dark green in aEF1A*GDP or aEF1A*GTP. Note the unwinding of helix A′ in aEF1A*GTP and similar orientation of helices A* in eEF1A2*GDP and aEF1A*GTP. (C) Tyr56 and Trp58 are responsible for an interaction of the A′ and A helices. Mutation of Tyr56 or Trp58 impairs the A′- A interaction during GTP binding and hydrolysis. (D) Superimposition of the domain II+III units of eEF1A2*GDP (pink), eEF1Ay (green) in the complex with eEF1Bα (not shown for sake of clarity) and aEF1A*GTP (blue) after alignment of domain I (not shown for sake of clarity) of all complexes. (E) Superimposition of the domain II+III units of bacterial EF-Tu in GDP (pink), GEF-induced (green) and GTP (blue) conformations after alignment of domain I (not shown for sake of clarity) of all complexes.
Thus, the positions of the A* and A′ helices are significantly different from the ‘intermediate’ state observed in the eukaryotic elongation factor complex with GEF (16).
A possible mechanism of the GDP-/GTP-dependent reversible changes in the A′-A* region may encompass the subdivision of A′ into two smaller helixes in the GTP form of aEF1A (14) (Figure 2A). Assuming the ‘GTP-conformations’ of eEF1A and aEF1A are similar, a comparison of the eEF1A2*GDP and aEF1A*GTP structures suggests the following mechanism. During the transition from the GDP to GTP conformation, the Tyr56-Trp58 segment of the helix A′ remains unchanged, whereas A′′ approaches the GTP-binding pocket, permitting Thr71 (Thr72 in eEF1A2) from the Switch I region to bind the γ-phosphate of GTP via Mg2+ (Figure 2A).
The role of conformational changes in the helix A′ is supported by recent findings that demonstrated the absolute importance of Tyr56 and Trp58 in eEF1Ay for yeast growth (28). There was no obvious reason for the lethality from the currently available data. However, the crystal structure of eEF1A2*GDP illustrates the importance of these residues in the interaction of the A′ and A helices. The aromatic ring of Tyr56 situated in the helix A′ forms a π-stacking interaction with the His26 imidazole ring, whereas the Nϵ1 atom of Trp58 forms a hydrogen bond with the hydroxyl group of Tyr29 in the helix A (Figure 2C). These interactions are probably needed to keep A′ and A helices together, leaving A′′ mobile and capable of promoting the Thr71/Thr72 interaction with GTP in aEF1A/eEF1A. Upon GTP hydrolysis, Thr71/Thr72 loses the Mg2+-mediated contact with the γ-phosphate, so the A′′ helix can move back and restore the integrity of the A′ helix. Thus, the strong fixation of A′ by the A helix may provide a basis for the correct positioning of the A′ helix in eEF1A2*GDP. The A-A′ contacts are obviously absent in the ‘intermediate state’ conformation of eEF1Ay observed in the complex with eEF1Bα (16). Consequently, eEF1Bα is supposedly able to interact with aY56A W58A mutated eEF1A. However, it is unlikely that the mutated protein would achieve the correct ‘GTP conformation’ after dissociation of eEF1Bα. Interestingly, the prokaryotic EF-Tu*GDP shows a different secondary structure of the effector region. Instead of A′′ in eEF1A2, an unstructured region in the Thermus aquaticus or β-hairpin in Escherichia coli EF-Tu*GDP is present (29).
Rotation of the domains (II+III) structural unit relative to domain I is different in eEF1A2*GDP and eEF1Ay bound to eEF1Bα
A comparison of the domain organization in eEF1A2*GDP, eEF1Ay bound to eEF1Bα and aEF1A*GTP illustrates the domain (II+III) unit movements relative to domain I during GDP/GTP exchange. Domains II and III of the factors were reported to move as a single body in molecular dynamic simulations (30). Evidently, eEF1Bα binding induces a 56° switch in position of the domain (II+III) unit relative to domain I (Figure 2D). The rearrangements of the nucleotide binding site and subsequent GTP binding lead to an increased angle of the domain II+III unit rotation (Figure 2D). A backward movement of the domain (II+III) unit is permitted only after GTP hydrolysis and produced the GDP-bound conformation. Notably, the bacterial and eukaryotic GEFs induce rotations of domain II+III of counterparts in opposite directions whereas subsequent GTP binding produces unidirectional rotation of aEF1A, and, possibly, eEF1A and EF-Tu domains (II+III) relative to domain I (Figure 2D and E).
The rotation of the domain (II+III) unit is accompanied by conformational changes in other regions connected with domain III. The interaction of GEF with domain II induces a change in the linker connecting domains II and III. Domain III is bound to domain I and cannot easily follow the domain II movement without rearrangement of the H-bond network. Indeed, in the eEF1Ay bound to eEF1Bα complex, the B and C helices acquire some novel contacts as compared to eEF1A2*GDP. Specifically, helix B binds to residues 422 and 430 instead of Arg381 and Arg382; and Thr142 of helix C contacts Ile343 and Val435 (Val433 in eEF1Ay) instead of Lys439 and Val437. Additionally, the Glu135–Lys386 salt bridge and the His136–Ser383 contact in eEF1A2*GDP are not observed in eEF1Ay because the conformational change induced by eEF1Bα enables Glu135 and His136 to contact the Switch II region.
Role of Mg2+ in GDP/GTP exchange in EF1A from evolutionary distinct organisms
The addition of homologous eEF1Bα to eEF1Ay or Artemia salina eEF1A accelerated the nucleotide exchange process in vitro (31,32). In eEF1Ay, the acceleration was hypothesized to be induced mainly by the forced exit of Mg2+ from the nucleotide binding site by GEF. In particular, Mg2+ displacement by Lys205 from eEF1Bα was suggested to destabilize and facilitate the dissociation of GDP from eEF1Ay (16–18).
Conversely, although Mg2+ was visible and functionally important for GTP hydrolysis in aEF1A*GTP (14), it was observed only under highly artificial conditions in aEF1A*GDP (33). Consequently, an alternative hypothesis was that the GDP/GTP exchange in aEF1A and eEF1A may occur solely due to the protein structure rearrangements, which are not necessarily coupled with the presence of Mg2+ (33).
Mg2+ had only a marginal effect on both spontaneous (Figure 3A) and GEF-dependent (Figure 3B) nucleotide exchanges in eEF1A2. The values of koff for spontaneous GDP/GDP exchange in eEF1A2 were 5.6 ± 0.3 .10−4 s−1 and 4.4 ± 0.5 .10−4 s−1, whereas the first-order rate constants for the GDP/GDP exchange catalyzed by eEF1Bα were 6.3 ± 0.3.10−3 s−1 and 10.0 ± 0.5.10−3 s−1 in the presence of 10 mM Mg2+ or EDTA, respectively. The spontaneous GDP/GDP exchange was observed to be independent of Mg2+ in eEF1Ay; however, the same study reported the importance of Mg2+ for the eEF1Bα-catalyzed nucleotide exchange process (18).
Figure 3.
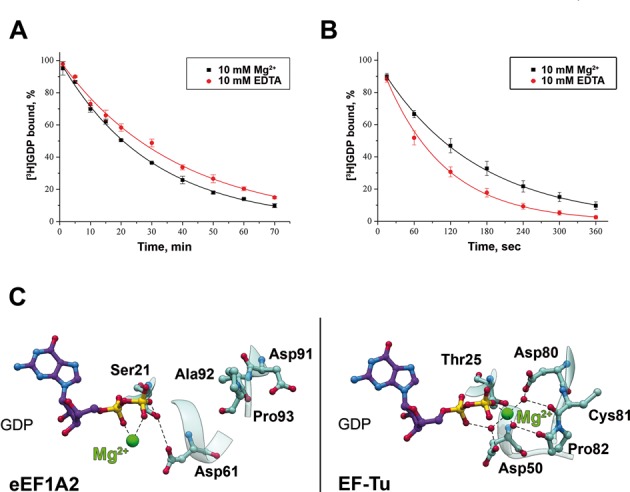
Mg2+ does not influence nucleotide exchange in eEF1A2. Mg2+ (black) and EDTA (red) do not have an impact on spontaneous (A) and eEF1Bα-catalyzed (B) nucleotide exchange process. The eEF1A2 concentration in the incubation mixture was 692 nM and eEF1Bα - 4 nM. The concentration of either Mg2+ or EDTA was 10 mM. Goodness (R2) of single exponential fits was calculated to be >0.999 for nucleotide exchange in the presence of both Mg2+ and EDTA. (C) Mg2+ contributes to GDP binding in EF-Tu rather than in eEF1A2.
In the eEF1A2*GDP structure Mg2+ is in contact with the α- and β-phosphates of GDP. In molecule A the distance from the Mg2+ ion to the Asp17 side chain is 2.7 Å, in molecule B it rises to 3.5 Å. Because molecule A is crystallized in more flexible conformation as evidenced by the unresolved electron density of Gly50 situated in the effector region we believe that approaching the Mg2+ ion by Asp17 occurs due to local intramolecular fluctuations and does not play any role in stabilization of the metal ion by eEF1A2. To our knowledge, the aspartate residue at the fourth position of a Walker A motif (GxxXxGKS/T) has never been described as a ligand in GTPases.
The affinity of eEF1A for GDP is much less than that of EF-Tu (reviewed in (34)). In EF-Tu*GDP (pdb–1EFC), Mg2+ is in contact with the β-phosphate and α-phosphate (12). However, Mg2+ is also linked to EF-Tu*GDP via direct contact with Thr25 and via water-mediated bonds with Asp51 and Asp81, whereas the corresponding residues in eEF1A2 (Asp61 and Asp91) are situated far from the magnesium binding site. There is also a direct Mg2+ contact with Thr25 which is conserved in all available EF-Tu*GDP structures. The side chain of corresponding Ser21 from the P-loop in eEF1A2 forms an H-bond with Asp61 instead. Thus, in eEF1A2*GDP the contacts of Mg2+ are limited by the α- and β-phosphates (Figure 3C). This finding, along with the kinetic data, favors the assumption that the presence of the magnesium ion cannot add any strength to the GDP stabilization in eEF1A2.
Molecular mechanism of the nucleotide exchange factor eEF1Bα function
Using the structures of both eEF1A2*GDP and eEF1Ay*GDP*eEF1Bα, we can deduce the GEF-mediated nucleotide exchange mechanism in eEF1A.
In order to destabilize GDP in eEF1A, the Switch I region has to be displaced, with a subsequent disruption of the (Glu68–His95, Arg69-Asp97, Glu68-Asp97) interaction network between Switch I and Switch II in the eEF1A2*GDP structure (Figure 4A and B). Such a rearrangement causes the loop 90–97 to move upwards and rotate. A key new observation is the orientation of Asp91, which is stabilized in eEF1A2*GDP by the interaction with His95, Asn101 and Ser107. In the presence of eEF1Bα, this residue makes a 180° flip to form a salt bridge with Lys20 and an H-bond with Ser21, which binds and stabilizes the β-phosphate in eEF1A2*GDP (Figure 4C and D). Ser21 also forms an H-bond with Lys205 of eEF1Bα (18). All of these events cause a destabilization of the β-phosphate position, which probably reflects its disorder in the eEF1Ay*eEF1Bα crystal structure (16). eEF1Bα clearly induces a rotamer change of His95 in eEF1A and permits the formation of H-bonds with Asp97 (eEF1A, helix B) and Val179 (eEF1Bα).
Figure 4.
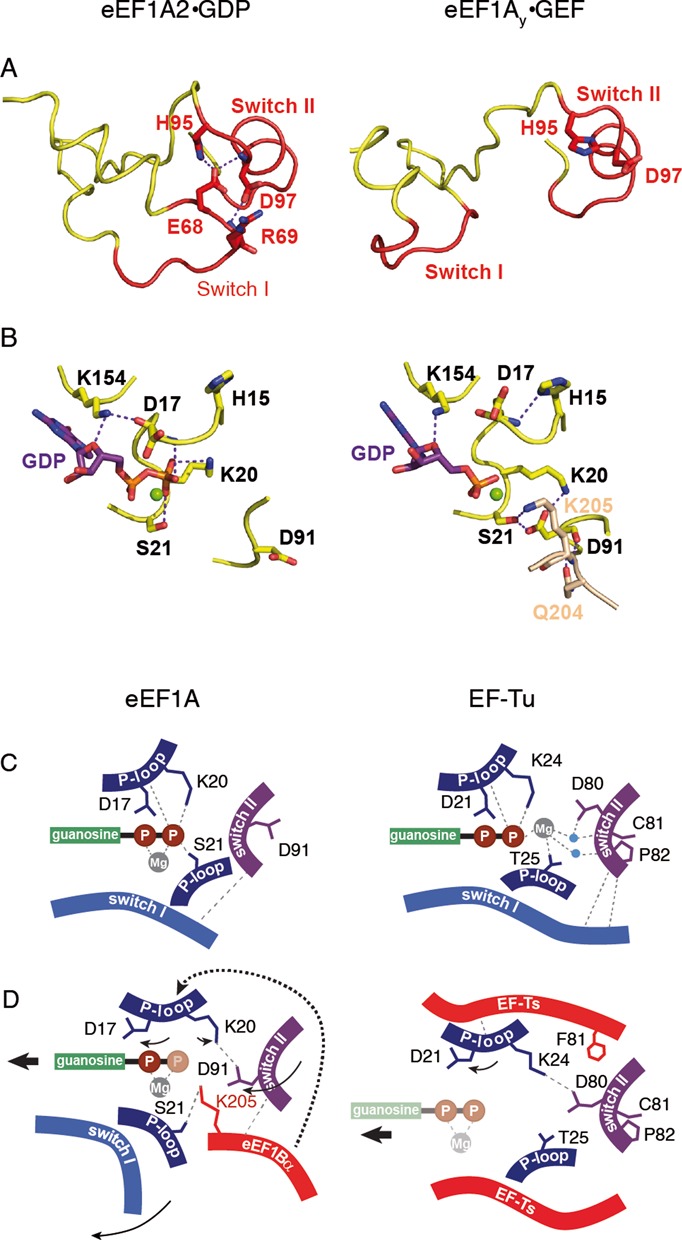
Mechanism of GEF-induced nucleotide exchange in eEF1A. (A) Conformation of the Switch I–Switch II region in eEF1A*GDP and in eEF1Ay-GEF. (B) Conformation of the GDP binding site in the absence and in the presence of GEF. Note the 180° rotation of Asp91. K205 and Q204 of eEF1Bα (numeration of yeast eEF1Bα) are shown in light brown. (C) Arrangement of the GDP-bound structures of eEF1A and EF-Tu. (D) Changes introduced in the GDP-bound structures of eEF1A and EF-Tu by corresponding GEFs. Eukaryotic GEF eEF1Bα directly disrupts contact of Ser21 with the β-phosphate, induces the conformational switch in P-loop leading to the disruption of Asp17 contact and prompts the conformational change in Switch II inducing 180° rotation of Asp91, with subsequent disruption of the Lys20 contact with the β-phosphate. Note the non-involvement of Mg2+ and GEF-induced rotation of Asp91 by 180°. Prokaryotic GEF EF-Ts causes conformational change in P loop, precluding contacts of Trp25 with Mg2+ linked to the β-phosphate, inducing rotation of Asp21 away from the β-phosphate and switch of Lys24 toward Asp81. Note the direct role of Mg2+ in the GDP stabilization, as well as similar positions of Asp81 in the GDP- and GEF-bound conformations. Water molecules are shown in blue. Magnesium ion is shown in gray. Compounds which are invisible in the structure, depicted as semi-transparent.
In the presence of eEF1Bα, the rotation of the domain (II+III) unit and the subsequent restoration of its interaction with helix B are observed. Such domain rearrangements decrease the distance between helices B and C. Consequently, the Arg96 side chain can reposition upwards and interact with Gln132 and Glu135 of helix C that disrupts the Gln132-His15 interaction. This results in His15 forming an H-bond with Asp17, which prevents it from interacting with the β-phosphate group and causes the peptide bond 16–17 to flip and reorient.
Upon interaction with eEF1Bα the H-bond between Asp17 and Lys154 is broken and amino group of the Lys154 residue flipped by about 70° when compared to eEF1A2*GDP alone (Figure 4C and D). Despite this rotation, Lys154 retains the H-bond with the oxygen atom of the ribose ring, but not Glu122. This leads to an alteration of the ribose and guanine ring orientation, whereas the α-phosphate remains in a similar position in the presence or absence of eEF1Bα.
DISCUSSION
Two models for the nucleotide exchange process in eEF1A exist. One implicates Lys205 from eEF1Bα inducing the Mg2+ removal and peptide flip in the P-loop of eEF1A for the dissociation of GDP and prevention of its re-binding (16,17). Another hypothesis suggests that Mg2+ is dispensable for GDP binding and dissociation, and instead emphasizes that the significant domain rotation destabilizes the interaction of the P-loop and the β-phosphate of GDP (33). The first model was based on the EF-Tu-GDP and eEF1Ay-eEF1Bα structures, and assumed that the GDP exchange mechanism is similar in pro- and eukaryotic elongation factors. The second model accounts for the structure of aEF1A*GDP and expands the available data from archaeal to eukaryotic elongation factors.
The structure of eEF1A2*GDP demonstrates no direct Mg2+-protein interactions. A spherical electron density observed between the α- and β-phosphates of GDP and the side chain of Asp17 was interpreted as the electrostatic interaction of the magnesium ion with the oxygen atoms of the phosphate groups situated ∼2.5 Å away. The distance from Mg2+ to the Asp17 side chain in the molecule A is about 2.7 Å, however, in the molecule B this distance increases to 3.5 Å which makes the formation of a bond between Asp17 and Mg2+ less probable. No interaction of Mg2+ with the corresponding Asp is found in all known X-ray structures of the translational GTPases. Furthermore, the fluctuations of Mg2+ and GDP in molecules A and B of the dimer coincide, which favors the notion that Mg2+ is stabilized in eEF1A2 via the GDP molecule. The coordination geometry for Mg2 was not determined, however, the water-mediated contacts, which are not visible at 2.7 Å resolution, are still possible. The kinetic experiments do not show any substantial Mg2+ effect on the nucleotide exchange rate in the presence or absence of eEF1Bα, which is consistent with the non-involvement of Mg2+ in the mechanism of GDP stabilization. The exclusion of Mg2+ resulted in the 3.6-fold acceleration of the eEF1Bα-catalyzed GDP dissociation from eEF1Ay (18) and in small 1.6-fold increase of koff for eEF1A2 (Figure 3B). Though one cannot entirely exclude some impact of Mg2+ upon the GDP release from eEF1A, the Mg2+ effect appears rather indirect and probably results from either subtle rearrangements of the GDP-binding site (35) induced by eEF1Bα or the increased stability of eEF1A*eEF1Bα complex in the presence of magnesium ions. Mg2+-induced constriction has been observed for other protein–protein complexes (36,37).
The insertion of the Gln-204 side chain from eEF1Bα between two antiparallel β-strands linking the Switch I and Switch II regions was previously claimed to force Asp91 away from Ser21 and a water molecule, contributing to GDP dissociation (16). This is inconsistent with our observations because the Asp91 of eEF1A2*GDP undergoes a 180° rotation away from the nucleotide binding site, and forms H-bonds with His95 and Asn101 from helix B of the Switch II region. However, in the GTP form of aEF1A (14) and in the GEF-complexed form of eEF1Ay (16) Asp91 is situated near the β-phosphate. Thus, eEF1Bα forces Asp91 back into the nucleotide-binding site rather than moving it out as was previously suggested (16) which may contribute to the dissociation of GDP as described in the Results section.
The 180° turn of Asp91 away from the GDP-binding site is detected also in aEF1A*GDP (13) and appears to be exclusive for non-bacterial elongation factors 1A. Interestingly, in the yeast termination factor eRF3, the corresponding Asp322 adopts the same as Asp91/90 in eEF1A2*GDP/aEF1A*GDP flipped position comparing to Asp81/Asp90 in EF-Tu*GTP/aEF1A*GTP correspondingly. However, contrary to the elongation factors, no nucleotide-dependent change in position of Asp322 was found in eRF3 (38).
The eEF1A2*GDP structure presented here contributes to the evolutionary understanding of the elongation factor 1 family. It is known that the structures of GDP-bound proteins display a large difference, while the GTP-bound forms of the G domain are mostly similar (39). The functional GTP form has the same configuration in EF-Tu and aEF1A and, possibly, in eEF1A, considering the principal common translation function of the factors. GTP forms of the elongation factors are very likely to preserve a universal mechanism of interaction with ribosomes in pro- and eukaryotes. However, the GDP-bound conformations of EF-Tu, aEF1A and eEF1A2 demonstrate essential differences. Thus, EF-Tu*GDP and aEF1A*GDP/eEF1A2*GDP represent various starting points toward achieving a universal GTP-bound conformation via different GEF-mediated mechanisms in bacterial and archaeal or eukaryotic cells. The bacterial and non-bacterial nucleotide exchange factors catalyze GDP/GTP exchange via distinct conformational changes, which results in similar GTP-bound conformations. Molecular details of the different mechanisms in prokaryotes and eukaryotes depicted as in (40) are summarized in Figure 4C and D.
The role for the domains’ rotation during the transition of ribosome-bound eEF1A from the GTP- to GDP-bound conformation has been assessed. The structure of the eEF1A*GTP*aminoacyl-tRNA complex in the 80S ribosome was modeled by superimposing the X-ray structures of the yeast 80S ribosomes (41) and the bacterial 70S ribosome with the aminoacyl-tRNA and EF-Tu*GDPCP (12). We observed that substitution of the GTP form of eEF1A with eEF1A*GDP, assuming that an interaction of eEF1A*GDP with tRNA persists for some time after GTP hydrolysis (8,9), caused domain I to rotate out of the ribosome while domains II and III remained in place (Supplementary Figure S1). This result means that after GTP hydrolysis, there is a disruption in the interaction between domain I of eEF1A and the ribosome. Consequently, eEF1A*GDP is retained on the ribosome mainly due to some domain (II+III) contacts and can dissociate easier. Thus, the domain rotation in eEF1A*GDP upon GTP hydrolysis is a first step of releasing the protein from the 80S ribosome, which resembles dissociation of EF-Tu from the 70S ribosome (reviewed in (1)).
The link between the A and A′ helices provided by the Tyr56-His26 and Trp58-Tyr29 interactions is suggested to play a vital role for adopting a specific GDP-bound conformation of eEF1A2. Otherwise the α-helical A*-A′ region would shift away from the nucleotide as seen in the Sulfolobus solfataricus aEF1A*GDP (13). As compared to eEF1A2, the archaeal protein has two Phe residues substituted for Tyr56 and Trp58 correspondingly, lacks the aromatic ring, equivalent to His26 and contains Met28 instead of Tyr29. Also, the A-A′ interaction may be important for the correct positioning of Thr71/Thr72 into the GTP nucleotide binding site of eEF1A. The mutation of either Trp58 or Tyr56 is lethal (28); therefore, the stabilization strength of each residue is probably not sufficient to maintain the interaction of the helix A and A′ during the unwinding of the helix A′ and shift of A′′ to the GTP-binding site. Moreover, we speculate that the link between A and A′, which forces these helices to move as a single unit, may be important for the GEF binding and dissociation mechanism. To reach the GDP binding site in eEF1A*GDP, GEF must separate the Switch I and Switch II regions. In eEF1A*GTP, the A-A′ unit apparently moves back into place and provides a mechanic force to dissociate GEF. This mechanism may be specific for the eukaryotic nucleotide exchange because the A-A′ link apparently cannot be formed in archaeal factors.
In higher vertebrates, there are two isoforms of eEF1A, which are 97% similar and are encoded by different genes. The expression of the isoforms is mutually exclusive: eEF1A2 appears in skeletal myocytes, cardiomyocytes and neurons; eEF1A1 is present in all remaining cells of the organism (42). Therefore, the role of isoforms during translation should be principally the same, whereas their tissue-specific localization suggests they may have different additional functions. Importantly, eEF1A2 is overexpressed in a number of human cancers (43,44) and was shown to have oncogene-like properties in some cases (45,46). The phosphorylation of Tyr29 has been reported in global phospho-proteomic cancer studies (47,48). This modification would prevent the Tyr29–Trp58 interaction in eEF1A2 (Figure 2C), which may affect the stability of the A-A′ helices linkage and result in an impairment of translation. The Tyr29-phosphorylated cellular pool could fulfill some other functional duty, such as signaling or actin bundling (49,50).
Phosphorylated Ser163 and Thr239 are highly conserved in both eEF1A1 and eEF1A2 of higher eukaryotes. Bioinformatics predicts that the phosphorylation of Ser163 in eEF1A1 and eEF1A2 (92% identical) is probably performed by different protein kinases, ataxia telangiectasia mutated or casein kinase 1, respectively, due to a local difference in the primary structures of the isoforms. Identification of a kinase for Thr239 is an intriguing task for the future. The local landscape near the phosphorylated Thr239 residue includes Lys146 and Lys244 which can be acetylated in vivo (51,52). Phosphorylated Ser163 is situated near Lys165 which is dimethylated in the A1 isoform (53) and trimethylated in the A2 isoform (6). Cross-talk between the modifications may provide a unique isoform-specific local landscape for eEF1A, which can be utilized, in particular, to differentially distribute and/or functionally modify the 97% similar isoforms.
A segment of the unstructured C-terminus was observed in molecule A of the eEF1A2 dimer. The C-terminal tail was previously proposed to participate in aminoacyl-tRNA binding by EF-Tu (54). However, the C-terminal tail of eEF1A2 adopts a somewhat different form compared to the EF-Tu conformation and is situated far from the putative tRNA-binding region. This region of eEF1A2 contains a number of lysine residues which may serve as a platform for binding other kinds of RNA (55,56).
eEF1A belongs to a family of G-proteins, which function as GTP hydrolysis-dependent molecular switches and are pivotal for cell life. Typically, the activity of G-proteins is regulated by GEF that stimulate dissociation of tightly bound GDP produced by GTP hydrolysis (57). Mechanism of GDP/GTP exchange usually involves Mg2+. The affinity of eEF1A for GDP and GTP is similar, contrary to its prokaryotic homolog EF-Tu. The data on X-ray structure of eEF1A2 explain the mechanics behind this, suggesting that a drop in the affinity for GDP is caused by exclusion of the magnesium ion role in the stabilization of GDP in eEF1A (Figure 3C). The independence from Mg2+, with consequent decrease in the GDP binding strength, seems to ensure the reliability of eEF1A functioning in case of GEF deficiency. In addition, eEF1A plays a number of non-orthodox roles and, therefore, is distributed evenly throughout the cell while the function and localization of its GEFs is mostly limited to the protein synthesis compartments (58). Similar affinity of eEF1A for GDP and GTP may help to maintain spontaneous GDP/GTP exchange in the GEF-deficient regions of cells where the eEF1A functions are not related to the protein synthesis. That is possible because the intracellular concentration of GTP is much higher than GDP.
The overall strategy of the eukaryotic ribosomal chain elongation is to ensure a high accuracy of the protein synthesis along with a reasonable rate. The change of the rate-limiting step from GDP dissociation in EF-Tu to GTP hydrolysis in eEF1A (32), which is a consequence of decreased affinity of the eukaryotic protein for GDP, apparently contributes to this function as the hydrolysis of GTP is directly triggered by the correct codon–anticodon recognition. We believe that the understanding of difference in the molecular mechanisms of nucleotide exchange in pro- and eukaryotic elongation factors will be helpful for the development of anti-bacterial drugs specifically targeted to EF-Tu.
The analysis of the crystal structure of eEF1A2*GDP together with the structures of eEF1Ay*eEF1Bα and aEF1A*GDP provides the important information on the conformational changes essential for the eEF1A function, refines existing and describes novel aspects of the mechanics of the nucleotide exchange process in the translation factors. The results provide background for comprehensive knowledge of the mechanism and evolution of the polypeptide chain elongation by the ribosome.
ACCESSION NUMBERS
Coordinates and structure factors have been deposited at the Protein Data Bank (pdb–4C0S).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr. Stephen Cusack (EMBL Grenoble Outstation, France) for continued interest and support of this work. The contribution of Oleksandra Novosylna and Nikolay Pydiura into the preparatory part of the work is greatly appreciated. We thank the staff of ESRF and of EMBL-Grenoble for their assistance and support in using beamlines ID14–4.
Footnotes
The authors wish it to be known that, in their opinion, the first three authors should be regarded as Joint First Authors.
FUNDING
France-Ukraine PICS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Voorhees R.M., Ramakrishnan V. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 2.Dever T.E., Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodnina M.V., Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr. Opin. Cell. Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Budkevich T.V., Timchenko A.A., Tiktopulo E.I., Negrutskii B.S., Shalak V.F., Petrushenko Z.M., Aksenov V.L., Willumeit R., Kohlbrecher J., Serdyuk I.N., et al. Extended conformation of mammalian translation elongation factor 1A in solution. Biochemistry. 2002;41:15342–15349. doi: 10.1021/bi026495h. [DOI] [PubMed] [Google Scholar]
- 5.Guzzo C.M., Yang D.C. Lysyl-tRNA synthetase interacts with EF1alpha, aspartyl-tRNA synthetase and p38 in vitro. Biochem. Biophys. Res. Commun. 2008;365:718–723. doi: 10.1016/j.bbrc.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 6.Kahns S., Lund A., Kristensen P., Knudsen C.R., Clark B.F., Cavallius J., Merrick W.C. The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res. 1998;26:1884–1890. doi: 10.1093/nar/26.8.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Negrutskii B.S., Shalak V.F., Kerjan P., El'skaya A.V., Mirande M. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J. Biol. Chem. 1999;274:4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 8.Petrushenko Z.M., Budkevich T.V., Shalak V.F., Negrutskii B.S., El'skaya A.V. Novel complexes of mammalian translation elongation factor eEF1A.GDP with uncharged tRNA and aminoacyl-tRNA synthetase. Implications for tRNA channeling. Eur. J. Biochem. 2002;269:4811–4818. doi: 10.1046/j.1432-1033.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- 9.Petrushenko Z.M., Negrutskii B.S., Ladokhin A.S., Budkevich T.V., Shalak V.F., El'skaya A.V. Evidence for the formation of an unusual ternary complex of rabbit liver EF-1alpha with GDP and deacylated tRNA. FEBS Lett. 1997;407:13–17. doi: 10.1016/s0014-5793(97)00242-1. [DOI] [PubMed] [Google Scholar]
- 10.Kjeldgaard M., Nissen P., Thirup S., Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 11.Nissen P., Thirup S., Kjeldgaard M., Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 12.Song H., Parsons M.R., Rowsell S., Leonard G., Phillips S.E. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution. J. Mol. Biol. 1999;285:1245–1256. doi: 10.1006/jmbi.1998.2387. [DOI] [PubMed] [Google Scholar]
- 13.Vitagliano L., Masullo M., Sica F., Zagari A., Bocchini V. The crystal structure of Sulfolobus solfataricus elongation factor 1alpha in complex with GDP reveals novel features in nucleotide binding and exchange. EMBO J. 2001;20:5305–5311. doi: 10.1093/emboj/20.19.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi K., Kikuno I., Kuroha K., Saito K., Ito K., Ishitani R., Inada T., Nureki O. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1alpha complex. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17575–17579. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi K., Saito K., Ishitani R., Ito K., Nureki O. Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex. Nucleic Acids Res. 2012;40:9319–9328. doi: 10.1093/nar/gks660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen G.R., Pedersen L., Valente L., Chatterjee I., Kinzy T.G., Kjeldgaard M., Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol. Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 17.Andersen G.R., Valente L., Pedersen L., Kinzy T.G., Nyborg J. Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex. Nat. Struct. Biol. 2001;8:531–534. doi: 10.1038/88598. [DOI] [PubMed] [Google Scholar]
- 18.Pittman Y.R., Valente L., Jeppesen M.G., Andersen G.R., Patel S., Kinzy T.G. Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha. J. Biol. Chem. 2006;281:19457–19468. doi: 10.1074/jbc.M601076200. [DOI] [PubMed] [Google Scholar]
- 19.Yaremchuk A., Shalak V.F., Novosylna O.V., Negrutskii B.S., Crepin T., El'skaya A.V., Tukalo M. Purification, crystallization and preliminary X-ray crystallographic analysis of mammalian translation elongation factor eEF1A2. Acta Crystallogr. F Struct .Biol. Cryst. Com. 2012;68:295–297. doi: 10.1107/S1744309112000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cans C., Passer B.J., Shalak V., Nancy-Portebois V., Crible V., Amzallag N., Allanic D., Tufino R., Argentini M., Moras D., et al. Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13892–13897. doi: 10.1073/pnas.2335950100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vonrhein C., Blanc E., Roversi P., Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z., Lasker K., Schneidman-Duhovny D., Webb B., Huang C.C., Pettersen E.F., Goddard T.D., Meng E.C., Sali A., Ferrin T.E. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunai F., Ando K., Ueno H., Numata O. Tetrahymena eukaryotic translation elongation factor 1A (eEF1A) bundles filamentous actin through dimer formation. J. Biochem. 2006;140:393–399. doi: 10.1093/jb/mvj169. [DOI] [PubMed] [Google Scholar]
- 27.Sanges C., Scheuermann C., Zahedi R.P., Sickmann A., Lamberti A., Migliaccio N., Baljuls A., Marra M., Zappavigna S., Reinders J., et al. Raf kinases mediate the phosphorylation of eukaryotic translation elongation factor 1A and regulate its stability in eukaryotic cells. Cell Death Dis. 2012;3:e276. doi: 10.1038/cddis.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belyi Y., Tartakovskaya D., Tais A., Fitzke E., Tzivelekidis T., Jank T., Rospert S., Aktories K. Elongation factor 1A is the target of growth inhibition in yeast caused by Legionella pneumophila glucosyltransferase Lgt1. J. Biol. Chem. 2012;287:26029–26037. doi: 10.1074/jbc.M112.372672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polekhina G., Thirup S., Kjeldgaard M., Nissen P., Lippmann C., Nyborg J. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure. 1996;4:1141–1151. doi: 10.1016/s0969-2126(96)00122-0. [DOI] [PubMed] [Google Scholar]
- 30.Kanibolotsky D.S., Novosyl'na O.V., Abbott C.M., Negrutskii B.S., El'skaya A.V. Multiple molecular dynamics simulation of the isoforms of human translation elongation factor 1A reveals reversible fluctuations between ‘open’ and ‘closed’ conformations and suggests specific for eEF1A1 affinity for Ca2+-calmodulin. BMC Struct. Biol. 2008;8:e4. doi: 10.1186/1472-6807-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gromadski K.B., Schummer T., Stromgaard A., Knudsen C.R., Kinzy T.G., Rodnina M.V. Kinetics of the interactions between yeast elongation factors 1A and 1Balpha, guanine nucleotides, and aminoacyl-tRNA. J. Biol. Chem. 2007;282:35629–35637. doi: 10.1074/jbc.M707245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen G.M., Moller W. Kinetic studies on the role of elongation factors 1 beta and 1 gamma in protein synthesis. J. Biol. Chem. 1988;263:1773–1778. [PubMed] [Google Scholar]
- 33.Vitagliano L., Ruggiero A., Masullo M., Cantiello P., Arcari P., Zagari A. The crystal structure of Sulfolobus solfataricus elongation factor 1alpha in complex with magnesium and GDP. Biochemistry. 2004;43:6630–6636. doi: 10.1021/bi0363331. [DOI] [PubMed] [Google Scholar]
- 34.Negrutskii B.S., El'skaya A.V. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- 35.Schummer T., Gromadski K.B., Rodnina M.V. Mechanism of EF-Ts-catalyzed guanine nucleotide exchange in EF-Tu: contribution of interactions mediated by helix B of EF-Tu. Biochemistry. 2007;46:4977–4984. doi: 10.1021/bi602486c. [DOI] [PubMed] [Google Scholar]
- 36.Yamazoe M., Onogi T., Sunako Y., Niki H., Yamanaka K., Ichimura T., Hiraga S. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873–5884. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira L., Cuervo A., Tavares P. Direct interaction of the bacteriophage SPP1 packaging ATPase with the portal protein. J. Biol. Chem. 2010;285:7366–7373. doi: 10.1074/jbc.M109.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong C., Ito K., Walsh M.A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 39.Vetter I.R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 40.Yang J., Zhang Z., Roe S.M., Marshall C.J., Barford D. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science. 2009;325:1398–1402. doi: 10.1126/science.1174468. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 42.Abbott C.M., Newbery H.J., Squires C.E., Brownstein D., Griffiths L.A., Soares D.C. eEF1A2 and neuronal degeneration. Biochem. Soc. Trans. 2009;37:1293–1297. doi: 10.1042/BST0371293. [DOI] [PubMed] [Google Scholar]
- 43.Pinke D.E., Kalloger S.E., Francetic T., Huntsman D.G., Lee J.M. The prognostic significance of elongation factor eEF1A2 in ovarian cancer. Gynecol. Oncol. 2008;108:561–568. doi: 10.1016/j.ygyno.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 44.Scaggiante B., Dapas B., Bonin S., Grassi M., Zennaro C., Farra R., Cristiano L., Siracusano S., Zanconati F., Giansante C., et al. Dissecting the expression of EEF1A1/2 genes in human prostate cancer cells: the potential of EEF1A2 as a hallmark for prostate transformation and progression. Br. J. Cancer. 2012;106:166–173. doi: 10.1038/bjc.2011.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anand N., Murthy S., Amann G., Wernick M., Porter L.A., Cukier I.H., Collins C., Gray J.W., Diebold J., Demetrick D.J., et al. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 2002;31:301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 46.Tomlinson V.A., Newbery H.J., Wray N.R., Jackson J., Larionov A., Miller W.R., Dixon J.M., Abbott C.M. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer. 2005;5:e113. doi: 10.1186/1471-2407-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 48.Rush J., Moritz A., Lee K.A., Guo A., Goss V.L., Spek E.J., Zhang H., Zha X.M., Polakiewicz R.D., Comb M.J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 49.Mateyak M.K., Kinzy T.G. eEF1A: thinking outside the ribosome. J. Biol. Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Negrutskii B., Vlasenko D., El'skaya A. From global phosphoproteomics to individual proteins: the case of translation elongation factor eEF1A. Expert Rev. Proteom. 2012;9:71–83. doi: 10.1586/epr.11.71. [DOI] [PubMed] [Google Scholar]
- 51.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 52.Wu X., Oh M.H., Schwarz E.M., Larue C.T., Sivaguru M., Imai B.S., Yau P.M., Ort D.R., Huber S.C. Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 2011;155:1769–1778. doi: 10.1104/pp.110.165852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dever T.E., Costello C.E., Owens C.L., Rosenberry T.L., Merrick W.C. Location of seven post-translational modifications in rabbit elongation factor 1 alpha including dimethyllysine, trimethyllysine, and glycerylphosphorylethanolamine. J. Biol. Chem. 1989;264:20518–20525. [PubMed] [Google Scholar]
- 54.Andersen G.R., Thirup S., Spremulli L.L., Nyborg J. High resolution crystal structure of bovine mitochondrial EF-Tu in complex with GDP. J. Mol. Biol. 2000;297:421–436. doi: 10.1006/jmbi.2000.3564. [DOI] [PubMed] [Google Scholar]
- 55.Miura P., Coriati A., Belanger G., De Repentigny Y., Lee J., Kothary R., Holcik M., Jasmin B.J. The utrophin A 5′-UTR drives cap-independent translation exclusively in skeletal muscles of transgenic mice and interacts with eEF1A2. Hum. Mol. Genet. 2010;19:1211–1220. doi: 10.1093/hmg/ddp591. [DOI] [PubMed] [Google Scholar]
- 56.Sikora D., Greco-Stewart V.S., Miron P., Pelchat M. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology. 2009;390:71–78. doi: 10.1016/j.virol.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 57.Wittinghofer A., Vetter I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 58.Sanders J., Brandsma M., Janssen G.M., Dijk J., Moller W. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1 beta gamma delta in the endoplasmic reticulum. J. Cell Sci. 1996;109:1113–1117. doi: 10.1242/jcs.109.5.1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


