Abstract
Human RecQL4 belongs to the ubiquitous RecQ helicase family. Its N-terminal region represents the only homologue of the essential DNA replication initiation factor Sld2 of Saccharomyces cerevisiae, and also participates in the vertebrate initiation of DNA replication. Here, we utilized a random screen to identify N-terminal fragments of human RecQL4 that could be stably expressed in and purified from Escherichia coli. Biophysical characterization of these fragments revealed that the Sld2 homologous RecQL4 N-terminal domain carries large intrinsically disordered regions. The N-terminal fragments were sufficient for the strong annealing activity of RecQL4. Moreover, this activity appeared to be the basis for an ATP-independent strand exchange activity. Both activities relied on multiple DNA-binding sites with affinities to single-stranded, double-stranded and Y-structured DNA. Finally, we found a remarkable affinity of the N-terminus for guanine quadruplex (G4) DNA, exceeding the affinities for other DNA structures by at least 60-fold. Together, these findings suggest that the DNA interactions mediated by the N-terminal region of human RecQL4 represent a central function at the replication fork. The presented data may also provide a mechanistic explanation for the role of elements with a G4-forming propensity identified in the vicinity of vertebrate origins of DNA replication.
INTRODUCTION
The RecQ-like helicase 4 (RecQL4) belongs to the well-conserved RecQ helicase family, which plays an important role for the maintenance of genome integrity. Whereas there is only one RecQ helicase in Escherichia coli, five different homologs have been identified in humans. Mutations in the genes of three of these are associated with diseases. Homozygous defects in the BLM (RecQL2) and in the WRN (RecQL3) gene lead to the Bloom and Werner syndrome, respectively, both of which are characterized by premature aging and an increased predisposition for cancer (1,2). Mutations of RecQL4 are linked to three different diseases: the Baller–Gerold syndrome, RAPADILINO and the Rothmund–Thomson syndrome (RTS) (3). All three syndromes display overlapping symptoms, including skeletal abnormalities, mental retardation, signs of premature aging and skin lesions. RTS patients also show an increased incidence of osteosarcomas (1,2,4).
Although RecQL4 shares the helicase domain that is characteristic for the family members, it has been identified rather lately as an active helicase (5–7). In vitro, the helicase activity appears to be easily masked by its strong DNA annealing activity (5–7).
RecQL4 is the only known human protein with homology to the yeast replication initiation factor Sld2 (8,9). The homology is restricted to the N-terminal 400 amino acids (aa) and particularly pronounced within the first 54 aa (10,11). Like Sld2 in yeast, the metazoan counterpart is essential for viability (8,12,13). While mice with disruptions within the helicase domain are viable and display an RTS phenotype (14,15), a truncated N-terminal gene product of only 200 amino acids (aa) leads to early embryonic lethality (13), indicating that the N-terminal domain of RecQL4 mediates essential functions, whereas the helicase performs an important albeit not essential role in the maintenance of genome stability (16). Therefore, it is not surprising that mutations in the N-terminal part of human RecQL4 are rare (3,17).
Current evidence indicates an important role of human RecQL4 during the initiation of DNA replication. It has been shown that RecQL4 is recruited to origins of replication at the late G1 phase of the cell cycle, but this association is lost after replication initiation has occurred (18). RecQL4 also interacts with several replication initiation factors, such as Mcm10 and TopBP1 (10,19), and it is necessary for the formation of the complex composed of Cdc45, MCM2–7 and GINS (CMG) that represents the eukaryotic replicative DNA helicase (20,21). RecQL4 promotes the assembly of replication initiation factors after pre-replicative complex formation in Xenopus laevis egg extracts (8,9), and after depletion from these extracts, an N-terminal fragment is sufficient to rescue chromatin binding of the DNA polymerases α and ϵ (9). Recent studies suggest additional functions of RecQL4, e.g. for maintaining the integrity of telomeres and the mitochondrial genome (22–27).
The RecQ helicases BLM and WRN are known to bind and dissolve G quadruplexes (G4) (28,29). G4 quadruplexes are DNA secondary structures formed by guanine-rich sequences. Four guanine bases self-assemble via Hoogsteen hydrogen bonds forming a structure that is further stabilized by the internalization of potassium. Recently, Biffi et al. (30) detected G4 structures in human cells and demonstrated that G4 formation is modulated during cell cycle progression. An accumulation of (predicted) potentially G4-forming sequences is also found in evolutionary conserved regions such as promoters and telomeres (reviewed in (31)). Most importantly, several recent studies suggest that the vast majority of human origins of replication is associated with potentially G4-forming motifs, which therefore might be of relevance for the initiation step of DNA replication (32–35).
Here, we dissected the biophysical and biochemical properties of the essential N-terminal domain of human RecQL4 and compared them to those of the full-length enzyme. We demonstrate that the N-terminal domain represents a largely intrinsically disordered entity that comprises multiple DNA-binding elements. These are responsible for the exceptionally strong DNA strand annealing and annealing-mediated strand exchange activity of this enzyme. Furthermore, the N-terminal part, but not the helicase region of RecQL4, binds tightly to G4 structures. These findings emphasize the importance of protein–DNA interactions of the N-terminal part of RecQL4 during the initiation of DNA replication.
MATERIALS AND METHODS
Random polymerase chain reaction screen
For the identification of soluble N-terminal fragments of human RecQL4, the random polymerase chain reaction (PCR) approach of Kawasaki and Inagaki (36) was modified. Stably expressed and correctly folded proteins gave rise to a green fluorescence when the randomly produced PCR products were combined with a green fluorescent protein (GFP) reporter (37) (Supplementary Figure S1). To this end, the vector pEGGsH6 was constructed by introducing a multiple cloning site, a tobacco etch virus (TEV) protease cleavage site, the open reading frame (ORF) of eGFP and a C-terminal hexahistidine (6xHis) tag in frame with the BamHI and the EcoRI sites of the vector pGEX1λT. Then, PCR reactions were performed on a template with the first 1280 bp of the ORF of RecQL4 (GeneBank NM_004260), using the sequence-specific forward primers RecQL4_159F (5′-GTACTCCGAGACGGATCCCAGGCCGGCGGCGGGCTC-3′) or RecQL4_178F (5′-GTACTCCGAGACGGATCCCGCAGCTCCGAGTCGCTCC-3′), corresponding to aa 54 and 60, respectively. This was necessary, since the extremely high GC content of the 5′ end of the human RECQL4 cDNA interfered with the random PCR. PCR was performed in 50 μl reactions with Phusion DNA polymerase and GC buffer (Finnzymes/Thermo). After 15 cycles of 15 s at 98°C, 30 s at 60°C and 45 s at 72°C in the presence of 20 μM forward primer and 5 ng template, the reverse primer Primer_CrandomXho 5′-TACCCGCAGCGTGAGCTCNNNNNNNNNNNNNNN-3′ was added at 200 μM, followed by two cycles of 15 s at 98°C, and 60 s at 80, 70, 60, 50, 40 and finally at 72°C. PCR products were purified with a PCR purification kit (Avegene) and amplified with primers B_Bamoutf 5′-GTACTCCGAGACGGATCC-3′ and D_Xhooutr 5′-TACCCGCAGCGTGAGCTC-3′ using Phusion polymerase with GC buffer and 25 cycles of 15 s at 98°C, 30 s at 60°C and 45 s at 72°C. PCR products were visible as smear without separate bands after agarose gel electrophoresis. Products corresponding to 300–1200 bp were excised and purified using a DNA extraction kit (Qiagen). After digestion with BamHI and XhoI (New England Biolabs), the fragment library was ligated into vector pEGGsH6 and plated on lysogeny broth (LB)/ampicillin plates. Colonies displaying fluorescence were replated and sequenced, and representative clones were subjected to test expressions to confirm the solubility of the GST-GFP fusions of the RecQL4 fragments. The RecQL4 fragments without GST- and GFP tags were subsequently recloned into the vector pRSFDuet-1 (Merck-Millipore) as described below.
Expression and purification of recombinant proteins
RecQL4_N54 was expressed and purified as described (10). All other fragments, except the full-length protein, were cloned as N- or C-terminal 6xHis fusions into the NcoI and XhoI restriction sites of the vector pRSFDuet-1. A pFastBac-1 plasmid containing the sequence for full-length RecQL4 with an N-terminal FLAG and 6xHis dual tag was a generous gift of Dr Yasushiro Furuichi, GeneCare Research Institute, Kamakura, Japan. A baculovirus was generated utilizing the Bac-To-Bac system (Life Technologies) according to the manufacturer's instructions. The N-terminal fragments were expressed and purified as follows: E. coli (BL21[DE3]) cells carrying the respective pRSF plasmid were grown at 37°C in 1 l LB medium supplemented with 30 μg/ml kanamycin until an OD595 of 0.5–0.7 was reached. After induction of protein expression with 1 mM isopropyl-β-D-thiogalactopyranoside for 5 h, cells were harvested, washed with phosphate-buffered saline (PBS) and resuspended into 40 ml PBS supplemented with 1 mg/ml lysozyme followed by incubation for 30 min on ice. Cells were disrupted by 40 × 10 s ultrasound pulses at 30% amplitude interrupted by 45 s incubations on ice, using a Branson W-450D Digital Sonifier. Triton X-100 was added to 3%, and after incubation at 4°C for 30 min and centrifugation at 25,000xg, the supernatant was added to 10 ml equilibrated DEAE sepharose (GE Healthcare). After 1 h incubation at 4°C, the supernatant was collected and added to 10 ml equilibrated S sepharose (GE Healthcare) and incubated at 4°C for 1 h. The sepharose was washed with 10 volumes of buffer A (50 mM Tris–HCl pH 8, 50 mM KCl, 3 mM β-mercaptoethanol [β-ME]) and eluted with buffer B (50 mM Tris–HCl pH 8, 600 mM KCl, 3 mM β-ME) in 5 ml portions. Protein-containing fractions were combined, diluted 1:3 in buffer A and loaded onto a 500 μl TALON® IMAC column (Clontech) equilibrated with buffer C (20 mM imidazole pH 8, 150 mM (NH4)2SO4, 3 mM β-ME). After binding for 1 h at 4°C and washing with 10 volumes of buffer C, the column was eluted with buffer D (350 mM imidazole, pH 8, 150 mM (NH4)2SO4, 3 mM β-ME). Protein-containing fractions were combined and applied to a Superose™ 12 column (GE Healthcare). Elution occurred in buffer E (20 mM potassium phosphate pH 7.4, 100 mM KCl, 10% glycerol). For purification of RecQL4_N388, gel filtration was omitted. For expression and purification of RecQL4_C464, the DEAE and S sepharose columns were omitted.
For the expression of full-length RecQL4, 250 ml High-Five™ insect cells were infected with the recombinant baculovirus and incubated in suspension culture for 48 h at 27°C. After harvest and a PBS wash, the cells were resuspended in lysis buffer (150 mM Tris–HCl pH 8, 150 mM NaCl, 10% glycerol, 0.5% NP-40, 3 mM β-ME) and incubated for 30 min on ice. After centrifugation at 25 000xg, the cell extract was added to 10 ml S sepharose equilibrated with buffer F (50 mM Tris–HCl pH 8, 50 mM KCl) for 1 h at 4°C. The sepharose was washed with 10 volumes of buffer F and eluted in 5 ml fractions with buffer G (50 mM Tris–HCl pH 8, 600 mM KCl). Protein-containing fractions were pooled, diluted 1:3 in buffer F and loaded onto 500 μl ANTI-FLAG® M2 Affinity Gel (Sigma-Aldrich). After incubation at 4°C for 1 h, the Affinity Gel was washed with 10 volumes of Tris-buffered saline (TBS) and eluted with 100 μg/ml FLAG peptide in TBS. Protein-containing fractions were combined and loaded onto 500 μl Ni-NTA resin (Qiagen) for 1 h at 4°C. The resin was washed with buffer H (50 mM Tris–HCl pH 8, 300 mM NaCl, 20 mM imidazole) and eluted with buffer I (50 mM Tris–HCl pH 8, 300 mM NaCl, 250 mM imidazole) in portions of 500 μl. Protein containing fractions were dialysed against buffer E.
Proteins were snap frozen with liquid nitrogen and stored at −80°C. Purity of the recombinant proteins was analyzed with sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and concentrations were determined by spectrophotometry at 280 nm wavelength using specific extinction coefficient that were derived from the respective amino acid composition.
Radioactive labeling and DNA substrate preparation
Oligonucleotides were 5′-end labeled with T4 polynucleotide kinase in reaction buffer A (Thermo) and γ-33P-ATP (5000 Ci/mmol; Hartmann Analytic, Braunschweig, Germany). Unincorporated material was removed using Microspin G25 columns (GE Healthcare). Oligonucleotides (listed in Supplementary Table S1) were purchased 3xHPLC-purified from Purimex (Grebenstein, Germany). Duplex, fork and bubble substrates were annealed by combining corresponding oligonucleotides in 30 mM Tris–HCl pH 8, 10 mM MgCl2, heating at 95°C for 5 min and cooling down slowly to room temperature. The G-quadruplex substrate was prepared before labeling. To this end, 500 pmol of the respective oligonucleotide was dissolved in 12.5 μl TE buffer and heated at 98°C for 5 min followed by 3-min chilling on ice. Then 25 μl of cold TE, containing 1.9 M NaCl and 0.1 M KCl, was added. After mixing, the solution was incubated for 60 h at 67°C.
Annealing assay
Annealing reactions were carried out by incubating radiolabeled LB2F and non-labeled LB2_R24 oligonucleotides (Supplementary Table S1) with protein in 10 μl annealing buffer (20 mM Tris acetate [Ac] pH 8, 0.5 mM MgAc, 50 mM KAc pH 7.3, 1 mM DTT, 50 μg/ml bovine serum albumin [BSA]) for the indicated times at 37°C. After addition of 5 μl stop buffer (2% SDS, 50 mM ethylenediaminetetraacetic acid [EDTA] pH 8, 40% glycerol, 400 nM unlabeled LB2F), the reaction products were resolved by neutral TBE-PAGE, visualized with a PhosphoImager (Typhoon Trio, GE Healthcare), and quantified with the ImageQuant software (GE Healthcare).
A fluorescence-based annealing assay was performed by mixing the 48-mer oligonucleotide Anneal-F (Supplementary Table S1) with protein and 200 nM 4′,6-diamidino-2-phenylindole (DAPI) in annealing buffer (38). After measuring and subtracting the basal fluorescence level, the complementary oligonucleotide Anneal-R was added and the solution was carefully mixed. The change in fluorescence (excitation: 358 nm, emission 461 nm, band width 5 nm) was measured at six time points per second over 10 min with a Cary Eclipse fluorescence spectrophotometer (Agilent).
Strand exchange assay
The indicated radiolabeled DNA substrates and proteins were incubated in 10 μl strand exchange buffer (20 mM Tris–HCl pH 7.5, 3.5 mM MgCl2, 100 μg/ml BSA, 5 mM DTT, 10% glycerol) for 30 min at 37°C. After addition of 10 μl stop buffer (2% SDS, 50 mM EDTA pH 8, 40% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol, 0.45 mg/ml proteinase K), the reaction products were resolved by neutral PAGE and visualized using a PhosphoImager (Typhoon Trio, GE Healthcare).
Electrophoretic mobility shift assay (EMSA)
The DNA-binding reactions were performed by incubation of the corresponding DNA with protein in 10 μl binding buffer (20 mM HEPES pH 7.4, 0.5 mM EDTA, 0.05% NP40, 10% glycerol) for 10 min at 25°C. For G4 reactions, the binding buffer was supplemented with 150 mM KCl. After addition of 5 μl loading buffer (0.1% bromophenol blue, 40% glycerol), the binding partners were analyzed on neutral PAGE. Products containing radiolabeled oligonucleotides were visualized using a PhosphoImager. Non-labeled DNA was stained in gel with SYBR Safe (Invitrogen) and scanned using Typhoon Trio. LB2F ss oligonucleotide was labeled with FAM. Quantification was done with ImageQuant software (GE Healthcare).
DNase I protection assay
10 μl reaction volumes contained the indicated radiolabeled DNA substrates and proteins in 20 mM Tris-HCl pH 7.5, 3.5 mM MgCl2, 3.5 mM ATP, 100 μg/ml BSA, 5 mM DTT and 10% glycerol. After addition of 5 units of DNase I, the reaction mixtures were incubated at 37°C for 10 min, followed by addition of 10 μl stop buffer (2% SDS, 50 mM EDTA pH 8, 40% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol, 0.45 mg/ml proteinase K) and resolved by neutral PAGE. Visualization occurred with a PhosphoImager.
RESULTS
A random PCR screen led to stably expressed N-terminal RecQL4 fragments
Initial attempts to express N-terminal fragments of human RecQL4 in E. coli failed since the recombinant proteins were either highly insoluble or became rapidly degraded during expression and purification. The exception was RecQL4_N388 covering the amino-terminal region up to the potential zinc knuckle (39). This observation was in line with structure and disorder predictions by the PSIPRED and DISOPRED servers (40,41), which suggested that the N-terminal region of human RecQL4 possesses very little regular secondary structure and a high propensity of intrinsic disorder. We therefore employed a random screen to identify N-terminal fragments for stable expression and purification. By preparing a fragment library of the N-terminus of RecQL4 (Supplementary Figure S1) in a vector containing a GFP folding reporter, the solubility of the expression products could be rapidly monitored by the fluorescence of the E. coli colonies on the plates (36). Fluorescence was observed when the inserted RecQL4 sequence was in frame with GFP and when the translated protein was natively folded (Figure 1). By sequencing, 23 independent protein fragments of the RecQL4 N-terminus were identified. Interestingly, we observed that the distribution of fragments was not random, but indicated an accumulation of fragments with specific lengths, in particularly around 170 aa. Based on test expressions of representative positive clones from the screen, we selected the N-terminal fragments indicated in Figure 2. The screen and our previous work (10) indicated boundaries of autonomously folding regions at around residues 54, 170, 250 and 388, respectively (Figure 2A). From those we also prepared N-terminal truncations. Out of these, two fragments starting at aa 54 and 251 proved suitable for expression and purification (Figure 2B). We also expressed and purified full-length RecQL4 (RecQL4_FL) in insect cells and a fragment comprising the complete helicase domain but lacking the N-terminal region and the putative zinc knuckle (C464). All seven constructs were expressed and purified to near homogeneity by a combination of ion exchange chromatography, affinity purification and gel filtration (Figure 2B); only N388 displayed minor signs of degradation during purification.
Figure 1.
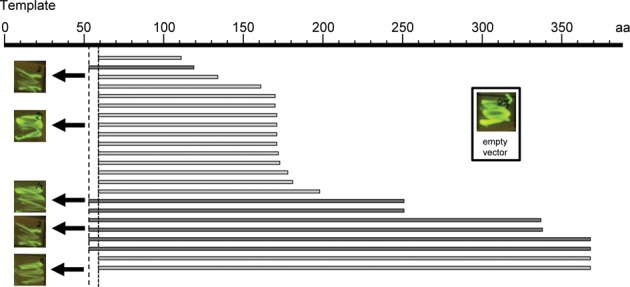
Stably expressed fragments from the N-terminus of RecQL4 as obtained from a random PCR and GFP fluorescence-based screen. Depicted is the coverage of independent clones retrieved from two experiments with fixed right primers covering RecQL4 from aa 54 (dark gray) or aa 60 (light gray), respectively. It is apparent that the distribution of clones is not random.
Figure 2.
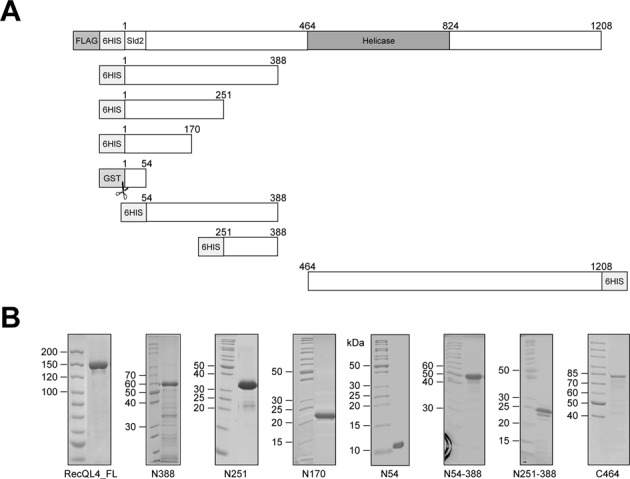
Purified fragments of RecQL4. (A) Schematic depiction of the full-length protein and the fragments including the tags used for purification. The C-terminal fragment contained the helicase domain. (B) Coomassie blue-stained SDS-PAGE gel of purified recombinant N- and C-terminal fragments expressed in E. coli and the full-length protein expressed in baculovirus-infected High-Five™ insect cells. Tags (FLAG, 6HIS, GST), the homeo-like N-terminal 54 residues homologous to the yeast Sld2 protein and the helicase domain are indicated.
Because of the apparent discrepancy between bioinformatics predictions suggesting a largely disordered structure and the results of our screen indicating the presence of several folding domains, we analyzed the structure of the N-terminal fragments by far UV CD spectroscopy (Supplementary Figure S2A). Both the spectra and their deconvolution indicated that apart from the α-helical homeo-like domain N54, our fragments contained little or no regular secondary structures such as α-helices or β-sheets. The content of secondary structure appeared to decrease in the order of N54 >> N170 > N251 > N54–388 ≈ N251–388. Uversky (42) showed that the folding state of a protein can be estimated by its position in a plot of [Θ]222 against [Θ]200 (Supplementary Figure S2B). By using this method, N54 was placed in the region typical for natively folded proteins, whereas the longer fragments N170 and N251 approached the region of proteins classified by Uversky as ‘pre-molten globule’ consistent with a very low content of regular secondary structure. The fragments N54-388 and N251-388 lacking the homeodomain-like region fell completely into the pre-molten globule cluster. This was also supported by [1H 15N] heteronuclear single quantum correlation (HSQC) nuclear magnetic resonance spectra of N170 and N251-388 (Supplementary Figure S3). Here, the well-folded homeo-like fold of N54 (10) was also discernible in the larger N170 construct (Supplementary Figure S3A). Only some amide resonances arising from the C-terminal end of N54 experienced a chemical shift perturbation in the context of the larger N170 construct. The remainder of the resonances for N170 exhibited low spectral dispersion and significant line broadening, consistent with a pre-molten globule-like folding state. The [1H,15N]-HSQC spectrum of N251-388 exhibited a severely reduced spectral dispersion and a significant variation in line width (Supplementary Figure S3B), suggesting a high conformational heterogeneity including local random-coil but also globule-like folding states. Proteins in a native pre-molten globule state contain more residual secondary structure and are more compact than intrinsically unfolded proteins (43). As typically for pre-molten globule proteins, the N-terminus of human RecQL4 was characterized by relatively low hydrophobicity and a high proportion of uncompensated, charged residues. This was experimentally supported by tryptophan fluorescence measurements. All N-terminal fragments except N54 showed after excitation at 295 nm an emission maximum of fluorescence at around 355 nm, indicating a predominantly hydrophilic environment of the tryptophans even under native conditions. Moreover, addition of guanidinium hydrochloride influenced the emission only marginally. Furthermore, all the fragments appeared to be completely denatured at concentrations below 1 M guanidinium hydrochloride (Supplementary Figure S4).
The N-terminus of RecQL4 catalyzed DNA annealing
It has previously been shown that RecQL4 has a strong DNA annealing activity that masks the helicase function of the protein (5–7). We therefore investigated, which part(s) of the protein mediate DNA annealing. To this end, we analyzed the kinetics by incubating a radiolabeled 24-mer with its complementary strand and our hRecQL4 fragments in the absence of ATP. The amount of formed heteroduplexes was quantified by employing native gel electrophoresis and autoradiography. In the absence of protein, 40% of the strands hybridized within 10 min (Figure 3A and B and Supplementary Figure S5), representing spontaneous annealing under assay conditions. The previously described N54 fragment (10) did not increase hybridization. In contrast, the same concentration of N388, which comprised the N-terminus of RecQL4 up to the putative zinc knuckle, converted the complementary single-stranded DNAs (ssDNAs) into double-stranded DNA (dsDNA) within 2 min. The same was true for fragment N54-388 confirming that the first 54 aa of human RecQL4 did not contribute to annealing. N170 and N251 displayed comparable hybridization activities with about 60% ssDNA being converted into dsDNA in 10 min. Hence, the region between aa 170 and 251 apparently did not or only little contribute to annealing. In a second approach, we determined annealing using a real-time assay, which yielded a better time resolution. The assay was based on the increased fluorescence yield of DAPI binding to dsDNA formed after annealing (38) (Figure 3C). Again, we observed extremely efficient annealing with little difference between N388 and N54-388, respectively (Figure 3D). We could also confirm that N170 and N251 possessed similar annealing activities. Importantly, fragment N251-388 was also able to anneal ssDNA approximately as efficient as N170 and N251. From this we conclude that there are two important regions responsible for strand annealing: one is located between aa 55 and 170 and the other one between aa 251 and 388.
Figure 3.
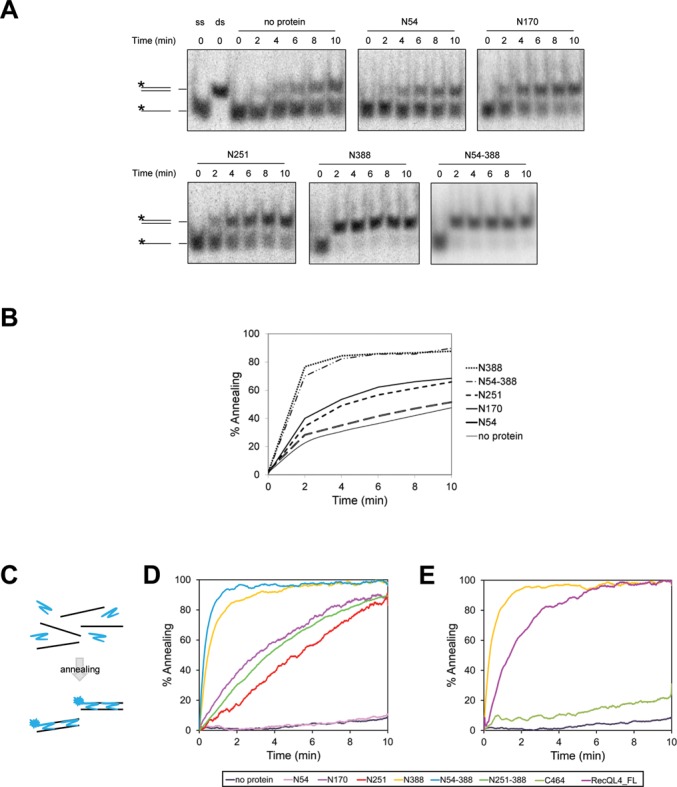
The strand annealing activity of RecQL4 was provided by the N-terminus. (A) DNA strand annealing assays were performed with 2 nM of LB2_R24 and complementary radiolabeled LB2F oligonucleotides and 20 nM protein. The reaction mixtures were incubated at 37°C for the time indicated. The radioactively labeled strands are marked by an asterisk. (B) Quantification of the DNA strand annealing assays as shown in (A). (C) Illustration of the fluorescence-based DNA strand annealing assay. In total, 2 nM of complementary Anneal-F and Anneal-R oligonucleotides were incubated with 200 nM DAPI at 22°C. After DNA annealing, the dsDNA was bound by DAPI leading to a fluorescence increase. (D) Fluorescence-based DNA strand annealing assays were done as described in (C) with 20 nM of N-terminal RecQL4 fragments or without protein (no protein). The oligonucleotides for the fluorescence-based annealing assay were longer than those used in (A) and showed less spontaneous annealing in the absence of protein. (E) The same as in (D). The diagram shows the annealing activity of the longest N-terminal fragment in comparison with the C-terminal fragment and the full-length protein (50 nM each). Protein concentrations were determined by UV absorbance as indicated in ‘Materials and Methods’.
Next, we studied whether the N-terminal 388 residues were also sufficient for explaining the annealing activity of full-length human RecQL4. We therefore measured annealing by equimolar amounts of fragment C464 (which included the helicase and the C-terminal domain of RecQL4, but lacked the N-terminus) and RecQL4_FL in the absence of ATP. The latter displayed a robust annealing activity that, however, was less active than that of equimolar amounts of N388 (Figure 3E). In contrast, C464 showed relatively little annealing activity. In the presence of ATP, both RecQL4_FL and C464 unwound Y-shaped substrate as expected (Supplementary Figure S6A and B). With the same assay, we failed to detect any ATP-dependent DNA helicase activity of N388 or other N-terminal fragments (data not shown), in agreement with (44), but in contrast to (5). These results reinforce the view that the N-terminus of RecQL4 is required and sufficient for the ATP-independent annealing activity of this protein, whereas it does not contribute to an ATP-dependent DNA helicase activity.
The N-terminal region of RecQL4 exhibited an annealing-dependent DNA strand exchange activity
DNA strand annealing is frequently associated with a DNA strand exchange activity. Therefore, we asked whether N170, N388 and RecQL4_FL possessed such an activity. In the absence of ATP, the fragments N170, N388 and the full-length protein induced an exchange of one DNA strand of a bubble with its respective complementary ssDNA, but were not able to mediate an exchange between a bubble and the corresponding dsDNA (Figure 4A). The N-terminal fragments catalyzed this reaction more effective than the full-length protein. DNA strand exchange was only initiated when the ssDNA sequence in the bubble and the acceptor strand were complementary, i.e. the reaction was initiated by annealing (compare Figure 4A and B). This interpretation was confirmed with a Y-shaped substrate and complementary ssDNA: all three protein constructs converted the Y-form DNA and a matching complementary strand into a dsDNA and the corresponding ssDNA product (Figure 4C). Consistently, no back reaction was detected when a dsDNA served as donor for a complementary ssDNA to form a Y (Figure 4D). Annealing-dependent strand exchange was mediated by the N-terminal domain of human RecQL4; the N-terminally deleted C464 fragment was not able to mediate strand exchange (Supplementary Figure S6C and D).
Figure 4.
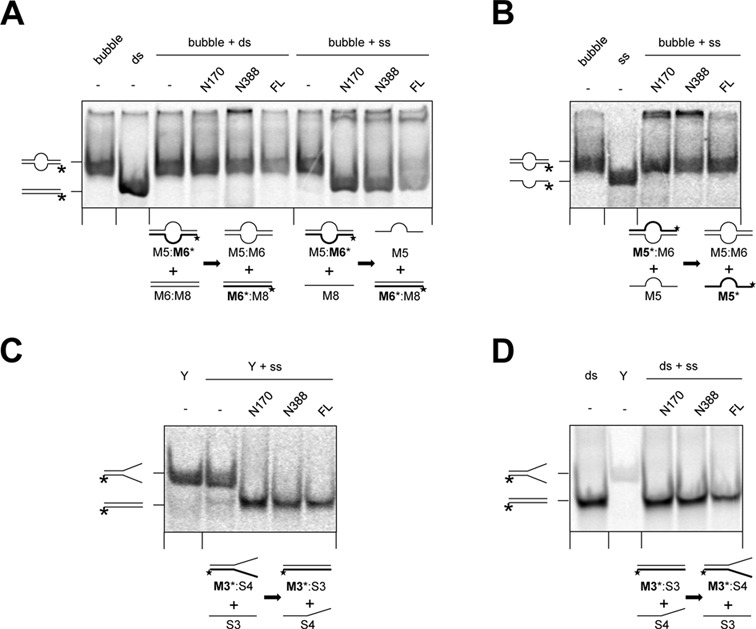
The N-terminus of RecQL4 possessed an ATP-independent DNA strand exchange activity. (A) Strand exchange occurred between a bubble and a complementary ssDNA, but not between a bubble and the corresponding dsDNA. DNA strand exchange assays were performed with 1 nM DNA and 2 μM RecQL4_N170 or 200 nM of RecQL4_N388 or RecQL4_FL, respectively, in the absence of ATP. A radiolabeled bubble DNA was incubated with the complementary ss- or dsDNA, respectively, for 30 min at 37°C. (B) Strand exchange required regions of complementary ssDNA. Reactions were performed as in (A), containing radiolabeled bubble DNA and an equal amount of the non-labeled, corresponding strand. (C) Strand exchange between a Y-shaped DNA and a complementary ssDNA. Reactions were performed as in (A), but contained radiolabeled Y-shaped DNA and a complementary ssDNA. (D) No strand exchange occurred in the reverse direction compared to (C). Reactions were performed as in (A), but contained radiolabeled dsDNA and a partly complementary ssDNA. Protein concentrations were determined by UV absorbance as indicated in ‘Materials and Methods’. The radioactively labeled strands are marked by an asterisk.
These results indicate that RecQL4 bears no DNA strand exchange activity in the classical sense. This would mediate an ATP-dependent exchange between an ssDNA acceptor and a complementary dsDNA donor (45). The here observed annealing-initiated exchange strongly resembles the strand exchange activities reported for several human RecQ helicases including RecQL4 (46,47). Importantly, the present experiments demonstrate that the N-terminus of RecQL4 lacking the RecQ helicase domain is sufficient to promote an at least limited passive branch migration reaction if two complementary ssDNA regions are available to initiate this reaction. It should be noted that this reaction required higher concentrations in vitro compared to the simple annealing reactions presented in Figure 3.
The N-terminus of RecQL4 consists of three independent DNA-binding domains
Next we evaluated the DNA-binding ability of the purified N-terminal fragments to determine potential preferences for certain DNA structures and to identify the DNA-binding regions within the protein. For a comparison of the DNA affinities of the different fragments, we employed electrophoretic mobility shift assays. As shown in Figure 5 and Supplementary Figure S7, all N-terminal fragments of human RecQL4 tested bind a 24-mer ssDNA, the corresponding dsDNA as well as a Y-shaped DNA consisting of a 16 base paired region and two 8 nt non-complementary overhangs, albeit with varying affinity. Under these conditions, we previously detected only weak binding by N54 (10), which possessed a low, sequence-independent binding affinity for ds- and Y-form DNA. Fragments N170 and N251 both bound ss-, ds- and Y-DNA considerably stronger than N54 (Figure 5, Supplementary Figure S7, and (10)). As there was no appreciable difference in the binding ability of the fragments N170 and N251, apparently there must have been another DNA-binding site between aa 55 and 170. Half of the DNA was bound in a reaction containing 1 μM of N388 (Figure 5B). Hence, there must have been at least one additional DNA-binding site between aa 251 and 388. This assessment was confirmed by binding assays with N251-388. This fragment contributed particularly to the binding of Y-DNA, but less to ss- and dsDNA binding. Next, we studied an N388 construct lacking the first homeodomain-forming 54 aa (10). This shortened fragment displayed a small impairment of DNA binding for all three DNA forms tested (Figure 5B), consistent with the low binding affinity of N54 (10). Finally, we compared the binding of N388 with RecQL4_FL (Figure 5C and Supplementary Figure S7E–G). There was no significant difference in the binding of the N-terminal fragment and the full-length enzyme to our three model DNAs. Yeast Sld2 has been reported to bind preferentially to a thymine-rich ssDNA region of the origin ARS1 (48). We therefore repeated the EMSAs utilizing 25-mer oligonucleotides with a high and low GC content. As can be seen in Figure 6 and Supplementary Figure S8, the oligonucleotide LB2_GC (76% GC content) was shifted significantly better by both N388 and RecQL4_FL than the AT-rich oligonucleotide LB2_AT (16% GC content). Apparently, LB2 (50% GC content) and LB2_GC (76% GC) were bound with similar affinities. This suggests that the N-terminus of human RecQL4 shows preference for GC-rich DNA, in difference to its homolog Sld2 from yeast.
Figure 5.
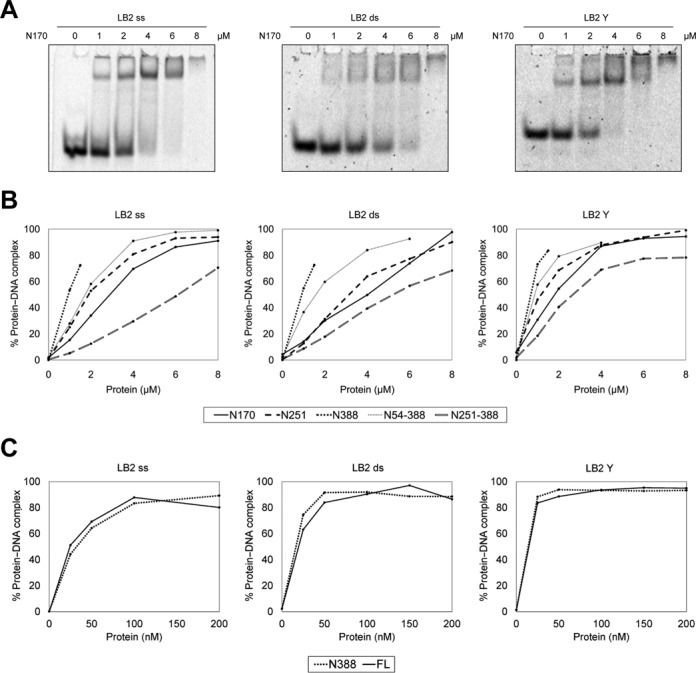
The N-terminus of RecQL4 favored binding to Y-shaped DNA. (A) EMSAs were performed with 1.3 μM of ss-, ds- or Y-shaped LB2-derived DNA and the indicated amounts of RecQL4_N170. (B) Quantification of EMSA assays of the N-terminal fragments and 1.3 μM of ss-, ds- or Y-shaped LB2-derived DNA. (C) Comparison of DNA binding of RecQL4_N388 and RecQL4_FL using the same substrates as in (A) and (B). All DNA-binding experiments and quantifications presented here were performed under identical conditions. All experiments were performed up to four times with minor variations in the concentration of the DNA substrates and produced in all cases comparable results.
Figure 6.
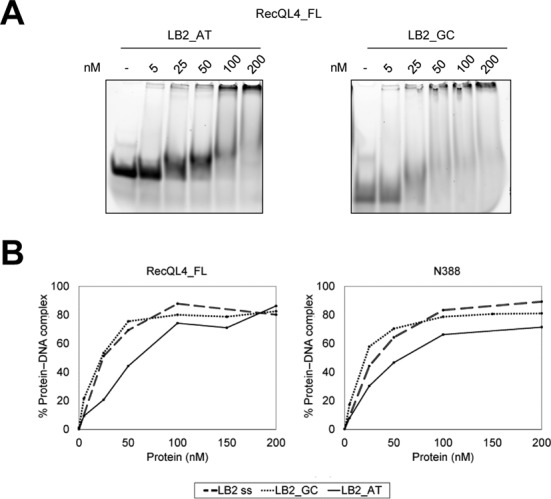
The N-terminus of RecQL4 favored binding to GC-rich over AT-rich ssDNA. (A) EMSAs were performed with 1.3 μM of 25-mers ssDNA. The sequence of these oligonucleotides is derived from a 232-bp human genomic fragment comprising the LB2 origin (Supplementary Table S1) (10). Compared to the genomic sequence, LB2_GC carries a base exchange to reduce secondary structure. LB2_GC and LB2_AT possess 76 and 16% G-C content, respectively. (B) Quantification of EMSA assays of (A).
Together, our results indicate the presence of multiple DNA-binding sites at the N-terminal region of RecQL4 that are decisive for the DNA-binding affinity of human RecQL4, first the homeodomain-like fold comprising aa 1–54 (10), followed by a second domain comprising aa 55–170 with importance for ssDNA binding (Figure 5B, left panel), and a third domain between aa 251–388 that mediated Y-DNA, but also ssDNA and dsDNA binding (Figure 5B). However, all fragments showed a higher affinity to Y-shaped DNA in comparison to ss- or dsDNA. Interestingly, the highly ordered first 54 aa contributed only little to overall DNA binding.
The N-terminus of RecQL4 bound G-quadruplexes much better than other DNA substrates
The majority of origins of replication are associated with G-rich sequences that have the propensity to form G-quadruplex (G4) DNA structures (32,34). Therefore, G4 structures might be important for the initiation of replication. Because other RecQ helicases have been shown to bind G4 structures (reviewed in (49)) and because RecQL4 performs largely unknown functions during the initiation of replication, we asked whether there is an affinity of RecQL4 for this non-canonical type of DNA structure. To this end, we compared the binding affinity of RecQL4_FL and the various N-terminal fragments described above to (intermolecular) tetraplexes by EMSA (Figure 7). These were compared with the affinities of the corresponding ss- and dsDNA of identical sequence. Surprisingly, we observed a substantial affinity to tetraplexes that exceeded binding to the corresponding ss- and ds- control DNAs by far. In the presence of 2 μM of N170, 250 nM N388 or 200 nM RecQL4_FL, the bands of the G4 structures were completely shifted, whereas only a fraction of the control DNAs migrated differently (Figure 7A). Thus, all three RecQL4 constructs displayed G4 binding, although a 10-fold higher concentration of N170 than RecQL4_FL was required to saturate binding to the tetraplex. In competition assays (Figure 7B, Supplementary Figure S9A and B) with 10 nM radiolabeled G4 and 200 nM RecQL4_FL, the addition of an up to 60-fold excess of the corresponding ss-, ds- or Y-form DNA had only marginal effects on the shift of the G4-DNA (Figure 7B). There was also no effect on G4 binding by N170 and N388 in reactions containing the competing ss-, ds- or Y-DNAs in an up to 60-fold molar excess (Supplementary Figure S9A and B, respectively). Only addition of a 30- to 50-fold molar excess of a 68-mer bubble DNA reverted the G4 shift, albeit not completely. G4 binding by RecQL4_FL could only be partly reverted by stepwise increasing the KCl concentration from 50 to 350 mM (Supplementary Figure S9C). Furthermore, the effect of ATP on G4 binding was studied (Supplementary Figure S10A and B). The presence of 5 mM ATP during the binding reaction did not affect binding of N388 or RecQL4_FL.
Figure 7.
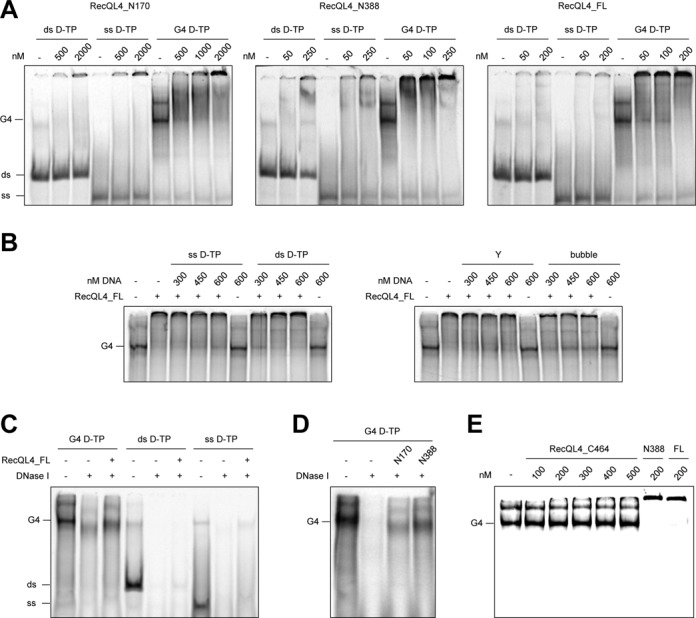
The N-terminus of RecQL4 bound G4 DNA with a high affinity. (A) EMSAs were performed with 5 nM of radiolabeled ds, ss or G4 DNA possessing the same DNA sequence and indicated amounts of RecQL4_N170, RecQL4_N388 or RecQL4_FL, respectively. (B) Competition assays were done like EMSAs with 10 nM of radiolabeled G4 DNA supplemented with the indicated amounts of unlabeled ss-, ds-, Y- or bubble DNA and 200 nM of RecQL4_FL. (C) Measurement of DNase I protection by RecQL4. The reaction mixtures contained 5 units of DNase I and 10 nM of G4-, ds- or ssDNA, and 200 nM of RecQL4_FL as indicated and they were incubated for 10 min at 37°C followed by deproteination in the presence of SDS and proteinase K. (D) Same as in (C), but the mixtures contained G4 DNA and 2 μM of RecQL4_N170 or 200 nM of RecQL4_N388. (E) The N-terminal, but not the helicase domain of human RecQL4 showed a high affinity for G4 DNA. The EMSA was performed with 5 nM of radiolabeled G4 DNA and the indicated amounts of RecQL4_C464, RecQL4_N388 or RecQL4_FL. The sequence and origin of the D-TP oligonucleotide is presented in Supplementary Table S1, and the preparation of a bi-molecular G4 DNA is described in detail in the Materials and Methods in the section ‘Radioactive labeling and DNA substrate preparation’. All gel shifts in panels (A), (B) and (E) were performed under identical conditions in DNA-binding buffer containing 150 mM KCl.
In principle, the observed G4 binding might also have been due to the ssDNA overhangs outside of the G4-forming sequence. By performing DNase I protection assays, we were able to demonstrate that this is not the case. In the presence of RecQL4_FL, G4 DNA and, to a lesser extent ss- and dsDNA, were protected against degradation catalyzed by DNase I (Figure 7C). A small increase in electrophoretic mobility of the G4 product in the reaction with RecQL4_FL and DNase I suggested that the ssDNA overhangs were not protected. Similarly, 2 μM N170 or 200 nM N388 also protected 10 nM of the G4 structure against DNase I under conditions, where G4 alone was almost completely degraded (Figure 7D).
Finally, as comparable concentrations of N388 and full-length RecQL4 were sufficient to achieve a complete G4 shift, we addressed if the N-terminal region alone was sufficient for G4 binding. Therefore, we analyzed the binding of G4-DNA to increasing concentrations of the C-terminal fragment C464. In the presence of 0.5 μM protein, no binding was detected, whereas N388 and RecQL4_FL resulted in a complete shift at 0.2 μM protein (Figure 7E). We also tested if RecQL4 exhibited further activities on G4 structures. Yet, neither any unwinding of G4 nor a promotion of G4 formation in reactions with or without ATP was apparent (Supplementary Figure S10C).
Taken together, we conclude that the N-terminus of RecQL4 is both necessary and sufficient for the high affinity binding of RecQL4 to G4 structures, but neither is involved in its formation or its dissolution.
DISCUSSION
RecQL4 is a multifunctional enzyme with pleiotropic roles in the cell. Whereas the N-terminal region is essential and apparently sufficient for its function(s) in the initiation of DNA replication (9,15,16,44), the RecQ helicase domain is important for the maintenance of genome integrity upon genotoxic stress (2). This division is most apparent in fungi, where the homologs of the two regions are expressed from two genes, SLD2 and HRQ1, whose products fulfill functions similar to human RecQL4 as independent entities (8,9,50–54).
Despite detailed cell biological analyses, still very little is known about the biochemical and structural properties of human RecQL4 (5–7). We therefore performed a detailed characterization of the N-terminal domain, as it represents the only known metazoan homolog of the essential yeast replication initiation factor Sld2 (8,9).
Various fragments of the N-terminus of human RecQL4 were expressed in E. coli and stable subspecies were identified using a random screen. The biophysical and structural characterization indicated that, apart from the amino-terminal homeo-like domain (10), the N-terminus was largely disordered and adopted a conformation reminiscent of a pre-molten globule. Moreover, the expression screen and the DNA-binding studies revealed that the N-terminus could be divided into at least three functional entities comprising residues 1–54, 55–170 and 251–388, respectively, which were connected by a linker region (aa 171–250) with little functional impact (Figure 8). Intrinsically disordered regions are common for eukaryotic proteins, and are particularly abundant in nuclear proteins, such as transcription factors (reviewed in (55–57)). These regions are very well adapted to mediate interactions with multiple nucleic acid and protein partners, and often adopt a non-random conformation upon binding (55). Considering the relatively high abundance of basic amino acids in the N-terminal region, the observed interactions with DNA are likely to be mediated to a significant extent by electrostatic contributions.
Figure 8.
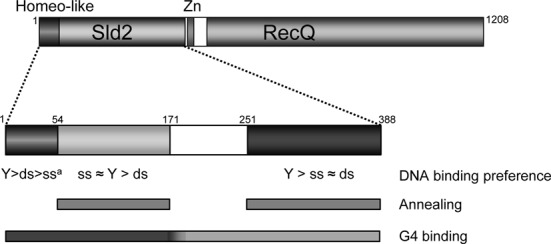
Schematic representation of the presumptive functional N-terminal domains of RecQL4. The regions implicated in DNA binding and their preferences for ss-, ds- and Y-shaped DNA, respectively, are shown. Below the region(s) contributing to G4 binding and annealing activity are indicated by bars. aBased on reference (10).
Sld2RecQL4 from yeast plays a prominent role in the initiation of DNA replication, where it is involved in the formation of the pre-initiation complex including Dpb11TopBP1 and Sld3Treslin. This step depends on timely-regulated phosphorylation events performed by the S phase kinases (58–60). Although RecQL4 appears to be also required for vertebrate initiation complex formation (8,9), the underlying regulation and interaction networks are different (9,10,19). Yeast Sld2 interacts directly with single-strand origin DNA (48). Importantly, this interaction competes with the binding of phosphorylated Mcm2–7 (61). Therefore, it seems that Sld2 mediates the assembly of the replicative helicase and the recruitment of GINS, which together with Cdc45 then unwind the origin (61,62). Can the biochemical properties of the N-terminus of human RecQL4 be connected to its essential function(s) during replication initiation? The multiple binding activities to ss-, ds- and Y-shaped DNA observed here, together with the previously reported interactions with various replication initiation factors, such as Mcm2–7, Mcm10 and TopBP1 (9,10,19,20), suggest a structural role both for origin unwinding and for the establishment of the replication fork. In particular, the preferential binding to Y-shaped DNA suggests a role in stabilization of the emerging initiation bubble. The strong annealing activity, which has also been seen for Sld2 (48), seems to contradict such a role. But strand annealing may counteract origin opening until the initiation complex has been correctly assembled. The strong annealing activity of metazoan RecQL4 is well documented (5,6,44,63), but this is not, as in other RecQ helicases, localized to the conserved helicase domain (reviewed in (64,65)). Nevertheless, it should be noted that the C464 helicase fragment catalyzes some strand annealing, though much less pronounced as compared to the N-terminal fragments of human RecQL4 (Figure 3E). It has been reported that there is an ATP-dependent strand exchange of RecQL4 fragments lacking the N-terminal region, but this activity is far lower than RecQL4_FL-mediated strand exchange in the absence of ATP (5).
Recent studies demonstrate that the vast majority of human origins of replication correlates with G-rich sequences displaying a propensity for G4 formation (32,34). More importantly, the G4 motifs are essential for origin function and their orientation determined the position of the replication start site for two model origins studied in chicken DT40 cells (35). Therefore, it is tempting to speculate that the high affinity binding of the N-terminus of RecQL4 to G4 structures is also involved in the replication initiation process. This principally differs from budding yeast, where Sld2 has been shown to bind to the thymine-rich ssDNA region of the origin (48). In fungi, origins are not in the vicinity of G4 motifs (reviewed in (33)). As others (44,66), we failed to detect any G4 forming or unwinding activity of RecQL4. Hence, G4 structures may snap out spontaneously, when an origin becomes unwound. This in turn may be further stabilized by the binding of RecQL4 (and perhaps ORC (67)) and then serve as an entry site for RPA and the polymerases α and ϵ (8,9), to further stabilize the newly opened bubble. The protection of a G4 structure by RecQL4 against DNase I digestion (Figure 7C), would be in agreement with such a scenario. This is further supported by the observation that RecQL4 protects G4-DNA from unwinding by the BLM helicase (66). Many proteins containing different motifs have been identified to interact with G4 structures (49,67,68). Among these are several helicases of the RecQ family where the ‘RecQ C-terminal’ (RQC) and the ‘helicase and RNaseD C-terminal’ (HRDC) domains were identified as G4 binders (69,70). In this study, we allocated G4 binding to the N-terminal 388 amino acids (Figure 7A and E). In fact, human RecQL4 lacks an HRDC domain and only possesses a poorly conserved RQC domain (39).
Many recent findings indicate that RecQL4 shuttles from the nucleus to the mitochondrion, where it preserves the DNA integrity and protects mitochondrial DNA against oxidative damage (24–27,71). As the mitochondrial genome contains potential G4-forming motifs at a 10-fold higher density compared to the nuclear genome (72), G4 binding may also be important for the mitochondrial function of RecQL4, even though it does not serve as a substrate for its helicase activity.
One of the intriguing properties of RecQL4_N388 was an unconventional ATP-independent strand-exchange activity (Figure 4). Usually, strand exchanges between homologous ss- and dsDNA, e.g. those catalyzed by RecA, are driven by ATP hydrolysis. We therefore do not expect that RecQL4_N388-mediated strand exchanges initiate strand invasion during homologous recombination. This activity rather resembles strand exchanges that are necessary for template switching at stalled replication forks (66,73).
Taken together, the present study demonstrates that beyond its role as a protein interaction platform, the N-terminus of human RecQL4 also acts as a complex moderator of DNA transactions that are mediated by multiple DNA-binding sites. These facilitate structure-specific DNA binding, strand annealing and annealing-initiated strand exchanges, which may be essential for the initiation of DNA replication in humans.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The cDNA for full-length human RecQL4 was a generous gift of Dr Yasushiro Furuichi, GeneCare Research Institute, Kamakura, Japan. We thank Annerose Gleiche for excellent technical assistance.
Footnotes
Present address: Kristin Kiosze, Institute of Biochemistry, Biocenter, Johann Wolfgang Goethe-University Frankfurt, Frankfurt, Germany.
FUNDING
Federal Government of Germany and the State of Thuringia [to Leibniz Institute for Age Research—Fritz Lipmann Institute [(FLI), member of Science Association ‘Gottfried Wilhelm Leibniz’ (WGL)]; Leibniz Graduate School on Ageing and Age-related Diseases (LGSA) [to H.K.]; Academy of Finland [251576 to J.E.S.]. Funding for open access charge: Leibniz Institute for Age Research—Fritz Lipmann Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chu W.K., Hickson I.D. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 2.Croteau D.L., Popuri V., Opresko P.L., Bohr V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siitonen H.A., Sotkasiira J., Biervliet M., Benmansour A., Capri Y., Cormier-daire V., Crandall B., Hannula-Jouppi K., Hennekam R., Herzog D., et al. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellermayer R. The versatile RECQL4. Genet. Med. 2006;8:213–216. doi: 10.1097/01.gim.0000214457.58378.1a. [DOI] [PubMed] [Google Scholar]
- 5.Xu X., Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capp C., Wu J., Hsieh T. Drosophila RecQ4 has a 3’-5’ DNA helicase activity that is essential for viability. J. Biol. Chem. 2009;284:30845–30852. doi: 10.1074/jbc.M109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T., Kohno T., Ishimi Y. DNA helicase activity in purified human RECQL4 protein. J. Biochem. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- 8.Sangrithi M.N., Bernal J.A., Madine M., Philpott A., Lee J., Dunphy W.G., Venkitaraman A.R. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Matsuno K., Kumano M., Kubota Y., Hashimoto Y., Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase α in the initiation of DNA replication. Mol. Cell. Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohlenschläger O., Kuhnert A., Schneider A., Haumann S., Bellstedt P., Keller H., Saluz H.P., Hortschansky P., Hänel F., Grosse F., et al. The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif. Nucleic Acids Res. 2012;40:8309–8324. doi: 10.1093/nar/gks591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaggioli V., Zeiser E., Rivers D., Bradshaw C.R., Ahringer J., Zegerman P. CDK phosphorylation of SLD-2 is required for replication initiation and germline development in C. elegans. J. Cell Biol. 2014;204:507–522. doi: 10.1083/jcb.201310083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J., Capp C., Feng L., Hsieh T.S. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev. Biol. 2008;323:130–142. doi: 10.1016/j.ydbio.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichikawa K., Noda T., Furuichi Y. Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nihon Yakurigaku Zasshi. 2002;119:219–226. doi: 10.1254/fpj.119.219. [DOI] [PubMed] [Google Scholar]
- 14.Mann M.B., Hodges C.A., Barnes E., Vogel H., Hassold T.J., Luo G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- 15.Hoki Y., Araki R., Fujimori A., Ohhata T., Koseki H., Fukumura R., Nakamura M., Takahashi H., Noda Y., Kito S., et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 2003;12:2293–2299. doi: 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- 16.Abe T., Yoshimura A., Hosono Y., Tada S., Seki M., Enomoto T. The N-terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells. Role of the N-terminal region of RECQL4 in cells. Biochim. Biophys. Acta. 2011;1813:473–479. doi: 10.1016/j.bbamcr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y. Rothmund-Thomson syndrome helicase, RECQ4: on the crossroad between DNA replication and repair. DNA Repair (Amst). 2010;9:325–330. doi: 10.1016/j.dnarep.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Thangavel S., Mendoza-maldonado R., Tissino E., Sidorova J.M., Yin J., Wang W., Monnat R.J., Falaschi A., Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X., Rochette P.J., Feyissa E.A., Su T.V., Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im J.S., Ki S.H., Farina A., Jung D.S., Hurwitz J., Lee J.K. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl Acad. Sci. U.S.A. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilves I., Petojevic T., Pesavento J.J., Botchan M.R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh A.K., Rossi M.L., Singh D.K., Dunn C., Ramamoorthy M., Croteau D.L., Liu Y., Bohr V.A. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012;287:196–209. doi: 10.1074/jbc.M111.295063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrarelli L.K., Popuri V., Ghosh A.K., Tadokoro T., Canugovi C., Hsu J.K., Croteau D.L., Bohr V.A. The RECQL4 protein, deficient in Rothmund-Thomson syndrome is active on telomeric D-loops containing DNA metabolism blocking lesions. DNA Repair (Amst). 2013;12:518–528. doi: 10.1016/j.dnarep.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croteau D.L., Rossi M.L., Canugovi C., Tian J., Sykora P., Ramamoorthy M., Wang Z.M., Singh D.K., Akbari M., Kasiviswanathan R., et al. RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell. 2012;11:456–466. doi: 10.1111/j.1474-9726.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De S., Kumari J., Mudgal R., Modi P., Gupta S., Futami K., Goto H., Lindor N.M., Furuichi Y., Mohanty D., et al. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J. Cell Sci. 2012;125:2509–2522. doi: 10.1242/jcs.101501. [DOI] [PubMed] [Google Scholar]
- 26.Chi Z., Nie L., Peng Z., Yang Q., Yang K., Tao J., Mi Y., Fang X., Balajee A.S., Zhao Y. RecQL4 cytoplasmic localization: implications in mitochondrial DNA oxidative damage repair. Int. J. Biochem. Cell Biol. 2012;44:1942–1951. doi: 10.1016/j.biocel.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta S., De S., Srivastava V., Hussain M., Kumari J., Muniyappa K., Sengupta S. RECQL4 and p53 potentiate the activity of polymerase γ and maintain the integrity of the human mitochondrial genome. Carcinogenesis. 2014;35:34–45. doi: 10.1093/carcin/bgt315. [DOI] [PubMed] [Google Scholar]
- 28.Mohaghegh P., Karow J.K., Brosh R.M., Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun H., Karow J.K., Hickson I.D., Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 30.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarsounas M., Tijsterman M. Genomes and G-quadruplexes: for better or for worse. J. Mol. Biol. 2013;425:4782–4789. doi: 10.1016/j.jmb.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 32.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.M., Lemaitre J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 33.Leonard A.C., Méchali M. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013;5:1–17. doi: 10.1101/cshperspect.a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cayrou C., Coulombe P., Puy A., Rialle S., Kaplan N., Segal E., Méchali M. New insights into replication origin characteristics in metazoans. Cell Cycle. 2012;11:658–667. doi: 10.4161/cc.11.4.19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valton A.L., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintomé C., Riou J.F., Prioleau M.N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawasaki M., Inagaki F. Random PCR-based screening for soluble domains using green fluorescent protein. Biochem. Biophys. Res. Commun. 2001;280:842–844. doi: 10.1006/bbrc.2000.4229. [DOI] [PubMed] [Google Scholar]
- 37.Waldo G., Standish B., Berendzen J., Terwilliger T. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 38.Sugiyama T., New J.H., Kowalczykowski S.C. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. U.S.A. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marino F., Vindigni A., Onesti S. Bioinformatic analysis of RecQ4 helicases reveals the presence of a RQC domain and a Zn knuckle. Biophys. Chem. 2013;177–178:34–39. doi: 10.1016/j.bpc.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Ward J., Sodhi J., McGuffin L., Buxton B., Jones D. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Buchan D., Minneci F., Nugent T., Bryson K., Jones D. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W340–W348. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uversky V. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uversky V.N., Gillespie J.R., Fink A.L. Why are ‘natively unfolded’ proteins unstructured under physiologic conditions. Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Rossi M.L., Ghosh A.K., Kulikowicz T., Croteau D.L., Bohr V.A. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst). 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 46.Popuri V., Croteau D.L., Brosh R.M., Bohr V.A. RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle. 2012;11:4252–4265. doi: 10.4161/cc.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Machwe A., Xiao L., Groden J., Matson S.W., Orren D.K. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 48.Kanter D.M., Kaplan D.L. Sld2 binds to origin single-stranded DNA and stimulates DNA annealing. Nucleic Acids Res. 2010;39:2580–2592. doi: 10.1093/nar/gkq1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murat P., Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 50.Pospiech H., Grosse F., Pisani F.M. The initiation step of eukaryotic DNA replication. Subcell. Biolchem. 2010;50:79–104. doi: 10.1007/978-90-481-3471-7_5. [DOI] [PubMed] [Google Scholar]
- 51.Groocock L.M., Prudden J., Perry J.J.P., Boddy M.N. The RecQ4 orthologue Hrq1 is critical for DNA interstrand cross-link repair and genome stability in fission yeast. Mol. Cell. Biol. 2012;32:276–287. doi: 10.1128/MCB.06184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bochman M.L., Paeschke K., Chan A., Zakian V.A. Hrq1, a homolog of the human RecQ4 helicase, acts catalytically and structurally to promote genome integrity. Cell Rep. 2014;6:346–356. doi: 10.1016/j.celrep.2013.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi D.H., Lee R., Kwon S.H., Bae S.H. Hrq1 functions independently of Sgs1 to preserve genome integrity in Saccharomyces cerevisiae. J. Microbiol. 2013;51:105–112. doi: 10.1007/s12275-013-3048-2. [DOI] [PubMed] [Google Scholar]
- 54.Barea F., Tessaro S., Bonatto D. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput. Biol. Chem. 2008;32:349–358. doi: 10.1016/j.compbiolchem.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Fink A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Nishikawa K. Natively unfolded proteins: an overview. Biophysics (Oxf). 2009;5:53–58. doi: 10.2142/biophysics.5.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pentony M.M., Jones D.T. Modularity of intrinsic disorder in the human proteome. Proteins. 2010;78:212–221. doi: 10.1002/prot.22504. [DOI] [PubMed] [Google Scholar]
- 58.Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr. Opin. Cell Biol. 2010;22:766–771. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 59.Zegerman P., Diffley J.F.X. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 61.Bruck I., Kanter D.M., Kaplan D.L. Enabling association of the GINS protein tetramer with the mini chromosome maintenance (Mcm)2–7 protein complex by phosphorylated Sld2 protein and single-stranded origin DNA. J. Biol. Chem. 2011;286:36414–36426. doi: 10.1074/jbc.M111.282822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruck I., Kaplan D.L. The replication initiation protein Sld2 regulates helicase assembly. J. Biol. Chem. 2014;289:1948–1959. doi: 10.1074/jbc.M113.532085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macris M.A., Krejci L., Bussen W., Shimamoto A., Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst). 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 64.Wu Y. Unwinding and rewinding: double faces of helicase. J. Nucleic Acids. 2012;2012 doi: 10.1155/2012/140601. doi:10.1155/2012/140601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bachrati C.Z., Hickson I.D. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 66.Singh D.K., Popuri V., Kulikowicz T., Shevelev I., Ghosh A.K., Ramamoorthy M., Rossi M.L., Janscak P., Croteau D.L., Bohr V.A. The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res. 2012;40:1–17. doi: 10.1093/nar/gks349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoshina S., Yura K., Teranishi H., Kiyasu N., Tominaga A., Kadoma H., Nakatsuka A., Kunichika T., Obuse C., Waga S. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J. Biol. Chem. 2013;288:30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sissi C., Gatto B., Palumbo M. The evolving world of protein-G-quadruplex recognition: a medicinal chemist's perspective. Biochimie. 2011;93:1219–1230. doi: 10.1016/j.biochi.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huber M.D., Duquette M.L., Shiels J.C., Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 70.Cahoon L.A., Manthei K.A., Rotman E., Keck J.L., Seifert H.S. Neisseria gonorrhoeae RecQ helicase HRDC domains are essential for efficient binding and unwinding of the pilE guanine quartet structure required for pilin antigenic variation. J. Bacteriol. 2013;195:2255–2261. doi: 10.1128/JB.02217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Croteau D.L., Singh D.K., Hoh Ferrarelli L., Lu H., Bohr V.A. RECQL4 in genomic instability and aging. Trends Genet. 2012;28:624–631. doi: 10.1016/j.tig.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Capra J.A., Paeschke K., Singh M., Zakian V.A. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Branzei D., Foiani M. Template switching: from replication fork repair to genome rearrangements. Cell. 2007;131:1228–1230. doi: 10.1016/j.cell.2007.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


