Abstract
We compared the nucleus accumbens (NAc) transcriptomes of generation 8 (G8), 34-day-old rats selectively bred for low (LVR) versus high voluntary running (HVR) behaviours in rats that never ran (LVRnon-run and HVRnon-run), as well as in rats after 6 days of voluntary wheel running (LVRrun and HVRrun). In addition, the NAc transcriptome of wild-type Wistar rats was compared. The purpose of this transcriptomics approach was to generate testable hypotheses as to possible NAc features that may be contributing to running motivation differences between lines. Ingenuity Pathway Analysis and Gene Ontology analyses suggested that ‘cell cycle’-related transcripts and the running-induced plasticity of dopamine-related transcripts were lower in LVR versus HVR rats. From these data, a hypothesis was generated that LVR rats might have less NAc neuron maturation than HVR rats. Follow-up immunohistochemistry in G9–10 LVRnon-run rats suggested that the LVR line inherently possessed fewer mature medium spiny (Darpp-32-positive) neurons (P < 0.001) and fewer immature (Dcx-positive) neurons (P < 0.001) than their G9–10 HVR counterparts. However, voluntary running wheel access in our G9–10 LVRs uniquely increased their Darpp-32-positive and Dcx-positive neuron densities. In summary, NAc cellularity differences and/or the lack of running-induced plasticity in dopamine signalling-related transcripts may contribute to low voluntary running motivation in LVR rats.
Introduction
Understanding the neuro-molecular basis for voluntary exercise behaviours is imperative. Accelerometry measurements suggest that over 90% of Americans who are 12 years old and older fail to meet US physical activity guidelines (Troiano et al. 2008). This statistic is troubling given that lifetime physical inactivity accelerates the secondary ageing of numerous organ systems, which leads to a diminished quality of life and an increased risk for chronic disease and premature mortality (Booth et al. 2011). A recent genome-wise association study of 772 twins found that additive genetic factors explained 47% of the variance for time spent performing moderate to vigorous intensity physical activity and 31% of the variance in the time spent being sedentary (Hoed et al. 2013). Thus, low and high levels of voluntary physical activity probably have a partial genetic basis.
The mesolimbic dopaminergic pathway in the midbrain and basal forebrain, specifically the nucleus accumbens (NAc), plays a major role in determining voluntary running behaviour in rodents (Waters et al. 2008; Knab et al. 2009, 2012; Knab & Lightfoot, 2010). Furthermore, Salamone & Correa (2012) contend that the NAc acts as a ‘gate’, ‘filter’ and/or ‘amplifier’ of information passing from various cortical or limbic areas to various motor areas of the brain, and suggest that the NAc participates in a variety of behavioural processes related to aspects of motivation. A selective breeding model was first developed by Swallow et al. (1998) to study the neurobiology of mice that voluntarily run high nightly distances (high voluntary running (HVR) mice) compared to control mice. Rhodes et al. (2003) examined brain activity patterns of HVR mice versus control lines through c-fos staining and determined that high neuron activation in the NAc differentiated running motivation between the HVR and control lines. Altered dopaminergic profiles also exist between the HVR lines and control lines (Rhodes et al. 2001; Rhodes & Garland, 2003; Mathes et al. 2010), which further suggest that NAc dopaminergic signalling is involved with voluntary exercise motivation. Beyond the HVR murine model of Garland and co-workers, other rodent data similarly suggest that NAc dopaminergic signalling plays an integral role in voluntary physical activity behaviours (Knab et al. 2009; Greenwood et al. 2011). Finally, genetic factors are known to be involved in both motivation and ability to engage in voluntary wheel running in rodents (Garland et al. 2011).
We recently developed a unique selective breeding model for rats that voluntarily run low (low voluntary running (LVR)) or high (HVR) nightly distances in voluntary running wheels (Roberts et al. 2013). In the current study, we sought to examine the NAc transcriptome in generation 8 (G8) 34-day-old rats selectively bred for low (LVR) versus high voluntary running (HVR) behaviours, and tested in subgroups of those that never ran (LVRnon-run and HVRnon-run), as well as after 6 days of voluntary wheel running (LVRrun and HVRrun). In addition, control wild-type Wistar (CTL) rats were tested. Importantly, the purpose of this ‘omics’ approach was to generate testable hypotheses as to which possible NAc features may be contributing to running motivation differences between lines. In the current study, RNA deep-sequencing (RNA-seq) and bioinformatics analyses from the G8 LVR and HVR rats directed hypothesis-driven follow-up immunohistochemistry experiments in G9–10 rats, which suggested that NAc neuronal maturation differences exist between LVR and HVR rats. Our newly generated hypothesis from these data is that LVR rats inherently possess less NAc neuron maturation than HVR rats, which, in turn, may suppress the development of voluntary wheel running reward. Likewise, while G9–10 HVR and LVR rats possessed similar NAc dopamine levels, the lack of running-induced plasticity in dopamine signalling-related mRNAs in the G8 LVR versus G8 HVR rats may be one culprit for low running motivation in the former line type. More in-depth discussion of these results is presented.
Methods
NAc RNA isolation from G8 HVR and LVR rats and cDNA library preparation for RNA-seq
All animal procedures outlined below were approved by the University of Missouri's Animal Care and Use Committee. Our selective breeding model to generate LVR and HVR rats is described elsewhere (Roberts et al. 2013). In brief, we reported that G8 HVR rats voluntarily ran about five times longer distances than G8 LVR rats, an effect that is mostly explained by between-line differences in time spent in running wheels versus running pace. In the current study, G8 LVR and HVR rats were weaned at 21 days of age, and then randomly divided into two groups at 28 days old, composed of: (a) those that voluntarily ran in wheels with bicycle computers for 6 days between the ages of 28 and 34 days (LVRrun and HVRrun); and (b) those that never ran in wheels (LVRnon-run and HVRnon-run). Wild-type Wistar rats that were never exposed to voluntary running wheels were purchased from Charles River Laboratories (Rayleigh, NC, USA) for this experiment. Food (Formulab Diet 5008, Purina) and water were provided ad libitum throughout the entirety of the experiment for all rats.
At 34 days of age between 17.00 and 19.00 h, up to 2 h prior to the dark cycle, rats were administered an i.p. injection of a lethal dose of sodium pentobarbital (60 mg kg−1 body mass). This time point was chosen as a ‘basal’ observational point to avoid running-induced line-type differences in NAc mRNAs that probably could exist during and in the hours following the dark cycle. Furthermore, 34 days is the time when rats end the 6-day period of voluntary wheel running for the selection process. Brains were quickly removed and NAc tissue was extracted from seven (4 male, 3 female) G8 HVRnon-run rats, eight (4 male, 4 female) G8 LVRnon-run rats, six G8 (3 male, 3 female) HVRrun rats, six (3 male, 3 female) G8 LVRrun rats and four (2 male, 2 female) CTL rats using a 2 mm punch tool and brain sectioning apparatus (Braintree Scientific, Braintree, MA, USA). Tissue plugs from 2 mm thick coronal brain slices, which were visibly identified as being from the NAc (Paxinos & Watson, 1998), were placed in Trizol and were stored at −80°C until processing. During tissue processing, samples were lysed in Trizol using a high speed shaking apparatus (Tissuelyser LT, Qiagen, Crawley, West Sussex, U.K.) with RNase-free stainless steel beads. RNA was subsequently separated according to the manufacturer's instructions, and isolated/DNase treated with columns (Macherey-Nagel, Bethlehem, PA, USA). High RNA integrity of each sample was confirmed using a BioAnalyzer 2100 automated electrophoresis system (Bio-Rad, Hercules, CA, USA) prior to cDNA library construction. cDNA library preparation was performed at the University of Missouri DNA Core as previously described (Roberts et al. 2013).
Illumina sequencing of NAc cDNA and statistical analyses of RNA-seq data
RNA-seq procedures were done at the University of Missouri DNA Core and are described in more detail elsewhere (Rustemeyer et al. 2011; Roberts et al. 2013). Differential gene expression patterns were analysed for annotated genes between the HVRrun and LVRrun rats using reads per kilobase per million mapped reads (RPKM) values. Our strategy to assess NAc mRNAs differentially expressed between G8 HVR and LVR rats is presented in Fig.1. We previously reported that 35 NAc transcripts exhibited line type specificity between G8 HVRnon-run and LVRnon-run rats (Roberts et al. 2013). However, it is possible that many of these transcripts could be normalized after 6 days of running, making these less probable line type-specific gene candidates (as in Fig.1C). Thus, we deemed an mRNA to be an inherent line type NAc candidate if it met the following thresholds: (a) the G8 HVRrun/G8 LVRrun fold change value was greater than ±1.25 (Student's t test P < 0.01); and (b) subsequent analysis of this mRNA candidate differed between the HVRnon-run and LVRnon-run groups at P < 0.05 regardless of fold change value. Transcripts that met the aforementioned HVRrun/LVRrun and HVRnon-run/LVRnon-run thresholds were considered more representative, intrinsic line type candidate genes (Fig.1B) and were also compared to control Wistar rats to further validate their line type specificity (not depicted in Fig.1).
Figure 1. Experimental strategy to assess influential NAc mRNAs differentially expressed between G8 HVR and LVR rats.
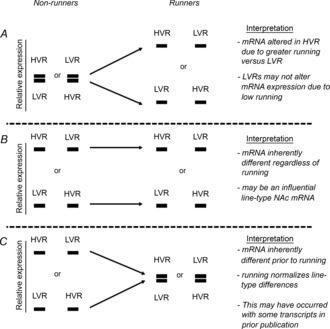
A, no inherent difference, but acquired difference with voluntary running. B, inherent difference is maintained after 6 days of voluntary running. C, inherent difference is lost after 6 days of voluntary running. NAc transcripts from HVR and LVR sitters (HVRnon-run and LVRnon-run) were also compared to age-matched control (CTL) Wistar rats to provide further evidence for line type differences.
Note that a false discovery rate threshold of q < 0.10 proved to be too stringent to detect line type NAc transcriptomic differences with and/or without running. While a ±1.25-fold cut-off (P < 0.01) threshold may seem statistically liberal compared to other publications using 1.5- to 2.0-fold change cut-offs to examine transcriptomic differences between treatments (Heruth et al. 2012; Song et al. 2012; Zhang et al. 2012), we contend that subtle mRNA differences exist within the NAc between the HVR and LVR rats given that: (a) our model is a physiological/in vivo model whereby rats are being observed in a ‘basal’ state, and (b) only eight transcripts differed at a ±1.5-fold change threshold in our previous publication when solely comparing the HVRnon-run versus LVRnon-run groups (Roberts et al. 2013). Of note, RNA-seq analyses were also performed in the current study at a relatively early generation of selective breeding to identify early NAc gene changes that occur prior to potential compensatory gene changes which may occur in later generations and, thus, could confound data interpretation.
Bioinformatics of RNA-seq data
Ingenuity Pathway Analysis (IPA; Ingenuity Systems Inc., Redwood, CA, USA) was used to examine NAc gene networks that differed between HVRrun and LVRrun rats, but were similar between HVRnon-run and LVRnon-run rats (line type differences probably due to running in Fig.1A and B differed between runner (HVRrun and LVRrun) as well as between non-runner (HVRnon-run and LVRnon-run) rats (inherent line type differences as in Fig.1B)). Past literature has also attributed differences in voluntary running behaviour to differences in NAc dopamine signalling (Knab & Lightfoot, 2010; Mathes et al. 2010; Greenwood et al. 2011; Knab et al. 2012; Roberts et al. 2012, 2013). Therefore, a list of dopamine signalling-related transcripts was constructed using the Gene Ontology (GO) Consortium database (http://www.geneontology.org) to examine if these gene expression patterns differed between LVR and HVR lines with or without 6 days of voluntary wheel running. Pathways for dopamine-related genes included the dopamine receptor-signalling pathway (GO ID: 0007212), adenylate cyclase-activating dopamine receptor pathway (GO ID: 0007191), adenylate cyclase-inhibiting dopamine receptor pathway (GO ID: 0007191) and/or the negative regulation of dopamine receptor signalling pathway (GO ID: 0060160).
Western blotting confirmation of line type-specific targets yielded by G8 RNA-seq analyses in female G10–G11 LVR and HVR rats
Western blotting of one of the gene candidates was performed in 35-day-old female G10–11 LVRrun/non-run and HVRrun/non-run rats (n = 6–7 per line and activity group). The Cadm4 gene was validated through Western blotting (Fig.2) due to it being the sole NAc inherent line type gene candidate that was most highly expressed on an RPKM basis and also associated with neuronal synaptogenesis, as discussed in the sections below.
Figure 2. Western blotting confirmation of NAc Cadm4 as a line type-specific gene.

All animals were female and were 35 days of age, and HVRrun and LVRrun animals spent 6 days in a voluntary running wheel (n = 6–7 per bar). *P < 0.05.
Briefly, 15–20 mg of NAc tissue was removed from brain and homogenized on ice in RIPA buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1% SDS, 1× protease inhibitor cocktail) using a Tissuelyser at 20 Hz for 1 min. The homogenate was centrifuged at 12,000 g for 10 min and the resultant supernatant was obtained for Western blotting. Protein concentrations were obtained using the BCA assay (Pierce Biotechnology, Rockford, IL, USA) and 60 μg of protein in loading buffer was loaded onto 18% SDS-PAGE gels. Proteins were transferred onto polyvinylidene difluoride membranes and all blots were incubated with Ponceau S (Sigma, St Louis, MO, USA) to verify equal loading in all lanes. Primary antibodies (rabbit polyclonal Cadm4 at 1:1000 (Abcam, Cambridge, MA, USA)) that had been diluted in Tris-buffered saline + Tween20 with 5% bovine serum albumin and applied to membranes overnight at 4°C, horseradish peroxidase-conjugated secondary antibody (1:2000; Cell Signaling Technology, Inc., Danvers, MA, USA), were applied for 1 h at room temperature, and ECL substrate (Pierce Biotechnology) was then applied for 5 min prior to exposure. Band densitometry was performed through the use of Kodak 4000R Imager and Molecular Imagery Software (Kodak Molecular Imaging Systems, New Haven, CT, USA) and statistical analyses on band densities were performed using a two-way (line type (LVR vs. HVR) × activity (run vs. non-run)) analysis of variance with Holm–Sidak post hoc analyses when appropriate.
Follow-up NAc tissue immunohistochemistry experiments in female G9–10 LVR and HVR rats from hypotheses derived from G8 RNA-seq analyses
Based upon resultant IPA gene network differences between lines of ‘cancer, cell cycle, cellular growth and proliferation’, we hypothesized that LVR rats might have fewer mature medium spiny neurons (MSNs). This hypothesis was tested by independent immunohistochemical (IHC) analyses for dopamine- and cAMP-regulated neuronal phosphoprotein (Darpp-32)-positive neurons, as well as for NAc doublecortin (Dcx)-positive neurons in female G9–10 LVR and HVR rats. Dcx is highly expressed by immature neurons in the adult cortex, and upon differentiation gradually decreases to undetectable levels (Brown et al. 2003). The stain was for total Darpp-32 protein that is prominently expressed in differentiated striatal MSNs, comprising 95% of neurons in the NAc (Arlotta et al. 2008), which was thus used as a molecular marker of mature/differentiated NAc MSNs in the current study.
Briefly, coronal brain slices from 39- to 40-day-old female G10 HVRrun (n = 7) and G10 LVRrun (n = 7) rats that voluntarily ran for 10–12 days were used for IHC. Coronal brain slices from age-matched G9 female LVRnon-run (n = 7) and G10 female HVRnon-run rats (n = 6) were also examined for Dcx- and Darpp-32-positive NAc neurons. Of note, brains were obtained 2–3 h into their dark/running cycle for the purpose of examining if distance ran on the night when they were killed correlated with c-fos-positive neurons in the NAc and other brain areas (data not included). This differed from the G8 NAc samples obtained for RNA-seq 2 h prior to the dark/running cycle. However, it is likely that NAc MSN and Dcx neuron densities do not transiently fluctuate between light and dark cycles.
Nore also that while these animals were 4–5 days older than the G8 animals interrogated for RNA-seq: (a) they were still similarly pre-pubescent as posited from previous literature (Zanato et al. 1994); and (b) Dcx protein expression is long lasting in premature neurons and gradually declines upon differentiation, as previously mentioned. Additionally, brains from the G8 animals were not preserved for IHC and, given that IHC experiments were done as a post hoc analysis, G9–10 animals that were ∼40 days of age were used due to convenience of sampling as opposed to 35 days of age. In spite of these minor differences, potential line type differences observed in NAc Dcx-positive and Darpp-32-positive neurons between the 39- to 40-day-old HVRrun and LVRrun rats logically provided us with a good representation of what probably occurred in G8 35-day-old HVRrun and LVRrun rats.
IHC procedures were similar to those performed by Rhodes et al. (2003). Briefly, rats were killed and were transcardially perfused with freshly made 4% paraformaldehyde (w/v) in phosphate-buffered saline (PBS, pH 7.4). Brains were subsequently removed, post-fixed in 4% paraformaldehyde overnight, and incubated in 30% sucrose (w/v) in PBS for 2 days. Thereafter, brains were removed from the 30% sucrose solution, wrapped in parafilm and stored until coronal slicing. Multiple frozen 40 μm coronal sections containing the NAc (Bregma + 1.00 mm) were obtained using a cryostat (Leica, Wetzlar, Germany) and each slice was placed in 24-well plates containing cryoprotectant solution (30% sucrose, 30% (w/v) ethylene glycol and 10% (w/v) polyvinylpryrrolidone in PBS, pH 7.4). Following long-term storage and prior to assaying, the free-floating sections were thoroughly washed in PBS and blocked in 10% normal goat serum for 2 h. IHC for Dcx (1:100 rabbit anti-Dcx, Abcam) and pan Darpp-32 (1:100 rabbit anti-Darpp-32, Cell Signaling) was subsequently performed on serial NAc sections in PBS containing 1.5% normal goat serum and 0.2% Triton X-100, and sections were incubated with the primary antibody solution for 2 days at 4°C. Sections were subsequently incubated with biotinylated goat anti-rabbit secondary antibody (1:400, Vector Labs, Burlingame, CA, USA) in PBS containing 0.2% Triton X-100 for 2 h at room temperature followed by incubation with ABC Elite kit reagents for 1 h (Vector Labs). Finally, sections were stained with diaminobenzidine + nickel solution through peroxidase reaction for 5 min. Section staining from all animals was performed simultaneously to avoid potential interassay variations in staining. Note that multiple coronal brain slices containing the NAc were collected per animal. As per the methods of Rhodes et al. (2003), one representative NAc section was immunostained. To standardize between-animal NAc sections, we ensured that there was morphological similarity of the following visual landmarks: (a) the orientation of the corpus collosum, (b) the initial presence of the lateral ventricle and (c) the presence of the anterior commissure which exists within the NAc core.
Micrographs were captured using an Olympus BX60 photomicroscope at 10× magnification (Olympus, Melville, NY, USA), and photographed with a Spot Insight digital camera (Diagnostic Instruments, Sterling Heights, MI, USA). Dcx-positive and Darpp-32-positive neurons from all samples were counted using Image J software (National Institutes of Health, Bethesda, MD, USA). Statistical analyses on Dcx-positive and Darpp-32-positive neurons were performed using a two-way (line type (HVR vs. LVR) × activity (run vs. non-run)) analysis of variance with Holm-Sidak post hoc analyses when appropriate.
NAc tissue dopamine in female G9–10 LVR and HVR rats as a follow-up to G8 RNA-seq analyses
To examine if dopamine concentrations were different between lines, NAc dopamine concentrations were determined in: (a) G10–11 LVRnon-run and HVRnon-run rats 1–2 h prior to their dark cycle; and (b) G10–11 LVRrun and HVRrun rats 1–2 h prior to their running dark cycle as well as 2–3 h into their dark cycle. Approximately 15–20 mg of NAc tissue was stereotactically removed from 35-day-old G10–11 LVRrun/non-run and HVRrun/non-run rats and homogenized on ice in 0.1 m perchloric acid and 0.001% EDTA (w/v) using a Tissuelyser at 20 Hz for 1 min. The homogenate was centrifuged at 12,000 g for 10 min and the resultant supernatant was obtained for dopamine analysis using high-performance liquid chromatography (HPLC) with electrochemical detection. Briefly, the system consisted of an ESA isocratic pump and a Thermo Separations Products Spectra system AS3500 auto sampler with a four-channel ESA Coularray Model 5600 detection system. Coularray settings were 25, 150, 250 and 800 mV. A Phenomenex 250 × 4.60 mm, 5 μm, Prodigy reversed phase column was used with a mobile phase consisting of 75 nm sodium hydrogen phosphate, 1.7 mm 1-octanesulfaonic acid, 100 μl l−1 triethylamine, 25 μm (500 μl of 100 mm EDTA/2 l, 5% acetonitrile adjusted to pH 3.0 with phosphoric acid) being pumped at 1 ml min−1. The system was controlled and data acquired and processed using the CoulArray software on a Pentium-based computer. Dopamine primary standard (1000 p.p.m., Sigma) was prepared in 1:1 acetonitrile/water, working dopamine standards (500 and 100 p.p.b.) were prepared in 1:1 acetonitrile/water, and dopamine retention was found to be at 8.3 min. Statistical analyses on NAc tissue dopamine content were performed using a two-way (line type (HVR vs. LVR) × activity (run vs. non-run vs. run during dark cycle)) analysis of variance with Holm–Sidak post hoc analyses when appropriate.
Results
Characterization of RNA-seq data from G8 LVR and HVR rats
The total number of reads as well as the percentage of reads aligned to the reference genome are presented in Table1. To ensure that there was uniform tiling across the reference genome, tiled reads from each sample were visualized using NexGen v2.2 software (data not shown).
Table 1.
RNA-seq reads mapped to reference library
| Group | Total reads | Reads aligned to reference library | Total annotated transcripts expressed* |
|---|---|---|---|
| LVRnon-run | 32,807,065 | 89.3% | 17,438 |
| HVRnon-run | 34,273,956 | 88.6% | 17,919 |
| LVRrun | 29,667,774 | 89.2% | 17,764 |
| HVRrun | 25,423,774 | 88.7% | 17,506 |
| CTL | 25,932,949 | 87.5% | 17,962 |
Chen et al. (2011) recently used laser capture microdissection to isolate NAc neurons for subsequent RNA-seq analysis, and sequencing results yielded high enrichments for glutamic acid decarboxylase 1 (Gad1) and Gad2 transcripts (RPKM, log2 values of 7–8), which encode enzymes in NAc MSNs to produce GABA. Our current RNA-seq data demonstrate that all samples were similarly enriched for Gad1 and Gad2, suggesting that NAc MSNs were indeed present within the assayed brain plugs (Fig.3A and B). The high correlation between RPKM values from a sampled rat versus the mean RPKM values from all of the 31 rats also demonstrates a high reliability of transcript detection using our current RNA-seq methods (Fig.3C). Specific differentially expressed transcripts were verified by RT-PCR (Table2).
Figure 3. Enrichment of an NAc MSN-specific mRNA marker in all groups as well as the reliability of RNA-seq measurements.
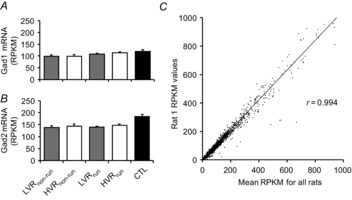
All groups presented high amounts of Grd1 (A) and Grd2 (B), which is indicative of NAc MSNs as previously shown by Chen et al. (2011). C, high correlation of RPKM values from a single rat compared to RPKM average values from all rats, demonstrating high reliability from the RNA-seq data.
Table 2.
Validation of selected RNA-seq transcripts by RT-PCR
| RNA-seq |
RT-PCR |
|||
|---|---|---|---|---|
| Transcript name* | Fold change | P | Fold change | P |
| Cadm4 | 2.1 | 0.007 | 3.5 | 0.009 |
| Ggcx | 1.5 | 0.000 | 1.6 | 0.001 |
| Oprm1 | 1.5 | 0.010 | 1.6 | 0.002 |
| Sgk1 | 1.5 | 0.009 | 1.7 | 0.04 |
Cell adhesion molecule 4 (Cadm4) promotes the formation of presynaptic terminals and induces functional synapses in the CNS by functioning in cell–cell adhesion. It was intrinsically different between LVR and HVR both in non-running and in voluntary running rats. It had RPKM > 34 for all group means. Gamma-glutamyl carboxylase (Ggcx) converts the reduced hydroquinone form of vitamin K to vitamin K epoxide. It was intrinsically different between LVR and HVR both in non-running and in voluntary running rats. It had RPKM > 2.5 for all group means. Opioid receptor mu 1 (Oprm1) is the principal target of endogenous opioid peptides and opioid analgesic agents, such as beta-endorphin and enkephalins, which exercise increases in the brain. It had RPKM > 1.3 for all group means. Serum- and glucocorticoid-inducible kinase-1 (Sgk1) expression plays an important role in cellular stress response, suggesting its involvement in the regulation of processes such as cell survival and neuronal excitability. It had RPKM > 27 for all group means.
NAc mRNA differences between G8 LVRrun vs. G8 HVRrun rats due to 6-day running differences
RNA-seq differences between G8 LVR and HVR rats with or without running are summarized in Fig.4.
Figure 4. Differentially expressed genes (DEGs) in HVRrun and LVRrun and similar or inherently different between HVRnon-run and LVRnon-run rats as well as CTL rats in RNA-seq analysis.
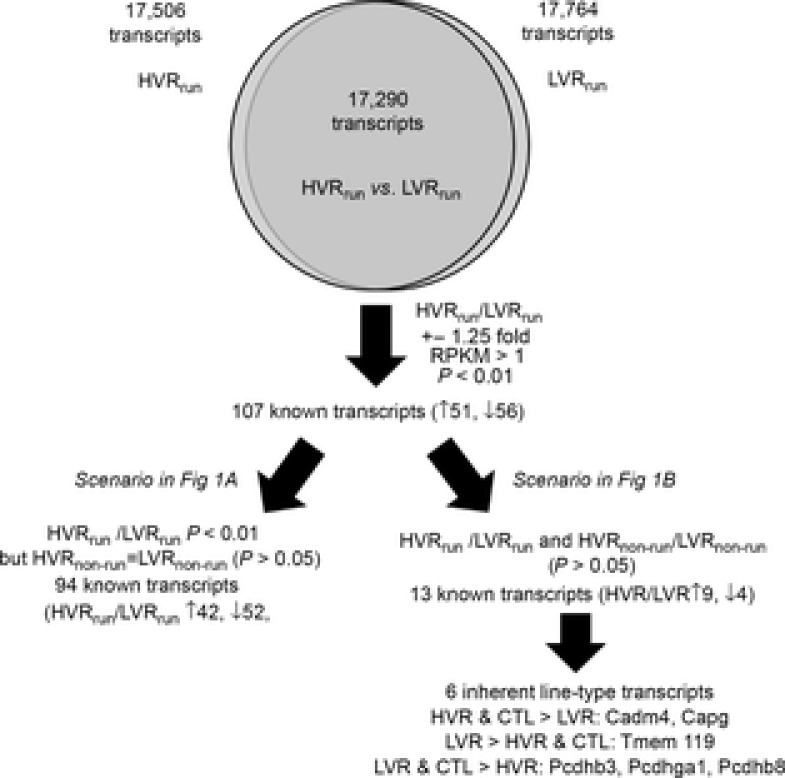
Total 6-day running distances (3.6 vs. 33.7 km) and times (121 vs. 1071 min) were significantly less in the LVRrun than HVRrun rats (P < 0.001; data not shown). Using the aforementioned thresholds (HVRrun/LVRrun > ±1.25-fold, unadjusted P < 0.01), we determined that 107 NAc mRNAs were differentially expressed between LVRrun and HVRrun rats. Of these 107 transcripts, only 13 were also differentially expressed between LVRnon-run and HVRnon-run rats, making them inherent/intrinsic line type NAc mRNA candidates (Fig.1B, discussed in the next section). Thus, the 94 remaining transcripts that were differentially expressed after 6 days of running between LVRrun and HVRrun rats (Fig.1A) were probably a consequence of drastic voluntary running differences (top 10 up- and down-regulated are presented in Table3). No sex differences were noted for any of the 107 transcripts (data not shown).
Table 3.
Top ten up- and down-regulated NAc transcripts between HVRrun and LVRrun but similar between HVRnon-run and LVRnon-run rats (scenario in Fig.1A)
| HVRrun | LVRrun | HVRrun/ | HVRnon-run | LVRnon-run | HVRnon-run/ | |||
|---|---|---|---|---|---|---|---|---|
| Transcript | RPKM avg | RPKM avg | LVRrun | P | RPKM avg | RPKM avg | LVRnon-run | P |
| Up-regulated in HVRrun and LVRrun but were similar between HVRnon-run and LVRnon-run | ||||||||
| Follistatin (Fst), mRNA | 2.30↑ vs. HVRnr | 1.37 | 1.68 | 0.002 | 1.41 | 1.59 | −1.13 | 0.449 |
| Serum/glucocorticoid regulated kinase 1 (Sgk1), mRNA | 41.20 | 26.97 | 1.53 | 0.009 | 31.52↑ vs. CTL | 27.59↑ vs. CTL | 1.14 | 0.380 |
| Dymeclin (Dym), mRNA | 2.46↑ vs. HVRnr | 1.67 | 1.47 | 0.000 | 1.81 | 1.89 | −1.04 | 0.678 |
| Protocadherin gamma subfamily A8 (Pcdhga8), mRNA | 3.71↑ vs. HVRnr | 2.56 | 1.45 | 0.002 | 2.76 | 2.64 | 1.05 | 0.520 |
| Protocadherin gamma subfamily A11 (Pcdhga11), mRNA | 2.39↑ vs. HVRnr | 1.65 | 1.44 | 0.000 | 1.89 | 1.83 | 1.04 | 0.693 |
| HAUS augmin-like complex, subunit 4 (Haus4), mRNA | 4.58 | 3.17 | 1.44 | 0.002 | 4.83 | 3.62 | 1.33 | 0.062 |
| Zinc finger protein 189 (Zfp189), mRNA | 11.66↑ vs. HVRnr | 8.34 | 1.40 | 0.009 | 8.03↑ vs. CTL | 7.95↑ vs. CTL | 1.01 | 0.938 |
| Solute carrier family 38, member 10 (Slc38a10), mRNA | 1.68 | 1.21 | 1.38 | 0.004 | 1.41 | 1.38 | 1.02 | 0.818 |
| Leucine rich repeat containing 8 family, member C (Lrrc8c), mRNA | 6.09↑ vs. HVRnr | 4.47 | 1.36 | 0.004 | 4.80 | 4.62 | 1.04 | 0.778 |
| Transmembrane protein 38A (Tmem38a), mRNA | 2.21↑ vs. HVRnr | 1.63 | 1.36 | 0.006 | 1.71 | 1.46 | 1.17 | 0.073 |
| Down-regulated in HVRrun and LVRrun but similar in HVRnon-run and LVRnon-run | ||||||||
| Steroid receptor RNA activator 1 (Sra1), non-coding RNA | 1.93↓ vs. HVRnr | 2.67 | −1.39 | 0.001 | 2.60 | 2.73 | −1.05 | 0.706 |
| Bcl2 modifying factor (Bmf), mRNA | 3.02↓ vs. HVRnr | 4.21 | −1.39 | 0.002 | 4.44 | 4.14 | 1.07 | 0.538 |
| Zinc finger protein 408 (Znf408), mRNA | 1.56↓ vs. HVRnr | 2.22 | −1.42 | 0.001 | 2.16 | 2.04 | 1.06 | 0.357 |
| PREDICTED: Rattus norvegicus zinc finger CCCH type, antiviral 1-like (LOC100365858), miscRNA | 1.23↓ vs. HVRnr | 1.77 | −1.43 | 0.003 | 1.66 | 1.73 | −1.04 | 0.748 |
| PREDICTED: Rattus norvegicus SRY-box containing gene 9 (LOC100361122), mRNA | 2.53 | 3.66 | −1.45 | 0.007 | 3.31 | 3.40 | −1.03 | 0.844 |
| Patatin-like phospholipase domain containing 7 (Pnpla7), mRNA | 1.49↓ vs. HVRnr | 2.17 | −1.45 | 0.004 | 2.05 | 1.94 | 1.05 | 0.327 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 4 (Adamts4), mRNA | 12.38 | 18.08 | −1.46 | 0.005 | 15.08 | 16.99 | −1.13 | 0.317 |
| RTEL1-TNFRSF6B readthrough, non-coding RNA | 2.07 | 3.83 | −1.85 | 0.006 | 2.92 | 3.43 | −1.17 | 0.311 |
| 3-Hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) (Hmgcs2), mRNA | 3.15↓ vs. HVRnr | 5.83 | −1.85 | 0.002 | 4.57↓ vs. CTL | 5.34 | −1.17 | 0.204 |
| Keratin 2 (Krt2), mRNA | 1.10 | 2.05 | −1.87 | 0.002 | 1.29 | 1.77 | −1.37 | 0.093 |
These transcripts were the top 10 up- and down-regulated NAc transcripts from the 94 transcripts that were differentially expressed in HVRrun versus LVRrun rats, probably a result of more running in HVRrun rats. Abbreviations: HVRrun = HVR 34-day-old 6-day runners, LVRrun = LVR 34-day-old 6-day runners, HVRnon-run = HVR 34-day-old non-runners, LVRnon-run = LVR 34-day-old non-runners, CTL = control Wistar rats. Symbols: ↑ vs. CTL = transcript in HVRnon-run or LVRnon-run rats was higher than in CTL rats (P < 0.05); ↓ vs. CTL = transcript in HVRnon-run or LVRnon-run rats was lower than in CTL rats (P < 0.05); ↑ vs. HVRnon-run or LVRnon-run = transcript in HVRrun or LVRrun is greater than HVRnon-run or LVRnon-run rats, respectively (P < 0.05); ↓ vs. HVRnon-run or LVRnon-run = transcript in HVRrun is less than HVRnon-run rats (P < 0.05). These significant differences are in bold type.
Inherent line type NAc mRNA candidates exclusive to the G8 LVR or G8 HVR rats with or without running when compared to CTL rats
As mentioned in the preceding paragraph, 13 differentially expressed NAc mRNAs existed between LVR and HVR rats regardless of running (Table4). While most of the 13 transcripts had RPKM values that were relatively low (<2.0), Cadm4, Retsat, Slc37a4 and Tmem119 were relatively highly expressed (>8.0) in both lines. Importantly, only 6 of these 13 NAc mRNAs were exclusive from CTL rats to either the LVR or the HVR line type (HVR & CTL > LVR: Cadm4, Capg; LVR > HVR & CTL: Tmem119; LVR & CTL > HVR: Pcdhb3, Pcdhga1, Pcdhb8). This list is distinguished from the 35 previous candidates in our recent publication (Roberts et al. 2013) only examining HVRnon-run and LVRnon-run rats mainly due to the fact that most of the previously reported candidates were normalized between lines after 6 days of running.
Table 4.
Differentially expressed NAc transcripts between HVRrun and LVRrun as well as between HVRnon-run and LVRnon-run rats
| HVRrun | LVRrun | HVRrun/ | HVRnon-run | LVRnon-run | HVRnon-run/ | |||
|---|---|---|---|---|---|---|---|---|
| Transcript | RPKM avg | RPKM avg | LVRrun | P | RPKM avg | RPKM avg | LVRnon-run | P |
| PREDICTED: Rattus norvegicus EG212225 protein-like (LOC100365310), miscRNA | 2.60 | 1.01 | 2.57 | 0.002 | 3.06 | 1.41 | 2.17 | 0.004 |
| PREDICTED: Rattus norvegicus EG212225 protein-like (LOC100360855), miscRNA | 3.96 | 1.61 | 2.47 | 0.003 | 4.93 | 2.07 | 2.39 | 0.002 |
| Cell adhesion molecule 4 (Cadm4), mRNA | 70.10 | 33.84 | 2.07 | 0.007 | 65.95 | 40.11↓ vs. CTL | 1.64 | 0.034 |
| Capping protein (actin filament), gelsolin-like (Capg), mRNA | 2.10 | 1.20 | 1.76 | 0.004 | 2.24 | 1.31↓ vs. CTL | 1.71 | 0.011 |
| PREDICTED: Rattus norvegicus DEAD/H (Asp–Glu–Ala–Asp/His) box polypeptide 11 (CHL1-like helicase homologue, S. cerevisiae) (Ddx11), mRNA | 2.34 | 1.41 | 1.66 | 0.007 | 2.69 | 1.38 | 1.95 | 0.001 |
| Gamma-glutamyl carboxylase (Ggcx), mRNA | 3.71 | 2.49 | 1.49 | 0.000 | 3.33 | 2.71 | 1.23 | 0.048 |
| Retinol saturase (all trans retinol 13,14 reductase) (Retsat), mRNA | 23.89 | 16.25 | 1.47 | 0.001 | 23.82 | 17.91 | 1.33 | 0.045 |
| PREDICTED: Rattus norvegicus similar to RIKEN cDNA 4632415K11 (RGD1308461), mRNA | 3.61 | 2.70 | 1.34 | 0.003 | 3.25 | 2.86 | 1.13 | 0.048 |
| Solute carrier family 37 (glucose 6-phosphate transporter), member 4 (Slc37a4), mRNA | 11.50↑ vs. HVRnr | 8.80 | 1.31 | 0.001 | 9.94 | 8.80 | 1.13 | 0.030 |
| Protocadherin beta 3 (Pcdhb3), mRNA | 1.80 | 2.59 | −1.44 | 0.008 | 1.79↓ vs. CTL | 2.38 | −1.33 | 0.001 |
| Transmembrane protein 119 (Tmem119), mRNA | 11.06 | 16.27 | −1.47 | 0.002 | 10.70 | 14.28↑ vs. CTL | −1.33 | 0.007 |
| Protocadherin gamma subfamily A1 (Pcdhga1), mRNA | 1.03 | 1.54 | −1.50 | 0.000 | 1.12↓ vs. CTL | 1.61 | −1.44 | 0.006 |
| Protocadherin beta 8 (Pcdhb8), mRNA | 1.91 | 3.03 | −1.58 | 0.001 | 1.72↓ vs. CTL | 2.66 | −1.54 | 0.000 |
Abbreviations: HVRrun = HVR 34-day-old 6-day runners, LVRrun = LVR 34-day-old 6-day runners, HVRnon-run = HVR 34-day-old non-runners, LVRnon-run = LVR 34-day-old non-runners, CTL = control Wistar rats. Symbols: ↑ vs. CTL = transcript in HVRnon-run or LVRnon-run rats was higher than in CTL rats (P < 0.05); ↓ vs. CTL = transcript in HVRnon-run or LVRnon-run rats was lower than in CTL rats (P < 0.05); ↑ vs. HVRnr or LVRnr = transcript in HVRrun or LVRrun is greater than HVRnon-run or LVRnon-run rats, respectively (P < 0.05); ↓ vs. HVRnr or LVRnr = transcript in HVRrun is less than HVRnon-run rats (P < 0.05). These significant differences are in bold type.
Western blotting confirmation of Cadm4 as line type-specific gene in G10–11 rats
Due to the relatively high NAc enrichment of Cadm4 on an RPKM basis, and its role in synaptogenesis, this protein was interrogated between lines in later generations to confirm our RNA-seq findings at the protein level. Remarkably, NAc Cadm4 protein was expressed to a lesser extent in LVRnon-run rats than in HVRnon-run rats (P = 0.03), and tended to be lower in the LVRrun versus HVRrun rats from G10–11 (P = 0.065; Fig.2), supporting its differential directionality in RNA-seq (Table4). Thus, this finding gave us further confidence in interpreting our RNA-seq data for hypotheses-generating purposes.
Hypothesis generation through bioinformatics of G8 LVR and HVR NAc transcriptome differences
Interestingly, the top associated gene network, as identified by IPA, as down-regulated in LVRrun versus HVRrun rats from the 94 differentially expressed NAc mRNAs included ‘cellular assembly and organization, cellular function and maintenance, cell cycle’. When considering the 13 transcripts that differed between lines independent of voluntary wheel running, the top associated down-regulated network in LVRnon-run versus HVRnon-run rats included ‘cancer, cell cycle, cellular growth and proliferation’ (network score: 20, 7/35 molecules differentially expressed: ↑Cadm4, ↑Capg, ↑Ggcx, ↑Restat, ↑Slc37a4, ↓Pcdhga1, ↓Tmem119). Others have previously found that voluntary running increases cell proliferation and survival of new cells within the subgranular zone of the mouse hippocampus (van Praag et al. 1999; Olson et al. 2006). However no reports, to our knowledge, suggest that voluntary running affects neurogenesis in the NAc. Thus, we hypothesized that these resultant networks may indicate that NAc neuron maturation differed between lines, and follow-up experiments described below were performed to test our newly generated hypotheses.
RNA-seq-guided IHC experiments suggest LVRnon-run rats possess fewer immature and medium spiny neurons in the NAc compared to HVRnon-run rats
Following the aforementioned line type differences discovered through IPA, we next performed IHC to assay the number of Dcx-positive NAc neurons in G9–10 LVRrun/non-run and HVRrun/non-run rats to determine if there was a line type difference in the number of immature neurons. Remarkably, LVRnon-run rats possessed 3.8-fold fewer Dcx-positive NAc neurons than HVRnon-run rats (P < 0.001; Fig.5A). LVRnon-run rats also possessed 6.3-fold fewer Darpp-32-positive NAc neurons than HVRnon-run rats (P < 0.001; Fig.5B).
Figure 5. Differences in NAc immature neurons (Dcx+) and MSNs (Darpp-32+) between G9–10 LVR and HVR rats.
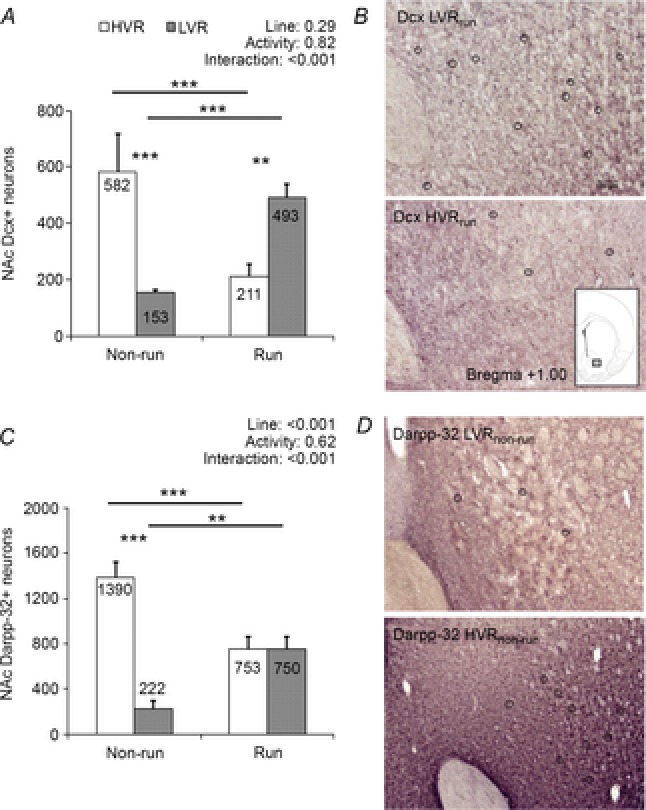
Line-type differences in NAc Dcx-positive (A) and Darpp-32-positive neurons (C), which are indicative of immature neurons and mature/differentiated MSNs, respectively. B and D, representative NAc micrographs of Dcx-positive and Darpp-32-positive cell staining, respectively. All animals were female and were 39–40 days of age, and HVRrun and LVRrun animals spent ∼10–12 days in a voluntary running wheel (n = 6–7 per bar). **P < 0.01, ***P < 0.001.
Contrary to the non-runner rats, LVRrun rats possessed 2.3-fold more Dcx-positive NAc neurons than HVRrun rats (P < 0.001; Fig.5A). Likewise, LVRrun rats possessed a similar number of Darpp-32-positive NAc neurons as HVRrun rats (P < 0.001; Fig.5B). HVRrun rats possessed significantly fewer Dcx-positive and Darpp-32-positive cells than HVRnon-run rats, respectively (P < 0.05; Fig.5A/B). Conversely, LVRrun rats possessed significantly more Dcx-positive and Darpp-32-positive cells than LVRnon-run rats (P < 0.05, Fig.3A and B).
Given that Dcx and Darpp-32 represent markers of immature neurons and mature/differentiated MSNs, respectively, these findings suggest that substantial differences in NAc neuronal maturation exist between the LVR and HVR lines types depending upon running status; this is discussed in greater detail below.
Of the HVR and LVR runners analysed for NAc MSNs, we find correlations exist for the following relationships.
HVRs: more NAc MSNs are associated with less total distances run prior to death (r = −0.58 when excluding one outlier; Fig.6A)
LVRs: more NAc MSNs are strongly associated with less total distances run prior to death (r = −0.92; Fig.6B)
Figure 6. Differences in NAc immature neurons (Dcx+) and MSNs (Darpp-32+) between G9–10 LVR and HVR rats Correlations between NAc IHC measures and 10-day running distance in G9–10 LVR and HVR runner rats.
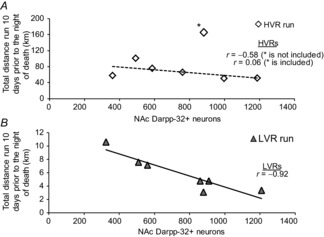
Negative associations exist between 10-day running distances and NAc MSN density in HVR (A) and LVR runners (B). Note that one HVR runner was excluded from analysis due to it being an outlier (* in A). However, r values are presented for both the inclusion and the exclusion of this animal.
Running induces plasticity in dopamine-related transcripts in HVR vs. LVR rats despite line type similarities in NAc tissue dopamine content
We had hypothesized prior to our RNA-seq analysis that more dopamine-related genes would be up-regulated within the NAc of LVR (LVRrun and LVRnon-run) and CTL rats versus HVR (HVRrun and HVRnon-run) rats. Only three of the 30 mRNAs designated by GO as dopamine-related transcripts statistically differed (P < 0.05) between LVRnon-run and HVRnon-run rats (LVR > HVR: Gna11, Oprd1; HVR > LVR: Slc6a3; Table5). Similarly, only four of the 30 dopamine-related transcripts statistically differed between LVRrun and HVRrun rats after 6 days of running (LVR > HVR: Adcy6, Arpp19; HVR > LVR: Oprd1, Oprm1; Table5). Interestingly, 6 days of running altered 16 of these transcripts within the HVR line (HVRrun vs. HVRnon-run rats, Table5), while only 1 of these 30 transcripts (Huntingtin; Htt) was altered between LVRrun versus LVRnon-run rats, suggesting a running-acquired plasticity in dopamine-related NAc mRNAs in HVR versus LVR rats.
Table 5.
Dopamine signalling-related NAc mRNA expression differences between HVR and LVR lines
| HVRrun | LVRrun | HVRrun/ | HVRnon-run | LVRnon-run | HVRnon-run/ | |||
|---|---|---|---|---|---|---|---|---|
| mRNA | RPKM avg | RPKM avg | LVRrun | P | RPKM avg | RPKM avg | LVRnon-run | P |
| Adcy5 | 26.21↑ vs. HVRnr | 21.21 | 1.24 | 0.08 | 20.30 | 21.80 | −1.08 | 0.36 |
| Adcy6 | 4.07 | 4.68 | −1.15 | 0.02 | 4.58 | 4.84 | −1.06 | 0.55 |
| Arpp19 | 107.05↑ vs. HVRnr | 82.12 | 1.30 | 0.03 | 76.93 | 82.71↓ vs. CTL | −1.08 | 0.54 |
| Cav2 | 7.02↓ vs. HVRnr | 8.51 | −1.22 | 0.06 | 8.53 | 7.87 | 1.08 | 0.28 |
| Caly | 142.02 | 142.84 | −1.01 | 0.90 | 138.85↓ vs. CTL | 135.83↓ vs. CTL | 1.02 | 0.71 |
| Cdnf | 0.36 | 0.38 | −1.08 | 0.83 | 0.23 | 0.35 | −1.56 | 0.22 |
| Drd1 | 81.54↑ vs. HVRnr | 65.53 | 1.24 | 0.06 | 58.32 | 60.44↓ vs. CTL | −1.03 | 0.77 |
| Drd2 | 59.03↑ vs. HVRnr | 50.26 | 1.17 | 0.15 | 45.29↓ vs. CTL | 47.72 | −1.06 | 0.53 |
| Drd3 | 0.54 | 0.64 | −1.20 | 0.34 | 0.71 | 0.54 | 1.30 | 0.32 |
| Drd4 | ND | ND | ND | ND | ND | ND | ND | ND |
| Drd5 | 1.22↑ vs. HVRnr | 1.07 | 1.13 | 0.41 | 0.92 | 0.99 | −1.09 | 0.56 |
| Dtnbp1 | 11.64 | 11.66 | −1.01 | 0.97 | 11.65 | 11.46 | 1.01 | 0.62 |
| Flna | 1.59↓ vs. HVRnr | 1.98 | −1.25 | 0.07 | 2.80 | 2.10 | 1.32 | 0.15 |
| Gna11 | 15.95↑ vs. HVRnr | 15.92 | 1.00 | 0.97 | 12.86 | 15.37 | −1.20 | 0.02 |
| Gna12 | 72.63↓ vs. HVRnr | 77.26 | −1.06 | 0.16 | 80.81 | 83.07 | 1.03 | 0.57 |
| Gna15 | 2.04 | 2.11 | −1.03 | 0.80 | 2.05 | 2.10 | −1.02 | 0.86 |
| Gnal | 71.17↑ vs. HVRnr | 61.86 | 1.15 | 0.07 | 57.67 | 59.44 | 1.03 | 0.74 |
| Gnaq | 33.91 | 33.15 | 1.02 | 0.49 | 31.73 | 33.53 | 1.05 | 0.24 |
| Gnas | 54.31 | 57.70 | −1.06 | 0.44 | 55.54↓ vs. CTL | 56.46 | −1.02 | 0.71 |
| Gnao1 | 105.13↑ vs. HVRnr | 100.23 | 1.05 | 0.30 | 89.96 | 102.82 | −1.15 | 0.24 |
| Htt | 0.80 | 0.69↓ vs. LVRnr | 1.15 | 0.06 | 0.72 | 0.84 | 1.16 | 0.20 |
| Klf16 | 41.93↑ vs. HVRnr | 40.12 | 1.05 | 0.45 | 36.45↓ vs. CTL | 38.36↓ vs. CTL | −1.05 | 0.37 |
| Lrrk2 | 17.70↑ vs. HVRnr | 14.76 | 1.20 | 0.09 | 13.34 | 14.75 | −1.11 | 0.31 |
| Nsg1 | 177.37 | 170.84 | 1.04 | 0.31 | 161.51↓ vs. CTL | 161.57↓ vs. CTL | 1.00 | 0.99 |
| Oprd1* | 3.10↑ vs. HVRnr | 2.50 | 1.24 | 0.02 | 2.03 | 2.64 | −1.32 | 0.01 |
| Oprm1 | 2.05 | 1.35 | 1.52 | 0.01 | 1.88 | 1.48 | 1.27 | 0.16 |
| Palm | 82.97 | 84.47 | −1.02 | 0.63 | 88.52 | 84.94 | 1.04 | 0.64 |
| Rgs9 | 129.27↑ vs. HVRnr | 107.27 | 1.21 | 0.16 | 97.37 | 101.51 | 1.04 | 0.69 |
| Slc6a3 | 0.10↓ vs. HVRnr | 0.09 | 1.10 | 0.63 | 0.16 | 0.10 | 1.58 | 0.03 |
| Slc9a3r1 | 20.79 | 23.78 | −1.15 | 0.07 | 21.83 | 23.35 | −1.08 | 0.49 |
This gene list was constructed according to the Gene Ontology (GO) dopamine receptor signalling pathway (GO ID: 0007212), adenylate cyclase-activating dopamine receptor pathway (GO ID: 0007191), adenylate cyclase-inhibiting dopamine receptor pathway (GO ID: 0007191), and/or the negative regulation of dopamine receptor signalling pathway (GO ID: 0060160) lists. Abbreviations: HVRrun = HVR 34-day-old 6-day runners, LVRrun = LVR 34-day-old 6-day runners, HVRnon-run = HVR 34-day-old non-runners, LVRnon-run = LVR 34-day-old non-runners, CTL = control Wistar rats; ND = not detected.
Symbols: cited as different between HVRnon-run and LVRnon-run rats in prior publication (Roberts et al. 2013); ↑ vs. CTL = transcript in HVRnon-run or LVRnon-run rats was higher than in CTL rats (P < 0.05); ↓ vs. CTL = transcript in HVRnon-run or LVRnon-run rats was lower than in CTL rats (P < 0.05); ↑ vs. HVRnon-run or LVRnon-run = transcript in HVRrun or LVRrun is greater than HVRnon-run or LVRnon-run rats, respectively (P < 0.05); ↓ vs. HVRnon-run or LVRnon-run = transcript in HVRrun is less than HVRnon-run rats (P < 0.05). These significant differences are in bold type.
Interestingly, NAc tissue dopamine content was similar between lines when examining G10–11 LVR versus HVR rats prior to or 2–3 h during the dark cycle; this finding suggests that running-induced alterations in dopamine-related transcripts within the HVR line were independent of running-induced increases in NAc dopamine content at the time period measured (Fig.7).
Figure 7. NAc tissue dopamine content between G10–11 LVR and HVR lines.
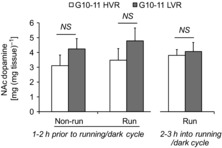
No significant differences (NS) existed between LVRnon-run and HVRnon-run rats 1–2 h prior to the running/dark cycle (P = 0.28), LVRrun and HVRrun rats 1–2 h prior to the running/dark cycle (P = 0.28), or LVRrun and HVRrun rats 2–3 h into the running/dark cycle (P = 0.72). All animals were female and were 35 days of age, and HVRrun and LVRrun animals spent 6 days in a voluntary running wheel (n = 6–7 per bar).
Discussion
RNA-seq provides an unbiased measure of the presence and prevalence of transcripts from known and unknown genes (Mortazavi et al. 2008). Therefore, our experimental approach consisted of two phases: (a) using RNA-seq and bioinformatics to develop hypotheses that delineate NAc characteristics which may differentiate running motivations between our unique model of LVR and HVR rats; and (b) performing follow-up experiments in later generations of these rats to test these hypotheses. Our newly generated hypotheses from these experiments are illustrated in Fig.8, and more in-depth discussions of our findings are presented below.
Figure 8. Summary figure of new hypotheses developed from current observations between the LVR and HVR lines.
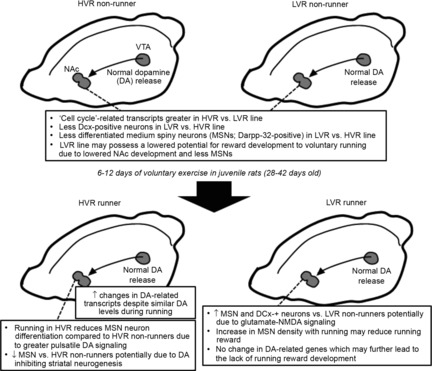
Hypotheses on neuron development between lines: the inherent up-regulation of ‘cell-cycle’-related transcripts in the HVR and LVR line and the higher densities of Dcx- and Darpp-32- positive neurons indicates that MSN development is inherently greater in the HVR line. However, voluntary running reverses these trends (MSNs: HVR = LVR; Dcx-positive neurons: LVR > HVR). Methamphetamine administration to rodents, which increases striatal dopamine levels, has been shown to decrease striatal neurogenesis. Hence, high pulsatile dopamine secretions into the NAc on a nightly basis due to high running may be the mechanism whereby HVRs experience a decrease in MSN density. Conversely, glutamate–NMDA receptor signalling is linked to an increase in striatal neurogenesis. Hence, this may be the mechanism whereby voluntary running in LVRs increases NAc Dcx- and Darpp-32-positive neurons. Hypotheses on MSN function between lines: initially, we hypothesized that lower inherent MSN density in LVR versus HVR non-runners may lead to the lack of voluntary running reward. However, there were negative associations between running distance and Darpp-32-positive neurons in the HVR and LVR runners (Fig.4A and B). Hence, we speculate that a greater MSN density may inhibit efferent targets that lead to a decreased motivation for voluntary running. Abbreviations: DA, dopamine; MSN, medium spiny neuron.
NAc medium spiny neurons are lower in LVRnon-run vs. HVRnon-run rats, whereas minimal running in the LVR line normalizes these differences
IHC experiments were performed in later LVR and HVR generations as a result of IPA suggesting that the top down-regulated NAc gene network in LVRnon-run versus HVRnon-run rats included ‘cancer, cell cycle, cellular growth and proliferation’. A novel observation was that LVRnon-run rats possessed 6.3-fold fewer mature NAc MSNs with the Darpp-32 marker and 3.8-fold fewer immature NAc MSNs with the Dcx marker, as compared to HVRnon-run rats. Thus, MSN density is innately diminished in the LVR versus HVR line, and fewer Dcx-positive neurons are able to potentially differentiate into MSNs. Lee et al. (2006) have shown that increases in dendritic spine density in dopaminoceptive NAc MSNs are linked to long lasting addictive behaviours. Hence, LVR rats not exposed to voluntary running may possess a diminished motivation to voluntarily run due to fewer NAc MSNs.
However, minimal running in the LVRrun versus HVRrun rats normalized the aforementioned line type differences that existed in non-runners by: (a) normalizing MSN density between lines, and (b) increasing the number of Dcx-positive neurons in LVRrun compared to HVRrun rats. Indeed, these findings are now difficult to reconcile. However, we posit that allowing LVR rats to partake in minimal running at a young age is associated with promotion of MSN differentiation and/or stimulation of striatal cytogenesis. Striatal cytogenesis has been shown to occur in juvenile rats weighing 200–250 g (Mao & Wang, 2001), these being older than the rats tested herein. Interestingly, glutamate release in the striatum has been shown to occur in rodents during treadmill exercise (Meeusen et al. 1997), and glutamate–N-methyl-d-aspartate receptor signalling has been linked to promoting striatal neurogenesis (Luk et al. 2003). Hence, this may be the underlying mechanism whereby voluntary running potentially increases NAc MSN and Dcx neuron densities in the LVR line.
HVRrun rats, after 6 days of voluntary running, possessed significantly fewer Darpp-32-positive cells relative to HVRnon-run rats, indicating decreased mature MSNs. In contrast to running-induced increases in Dcx-positive and Darpp-32-positive cells within the LVR line, the high volumes of running in HVRrun rats decreased these markers for neuronal developmental stage relative to HVRnon-run rats. A recent review by Corty & Freeman (2013) gives one potential explanation. They describe that during neuronal development, transient and unnecessary neuronal–neuronal connections often occur. They indicate that selective axonal and dendritic pruning takes place to achieve neural circuit refinement, and as a result, programmed cell death follows. Thus, our speculation is that programmed cell death could be one potential cause for the decrease in MSN density in the NAc of our HVR rats after 6 days of running from 28 to 34 days of age, with the LVR line lagging in their relative neuronal development.
When examining correlations between Darpp- 32-positive neurons and running distances in HVR and LVR rats, we report that modest to strong negative associations exist between lower total distances run prior to death and a greater density of Darpp-32-positive neurons in HVR (r = −0.58 with the exclusion of one outlier) and LVR rats (r = −0.92). While these correlations were performed on a limited number of rats, these findings may suggest one of multiple possibilities: (a) a possible inhibitory effect of MSNs on voluntary running distances in the LVR line; and/or (b) an anti-differentiation and/or apoptotic effect of running on MSNs; note, however, that the latter is precluded by the lack of up-regulated pro-apoptosis pathways in the LVR line as per the RNA-seq analysis. With regards to the former, NAc MSNs may send inhibitory signals to the globus pallidus, which is a known regulator of voluntary movement (Smith & Bolam, 1990). Hence, future experiments manipulating NAc MSN number in our HVR and LVR animals, via localized neurotoxin injections, will yield critical information on this possible relationship.
Expression of several dopamine-related mRNAs is amplified between HVRrun and HVRnon-run rats, but is mostly similar between LVRrun and LVRnon-run rats despite similarities in NAc dopamine tissue content between lines
A relationship between striatal dopaminergic signalling and voluntary running behaviour is well established (Garland et al. 2011), although transcriptomics analyses documenting this relationship have been previously limited to the examination of a few select NAc mRNAs, the sole exception being the recent publication by Mathes et al. (2010) where only HVR and control mouse lines were compared. Our current transcriptomics analyses unveil additional numerous dopamine-related transcript differences that exist between and within the LVR and HVR lines with or without 6 days of voluntary running. Only three dopamine-related transcripts innately differed between LVRnon-run and HVRnon-run rats. However, 16/30 dopamine-related mRNAs differed between HVRrun and HVRnon-run rats compared to only 1/30 between LVRrun and LVRnon-run rats. This finding confirms and expands what Knab et al. (2009), Greenwood et al. (2011) and our own data examining Drd1/2/5 NAc mRNA expression patterns in G5–6 HVRrun versus LVRrun rats via real-time PCR (unpublished findings) have similarly reported, which is that high levels of wheel running in rodents produces plasticity in the mRNA expression of dopamine-related genes within the NAc. We posit that upregulation of the dopaminergic signalling pathway at the transcriptome level could play a positive reinforcer for greater voluntary running. However, according to Mathes et al. (2010) and Swallow et al. (1998), replication of these results in additional LVR and HVR lines are potentially required to differentiate between an actual selection response from possible effects of genetic drift.
Unexpectedly, and contrary to the data of Mathes et al. (2010) who reported that HVR versus control mice expressed more NAc tissue dopamine, which drove the increased expression of dopamine-related transcripts, NAc dopamine differences did not exist between our LVR and HVR lines. Potential explanations could be that the running-induced plasticity of dopamine-related transcripts within our HVR line may be due to: (a) a given genetic architecture within the HVR line which predisposes them to differentially regulate dopamine-related transcripts in response to voluntary running without differences in dopamine levels, or (b) a species difference between our animals and the Garland's HVR mice. Notwithstanding this, the plasticity of dopamine-related genes within the HVR line could play some role in the progressive increase in daily distances of voluntary running that occurs in the initial weeks of accessibility to running wheels.
Conclusions
While the current study provides a plethora of data which potentially explain differences in voluntary exercise motivation, a limitation to the current study is that the HVR and LVR transcriptomes were analysed in a snapshot fashion early in the rodents’ lives.
Nonetheless, the current study illustrates novel and potentially important concepts including: (a) that NAc MSN maturation differences may partially be responsible for differences in voluntary running motivation between the HVR and LVR lines; and (b) that voluntary running early in life is able to induce plasticity in neuron populations and mRNA expression profiles. Specifically, LVR rats exhibit a lack of plasticity in dopamine-related transcripts, which could also interfere with the acquisition of voluntary exercise reward in these animals with low voluntary running activity.
Acknowledgments
We thank Dr Charlotte Tate for her past conversations with F.W.B., which helped guide the development of the HVR and LVR model.
Glossary
- CTL
control Wistar rats
- Dcx
doublecortin
- G
generation
- Gad
glutamic acid decarboxylase
- GO
Gene Ontology
- HVRnon-run
HVR 34-day-old non-runners
- HVRrun
HVR 34-day-old 6-day runners
- IHC
immunohistochemistry
- IPA
Ingenuity Pathway Analysis
- LVRnon-run
LVR 34-day-old non-runners
- LVRrun
LVR 34-day-old 6-day runners
- MSN
medium spiny neuron
- NAc
nucleus accumbens
- RPKM
reads per kilobase per million mapped reads
- RNA-seq
RNA deep-sequencing
Key points
Selective breeding experiments with laboratory rodents have demonstrated the heritability of voluntary exercise.
We performed RNA sequencing and bioinformatics analyses of the reward and pleasure hub in the brain – the nucleus accumbens – in rats selectively bred for low voluntary running (LVR) versus high voluntary running (HVR).
The discovery of unique genes and ‘cell cycle’-related gene pathways between lines guided our hypothesis that neuron maturation may be lower in LVR rats.
Testing of this hypothesis revealed that the LVR line inherently possessed fewer mature medium spiny neurons and fewer immature neurons than their HVR counterparts. However, minimal running in LVR rats appeared to rescue and/or reverse these effects.
Neuron maturation in the nucleus accumbens is related to low running voluntary behaviour in our model; this allows researchers to understand the potential neural mechanisms that underlie the motivations for low physical activity behaviour.
Additional information
Competing interests
All authors disclose no competing interests.
Author contributions
M.D.R. outlined the experiments, helped procure funding, performed bioinformatics and drafted the manuscript. F.W.B. procured funding, conceived and maintained the selective breeding model and helped draft the manuscript. R.G.T., J.M.C. and K.D.W. critically assisted with RNA-seq bioinformatics and helped draft the manuscript. G.E.R. performed HPLC and helped draft the manuscript. A.J.H. and C.Z. critically assisted with IHC and writing of the manuscript. R.G.T., J.M.C., J.D.B. and T.E.C. assisted in tissue collection, data analysis, and/or writing of the manuscript.
Funding
Partial funding for this project was obtained from a grant awarded to F.W.B. by the College of Veterinary Medicine at the University of Missouri, NIH T32-AR048523 (M.D.R.) and AHA 11PRE7580074 (J.M.C.). This project was also largely supported by funds donated through the College of Veterinary Medicine's Development Office.
Translational Perspective
Understanding the neural and molecular mechanisms that regulate voluntary exercise motivation is crucial given that most adolescents and adults do not engage in regular physical activity (Troiano et al. 2008), and therapies are needed to correct this physical activity motivational deficiency. While the current study offers no immediate ‘cure’ to lowered voluntary exercise motivation, the transcriptomics evaluations and follow-up experiments comparing the LVR and HVR lines herein continue to contribute to our understanding of the neuro-molecular basis for voluntary exercise behaviours.
References
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y. Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth FW, Laye MJ. Roberts MD. Lifetime sedentary living accelerates some aspects of secondary aging. J Appl Physiol. 2011;111:1497–1504. doi: 10.1152/japplphysiol.00420.2011. [DOI] [PubMed] [Google Scholar]
- Brown JP, Couillard-Despres S, Cooper-Kuhn CM, Winkler J, Aigner L. Kuhn HG. Transient expression of doublecortin during adult neurogenesis. J Comp Neurol. 2003;467:1–10. doi: 10.1002/cne.10874. [DOI] [PubMed] [Google Scholar]
- Chen H, Liu Z, Gong S, Wu X, Taylor WL, Williams RW, Matta SG. Sharp BM. Genome-wide gene expression profiling of nucleus accumbens neurons projecting to ventral pallidum using both microarray and transcriptome sequencing. Front Neurosci. 2011;5:98. doi: 10.3389/fnins.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM. Freeman MR. Cell biology in neuroscience: Architects in neural circuit design: glia control neuron numbers and connectivity. J Cell Biol. 2013;203:395–405. doi: 10.1083/jcb.201306099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Jr, Schutz H, Chappell MA, Keeney BK, Meek TH, Copes LE, Acosta W, Drenowatz C, Maciel RC, van Dijk G, Kotz CM. Eisenmann JC. The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J Exp Biol. 2011;214:206–229. doi: 10.1242/jeb.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood BN, Foley TE, Le TV, Strong PV, Loughridge AB, Day HE. Fleshner M. Long-term voluntary wheel running is rewarding and produces plasticity in the mesolimbic reward pathway. Behav Brain Res. 2011;217:354–362. doi: 10.1016/j.bbr.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heruth DP, Gibson M, Grigoryev DN, Zhang LQ. Ye SQ. RNA-seq analysis of synovial fibroblasts brings new insights into rheumatoid arthritis. Cell Biosci. 2012;2:43. doi: 10.1186/2045-3701-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoed M, Brage S, Zhao JH, Westgate K, Nessa A, Ekelund U, Spector TD, Wareham NJ. Loos RJ. Heritability of objectively assessed daily physical activity and sedentary behavior. Am J Clin Nutr. 2013;98:1317–1325. doi: 10.3945/ajcn.113.069849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knab AM, Bowen RS, Hamilton AT, Gulledge AA. Lightfoot JT. Altered dopaminergic profiles: implications for the regulation of voluntary physical activity. Behav Brain Res. 2009;204:147–152. doi: 10.1016/j.bbr.2009.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knab AM, Bowen RS, Hamilton AT. Lightfoot JT. Pharmacological manipulation of the dopaminergic system affects wheel-running activity in differentially active mice. J Biol Regul Homeost Agents. 2012;26:119–129. [PMC free article] [PubMed] [Google Scholar]
- Knab AM. Lightfoot JT. Does the difference between physically active and couch potato lie in the dopamine system? Int J Biol Sci. 2010;6:133–150. doi: 10.7150/ijbs.6.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Kim Y, Kim AM, Helmin K, Nairn AC. Greengard P. Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proc Natl Acad Sci U S A. 2006;103:3399–3404. doi: 10.1073/pnas.0511244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kennedy TE. Sadikot AF. Glutamate promotes proliferation of striatal neuronal progenitors by an NMDA receptor-mediated mechanism. J Neurosci. 2003;23:2239–2250. doi: 10.1523/JNEUROSCI.23-06-02239.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L. Wang JQ. Gliogenesis in the striatum of the adult rat: alteration in neural progenitor population after psychostimulant exposure. Brain Res Dev Brain Res. 2001;130:41–51. doi: 10.1016/s0165-3806(01)00195-x. [DOI] [PubMed] [Google Scholar]
- Mathes WF, Nehrenberg DL, Gordon R, Hua K, Garland T., Jr Pomp D. Dopaminergic dysregulation in mice selectively bred for excessive exercise or obesity. Behav Brain Res. 2010;210:155–163. doi: 10.1016/j.bbr.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen R, Smolders I, Sarre S, de Meirleir K, Keizer H, Serneels M, Ebinger G. Michotte Y. Endurance training effects on neurotransmitter release in rat striatum: an in vivo microdialysis study. Acta Physiol Scand. 1997;159:335–341. doi: 10.1046/j.1365-201X.1997.00118.x. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L. Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Olson AK, Eadie BD, Ernst C. Christie BR. Environmental enrichment and voluntary exercise massively increase neurogenesis in the adult hippocampus via dissociable pathways. Hippocampus. 2006;16:250–260. doi: 10.1002/hipo.20157. [DOI] [PubMed] [Google Scholar]
- Paxinos G. Watson C. The Rat Brain in Stereotaxic Coordinates. New York: Academic Press; 1998. [Google Scholar]
- Rhodes JS. Garland T. Differential sensitivity to acute administration of Ritalin, apomorphine, SCH 23390, but not raclopride in mice selectively bred for hyperactive wheel-running behavior. Psychopharmacology (Berl) 2003;167:242–250. doi: 10.1007/s00213-003-1399-9. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Garland T., Jr Gammie SC. Patterns of brain activity associated with variation in voluntary wheel-running behavior. Behav Neurosci. 2003;117:1243–1256. doi: 10.1037/0735-7044.117.6.1243. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Hosack GR, Girard I, Kelley AE, Mitchell GS. Garland T., Jr Differential sensitivity to acute administration of cocaine, GBR 12909, and fluoxetine in mice selectively bred for hyperactive wheel-running behavior. Psychopharmacology (Berl) 2001;158:120–131. doi: 10.1007/s002130100857. [DOI] [PubMed] [Google Scholar]
- Roberts MD, Brown JD, Company JM, Oberle LP, Heese AJ, Toedebusch RG, Wells KD, Cruthirds CL, Knouse JA, Ferreira JA, Childs TE, Brown M. Booth FW. Phenotypic and molecular differences between rats selectively bred to voluntarily run high vs. low nightly distances. Am J Physiol Regul Integr Comp Physiol. 2013;304:R1024–1035. doi: 10.1152/ajpregu.00581.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MD, Gilpin L, Parker KE, Childs TE, Will MJ. Booth FW. Dopamine D1 receptor modulation in nucleus accumbens lowers voluntary wheel running in rats bred to run high distances. Physiol Behav. 2012;105:661–668. doi: 10.1016/j.physbeh.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Rustemeyer SM, Lamberson WR, Ledoux DR, Wells K, Austin KJ. Cammack KM. Effects of dietary aflatoxin on the hepatic expression of apoptosis genes in growing barrows. J Anim Sci. 2011;89:916–925. doi: 10.2527/jas.2010-3473. [DOI] [PubMed] [Google Scholar]
- Salamone JD. Correa M. The mysterious motivational functions of mesolimbic dopamine. Neuron. 2012;76:470–485. doi: 10.1016/j.neuron.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD. Bolam JP. The neural network of the basal ganglia as revealed by the study of synaptic connections of identified neurones. Trends Neurosci. 1990;13:259–265. doi: 10.1016/0166-2236(90)90106-k. [DOI] [PubMed] [Google Scholar]
- Song HK, Hong SE, Kim T. Kim do H. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS One. 2012;7:e35552. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow JG, Carter PA. Garland T., Jr Artificial selection for increased wheel-running behavior in house mice. Behav Genet. 1998;28:227–237. doi: 10.1023/a:1021479331779. [DOI] [PubMed] [Google Scholar]
- Troiano RP, Berrigan D, Dodd KW, Masse LC, Tilert T. McDowell M. Physical activity in the United States measured by accelerometer. Med Sci Sports Exerc. 2008;40:181–188. doi: 10.1249/mss.0b013e31815a51b3. [DOI] [PubMed] [Google Scholar]
- van Praag H, Kempermann G. Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- Waters RP, Renner KJ, Pringle RB, Summers CH, Britton SL, Koch LG. Swallow JG. Selection for aerobic capacity affects corticosterone, monoamines and wheel-running activity. Physiol Behav. 2008;93:1044–1054. doi: 10.1016/j.physbeh.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanato VF, Martins MP, Anselmo-Franci JA, Petenusci SO. Lamano-Carvalho TL. Sexual development of male Wistar rats. Braz J Med Biol Res. 1994;27:1273–1280. [PubMed] [Google Scholar]
- Zhang LQ, Cheranova D, Gibson M, Ding S, Heruth DP, Fang D. Ye SQ. RNA-seq reveals novel transcriptome of genes and their isoforms in human pulmonary microvascular endothelial cells treated with thrombin. PLoS One. 2012;7:e31229. doi: 10.1371/journal.pone.0031229. [DOI] [PMC free article] [PubMed] [Google Scholar]


