Abstract
Importance
Newer sequencing technologies in combination with traditional gene mapping techniques, such as linkage analysis, can help identify the genetic basis of disease for patients with rare disorders of uncertain etiology. This approach may expand the phenotypic spectrum of disease associated with those genetic mutations.
Objective
To elucidate the molecular cause of a neuromuscular disease among a family in which 4 members, a mother and her 3 sons, were affected.
Design, Setting, and Participants
Two of 4 affected members manifested nemaline myopathy, a common subtype of congenital myopathy, while the other 2 had a nonspecific myopathy. Single-nucleotide polymorphism–based linkage analysis was performed on DNA samples from the 4 affected family members, and whole-genome sequencing was performed in the proband. Real-time quantitative reverse transcription–polymerase chain reaction, immunofluorescence, and Western blot analysis were performed on muscle biopsy specimens.
Main outcomes and Measures
Whole-genome sequencing and linkage analysis identified a variant in a gene that explains the phenotype.
Results
We identified a novel neurofilament light polypeptide (NEFL) nonsense mutation in all affected members. NEFL mutations have been previously linked to Charcot-Marie-Tooth disease in humans. This led us to reevaluate the diagnosis, and we recognized that several of the findings, especially those related to the muscle biopsy specimens and electromyography, were consistent with a neurogenic disease.
Conclusions and Relevance
NEFL mutations are known to cause Charcot-Marie-Tooth disease in humans and motor neuron disease in mice. We report the identification of an NEFL mutation in a family clinically manifesting congenital myopathy. We also describe potential overlap between myopathic and neurogenic findings in this family. These findings expand the phenotypic spectrum of diseases associated with NEFL mutations. This study is an example of the power of genomic approaches to identify potentially pathogenic mutations in unsuspected genes responsible for heterogeneous neuromuscular diseases.
Neuromuscular disorders include diseases related to skeletal muscle, neuromuscular junction, peripheral nerves, and anterior horn cells. Primary defects in any of these tissues lead to weakness as a common outcome. Neurogenic and myopathic conditions often manifest with distinct sets of clinicopathological findings, but overlapping findings sometimes make determination of the primary etiology difficult.1-4 In such situations, a molecular diagnosis may clarify the pathogenesis and lead to a revised diagnosis that reflects the primary cause of a patient's weakness.1,2
Congenital myopathy (CM) is characterized by early-onset muscle weakness due to primary skeletal muscle dysfunction.5 Patients with nemaline myopathy (NM), the most common type of CM, typically present with proximal muscle weakness and the presence of threadlike structures (nemaline rods) within skeletal muscle fibers seen on light microscopy using Gömöri trichrome staining.6,7 To date, mutations in 9 different genes (TPM3, NEB, ACTA1, TNNT1, TPM2, CFL2, KBTBD13, KLHL40, and KLHL41) have been reported to cause NM or one of its variants.6,8-10 In addition to myopathies, nemaline rods are observed in other conditions, including peripheral neuropathies,11 mitochondrial defects,12 human immunodeficiency virus infection,13 and polymyositis.14 Furthermore, patients with NM sometimes are seen with findings suggestive of a neurogenic etiology,15 and mutations in some genes (eg, DNM2) may cause a primary skeletal myopathy, such as centronuclear myopathy, or a neuropathy, such as Charcot-Marie-Tooth (CMT) disease.16
In this article, we describe a family with 4 members identified as having CM, including a mother (the proband) and her 3 sons, with variable clinicopathological findings. Two individuals were diagnosed as having NM, one had a nonspecific CM, and one manifested neurogenic atrophy with myopathic features. Although appearing to be a primary skeletal myopathy, some of the findings were consistent with a neurogenic process. To determine the genetic basis of their disease, linkage analysis was performed on DNA samples from the 4 affected members, and whole-genome sequencing was conducted on the proband's DNA. Whole-genome sequencing identified a unique heterozygous variant in the neurofilament light polypeptide gene (NEFL) (Online Mendelian Inheritance in Man 162280), a gene whose mutations were previously shown to cause CMT disease, a heterogeneous neuropathy affecting the peripheral nervous system.17,18 The NEFL variant was present in all affected members. Additional experiments were performed to determine the pathogenicity of the identified mutation.
Methods
Patient Enrollment
The mother (proband 20-1), father (20-5), and their 3 sons (20-2, 20-3, and 20-4)were enrolledin an institutional review board–approved study at Boston Children's Hospital (Figure 1A). The family gave written informed consent and was enrolled in the Beggs laboratory CM registry and research study. Blood samples were collected, and DNA was extracted.
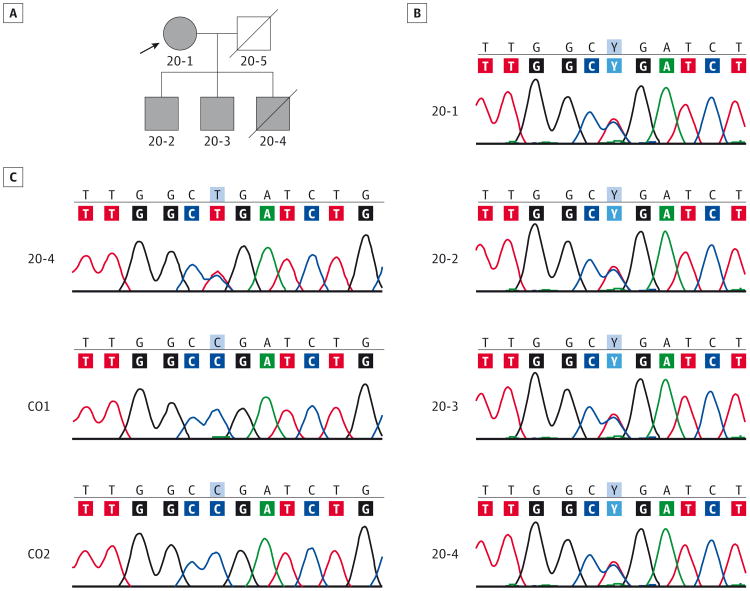
Figure 1. Genetic Analysis and Confirmation of an NEFL Mutation in the Family With a Congenital Myopathy Diagnosis.
A, Shown is the affected proband mother (20-1) and her 3 affected sons (20-2, 20-3, and 20-4). The square represents a male individual; a circle, a female individual; the arrow; the proband; and a slash mark, a deceased individual. B, Chromatograms from Sanger sequencing of genomic polymerase chain reaction products in the proband and her 3 affected sons show the heterozygous c.1261C>T change. C, Chromatograms of the sequenced real-time quantitative reverse transcription–polymerase chain reaction products derived from muscle biopsy samples in patient 20-4 and 2 controls (CO1 and CO2) show a mutant transcript peak of similar height to that of the wild type, suggesting no significant nonsense-mediated decay of the mutated NEFL messenger RNA. NEFL, neurofilament light polypeptide gene.
Muscle Biopsy
The slides and muscle biopsy specimens for 3 affected members of the family (unavailable for 20-3) were made available for study courtesy of several institutions. The slides stained with hematoxylin-eosin, nicotinamide adenine dinucleotide– tetrazolium reductase, and modified Gömöri trichrome were reevaluated by 2 of us who specialize in neuropathology (P.D-.S.C.C. and U.D.G.).
Electromyography and Nerve Conduction Studies
Electromyography (EMG) and nerve conduction studies were performed in the proband. Standardized techniques were used.
Single-Nucleotide Polymorphism–Based Linkage Analysis, Whole-Genome Sequencing, and Sanger Sequencing
Single-nucleotide polymorphism (SNP)–based linkage analysis was performed on DNA samples from the 4 affected family members (20-1, 20-2, 20-3, and 20-4) using an array (genome-wide human SNP 6.0; Affymetrix). Linkage and copy number variation analysis was performed using a pipeline developed by 2 of us (K.S.A. and K.M.) who specialize in informatics. Whole-genome sequencing was performed on the proband's DNA using an available platform (Complete Genomics Incorporated) as described previously.19 Nonpathogenic variants were filtered using dbSNP131, and potential disease-causing variants were confirmed using Sanger sequencing. Primer sequences for genomic polymerase chain reaction (PCR) and Sanger sequencing to detect the NEFL mutation were NEFL_F: ACCCGACTCAGTTTCACCAG and NEFL_R: TTCCTCCACTTC-GATCTGCT.
Real-time Quantitative Reverse Transcription–PCR and Complementary DNA Sequencing
Total RNA was extracted from muscle biopsy tissue from patient 20-4 and age-matched control subjects using a fibrous tissue minikit (RNeasy; Qiagen). Extracted messenger RNA was converted to complementary DNA using a synthesis system (SuperScript III First-Strand; Invitrogen). Sanger sequencing was performed using primers NEFL_F and NEFL_R (as above) flanking the mutant transcript.
A real-time quantitative reverse transcription–PCR assay was performed to measure NEFL transcript levels 5′ and 3′ to the mutation site. A probe (Taqman Hs04187794; Life Technologies) measured NEFL transcript levels 5′ to the mutation site with dye-labeled (VIC; Applied Biosystems) glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (catalog No. 4310884E; Life Technologies) as an internal control. A dye (SYBR Green; Bio-Rad) was used to measure the levels of the NEFL transcript 3′ to the mutation using the following primers: NEFL_Distal_1F: GATCGAAGTGGAGGAAACCA and NEFL_Distal_1R: GGCCTCTTCCTTGTCCTTCT. GAPDH transcript levels were evaluated as an internal control. Real-time quantitative reverse transcription–PCR was performed in duplicate and analyzed using a real-time quantitative reverse transcription–PCR system (7300; Applied Biosystems) with software (7500 Real-Time PCR System Sequence Detection Software, version 1.4; Applied Biosystems). The PCR program used was 50°C for 2 minutes, 95°C for 10 minutes, and 40 repetitions of 95.8°C for 15 seconds and 60°C for 1 minute. The relative amount of NEFL expression in the patient was calculated as a fold-change relative to an age-matched control after normalizing for GAPDH expression levels.
Western Blot and Immunofluorescence
Tissues from muscle biopsy specimens were frozen and stored at -80°C until analysis. Protein isolation and Western blot procedures were performed as described previously.20 Transferred proteins were detected with antibodies against NEFL (PA1-32240, 1:250 dilution; Thermo Scientific) and GAPDH (FL-335, 1:1000 dilution; Santa Cruz Biotechnology) and visualized using enhanced chemiluminescence. Quantification of protein levels was normalized to GAPDH using a software program (Quantity One 4.2.1; Bio-Rad) on an image station (440 Kodak DS; Eastman Kodak Co).
For immunofluorescence studies, 8-μm frozen transverse sections of muscle biopsy specimens from patients and age-matched controls were stained with rabbit polyclonal anti-NEFL (PA1-32240, 1:250 dilution; Thermo Scientific) and a-bun-garotoxin AlexaFluor 594 conjugate (B13423, 1:40 dilution; Invitrogen) antibodies. Alexa Fluor-conjugated antirabbit IgG (1: 100; Molecular Probes) was used as a secondary antibody. Coverslips were mounted using mounting medium with 4′,6-diamidino-2-phenylindole dihydrochloride (Vectashield; Vector Laboratories). Staining was evaluated using a microscope (Eclipse 90i; Nikon) with software (NIS-Elements AR; Nikon).
Results
Heterogeneous Clinicopathological Findings in Affected Members of the Family With Diagnosis of CM
The clinical findings in the family are summarized in Table 1. The mother (proband) and her 3 sons had manifested hypotonia since early infancy but without feeding or respiratory difficulties. All the sons had undescended testes. Delayed motor milestones of varying severity were present in all 4 members. They had no contractures at birth but developed them over time involving various joints. Footdrop had been noted in all 4 affected members during childhood. They became wheelchair dependent at different times of their lives. Facial and body muscle weakness, especially of the shoulders and distal lower extremities, and scoliosis were present. The proband underwent surgery for hammertoe deformity, and her 3 sons required multiple surgical procedures, including heel cord lengthening and spinal fusion. One of the affected sons died at age 35 years, likely due to respiratory insufficiency. The remaining affected family members are alive at ages 72,49, and 47 years. Coincidentally, the father had developed an autoimmune disease associated with scleroderma and polymyositis late in adult life and died. Serum creatine kinase levels were within normal limits in all affected individuals.
Table 1. Clinical Features of Patients with the NEFL Mutation.
| Variable | 20-1 | 20-2 | 20-3 | 20-4 |
|---|---|---|---|---|
| Age, y/sex | Alive, 72/F | Alive, 49/M | Alive, 47/M | Died at 35/M |
| Onset | Birth, floppy | Birth; floppy, facial weakness | Birth, floppy | Birth, facial weakness |
| Delayed motor milestones | Mild delay, slow runner, delayed speech | ++, Never walked, high-pitched and soft speech | Mild delay, walked before 2 y, slow speech | Mild delay by 6 mo, never could run, no speech issues |
| Facial involvement | +, Mild ptosis | + | +, Lazy eyes as child, needed patching | + |
| High-arched palate | + | + | − | + |
| Footdrop | +, Early childhood | +, Early childhood | +, Early childhood | +, Early childhood |
| Wristdrop | − | − | +, Severe | − |
| Muscle weakness | No obvious asymmetry | Distal leg and shoulder muscles more affected | Distal leg and shoulder muscles more affected | Distal leg and shoulder muscles more affected |
| Contractures | Knees, ankles | Ankles | Hips, knees, ankles, elbows | Hips, knees, jaw, ankles, fingers |
| Scoliosis | ++ | ++ | + | ++ |
| Deep tendon reflexes | Upper limbs present and lower limbs absent | ↓↓ | ↓ | Absent |
| Wheelchair dependence | From mid-50s | From early childhood | From high school | From early adolescence |
| Respiratory status affected | +, Nighttime bilevel positive airway pressure late in life | +, Shortness of breath | +, Recurrent pneumonia | +, Shortness of breath, sleep apnea |
| Surgical interventions | Hammertoe surgery | Spinal fusion surgery, heel cord lengthening | Rod placement for neck and upper back, spinal fusion, heel cord lengthening | Spinal fusion, heel cord lengthening |
Abbreviations: F, female; M, male; NEFL, neurofilament light polypeptide gene; +, present; ++, 2 or more areas; −, absent; ↓, decreasing; ↓↓, greatly decreasing.
The histopathogical findings in muscle biopsy specimens from affected family members are summarized in Table 2. The family was diagnosed as having NM because nemaline bodies were present in the proband and one son (20-3). Nemaline bodies were not evident in the muscle biopsy specimens of the other 2 sons (20-2 and 20-4). Patients 20-2 and 20-3 had additional findings suggestive of a neurogenic process, including type II fiber predominance, grouped fiber atrophy, and small angular fibers (Figure 2A-I).
Table 2. Histopathological Findings.
| Variable | 20-1 | 20-2 | 20-3 | 20-4 |
|---|---|---|---|---|
| Overall interpretation | Nemaline myopathy | Neurogenic atrophy With myopathic features | Nemaline myopathy with neurogenic atrophy | Nonspecific congenital myopathy |
| Fiber type predominance | Type I | Type II | Type II | No predominance |
| Fiber size variation | + | + | + | + |
| Nemaline rods | +, Occasional | − | + (1%-5% Fibers)- | − |
| Lipid accumulation | NA | + | + | + |
| Endomysial connective tissue | ↑ | ↑ | ↑ | ↑ |
| Myofiber degeneration/regeneration | + | − | − | + |
| Additional findings | Ring and atrophic fibers, internalized nuclei present | First muscle biopsy (in infancy) consistent with neurogenic atrophy; second (at age 11 y) showed grouped atrophy, round and angulated fibers, increased internalized nuclei | Grouped atrophy, angulated and atrophic fibers, nuclear chains | NA |
| Electron microscopy findings | Few nemaline rods, focal myofibrillar disruption, Z-band smearing | NA | Nemaline rods seen in continuity with Z-bands often associated with disruption of myofibrillar apparatus | NA |
Abbreviations: NA, not available; +, present; −, absent; ↑, increasing.
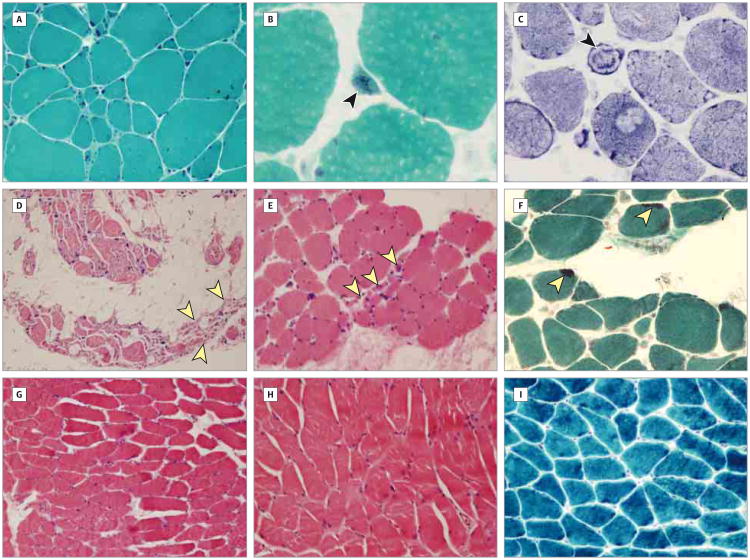
Figure 2. Histopathological Findings in Muscle Biopsy Specimens From Affected Members of the Family.
A-C. Proband 20-1. Deltoid muscle biopsy was performed at age 42 years. Marked excess variability was observed in fiber size, with increased number of internalized nuclei and increased endomysial connective tissue (A). Occasional fibers contained nemaline bodies (arrowhead) (B), and occasional ring fibers (arrowhead) were also noted (C) (modified Gömöri trichrome, original magnification ×200 [A] and ×600 [B]; nicotinamide adenine dinucleotide– tetrazolium reductase, original magnification ×400 [C]). D-F. Patient 20-3. Quadriceps muscle biopsy was performed at age 32 years. The biopsy specimen revealed group atrophy (arrowheads) and marked adipose tissue replacement (D). Small group atrophy was present (arrowheads) even in more well-preserved areas (E). Some of the fibers contained nemaline bodies (arrowheads) (F) (hematoxylin-eosin, original magnification ×100 [D] and ×200 [E]; modified Gömöri trichrome, original magnification ×200 [F]). G-I. Patient 20-4. Quadriceps muscle biopsy was performed at age 11 years. Mild variability in fiber size was present and there was no significant increase in endomysial connective tissue (G and H). Nemaline bodies were not identified (I) (hematoxylin-eosin, original magnification ×100 [G] and ×200 [G]; modified Gömöri trichrome, original magnification ×200 [I]).
The EMG and nerve conduction data were available only for the proband, performed at age 42 years. The motor and sensory nerve conduction study results were normal in the left upper and lower extremities except for a moderately low tibial motor amplitude and a slightly low median motor and sensory amplitude with prolonged distal latencies (eTable in the Supplement). The needle EMG revealed diffusely distributed abnormal spontaneous activity in the form of positive sharp waves, fibrillation potential activity, and complex repetitive discharges, as well as abnormal (giant amplitude, long duration, and polyphasic) motor unit potentials (Table 3) consistent with a primary chronic neurogenic disorder.
Table 3. Electromyography Findings in the Proband (20-1).
| Muscle, Left Side | Spontaneous | Voluntary Activity Comment on Motor Unit Potentials | |||
|---|---|---|---|---|---|
| Fibrillation Potentials/Positive Shar Waves | Fasciculation Potentials | Miscellaneous | |||
| Tibialis anterior | +++ | None | CRD | High amplitude, up to 5 mV; polyphasic and complex (some short) | |
| Extensor digitorum brevis | ++ | None | None | Reduced number; polyphasic, large and complex | |
| Vastus lateralis | ++ | None | None | Reduced number; long duration; high amplitude, up to 8 mV; polyphasic and complex | |
| First dorsal interosseus | None | None | None | Large; high amplitude, up to 10 mV | |
| Biceps | None | None | None | Polyphasic and complex | |
| Deltoid | + | None | CRD | Long duration, high amplitude, complex in form | |
| Infraspinatus | + | None | None | Large, polyphasic | |
| Paraspinal midthoracic | + | None | Myotonia | Not examined | |
| Paraspinal lumbar | ++ | None | None | Complex | |
| Paraspinal cervical | None | None | None | Large, complex | |
Abbreviations: CRD, complex repetitive discharge; +, more than 1 area; ++, 2 or more areas; +++, every area.
Mutated NEFL in the Affected Family Members
Linkage and copy number variation analyses were performed on DNA samples from the 4 affected members. No copy number variations were identified. Only 25.8% of the genome was consistent with linkage using a dominant transmission model, with a maximum logarithm of the odds score of 0.59. Whole-genome sequencing on the proband's DNA identified 17 872 exonic variants, 936 of which were unique and not present in db-SNP131. Of those 936 variants, only 273 were present in the linked region. The 273 variants were evaluated by gene expression and function, followed by Sanger sequencing. We sequenced 5 candidate variants in all 4 affected family members and identified a single heterozygous nonsense NEFL mutation (c.1261C>T; p.R421X). In addition, we screened the 273 linked variants for 333neuromuscular disease–associated genes, which include genes mutated in myopathies, muscular dystrophies, motor neuron diseases, ataxias, and neuropathies.21 This narrowed the list of variants to 6, which were then filtered using the National Heart, Lung, and Blood Institute Exome Sequencing Project database (http://evs.gs.washington.edu/EVS/) with frequency less than 0.005, Poly-Phen-2 (http://genetics.bwh.harvard.edu/pph2/) score greater than 0.1, and dominant inheritance pattern. We again identified the same single nonsense NEFL mutation (c.1261C>T; p.R421X), present in all 4 affected family members. This mutation was not present in the National Heart, Lung, and Blood Institute Exome Sequencing Project database, 1000 Genomes Project (http://browser.1000genomes.org), Genome Variant Database for Human Disease at the University of Miami (Florida) Miller School of Medicine (http://genomics.med.miami.edu), or dbSNP build 139.
Relative Stability of the Nonsense Mutation–Containing NEFL Transcript
Because nonsense mutations often lead to nonsense-mediated decay and loss of mutant transcripts, the stability of NEFL c.1261C>T transcripts was evaluated using messenger RNA extracted from patient 20-4 and control quadriceps muscles. Two primer pairs, one proximal and one distal to the site of mutation, were used to evaluate NEFL transcript levels. Real-time quantitative reverse transcription–PCR analysis showed that NEFL transcript levels in the patient were normal or slightly elevated at 1.32 fold (range, 1.03-1.70 fold) and 2.02 fold (range, 1.97-2.08 fold) higher compared with the control when measured at the 5′ and 3′ sites, respectively. Furthermore, PCR and sequence analysis of NEFL complementary DNA in the quadriceps muscle complementary DNA of patient 20-4 showed that the mutation was easily detectable at levels comparable to those of the wild-type allele. In this patient, the mutant T and the wild-type C peaks were clearly present, of similar amplitudes (Figure 1C), and comparable to the results of genomic DNA sequencing (Figure 1B). These findings suggest that mutant transcripts are stable and not undergoing significant nonsense-mediated decay.
No Obvious Differences in Protein Amounts on Western Blot and Immunofluorescence
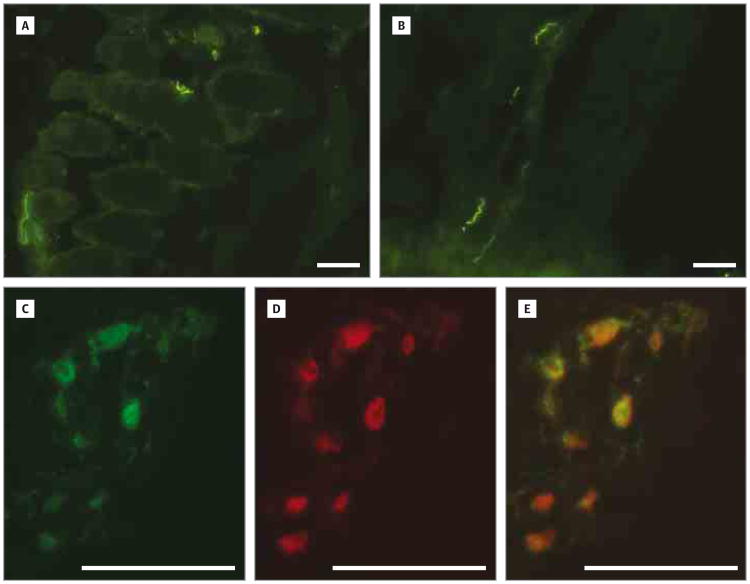
Western blot and indirect immunofluorescence analyses were under taken to look for gross alterations in NEFL protein size, levels, or localization. Muscle biopsy specimens were available from patients 20-2 and 20-4 for further investigation. Indirect immunofluorescence demonstrated that NEFL protein was present and localized appropriately to what appeared to be nerve terminals of the affected family member (20-2) and control (Figure 3A and B). Double-label indirect immunofluorescence for NEFL and a-bungarotoxin to label acetylcholine receptors confirmed that these areas were motor end plates (Figure 3C-E). Western blot analysis of muscle proteins from 2 affected members (20-2 and 20-4) and 2 age-matched controls detected faint bands corresponding to full-length NEFL protein in all the specimens (data not shown). The presence of a truncated NEFL protein related to the p.R421X mutation could not be evaluated because of the low overall abundance of NEFL in skeletal muscle and a nonspecific or cross-reacting band at that molecular weight.
Figure 3. Normal Localization of NEFL at Motor End Plates in Affected Muscle.
A and B, Indirect immunofluorescence shows similar localization of NEFL protein in transverse sections of control (A) and patient 20-2 (B) skeletal muscle biopsy specimens. C-E, Shown is double-label indirect immunofluorescence of a longitudinal section of myofiber from patient 20-2 stained for NEFL (green in C) and a-bungarotoxin (red in D). The merged image in E identifies NEFL-positive structures as motor end plates (scale bars are 50μm).
Discussion
NEFL encodes the neurofilament light polypeptide that, together with neurofilament medium and heavy polypeptides, forms type IV intermediate filament heteropolymers, a major component of the neuronal cytoskeleton.17 Mutations in NEFL are responsible for 2 subtypes of CMT disease, CMT1F and CMT2E.22 Motor neuron diseases, such as amyotrophic lateral sclerosis, are also associated with neurofilament protein defects, with misaccumulations of neurofilament proteins in motor neuron cell body and proximal axons, hallmarks of sporadic and familial amyotrophic lateral sclerosis.23 Furthermore, a transgenic mouse model of a mutant NEFL protein (p.L394P) leads to a selective loss of ventral horn motor neurons and denervation atrophy of the skeletal muscle that suggests it might be a candidate gene for motor neuron diseases.24
Lack of evidence from whole-genome sequencing for a mutation in genes associated with NM or another CM, as well as the identification of the NEFL c.1261C>T mutation in all affected members of this family, suggested an underlying neurogenic etiology and led us to reevaluate the clinicopathological findings. While the clinical presentations of affected family members were typical of a CM, the muscle biopsy and EMG findings revealed overlapping neurogenic and myopathic characteristics. The EMG findings of diffusely distributed giant amplitude and polyphasic motor unit potentials, positive sharp waves, and fibrillation potentials in the proband pointed to a neurogenic process, although similar neuropathic changes occasionally can be seen in a chronicmyopathic process.15,25 Furthermore, muscle biopsy specimens from the various family members disclosed features consistent with both myopathic and neurogenic processes. The myopathic findings included myofiber size variation, rounding of fibers, ring fibers, and nemaline bodies, while the neurogenic features included grouped fiber atrophy and multiple angulated fibers. Nemaline bodies, although often considered diagnostic of NM, are not pathognomonic and have been reported in patients with neurogenic disorders, such as CMT disease and progressive spinal muscular atrophy.11,26 Similarly, various mutations in an unrelatedgene, DNM2, can cause either a peripheral neuropathy, CMT2B, or an autosomal dominant centronuclear myopathy, a type of CM; however, DNM2 is expressed in both neuronal and muscle tissues, so the pleiotropic affects may be related to disruption of different tissue-specific functions of the protein.16 In the family described herein, because NEFL is not known to be expressed at appreciable levels in skeletal muscle, we are left with the conclusion that the muscle pathology and weakness are a secondary consequence of neuronal dysfunction as a result of a disrupted neuronal cytoskeleton. The absence of neuronal tissue for examination and the limited muscle biopsy specimens from only some affected members of the family precluded any studies to examine the basis of differential myopathic findings in some individuals, but differences might be related to distinct genetic backgrounds, variable environmental influences, or even sampling differences between different muscle groups at different ages and at different physiological and disease states.
It has been postulated that NEFL mutations cause a dominant phenotype due to a dominant-negative effect.27 Our experiments on muscle biopsy specimens have shown the stability of the mutant transcript in this family. Although no nervous system tissue or skin fibroblasts were available for further testing, the dominant nature of this mutation leads us to hypothesize that the translated truncated protein may be stable, potentially interfering with the normal functioning of wild-type protein in mature neurofilament heteropolymers.
Conclusions
In conclusion, we report a dominant nonsense NEFL mutation previously described exclusively in neurogenic disorders in a family of 4 with the clinical presentation of a chronic CM. The histopathological results and EMG demonstrated significant overlap between findings characteristic of a primary myopathy and a neurogenic disease. The pathogenic mechanisms of disease secondary to the NEFL mutation remain to be elucidated, and creation of a knockin mouse model of the human mutation may aid our understanding of these findings.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by grants K08 AR055072 (Dr Agrawal) and R01 AR044345 (Dr Beggs) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health, by grant MDA201302 from the Muscular Dystrophy Association (Dr Beggs), and by the Jean-Pierre and Nancy Boespflug Myopathic Research Foundation.
Role of Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Agrawal and Beggs had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Agrawal, Beggs.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Agrawal, Joshi, Marinakis, Ciarlini, Chad, Beggs.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Schmitz-Abe, Markianos.
Obtained funding: Beggs.
Administrative, technical, or material support: Agrawal, Beggs.
Study supervision: Agrawal, Beggs.
Conflict of Interest Disclosures: None reported.
Additional Contributions: We thank the members of this unique family, whose active participation made this study possible.
References
- 1.Komlósi K, Hadzsiev K, Garbes L, et al. Exome sequencing identifies Laing distal myopathy MYH7 mutation in a Roma family previously diagnosed with distal neuronopathy. Neuromuscul Disord. 2014;24(2):156–161. doi: 10.1016/j.nmd.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Igari R, Wada M, Sato H, Hayashi KY, Nishino I, Kato T. A case of inclusion body myopathy with Paget's disease of bone and frontotemporal dementia (IBMPFD) showing clinical features of motor neuron disease. Rinsho Shinkeigaku. 2013;53(6):458–464. doi: 10.5692/clinicalneurol.53.458. in Japanese. [DOI] [PubMed] [Google Scholar]
- 3.Kudryashova E, Wu J, Havton LA, Spencer MJ. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum Mol Genet. 2009;18(7):1353–1367. doi: 10.1093/hmg/ddp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer JG, Ferrier A, Kothary R. More than a bystander: the contributions of intrinsic skeletal muscle defects in motor neuron diseases. Front Physiol. 2013;4:356. doi: 10.3389/fphys.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.North KN, Wang CH, Clarke N, et al. International Standard of Care Committee for Congenital Myopathies Approach to the diagnosis of congenital myopathies. Neuromuscul Disord. 2014;24(2):97–116. doi: 10.1016/j.nmd.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanoudou D, Beggs AH. Clinical and genetic heterogeneity in nemaline myopathy: a disease of skeletal muscle thin filaments. Trends Mol Med. 2001;7(8):362–368. doi: 10.1016/s1471-4914(01)02089-5. [DOI] [PubMed] [Google Scholar]
- 7.North KN. Clinical approach to the diagnosis of congenital myopathies. Semin Pediatr Neurol. 2011;18(4):216–220. doi: 10.1016/j.spen.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Sambuughin N, Yau KS, Olivé M, et al. Dominant mutations in KBTBD13, a member of the BTB/Kelch family, cause nemaline myopathy with cores. Am J Hum Genet. 2010;87(6):842–847. doi: 10.1016/j.ajhg.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravenscroft G, Miyatake S, Lehtokari VL, et al. Mutations in KLHL40 are a frequent cause of severe autosomal-recessive nemaline myopathy. Am J Hum Genet. 2013;93(1):6–18. doi: 10.1016/j.ajhg.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta VA, Ravenscroft G, Shaheen R, et al. Identification of KLHL41 mutations implicates BTB-Kelch–mediated ubiquitination as an alternate pathway to myofibrillar disruption in nemaline myopathy. Am J Hum Genet. 2013;93(6):1108–1117. doi: 10.1016/j.ajhg.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danon MJ, Sarpel G, Manaligod JR. Nemaline rod myopathy and Charcot-Marie-Tooth disease: report of a case in a 10-year-old girl. Arch Neurol. 1980;37(2):123–127. doi: 10.1001/archneur.1980.00500510081021. [DOI] [PubMed] [Google Scholar]
- 12.Kornfeld M. Mixed nemaline-mitochondrial “myopathy”. Acta Neuropathol. 1980;51(3):185–189. doi: 10.1007/BF00687385. [DOI] [PubMed] [Google Scholar]
- 13.Dalakas MC, Pezeshkpour GH, Flaherty M. Progressive nemaline (rod) myopathy associated with HIV infection. N Engl J Med. 1987;317(25):1602–1603. doi: 10.1056/NEJM198712173172511. [DOI] [PubMed] [Google Scholar]
- 14.Cape CA, Johnson WW, Pitner SE. Nemaline structures in polymyositis: a nonspecific pathological reaction of skeletal muscles. Neurology. 1970;20(5):494–502. doi: 10.1212/wnl.20.5.494. [DOI] [PubMed] [Google Scholar]
- 15.North KN, Laing NG, Wallgren-Pettersson C. ENMC International Consortium and Nemaline Myopathy. Nemaline myopathy: current concepts. J Med Genet. 1997;34(9):705–713. doi: 10.1136/jmg.34.9.705. published correction appears in J Med Genet. 1997;34(10):879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Böhm J, Biancalana V, Dechene ET, et al. Mutation spectrumin the large GTPase dynamin 2, and genotype-phenotype correlation in autosomal dominant centronuclear myopathy. Hum Mutat. 2012;33(6):949–959. doi: 10.1002/humu.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordanova A, De Jonghe P, Boerkoel CF, et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain. 2003;126(pt 3):590–597. doi: 10.1093/brain/awg059. [DOI] [PubMed] [Google Scholar]
- 18.Fabrizi GM, Cavallaro T, Angiari C, et al. Giant axon and neurofilament accumulation in Charcot-Marie-Tooth disease type 2E. Neurology. 2004;62(8):1429–1431. doi: 10.1212/01.wnl.0000120664.07186.3c. [DOI] [PubMed] [Google Scholar]
- 19.Touma M, Joshi M, Connolly MC, et al. Whole genome sequencing identifies SCN2A mutation in monozygotic twins with Ohtahara syndrome and unique neuropathologic findings. Epilepsia. 2013;54(5):e81–e85. doi: 10.1111/epi.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wattanasirichaigoon D, Swoboda KJ, Takada F, et al. Mutations of the slow muscle a-tropomyosin gene, TPM3, are a rare cause of nemaline myopathy. Neurology. 2002;59(4):613–617. doi: 10.1212/wnl.59.4.613. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan JC, Hamroun D. The 2013 version of the gene table of monogenic neuromuscular disorders (nuclear genome) Neuromuscul Disord. 2012;22(12):1108–1135. doi: 10.1016/j.nmd.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Bird TD. Charcot-Marie-Tooth neuropathy type 1. 1998 Aug 31. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 1993-2014. [Accessed on July 29, 2014]. updated February 20, 2014. Available from http://www.ncbi.nlm.nih.gov/books/NBK1205/ [Google Scholar]
- 23.Garcia ML, Singleton AB, Hernandez D, et al. Mutations in neurofilament genes are not a significant primary cause of non–SOD1-mediated amyotrophic lateral sclerosis. Neurobiol Dis. 2006;21(1):102–109. doi: 10.1016/j.nbd.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Lee MK, Marszalek JR, Cleveland DW. A mutant neurofilament subunit causes massive, selective motor neuron death: implications for the pathogenesis of human motor neuron disease. Neuron. 1994;13(4):975–988. doi: 10.1016/0896-6273(94)90263-1. [DOI] [PubMed] [Google Scholar]
- 25.Daube JR, Rubin DI. Needle electromyography. Muscle Nerve. 2009;39(2):244–270. doi: 10.1002/mus.21180. [DOI] [PubMed] [Google Scholar]
- 26.Konno H, Iwasaki Y, Yamamoto T, Inosaka T. Nemaline bodies in spinal progressive muscular atrophy: an autopsy case. Acta Neuropathol. 1987;74(1):84–88. doi: 10.1007/BF00688343. [DOI] [PubMed] [Google Scholar]
- 27.Abe A, Numakura C, Saito K, et al. Neurofilament light chain polypeptide gene mutations in Charcot-Marie-Tooth disease: nonsense mutation probably causes a recessive phenotype. J Hum Genet. 2009;54(2):94–97. doi: 10.1038/jhg.2008.13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.