Summary
In the intracellular pathogen Brucella abortus, the general stress response (GSR) signaling system determines survival under acute stress conditions in vitro, and is required for long-term residence in a mammalian host. To date, the identity of the Brucella sensor kinase(s) that function to perceive stress and directly activate GSR signaling have remained undefined. We demonstrate that the flavin-binding sensor histidine kinase, LovhK (bab2_0652), functions as a primary B. abortus GSR sensor. LovhK efficiently and specifically phosphorylates the central GSR regulator, PhyR, and activates transcription of a set of genes that closely overlaps the known B. abortus GSR regulon. Deletion of lovhK severely compromises cell survival under defined oxidative and acid stress conditions. We further show that lovhK is required for cell survival during the early phase of mammalian cell infection and for establishment of long-term residence in a mouse infection model. Finally, we present evidence that particular regions of primary structure within the two N-terminal PAS domains of LovhK have distinct sensory roles under specific environmental conditions. This study elucidates new molecular components of a conserved signaling pathway that regulates B. abortus stress physiology and infection biology.
Keywords: Brucella abortus, signal transduction, two-component system, LOV domain, stress response, alphaproteobacteria, response regulator, PhyR, virulence, ecfG
Introduction
The α-proteobacteria, Brucella spp., are intracellular pathogens that are particularly adept at establishing long-term interactions with a range of mammalian cell types (Moreno & Moriyon, 2006). The ability of these species to stably inhabit phagocytes and other host cells facilitates successful evasion of the immune response and underlies the development of chronic infection. We have previously shown that the general stress response (GSR) signaling pathway regulates Brucella abortus stress physiology, and chronic B. abortus persistence in a mouse infection model (Kim et al., 2013). In this study, we identify and characterize protein sensors that control activation and repression of the B. abortus GSR pathway.
In the α-proteobacteria, the GSR is controlled by a hybrid signaling system that integrates features of two-component signal transduction (TCS) and alternative σ factor regulation. This system is encoded from a conserved genetic locus that includes three main components: phyR, nepR, and an EcfG-family σ factor (annotated rpoE1 in B. abortus) (Figure 1A; Figure S1). PhyR is a core GSR regulator that contains a σ-like (SL) domain positioned N-terminal to a TCS receiver domain. The current model of PhyR function, which has now been investigated in a number of α-proteobacteria (Abromaitis & Koehler, 2013, Bastiat et al., 2010, Correa et al., 2013, Francez-Charlot et al., 2009, Gourion et al., 2008, Gourion et al., 2009, Herrou et al., 2010, Iguchi et al., 2013, Jans et al., 2013, Kaczmarczyk et al., 2011, Kim et al., 2013, Lourenco et al., 2011), is that stress-dependent phosphorylation of the PhyR receiver domain increases affinity of the SL domain for the anti-σ factor NepR (Francez-Charlot et al., 2009) (see Figure 1B). Thus, phosphorylation activates PhyR as an anti-anti-σ factor. Phospho-PhyR (PhyR~P) binding to NepR releases ECF σ (Francez-Charlot et al., 2009) (RpoE1, or σE1 in B. abortus) to directly regulate transcription.
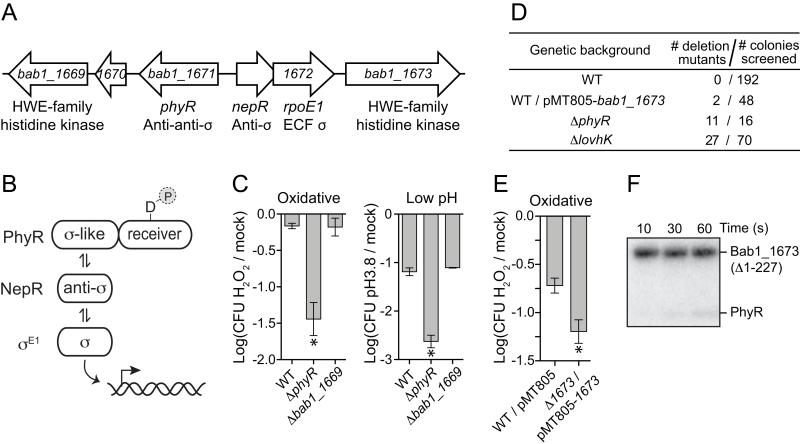
Figure 1.
Two HWE-type histidine kinases adjacent to the general stress response (GSR) genetic locus, bab1_1669 and bab1_1673, do not function as GSR activators.
(A) Arrangement of genes at the B. abortus GSR locus. (B) Core proteins of the alphaproteobacterial GSR regulatory system. PhyR is phosphorylated under stress conditions, and the general stress sigma factor, σE1, is activated when phospho-PhyR binds NepR and releases σE1. (C) Survival of B. abortus cells subjected to either 5 mM hydrogen peroxide or acid stress (pH 3.8) was assessed by enumerating colony forming units from both stressed and mock treated cultures. Data represent mean ± S.D. of three independent replicates (* = p<0.0006 based on one-way ANOVA followed by Dunnett’s multiple comparison test). (D) bab1_1673 is essential in strains with an intact GSR locus. Using a two-step double recombination strategy, we attempted to delete bab1_1673 from several genetic backgrounds including ΔphyR, ΔlovhK, and a background in which bab1_1673 was expressed from a complementing plasmid (pMT805-bab1_1673). After the second recombination step, colonies were screened by PCR to determine whether the chromosomal bab1_1673 locus was deleted or intact. The Table reports the number of recovered colonies containing a bab1_1673 deletion in a given genetic background. (E) Oxidative stress survival of cells overexpressing bab1_1673 compared to an empty plasmid control strain. Data presented as in C. (F) Assay of phosphoryl transfer from purified Bab1_1673~P to PhyR across a 60s timescale.
Though the interactions between PhyR, NepR and ECF σ are generally understood, the identities of the stress sensor protein (or proteins) that phosphorylate PhyR, and thus activate the GSR, remain largely uncharacterized. Exceptions include the PhyK sensor histidine kinase of Caulobacter crescentus, which is proposed to phosphorylate PhyR during stress (Foreman et al., 2012, Lourenco et al., 2011), the RsiC kinase of Sinorhizobium meliloti, which is proposed to exert positive and negative control over the phosphorylation state of two paralogous PhyR proteins (Sauviac & Bruand, 2014), and the LOV histidine kinase EL368, which phosphorylates two receiver domains in the marine bacterium, Erythrobacter litoralis, including PhyR (Correa et al., 2013). Additionally, PhyP of Sphingomonas str. FR1 (Kaczmarczyk et al., 2011) and the LovK kinase of C. crescentus (Foreman et al., 2012) function as negative regulators of the GSR system, likely by dephosphorylating PhyR~P. Notably, PhyK, RsiC, EL368, PhyP, and LovK are all related to an atypical subset of the histidine kinase family known as HWE kinases, which lack a recognizable F box and contain a signature HWE motif in the catalytic/ATPase domain (Karniol & Vierstra, 2004).
Given the emerging role of HWE-family kinases in the control of the α-proteobacterial GSR, we sought to test whether these proteins function as GSR regulators in B. abortus. We identified three HWE-related kinase genes in B. abortus, bab1_1669, bab1_1673, and lovhK (bab2_0652), and characterized their function in the GSR. Two of these genes, bab1_1673 and bab1_1669, directly flank the GSR locus on B. abortus chromosome 1 (Figure 1A). We show that bab1_1669 does not function in GSR under our assayed conditions; bab1_1673 is essential under standard culture conditions, and functions as a negative regulator of oxidative stress survival. We further demonstrate that the flavin-binding HWE kinase, LovhK (Swartz et al., 2007), functions as a primary B. abortus GSR sensor in vitro. Specifically, a) a ΔlovhK null strain has equivalent oxidative and acid stress survival phenotypes as a ΔphyR null, b) LovhK efficiently and specifically phosphorylates PhyR in vitro, and c) LovhK activates transcription of a set of genes that closely overlaps the known B. abortus GSR regulon. However, ΔlovhK, ΔphyR, and ΔrpoE1 null mutants have incongruent phenotypes in our animal and cell-based infection experiments: unlike ΔphyR and ΔrpoE1, B. abortus ΔlovhK exhibits reduced viability in THP-1 macrophage-like cells and is attenuated at an early stage of infection in a mouse model. We conclude that the regulatory output of B. abortus LovhK in the host may involve coordinate signal detection and integration by multiple histidine kinases that engage more than one downstream pathway. Indeed, our functional analysis of mutant strains with site directed changes in the PAS sensory domains of LovhK, or in genes regulated downstream of LovhK provides evidence that the B. abortus GSR system senses multiple environmental signals that can affect multiple adaptive responses.
Results
Functional analysis of two HWE-type kinases encoded at the B. abortus GSR genetic locus
α-proteobacteria commonly encode histidine kinases directly adjacent to, or near, the GSR genetic locus containing the phyR, nepR, and ecfG-family (ECF σ) genes (Figure S1). Three of these GSR locus-proximal genes have been characterized: phyP of Sphingomonas str. FR1, phyK of C. crescentus (Foreman et al., 2012, Kaczmarczyk et al., 2011, Lourenco et al., 2011) and rsiC of Sinorhizobium meliloti (Sauviac & Bruand, 2014). B. abortus bab1_1669 and bab1_1673 directly flank phyR-nepR-rpoE1 on chromosome 1, and each encode putative HWE-type sensor histidine kinases (Figure 1A). Using functional assays we previously established (Kim et al., 2013), we sought to test the role of these genes in stress adaptation. We successfully deleted bab1_1669 from the chromosome. Deletion of bab1_1673 required addition of an extra-chromosomal copy of bab1_1673 on a plasmid (Figure 1D). Thus bab1_1673 is essential under standard laboratory culture conditions. However, the essential nature of bab1_1673 requires a fully-intact set of GSR regulatory genes, as bab1_1673 could be deleted in strains lacking phyR or lovhK. We conclude that bab1_1673 genetically interacts with the GSR system; the functional implications of this result are detailed in the discussion.
A Δbab1_1669 null strain was not defective in oxidative or acid stress survival relative to wild-type and the stress-sensitive ΔphyR strain. (Figure 1C); the essential nature of bab1_1673 precluded assessment of its null phenotype. However, overexpression of bab1_1673 from its native promoter from a multi-copy replicating plasmid reduced survival of cells subjected to oxidative stress (Figure 1E). This suggests Bab1_1673 functions as a negative regulator of the B. abortus GSR system, analogous to Sphingomonas phyP or C. crescentus lovK. We further demonstrated that the purified kinase domain of Bab1_1673 (MBP-Bab1_1673(Δ1-227)) is an active autokinase in vitro (Figure 1F). While we do observe a very low level of phosphoryltransfer from Bab1_1673 to PhyR, the kinetics of this process are not of a magnitude consistent with a typical cognate kinase/receiver pair (Skerker et al., 2005). Attempts to radiolabel PhyR with [32P]acetyl phosphate were not successful, and we were thus unable to test activity of Bab1_1673 as a PhyR~P phosphatase. We conclude that bab1_1669 has no role in regulation of B. abortus GSR under our assayed in vitro conditions, while bab1_1673 genetically interacts with the GSR system and can function as a negative regulator of cellular stress survival.
LovhK is a PhyR kinase that regulates oxidative and acid stress survival in vitro
In C. crescentus, the flavin-binding HWE-kinase LovK functions together with the single domain receiver protein LovR to repress GSR signaling (Foreman et al., 2012), and activate cellular adhesion (Purcell et al., 2007, Fiebig et al., 2014). B. abortus also encodes a flavin-binding HWE kinase, LovhK, which has been previously shown to function as a blue-light regulated determinant of intracellular replication (Swartz et al., 2007). In Brucella melitensis, the ortholog of LovhK known as BM-LOV-HK has recently been shown to directly or indirectly modulate expression of genes involved in GSR, Type IV secretion, and quorum sensing (Gourley et al., 2014). However, the direct molecular signaling partner(s) of Brucella LovhK remain undefined. Inspection of the primary structure of the N-terminal sensory region of B. abortus LovhK revealed a second PAS sensory domain that is not present in C. crescentus LovK, which suggested that B. abortus LovhK may function differently than C. crescentus LovK. Nonetheless, we tested the role of LovhK as a regulator of the B. abortus GSR system as neither of the other two HWE-type kinases encoded in the genome (bab1_1669 and bab1_1673) appear to phosphorylate PhyR and thereby function as GSR activators.
A LovhK expression construct lacking the LOV domain (LovKΔ8-109) yields abundant, stable protein which has autokinase activity in vitro and rapidly transfers phosphoryl groups to equimolar PhyR; within 10 seconds we observed PhyR~P (Figure 2A and Figure S2). To test the specificity of LovhK for PhyR, we cloned, expressed, and purified the receiver domains of all 23 annotated response regulators encoded in the B. abortus genome. Incubation of phosphorylated LovhKΔ8-109 (LovhK~P) with equimolar concentrations of each of the 23 receiver domains revealed efficient phosphotransfer to a single substrate, PhyR, during a 30 second incubation (Figure 2B). Given that histidine kinases generally have a kinetic preference for phosphotransfer to their cognate substrate(s) (Skerker et al., 2005), these data support a model in which PhyR is the primary receiver for LovhK in B. abortus.
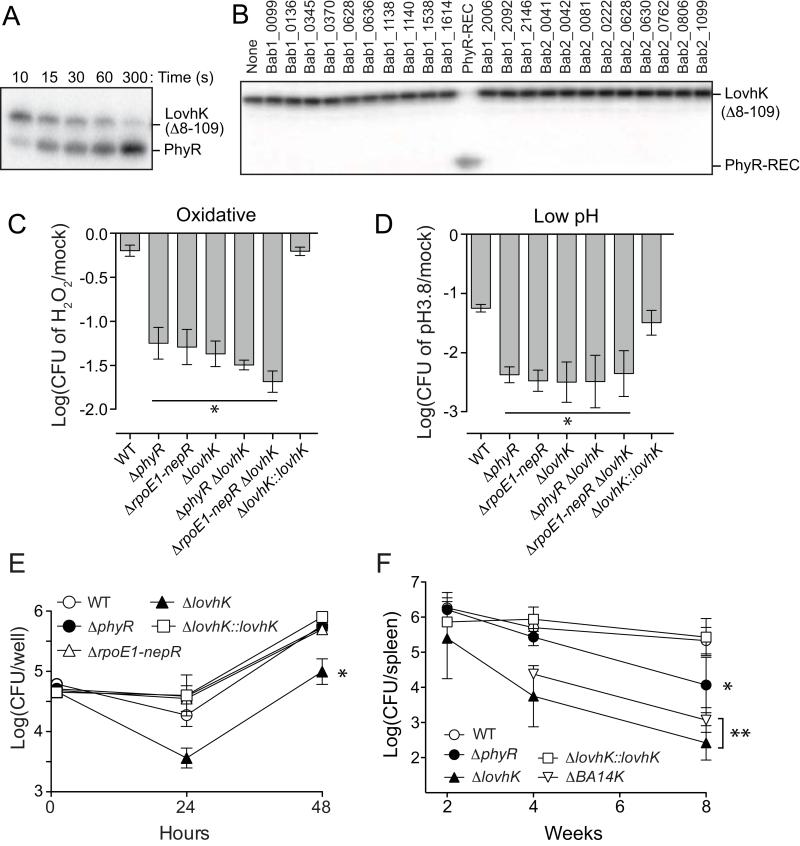
Figure 2.
LovhK functions as a PhyR kinase and activates B. abortus GSR.
(A) Time course of in vitro phosphotransfer between phospho-LovhK and PhyR between 10 and 300s. The bands corresponding to the phosphorylated proteins are marked. The stable LovhK(Δ8-109) variant, which lacks a LOV domain, was used in this phosphoryltransfer assay. (B) Phosphotransfer profile (Laub et al., 2007) of LovhK against each of the 23 response regulator receiver domains encoded in the B. abortus genome; incubation for 30 seconds. (C) Oxidative and (D) acid stress survival as in Figure 1 (* = p<0.001 and p<0.0005 based on one-way ANOVA followed by Dunnett’s multiple comparison test in oxidative stress and acid stress, respectively). (E) Replication of wild-type and mutant B. abortus strains (see key) within human macrophage-like THP-1 cells at 1 hour, 24 hours, and 48 hours post-infection. Data represent mean ± S.D. of three wells per time point; three independent replicates (* = p<0.001 at 24 and 48 hours based on one-way ANOVA followed by Dunnett’s multiple comparison test). (F) Kinetics of BALB/c mouse spleen colonization by wild-type and mutant B. abortus (see key) (* = p<0.001; ** p<0.0001 at 8 weeks based on one-way ANOVA followed by Dunnett’s multiple comparison test). Data indicate mean ± S.D. of five mice per time point. Data represent two independent experiments.
We next tested the functional role of lovhK in oxidative and acid stress survival in vitro. A ΔlovhK null strain exhibits reduced survival in both oxidative (5 mM H2O2) and acid stress (pH 3.8) conditions relative to mock treated controls; this survival defect is statistically equivalent to ΔrpoE1-nepR and ΔphyR null strains (Figure 2C and 2D). Replacement of the ΔlovhK allele with the full-length wild-type lovhK allele (noted as ΔlovhK::lovhK) restores cell survival to wild-type levels, confirming that this survival defect is due to the deletion of lovhK. Double deletion strains in which the ΔlovhK null mutation is combined with either ΔrpoE1-nepR or ΔphyR have equivalent survival phenotypes, providing genetic evidence that lovhK, phyR, and nepR-rpoE1 function in the same stress response pathway. From these data, we conclude that LovhK is a cognate sensor histidine kinase of PhyR, which functions as a stress sensor (and GSR pathway activator) under oxidative and acid stress conditions.
lovhK mediates cell survival inside THP-1 cells and is required for establishment of long-term infection in a mouse model
We have previously shown that GSR mutant strains lacking either rpoE1 or phyR are attenuated during the chronic phase of infection in BALB/c mice (> 1 month post-infection) but exhibit no survival or replication defect in primary murine macrophages (Kim et al., 2013). However, a B. abortus mutant lacking lovhK has been reported to exhibit a replication defect in a J774A.1 murine macrophage cell line (Swartz et al., 2007). Our data show that a ΔlovhK null mutant exhibits normal entry into terminally differentiated human THP-1 macrophage-like cells, but has a survival defect between 1 and 24 hours post-infection. After 24 hours, ΔlovhK replicates at a rate equivalent to wild-type (Figure 2E). Consistent with previous observations in primary murine macrophages (Kim et al., 2013), neither the ΔrpoE1-nepR nor ΔphyR strains exhibit any defect in growth or survival inside THP-1 cells.
To extend our analysis of the functional role of lovhK in B. abortus infection biology, we infected BALB/c mice and monitored bacterial load in the spleen over the course of 8 weeks. ΔlovhK was attenuated at an early stage of infection; viable colony forming units per spleen were 2 logs below wild-type by 4 weeks post-infection and 3 logs lower by 8 weeks. As observed previously (Kim et al., 2013), a ΔphyR null mutant exhibited no defect during the early stages of infection, and was not significantly attenuated until >1 month post-infection (Figure 2F). In both THP-1 infection experiments and mouse infection experiments, replacement of the ΔlovhK allele with the wild-type allele complemented the growth/replication defect of ΔlovhK (Figure 2E and 2F). These infection assays are distinct from our functional assays of these GSR mutants in defined in vitro stress conditions, where ΔlovhK, ΔphyR, and ΔrpoE1-nepR null strains have statistically equivalent cell survival phenotypes. The implications of these genetic results as they relate to GSR signaling complexity and specificity in B. abortus are discussed below.
lovhK and rpoE1 regulate a highly overlapped set of genes under in vitro stress conditions
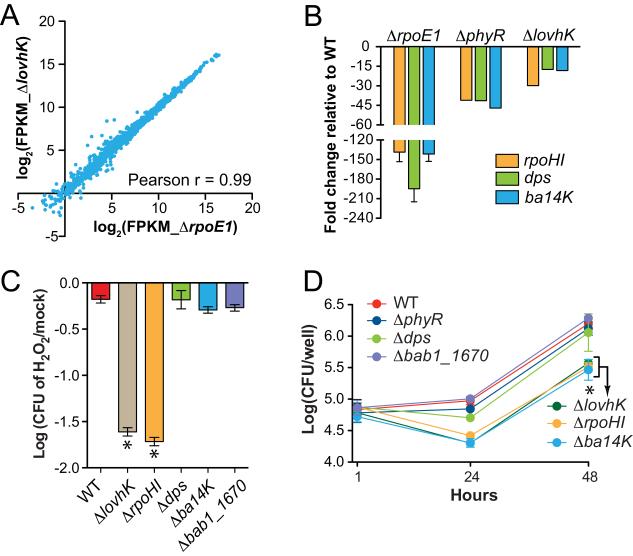
Given the clear biochemical and genetic connection between lovhK and the GSR regulators, phyR and rpoE1, we sought to define genes under direct and indirect transcriptional control of LovhK and to compare this gene set to that controlled by the GSR sigma factor, RpoE1 (σE1). To this end, we subjected wild-type B. abortus, ΔlovhK, and ΔrpoE1 strains to an equivalent oxidative stress (5 mM H2O2) and measured transcript levels in each strain by RNA-seq. Genes that show statistically significant differences in expression when comparing ΔlovhK or ΔrpoE1 to the wild-type parent strain are highly similar; 93% of genes exhibiting differential expression above the significance cutoff in the ΔlovhK dataset are above the significance cutoff in the ΔrpoE1 dataset (Table S1). A direct comparison of the ΔlovhK and ΔrpoE1 transcript levels revealed only five genes with statistically significant differential expression (Table 1 and Figure S3). However, all five of these genes are present in both the ΔlovhK and ΔrpoE1 regulons; it is simply the magnitude of expression that differs in the ΔlovhK and ΔrpoE1backgrounds. As σE1 is the direct transcriptional regulator in this system it is not surprising that a rpoE1 null show that largest magnitude changes in transcription. Expression patterns of three of these genes were further validated by qRT-PCR in ΔlovhK, ΔphyR and ΔrpoE1 (Figure 3B). Thus, the genes under direct and indirect transcriptional control of LovhK and σE1 during oxidative stress are, in statistical terms, nearly identical. The strong statistical overlap between the ΔlovhK and ΔrpoE1 RNA-seq datasets is further visualized in Figure 3A, in which log-transformed expression levels of all genes (quantified in fragments per kilobase pairs of transcript per million fragments mapped; FPKM) are presented. Together, these data support our model whereby LovhK directly phosphorylates PhyR to regulate the B. abortus GSR.
Table 1.
Genes differentially expressed between ΔlovhK and ΔrpoE1 null strains under in vitro oxidative stress conditions.
| Gene ID | FPKM |
Log2 (ΔlovhK/ ΔrpoE1) |
Annotation | ||
|---|---|---|---|---|---|
| WT | Δ rpoE1 | Δ lovhK | |||
| bab1_1670 | 7333 | 211 | 690 | 1.71 | Hypothetical protein |
| bab1_1775 | 662 | 5 | 48 | 3.16 | RNA polymerase factor sigma 32, rpoH1* |
| bab2_0505 | 5310 | 39 | 372 | 3.26 | Immunoreactive 14 kDa protein, ba14K* |
| bab2_0696 | 655 | 7 | 97 | 3.74 | Hypothetical protein |
| bab1_2150 | 4594 | 22 | 337 | 3.91 | DNA starvation/stationary phase protection protein, dps* |
Genes selected for confirmation with qRT-PCR. FPKM=Fragments Per Kilobase of transcript per Million mapped reads. Raw RNA-seq data from replicate experiments is available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), accession number GSE59513.
Figure 3.
LovhK regulates a set of genes that overlaps the σE1 GSR regulon.
(A) Transcriptome analysis of wild type, ΔrpoE1, and ΔlovhK deletion strains subjected to oxidative stress. Pairwise comparison of log2 transformed RNA-seq expression data (FPKM) for 3,414 genes between ΔrpoE1 and ΔlovhK mutants (Pearson r = 0.99). (B) Quantitative RT-PCR validation of expression levels of three transcripts (rpoH1, dps, and ba14k) in ΔrpoE1, ΔphyR, and ΔlovHK backgrounds relative to wild type. (C) Oxidative stress survival as in Figure 1 (* = p<0.0001 by one-way ANOVA followed by Dunnett’s multiple comparison test). (D) Intracellular replication of B. abortus strains in THP-1 cells at 1 hour, 24 hours, and 48 hours post-infection.
A functional analysis of genes regulated downstream of LovhK/σE1
To test the functional role of a subset of genes in the lovhK/rpoE1 regulon, we generated single deletion mutants of four different genes that are strongly induced under oxidative stress by LovhK and σE1 (Table S1, Figures 2 and 3). These genes include ba14k (bab2_0505) (Chirhart-Gilleland et al., 1998, Vemulapalli et al., 2006), rpoH1 (bab1_1775) (Delory et al., 2006), dps (bab1_2150), and bab1_1670. Only the ΔrpoH1 (σ32-family gene) null mutant was defective in oxidative stress survival; its survival phenotype is equivalent to ΔlovhK (Figure 3C). Mutants lacking ba14K or the alternative sigma factor genes rpoH1 exhibit intracellular survival phenotypes in THP-1 cells that are similar to the lovhK null (Figure 3D). Specifically, both of these single deletion mutants exhibit normal THP-1 entry, but have a survival defect between 1 and 24 hours post-infection. After 24 hours, Δba14K and ΔrpoH1 replicate at a rate equivalent to wild-type. Neither Δdps nor Δbab1_1670 exhibit defects in intracellular replication (Figure 3D). We further assessed one of these mutants, Δba14K, in an animal infection model. Deletion of the conserved hypothetical gene Δba14K results in significant attenuation in Balb/c mice as assessed by enumeration of CFU per spleen; the number of CFU/spleen is 1.5 logs lower than wild-type at 4 weeks and 2.5 logs lower than wild-type at 8 weeks post-infection (Figure 2F). This animal infection phenotype is intermediate between ΔlovhK and ΔphyR, and is consistent with previous reports of Δba14K attenuation in a mouse model (Vemulapalli et al., 2006), though the clearance kinetics and total bacterial load in the spleen differ. B. abortus Ba14K has been previously described as an immunoreactive protein (Chirhart-Gilleland et al., 1998) that affects LPS synthesis (Vemulapalli et al., 2006). Thus, Ba14K may play a role in modulation of B. abortus interface with the host immune system. Collectively, the disparate infection phenotypes of these null mutants provide evidence that genes regulated downstream of the lovhK-phyR-nepR-rpoE1 (i.e. GSR) system have diverse roles in the regulation of stress adaptation and host infection.
Evidence for distinct sensory roles for the two PAS domains of LovhK
B. abortus LovhK (Swartz et al., 2007) is a cytoplasmic HWE kinase that contains two sensory domains: an N-terminal flavin-binding PAS/LOV domain (Herrou & Crosson, 2011) followed by a second PAS domain that does not contain a clear cofactor binding sequence (Henry & Crosson, 2011) (Figure 4A). We hypothesized that these domains perceive distinct environmental stimuli or affect different regulatory outputs during specific stress or infection conditions. LovhK residue C43 (previously annotated C69 (Swartz et al., 2007)) is known to form a cysteinyl-C(4a) flavin adduct upon blue light absorption. Replacement of this residue with alanine abrogates light-dependent adduct formation (Rinaldi et al., 2012, Swartz et al., 2007) and light-regulated intracellular replication in a murine macrophage cell line (Swartz et al., 2007). To date, the function of the second PAS domain of LovhK has remained undefined.
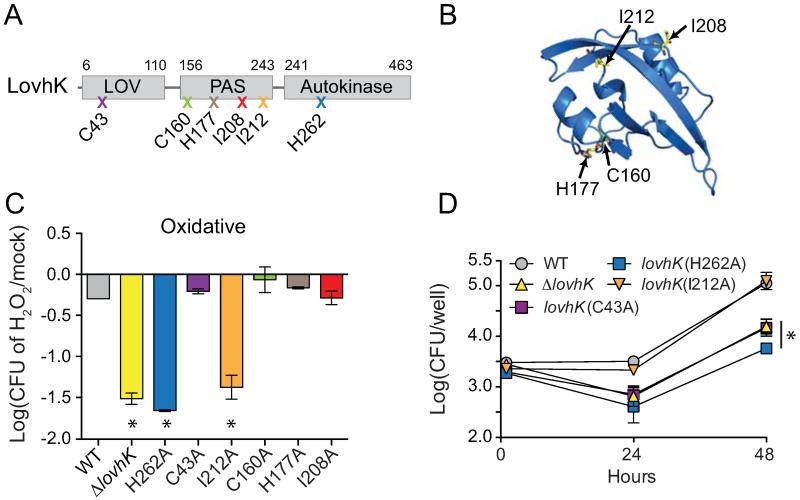
Figure 4.
Point mutations in LovhK sensory domains result in distinct stress survival and infection phenotypes.
(A) LovhK domain structure. Mutations were selected based on a structural homology model and are indicated by an X with the corresponding amino acid residue; color-coding of the marked mutation (X) corresponds to the graphs in panels C and D and in Figure 5B. (B) Model of LovhK PAS domain based on a homologous structure of FixL-PAS of Rhizobium meliloti (PDB: 1D06). Positions of mutated residues are marked. (C) Oxidative stress survival of strains bearing mutant lovhK alleles as in Figure 1 (* = p<0.0005 by one-way ANOVA followed by Dunnett’s multiple comparison test). (D) Intracellular survival and replication of wild type and lovhK mutant strains in THP-1 cells at 1 hour, 24 hours, and 48 hours post-infection. Error bars, S.D. Data represent three independent experiments with three wells per time point (* = p<0.001 by one-way ANOVA followed by Dunnett’s multiple comparison test).
To identify important functional residues in the second undefined PAS domain, we constructed a structural model based on an X-ray crystal structure of a homologous PAS domain of Rhizobium meliloti FixL (Miyatake et al., 2000) (Figure 4B). We generated LovhK point mutants in which we mutated residues in the putative ligand-binding pocket, C160, H166, I208, and I212, to alanine (Figure 4A and B). We then replaced the wild-type allele of lovhK with mutant alleles harboring these single point mutations, and assayed oxidative stress survival of these lovhK(PAS mutant) strains. Of the five assayed PAS point mutants, only lovhK(I212A) exhibited significantly reduced survival under oxidative stress (Figure 4C). We further assayed this mutant in a THP-1 cell-based infection assay, but observed no difference in infection, survival or replication relative to wild-type (Figure 4C and 4D).
We next replaced the wild-type allele of lovhK with a point mutant allele in which the conserved site of histidine phosphorylation was changed to alanine (lovhK(H262A)). As expected, an intact LovhK phosphorylation site is required for oxidative stress adaptation and intracellular survival and replication in THP-1 macrophage-like cells. Notably, mutation of the photo-responsive C43 residue to alanine in the LovhK PAS/LOV domain (lovhK(C43A)) has no effect on oxidative stress survival. However, consistent with a previous report (Swartz et al., 2007), this mutation does reduce intracellular survival in THP-1 between 1 and 24 hours post-infection (Figure 4C and D). Thus the defect in survival of the lovhK(C43A) in THP-1 is likely not due to oxidative stress encountered during the early phase of infection and intracellular trafficking.
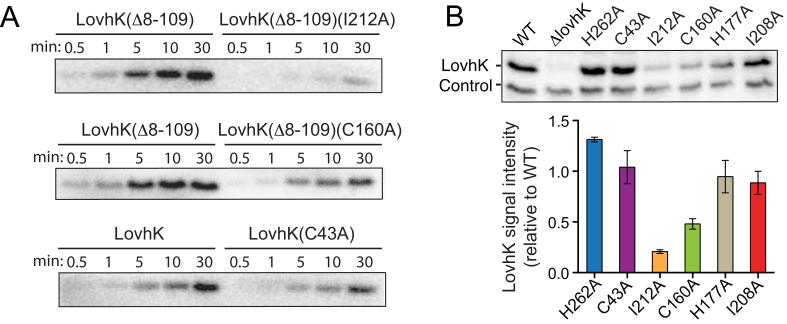
To more clearly interpret our lovhK mutagenesis data, we assayed steady-state levels of full-length LovhK and LOV, PAS, and kinase domain mutants in B. abortus cells by Western blot using polyclonal LovhK antiserum. All LovhK variants except for LovhK(C160A) and LovhK(I212A) were expressed, within error, to wild-type levels (Figure 5B). While it is possible that the oxidative stress survival defect of I212A is due to low expression or poor stability, we note that C160A expression is also reduced yet this mutant exhibits no defect in oxidative stress survival. Moreover, I212A is not defective in a THP-1 infection model, which provides evidence that the level of expressed LovhK(I212A) is functional in the context of LovhK signaling inside a mammalian host cell. These results demonstrate that perturbation of regions of LOV and PAS sensor domain structure in B. abortus LovhK affect specific features of either stress physiology and/or infection biology.
Figure 5.
Mutations in LovhK sensor domains affect in vitro kinase activity and steady state protein levels in B. abortus cells.
(A) In vitro autophosphorylation assays with purified full-length LovhK and variants carrying mutations in the N-terminal LOV (C43A) or PAS (C160A; I212A) domains (proteins labeled above each autoradiograph). (B) (top) Western blot of wild-type and mutant LovhK alleles expressed from the native locus on the B. abortus chromosome. Control is a non-specific band that cross-reacts with LovhK antiserum. (bottom) Mean signal intensity (±S.D.) of LovhK mutant variants normalized to wild-type LovhK from replicate Western blots.
LovhK sensor domain mutations can affect autophosphorylation activity
Given the oxidative stress survival phenotype of lovhK(I212A), we postulated that this sensor domain mutation may affect kinase activity. We assayed in vitro autophosphorylation of purified LovhK(I212A) utilizing the active and stable lovhKΔ8-109 allele as the parent construct. The steady-state phosphorylation level of LovhKΔ8-109(I212A) is significantly reduced relative to LovhKΔ8-109 (5 μM concentration of each protein was assayed) (Figure 5A). The I160A mutant, which has no defect under oxidative stress conditions (Figure 4C), is more active than I212A, though steady-state phosphorylation is still diminished relative to the parent protein LovhKΔ8-109 (5 μM concentrations; Figure 5A). We further demonstrate that autophosphorylation of LovhK(C43A) is lower than wild-type (full-length) LovhK (purified under dim red light), which is consistent with previous data (Swartz et al., 2007). We conclude that particular lesions in the sensory domains of LovhK can affect both cellular LovhK levels and autophosphorylation activity. However, we acknowledge that the effects of these mutations on protein tertiary and quaternary structure remain undefined.
Discussion
HWE kinases are commonly located adjacent to α-proteobacterial GSR loci (Figure S1), and have been recently implicated in GSR regulation in several species (Correa et al., 2013, Foreman et al., 2012, Kaczmarczyk et al., 2011, Lourenco et al., 2011, Sauviac & Bruand, 2014). Here, we report the characterization of the three HWE-type histidine kinases encoded in the genome of the intracellular pathogen, B. abortus. We have functionally and genetically linked two of these kinases, Bab1_1673 and LovhK, to B. abortus GSR regulation.
Bab1_1673 can only be deleted from the chromosome in the presence of a complementing copy of the gene on a plasmid, or in a genetic background in which either phyR or lovhK are missing. When overexpressed, bab1_1673 functions as an inhibitor of stress survival (Figure 1E). The genetic properties of Bab1_1673 are similar to Sphingomonas str. FR1 phyP (Kaczmarczyk et al., 2011). PhyP is an essential HWE-like kinase encoded from a gene adjacent to the Sphingomonas str. FR1 GSR locus; PhyP likely functions to remove phosphoryl groups from PhyR~P, and thus repress GSR signaling (Kaczmarczyk et al., 2011). Notably, PhyP and Bab1_1673 are reciprocal top BLAST hits; overall sequence identity in the HWE-like domain is approximately 30%. It remains to be determined 1) why repressive HWE kinase genes such as bab1_1673 and phyP are essential in the context of an intact GSR signaling system, and 2) the biochemical mechanism by which these proteins function to repress stress adaptation. We note that the primary genetic repressor of GSR signaling, the anti-σ factor nepR, also cannot be deleted from a wild-type background in other α-proteobacteria (Campagne et al., 2012, Herrou et al., 2010, Herrou et al., 2012, Kaczmarczyk et al., 2011). It therefore seems that de-repression (or hyperactivation) of the GSR in α-proteobacteria strongly inhibits cell growth or division.
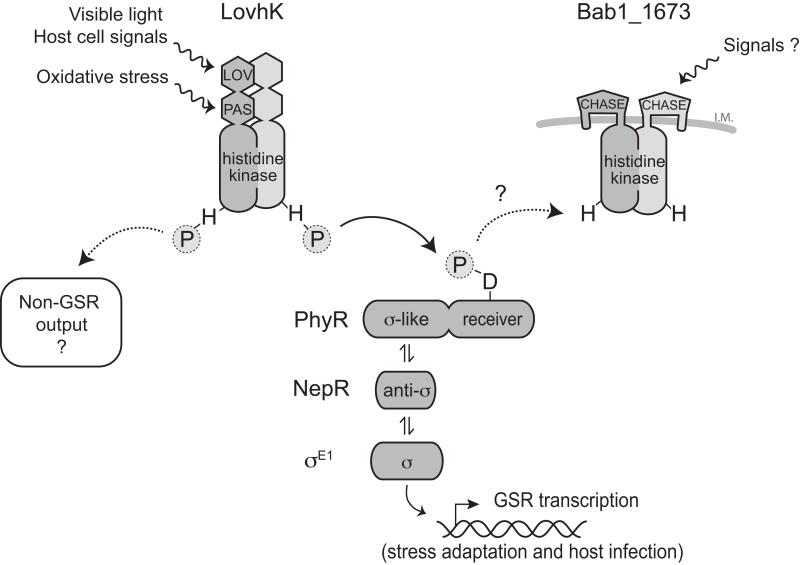
The B. abortus HWE histidine kinase, LovhK, has been previously reported to function as a light-regulated kinase that controls intracellular replication in a light-dependent manner (Swartz et al., 2007). To date, the molecular pathway(s) through which this protein directly functions have remained undefined. We present genetic and biochemical evidence that LovhK is a cognate sensor histidine kinase of PhyR that functions as a stress sensor (and GSR pathway activator) in B. abortus (Figure 2). Phosphotransfer experiments show that LovhK stably phosphorylates only 1 of the 23 response regulator receiver domains encoded in B. abortus: PhyR (Figure 2), though we cannot eliminate the possibility that LovhK may phosphorylate other substrates to a level that is not detectable with our assay. Indeed, it is established that the LOV histidine kinase, EL368, of E. litoralis phosphorylates two different receiver domains, including PhyR (Correa et al., 2013). Nonetheless, the ΔlovhK null phenotypes defined under in vitro stress conditions match the null phenotypes of ΔphyR and ΔrpoE1 mutants, and the LovhK and σE1 transcriptional regulons defined under oxidative stress are nearly equivalent (Figure 3), which suggests a linear signaling model in which LovhK functions as the upstream molecular sensor of environmental stress that phosphorylates PhyR and releases σE1 to drive transcription (see Figure 1B). However, animal and cell-based infection experiments reported herein reveal additional complexity in LovhK signaling. Specifically, the phenotypes of single ΔlovhK, ΔphyR and ΔrpoE1 null mutants are not congruent between animal and cell-based infection models: ΔlovhK survival is attenuated in THP-1 cells, while ΔphyR and ΔrpoE1 strains have no survival or replication defect in primary murine macrophages (Kim et al., 2013) or THP-1 cells (Figure 2E). Moreover, ΔlovhK is cleared from a murine host earlier than ΔphyR and ΔrpoE1 (Figure 2F). These results suggest that LovhK interfaces with another pathway in the context of a mammalian host cell, or that signals present or absent in the host, e.g. light (Bonomi et al., 2012, Correa et al., 2013, Purcell et al., 2007, Swartz et al., 2007) or other physicochemical differences inside the host, modulate the signaling output of LovhK in unexpected ways. Importantly, we show that regions of primary structure in each of the two PAS sensory domains of LovhK have specific roles in signal perception and kinase regulation (Figures 4 5 and 6). It is possible to envision models in which multiple environmental signals can be integrated via LovhK; the complexity of possible signaling logic is further increased when one considers the role of the HWE sensor kinase Bab1_1673 as a pathway repressor (and perhaps Bab1_1669, which may have functions in B. abortus GSR under conditions that have not yet been explored) (Figure 6).
Figure 6.
Molecular model of the B. abortus General Stress Response (GSR) regulatory system. CHASE is the predicted sensor domain (Zhulin et al., 2003) contained at the amino terminus of Bab1_1673.
It is not clear what role the photo-responsive LOV domain of LovhK plays in GSR activation under stress, or in natural B. abortus transmission and infection in the wild. Residue C43 within the N-terminal LOV domain is certainly required for light-dependent cysteinyl-flavin adduct formation and LovhK photoactivation in vitro (Swartz et al., 2007) and for robust infection of mammalian phagocytic cells (Figure 4D) (Swartz et al., 2007). However, this key LOV domain residue is not required for oxidative stress survival of B. abortus in vitro (Figure 4C). In short, lovhK is required for oxidative stress survival, but LOV domain photoactivity is not required in this in vitro context. There is evidence that LOV domains may have a more complex sensory role in which both cellular redox state and/or light are functional input signals (Bury & Hellingwerf, 2014, Metz et al., 2012, Noll et al., 2007, Purcell et al., 2010). This putative redox-sensing feature of LOV domains may also be involved in stress response regulation by the YtvA-family of LOV proteins, which control σB in Gram positive bacteria (Akbar et al., 2001, Avila-Perez et al., 2006, Gaidenko et al., 2006, Suzuki et al., 2007, Tiensuu et al., 2013), though this area has not been fully explored. To understand the role of the B. abortus GSR system in infection biology, it will be necessary to identify relevant host signals to which LovhK responds, and to elucidate how LovhK interfaces with Bab1_1673 in cell signaling.
An additional layer of regulatory complexity to consider when interpreting infection phenotypes of GSR pathway mutants is that low molecular weight phosphodonors such as acetyl phosphate may participate in B. abortus GSR signaling inside the mammalian host. In such a scenario, changes in the intracellular levels of acetyl phosphate could affect GSR signaling in the absence of the lovhK gene via direct phosphorylation of the PhyR receiver domain (Wolfe, 2005). Indeed, it is known that the B. abortus GSR system strongly controls transcription of genes that are predicted to control acetyl phosphate synthesis (Kim et al., 2013).
Overall, our study underscores the importance of assaying multiple genetic-environment combinations when assessing the function of environmental regulatory systems. Such experiments on the B. abortus GSR regulatory system have shed light on the molecular machinery required for adaptation to environmental stress conditions relevant to infection biology, and for adaptation to the complex mammalian host environment. We have identified key sensory residues in LovhK that are required for adaptation and survival in vitro and in vivo and conclude that B. abortus utilizes LovhK and the GSR system in nuanced ways to respond to specific conditions (and combinations of conditions) encountered inside and outside its host.
Experimental Procedures
Bacterial strain culture and reagents
Escherichia coli strains used for cloning were maintained in Luria–Bertani (LB) agar at 37°C. Liquid cultures were grown in LB broth (Fisher scientific). Brucella abortus 2308 and its derivative strains were grown in Brucella broth (BD Difco) or on Schaedler agar (BD Difco) supplemented with 5% defibrinated bovine blood (SBA) at 37°C with 5% CO2. Antibiotics were added as required at the following concentrations: 50 μg ml-1 of kanamycin (Kan), 100 μg ml-1 of ampicillin (Amp). All studies on live B. abortus were performed at biosafety level 3 (BSL3) as per CDC select agent regulations and standard operating procedures at the University of Chicago Howard Taylor Ricketts Laboratory.
Plasmid and Strain Construction
To create gene deletion alleles, 500 to 700 bp regions flanking the target genes were PCR-amplified from B. abortus 2308 using primers listed in Table S3. To create point mutation alleles 500-700 bp regions flanking the site of interest were amplified with primers containing the point mutations of interest. The flanking regions were joined by overlap extension PCR to generate null or point mutant alleles which were then ligated into the suicide plasmid pNPTS138. This plasmid encodes a kanamycin resistance marker to select primary integrants and sacB for counterselection on 10% sucrose to identify clones in which the plasmid has been lost in a second recombination event. Clones were screened by PCR to identify those carrying the null alleles. Sequencing the PCR products identified clones bearing point mutant alleles. All other plasmids were constructed by PCR amplification of the target sequence, restriction digestion and ligation into the desired plasmid. All plasmids were confirmed by sequencing. Plasmids were transformed into B. abortus 2308 by electroporation. Strains, plasmids, and oligonucleotides used in this study are listed in Tables S2 and S3.
Oxidative and acid stress assays
Stress assays were performed as previously described (Kim et al., 2013). Briefly, B. abortus cells grown on SBA were harvested into Gerhardt’s minimal medium (pH 6.8) and then adjusted to a cell density of 108 cfu/ml. Serial dilution of cultures after 60 or 90 min treatments of H2O2 (5 mM) or pH 3.8 were plated on SBA to measure viable colony forming units.
Cell culture and macrophage infection assay
Human monocytic THP-1 cells were cultured in RPMI 1640 medium supplemented with 10% heat-treated fetal bovine serum (FCS) and 2 mM L-glutamine, and were differentiated to macrophages by the addition of 40 nM phorbol 12-myristate 13-acetate (PMA) (Sigma) for 24 h at 37°C in 5% CO2. For infection assays, 5 × 104 macrophages were infected with 5 × 105 brucellae to achieve a multiplicity of infection (MOI) of 1:100 in 96-well plates for 1 h in triplicate. Following infection, extracellular bacteria were removed by treatment with 50 μg/ml gentamycin for 15 min. To determine the numbers of intracellular brucellae, cells were washed three times with PBS and lysed with 0.1 ml of 0.1% Triton X-100. The lysate was serially diluted, and plated on Tryptic Soy agar.
Experimental infection of BALB/c mice
Female BALB/c mice (4–5 weeks old) were purchased from Harlan Laboratories, Inc. Animal studies on wild-type and mutant strains were conducted at the University of Chicago under protocols approved by the institutional animal care and use committee and the institutional biosafety committee. Specifically, animals used in this study were housed under pathogen-free conditions in a BSL3 animal facility at the University of Chicago Howard Taylor Ricketts Laboratory. Six-week-old, female BALB/c mice were infected with 5 × 104 cfu of wild-type or mutant strains via the intraperitoneal route. Mouse spleens were removed aseptically and homogenized in 1 ml of PBS, and serial dilutions of homogenized spleens were plated on Schaedler blood agar to enumerate bacterial CFU at 2, 4, and 8 weeks postinfection.
RNA isolation and qRT-PCR
Total RNA was extracted from cells growing exponentially (OD600=0.15–0.2) in Brucella broth at 37°C after 10 min of 5 mM H2O2 exposure with a modified hot phenol-SDS extraction method (Chuang et al., 1993) and the RNeasy kit (Qiagen). Reverse transcription was performed with Superscript III cDNA kit (Invitrogen) and qPCR was carried out with SYBR Green PCR kits (Applied Biosystems). Relative quantification of specific gene expression was calculated with the 2−ΔΔCt method, with rplK as the housekeeping reference, and normalized against the B. abortus 2308 wild type. Each assay was performed at least in triplicate on three independent cultures.
Transcriptome profiling by RNA-seq
For RNA sequencing analysis, identical culture conditions and RNA purification procedures were used. rRNA was depleted using Ribo-Zero rRNA Removal (Gram-negative bacteria) Kit (Epicentre). Libraries were prepared with Illumina TruSeq RNA kit according to manufacturer’s instructions and were then quantified using a 2100 Bioanalyzer (Agilent) and sequenced on a HiSeq2500 (Illumina). The obtained RNA-seq reads were aligned to the genome sequence of B. abortus 2308 (RefSeq AM040265) using the short-read aligner Bowtie 2 (Langmead & Salzberg, 2012). Alignments were visualized using Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) (Robinson et al., 2011). To calculate transcript abundances, Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values were assessed by using Cufflinks (http://cufflinks.cbcb.umd.edu/) with appropriate parameters set for quartile normalization (Trapnell et al., 2012). Differential expression analysis was carried out using Cuffdiff, with quartile normalization. A fold change of greater than 1.5-fold and false-discovery rate cutoff of 0.05 was used to determine significant differential expression between two different conditions. RNA-seq datasets are available at PATRIC (Pathosystems Resource Integration Center, http://patricbrc.org/) and the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) at accession number GSE59513.
Recombinant protein purification
Genes to be overexpressed and purified were amplified and cloned into pET overexpression vectors. Expression of N-terminally His-tagged proteins was induced in E. coli BL21 (DE3) by addition of 250 μM of isopropyl β-D-1-thiogalactopyranoside (IPTG) at 30° C. After cell lysis in a French pressure cell and centrifugation, soluble recombinant proteins were affinity-purified by using Ni2+ Chelating Resin (GE Amersham Pharmacia) using a 250 to 500 mM imidazole gradient in 20 mM Tris buffer pH 7.5, 150 mM NaCl. Purity of the proteins was assessed to be ≥95% by SDS/PAGE with Coomassie blue staining.
In vitro kinase and phosphoyltransfer assays
All kinase and phosphotransfer reactions were performed following previously published methods (Willett & Kirby, 2012). For kinase reaction, 5 μM of purified LovhKΔ8-109 and its variants was incubated in kinase buffer (25 mM Tris pH 7.5, 50 mM KCl, 1 mM of CaCl2 MgCl2, MnCl2, and DTT) and ATP mix (250 μM ATP, 0.3 μM [γ-32P]ATP). Reactions were quenched by addition of SDS/PAGE sample buffer. For phosphotransfer from LovhK(Δ8-109) and MBP-1673(Δ1-227) to PhyR or the REC domain proteins of 23 Brucella response regulators, the 32P-LovhK(Δ8-109) and 32P-MBP-1673(Δ1-227) were first generated by a 30-min autophosphorylation reactions and then mixed with equimolar (5 μM) of the response regulator proteins. 32P-labeled products on SDS/PAGE gels were quantified by using a Typhoon Imager.
Protein immunoblot analysis
Rabbit LovhK polyclonal antiserum (Josman, LLC) was raised against His6-LovHK. Rabbits were immunized with purified LovhK on days 0, 21, 35, 49, and 63. Total B. abortus protein from liquid cell culture was concentrated by precipitation with 10% trichloroacetic acid, separated by 12 %fThe Brucella abortus virulence regulator, LovhK, is a sensor kinase in the general stress response signaling pathway SDS/PAGE, and then transferred to a PVDF membrane (Millipore). The membrane was probed with the primary polyclonal anti-LovhK antiserum (1:1,000) followed by HRP-conjugated goat anti-rabbit secondary antibodies (1:10,000) (Thermo Scientific). Blots were developed using SuperSignal West Femto Substrate (Thermo Scientific). The signal intensity was measured using Image Lab™ software in the ChemiDoc MP system (Bio-Rad).
Supplementary Material
Acknowledgements
We thank members of the Crosson Lab for discussions and guidance for this study. Robert Foreman generated plasmids to produce B. abortus ΔrpoE1, ΔrpoH1, and Δdps mutant strains. This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), grant numbers U19AI107792 and R01AI107159 to S.C. J.W.W. is supported by a NIH Ruth Kirschstein Postdoctoral fellowship (F32GM109661).
Footnotes
The authors of this study have no conflict of interest to declare.
References
- Abromaitis S, Koehler JE. The Bartonella quintana extracytoplasmic function sigma factor RpoE has a role in bacterial adaptation to the arthropod vector environment. J Bacteriol. 2013;195:2662–2674. doi: 10.1128/JB.01972-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar S, Gaidenko TA, Kang CM, O’Reilly M, Devine KM, Price CW. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis. J Bacteriol. 2001;183:1329–1338. doi: 10.1128/JB.183.4.1329-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Perez M, Hellingwerf KJ, Kort R. Blue light activates the sigmaB-dependent stress response of Bacillus subtilis via YtvA. J Bacteriol. 2006;188:6411–6414. doi: 10.1128/JB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiat B, Sauviac L, Bruand C. Dual control of Sinorhizobium meliloti RpoE2 sigma factor activity by two PhyR-type two-component response regulators. J Bacteriol. 2010;192:2255–2265. doi: 10.1128/JB.01666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomi HR, Posadas DM, Paris G, Carrica Mdel C, Frederickson M, Pietrasanta LI, Bogomolni RA, Zorreguieta A, Goldbaum FA. Light regulates attachment, exopolysaccharide production, and nodulation in Rhizobium leguminosarum through a LOV-histidine kinase photoreceptor. Proc Natl Acad Sci U S A. 2012;109:12135–12140. doi: 10.1073/pnas.1121292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury A, Hellingwerf KJ. On the in vivo redox state of flavin-containing photosensory receptor proteins. Methods Mol Biol. 2014;1146:177–190. doi: 10.1007/978-1-4939-0452-5_9. [DOI] [PubMed] [Google Scholar]
- Campagne S, Damberger FF, Kaczmarczyk A, Francez-Charlot A, Allain FH, Vorholt JA. Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria. Proc Natl Acad Sci U S A. 2012;109:E1405–1414. doi: 10.1073/pnas.1117003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirhart-Gilleland RL, Kovach ME, Elzer PH, Jennings SR, Roop RM., 2nd Identification and characterization of a 14-kilodalton Brucella abortus protein reactive with antibodies from naturally and experimentally infected hosts and T lymphocytes from experimentally infected BALB/c mice. Infect Immun. 1998;66:4000–4003. doi: 10.1128/iai.66.8.4000-4003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SE, Daniels DL, Blattner FR. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa F, Ko WH, Ocasio V, Bogomolni RA, Gardner KH. Blue light regulated two-component systems: enzymatic and functional analyses of light-oxygen-voltage (LOV)-histidine kinases and downstream response regulators. Biochemistry. 2013;52:4656–4666. doi: 10.1021/bi400617y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delory M, Hallez R, Letesson JJ, De Bolle X. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J Bacteriol. 2006;188:7707–7710. doi: 10.1128/JB.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet. 2014;10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman R, Fiebig A, Crosson S. The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J Bacteriol. 2012;194:3038–3049. doi: 10.1128/JB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A, Frunzke J, Reichen C, Ebneter JZ, Gourion B, Vorholt JA. Sigma factor mimicry involved in regulation of general stress response. Proc Natl Acad Sci U S A. 2009;106:3467–3472. doi: 10.1073/pnas.0810291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidenko TA, Kim TJ, Weigel AL, Brody MS, Price CW. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J Bacteriol. 2006;188:6387–6395. doi: 10.1128/JB.00691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourion B, Francez-Charlot A, Vorholt JA. PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J Bacteriol. 2008;190:1027–1035. doi: 10.1128/JB.01483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourion B, Sulser S, Frunzke J, Francez-Charlot A, Stiefel P, Pessi G, Vorholt JA, Fischer HM. The PhyR-sigma(EcfG) signalling cascade is involved in stress response and symbiotic efficiency in Bradyrhizobium japonicum. Mol Microbiol. 2009;73:291–305. doi: 10.1111/j.1365-2958.2009.06769.x. [DOI] [PubMed] [Google Scholar]
- Gourley CR, Petersen E, Harms J, Splitter G. Decreased in vivo virulence and altered gene expression by a Brucella melitensis light-sensing histidine kinase mutant. Pathog Dis. 2014 doi: 10.1111/2049-632X.12209. doi: 10.1111/2049-1632X.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J, Foreman R, Fiebig A, Crosson S. A structural model of anti-anti-sigma inhibition by a two-component receiver domain: the PhyR stress response regulator. Mol Microbiol. 2010;78:290–304. doi: 10.1111/j.1365-2958.2010.07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J, Rotskoff G, Luo Y, Roux B, Crosson S. Structural basis of a protein partner switch that regulates the general stress response of alpha-proteobacteria. Proc Natl Acad Sci U S A. 2012;109:E1415–1423. doi: 10.1073/pnas.1116887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi H, Sato I, Yurimoto H, Sakai Y. Stress resistance and C1 metabolism involved in plant colonization of a methanotroph Methylosinus sp. B4S. Arch Microbiol. 2013;195:717–726. doi: 10.1007/s00203-013-0922-6. [DOI] [PubMed] [Google Scholar]
- Jans A, Vercruysse M, Gao S, Engelen K, Lambrichts I, Fauvart M, Michiels J. Canonical and non-canonical EcfG sigma factors control the general stress response in Rhizobium etli. Microbiologyopen. 2013;2:976–987. doi: 10.1002/mbo3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarczyk A, Campagne S, Danza F, Metzger LC, Vorholt JA, Francez-Charlot A. Role of Sphingomonas sp. strain Fr1 PhyR-NepR-sigmaEcfG cascade in general stress response and identification of a negative regulator of PhyR. J Bacteriol. 2011;193:6629–6638. doi: 10.1128/JB.06006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Caswell CC, Foreman R, Roop RM, 2nd, Crosson S. The Brucella abortus general stress response system regulates chronic mammalian infection and is controlled by phosphorylation and proteolysis. J Biol Chem. 2013;288:13906–13916. doi: 10.1074/jbc.M113.459305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Biondi EG, Skerker JM. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 2007;423:531–548. doi: 10.1016/S0076-6879(07)23026-5. [DOI] [PubMed] [Google Scholar]
- Lourenco RF, Kohler C, Gomes SL. A two-component system, an anti-sigma factor and two paralogous ECF sigma factors are involved in the control of general stress response in Caulobacter crescentus. Mol Microbiol. 2011;80:1598–1612. doi: 10.1111/j.1365-2958.2011.07668.x. [DOI] [PubMed] [Google Scholar]
- Metz S, Jager A, Klug G. Role of a short light, oxygen, voltage (LOV) domain protein in blue light- and singlet oxygen-dependent gene regulation in Rhodobacter sphaeroides. Microbiology. 2012;158:368–379. doi: 10.1099/mic.0.054700-0. [DOI] [PubMed] [Google Scholar]
- Miyatake H, Mukai M, Park SY, Adachi S, Tamura K, Nakamura H, Nakamura K, Tsuchiya T, Iizuka T, Shiro Y. Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies. J Mol Biol. 2000;301:415–431. doi: 10.1006/jmbi.2000.3954. [DOI] [PubMed] [Google Scholar]
- Moreno E, Moriyon I. In: The Genus Brucella. In: The prokaryotes: a handbook on the biology of bacteria. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. Springer; New York: 2006. pp. 315–456. [Google Scholar]
- Noll G, Hauska G, Hegemann P, Lanzl K, Noll T, von Sanden-Flohe M, Dick B. Redox properties of LOV domains: chemical versus photochemical reduction, and influence on the photocycle. Chembiochem. 2007;8:2256–2264. doi: 10.1002/cbic.200700304. [DOI] [PubMed] [Google Scholar]
- Purcell EB, McDonald CA, Palfey BA, Crosson S. An analysis of the solution structure and signaling mechanism of LovK, a sensor histidine kinase integrating light and redox signals. Biochemistry. 2010;49:6761–6770. doi: 10.1021/bi1006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi J, Gallo M, Klinke S, Paris G, Bonomi HR, Bogomolni RA, Cicero DO, Goldbaum FA. The beta-scaffold of the LOV domain of the Brucella light-activated histidine kinase is a key element for signal transduction. J Mol Biol. 2012;420:112–127. doi: 10.1016/j.jmb.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauviac L, Bruand C. A Putative Bifunctional Histidine Kinase/Phosphatase of the HWE Family Exerts Positive and Negative Control on the Sinorhizobium meliloti General Stress Response. J Bacteriol. 2014;196:2526–2535. doi: 10.1128/JB.01623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Takaya N, Hoshino T, Nakamura A. Enhancement of a sigma(B)-dependent stress response in Bacillus subtilis by light via YtvA photoreceptor. J Gen Appl Microbiol. 2007;53:81–88. doi: 10.2323/jgam.53.81. [DOI] [PubMed] [Google Scholar]
- Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- Tiensuu T, Andersson C, Ryden P, Johansson J. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol Microbiol. 2013;87:909–924. doi: 10.1111/mmi.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemulapalli TH, Vemulapalli R, Schurig GG, Boyle SM, Sriranganathan N. Role in virulence of a Brucella abortus protein exhibiting lectin-like activity. Infect Immun. 2006;74:183–191. doi: 10.1128/IAI.74.1.183-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett JW, Kirby JR. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet. 2012;8:e1003084. doi: 10.1371/journal.pgen.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J Bacteriol. 2003;185:285–294. doi: 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.