Summary
The catalytic A1 subunit of cholera toxin (CTA1) has a disordered structure at 37°C. An interaction with host factors must therefore place CTA1 in a folded conformation for the modification of its Gsα target which resides in a lipid raft environment. Host ADP-ribosylation factors (ARFs) act as in vitro allosteric activators of CTA1, but the molecular events of this process are not fully characterized. Isotope-edited Fourier transform infrared spectroscopy monitored ARF6-induced structural changes to CTA1, which were correlated to changes in CTA1 activity. We found ARF6 prevents the thermal disordering of structured CTA1 and stimulates the activity of stabilized CTA1 over a range of temperatures. Yet ARF6 alone did not promote the refolding of disordered CTA1 to an active state. Instead, lipid rafts shifted disordered CTA1 to a folded conformation with a basal level of activity that could be further stimulated by ARF6. Thus, ARF alone is unable to activate disordered CTA1 at physiological temperature: additional host factors such as lipid rafts place CTA1 in the folded conformation required for its ARF-mediated activation. Interaction with ARF is required for in vivo toxin activity, as enzymatically active CTA1 mutants that cannot be further stimulated by ARF6 fail to intoxicate cultured cells.
Keywords: ADP-ribosylation factor, cholera toxin, Fourier transform infrared spectroscopy, host-pathogen interactions, protein folding
Introduction
Cholera toxin (CT) travels from the cell surface to the endoplasmic reticulum (ER) as an AB holotoxin (Wernick et al., 2010). In the ER, the catalytic CTA1 subunit is displaced from the rest of the toxin (Taylor et al., 2011b; Taylor et al., 2014; Tsai et al., 2001). The free CTA1 polypeptide is an unstable protein, so it shifts to an unfolded conformation upon its release from the holotoxin at the physiological temperature of 37°C (Pande et al., 2007). This identifies the dissociated CTA1 subunit as a misfolded protein for export to the cytosol via the quality control system of ER-associated degradation (ERAD) (Banerjee et al., 2010; Massey et al., 2009; Taylor et al., 2011a; Teter and Holmes, 2002; Teter et al., 2003). Most exported ERAD substrates are appended with poly-ubiquitin chains and consequently degraded by the 26S proteasome (Vembar and Brodsky, 2008). CTA1 escapes ubiquitin-dependent proteasomal degradation because it has an arginine-over-lysine amino acid bias which restricts the number of potential sites for ubiquitination (Hazes and Read, 1997; Rodighiero et al., 2002). The disordered conformation of CTA1 renders it susceptible to ubiquitin-independent proteasomal degradation by the core 20S proteasome, but the relatively slow turnover of cytosolic CTA1 (t½ = 2 hr) allows the toxin to persist long enough to elicit a cytopathic effect (Pande et al., 2007). Thus, the structure of the CTA1 polypeptide allows it to both exploit and evade ERAD.
CTA1 enters the cytosol in an unfolded state as it passes through one or more protein-conducting channels in the ER membrane (Bernardi et al., 2008; Bernardi et al., 2010; Dixit et al., 2008; Saslowsky et al., 2010; Schmitz et al., 2000). The translocated pool of CTA1 must therefore regain a folded conformation in order to act upon its Gsα target which is located in lipid rafts at the cytoplasmic face of the plasma membrane (Allen et al., 2007; Kamata et al., 2008; Oh and Schnitzer, 2001). Renaturation will not occur spontaneously in the absence of cellular factors, and CTA1 alone has little to no in vitro activity at 37°C (Murayama et al., 1993; Ray et al., 2012). We recently demonstrated that lipid rafts exhibit a chaperone-like activity that places disordered CTA1 in a folded conformation with catalytic activity at 37°C (Ray et al., 2012). Moreover, intact lipid rafts were required for the in vivo activity of cytosolic CTA1. Lipid rafts thus represent a host factor that allows cytosolic CTA1 to attain an ordered, active structure. Yet lipid rafts did not act through allostery: addition of lipid rafts to the folded CTA1 polypeptide at 25°C did not enhance toxin activity. The raft-induced gain of toxin function at 37°C thus appeared to result from a simple gain-of-structure which placed CTA1 in a folded conformation with basal activity. Optimal stimulation of CTA1 likely requires an allosteric interaction with host ADP-ribosylation factors (ARFs) (Welsh et al., 1994).
ARF proteins are small molecular weight GTPases that were originally isolated from cell extracts as in vitro allosteric activators of CTA1 (Kahn and Gilman, 1984, 1986; Tsai et al., 1987; Tsai et al., 1988). There are six members of the ARF family grouped in three classes, and each class can activate CTA1 (Moss and Vaughan, 1995; Price et al., 1992; Welsh et al., 1994). ARF6 can bind to CTA1 over a range of temperatures in either the absence or presence of GTP (Pande et al., 2007), but only ARF/GTP can stimulate the ADP-ribosyltransferase activity of CTA1 (Bobak et al., 1990; Kahn and Gilman, 1984, 1986; Price et al., 1992; Tsai et al., 1987). A crystal structure of ARF6/GTP bound to CTA1 at 25°C indicated that the stimulation of toxin activity results from an ARF-induced change to the conformation of CTA1 which allows NAD (the donor molecule for the ADP-ribosylation reaction) to access the toxin active site (O’Neal et al., 2005). The importance of CTA1-ARF interactions for in vivo toxin activity has yet to be established, however.
The allosteric activation of CTA1 by ARF is well-established. Yet it is unclear whether ARF6 can activate a disordered CTA1 polypeptide, or if ARF6 can only activate a structured CTA1 subunit such as one present in the 25°C co-crystal of ARF6 and CTA1 (O’Neal et al., 2005). The standard in vitro assay for CTA1 activity is performed at 25°C or 30°C rather than the physiological temperature of 37°C (Bobak et al., 1990; Haun et al., 1993; Kahn and Gilman, 1984, 1986; Lai et al., 1983; Teter et al., 2006; Tsai et al., 1987; Tsai et al., 1988). This is because the isolated CTA1 polypeptide has a disordered conformation with minimal enzymatic activity against Gs or synthetic substrates at 37°C (Murayama et al., 1993; Ray et al., 2012). When CTA1-ARF interactions have been examined at 37°C, CTA1 and ARF are typically added together before incubation at 37°C (Murayama et al., 1993; Pande et al., 2007). This protocol allows ARF6 to stabilize the folded conformation of CTA1 initially present at low temperature, but it does not determine whether ARF6 can activate the disordered CTA1 subunit. In this paper we examined whether ARF6 can activate a disordered, as well as a structured, CTA1 polypeptide. A biochemical and biophysical analysis of CTA1 structure / function indicated that ARF6 cannot promote the refolding of disordered CTA1 to an active state. Instead, lipid rafts shifted the disordered, 37°C structure of CTA1 to a folded conformation with a basal level of activity that could be further stimulated by ARF6. In vivo toxin activation by ARF is apparently essential for productive intoxication, as enzymatically active CTA1 mutants that cannot be further stimulated by ARF6 fail to elicit a cytopathic effect from cultured cells. These observations provide new molecular details for the role of ARF6 in CTA1 activation and indicate additional host factors such as lipid rafts must place CTA1 in the folded conformation required for ARF-stimulated toxin activity.
Results
Lipid rafts but not ARF6 shift disordered CTA1 from a protease-sensitive to protease-resistant conformation
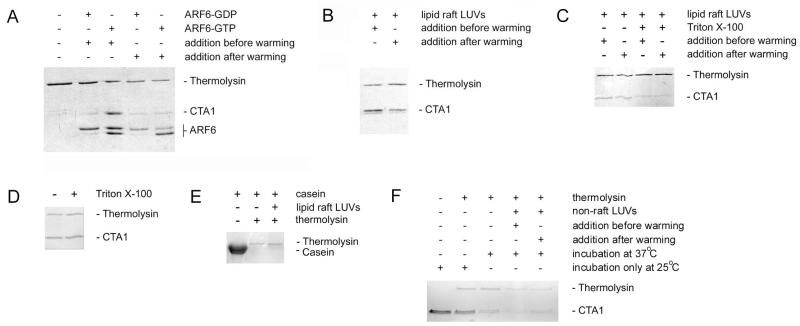
CTA1-ARF6 interactions were initially examined with a protease sensitivity assay (Fig. 1). The isolated CTA1 polypeptide shifts from a protease-resistant conformation to a protease-sensitive conformation with increasing temperature, which is consistent with its intrinsic thermal instability (Pande et al., 2007). ARF6/GTP but not ARF6/GDP prevented the temperature-induced transition of CTA1 to a protease-sensitive conformation (Fig. 1A). This confirmed our previous report (Pande et al., 2007) and demonstrated the active form of ARF6 can stabilize the folded, protease-resistant conformation of CTA1. However, our previous study did not determine whether ARF6/GTP could protect CTA1 from proteolysis when added to the unfolded toxin. To address this issue, we heated CTA1 to 37°C before the addition of ARF6. Under this condition, neither ARF6/GDP nor ARF6/GTP protected CTA1 from proteolysis. The presence of active ARF6 was confirmed by the generation of an ARF6 doublet which, as previously reported (Pande et al., 2007), results from proteolytic nicking of ARF6/GTP but not ARF6/GDP. The same GTP-bound ARF6 protein thus produced a different result based on the starting temperature of the experiment: ARF6/GTP protected CTA1 from proteolysis when added at a low temperature (25°C) that maintained CTA1 in a folded conformation, but ARF6/GTP did not protect CTA1 from proteolysis when added at a higher temperature (37°C) that generated an unfolded toxin conformation. This observation provided an internal control demonstrating ARF6/GTP does not directly inhibit the activity of the thermolysin protease, as CTA1 was degraded by thermolysin in the presence of ARF6/GTP when ARF6/GTP was added to CTA1 after the toxin had been warmed to 37°C. Our results suggested ARF6/GTP can stabilize a folded CTA1 polypeptide but cannot induce a gain-of-structure in the disordered CTA1 polypeptide. The inactive, GDP-bound form of ARF6 could neither stabilize folded CTA1 nor induce a gain-of-structure in disordered CTA1. The different effects of ARF6/GDP vs. ARF6/GTP on CTA1 structure could not be attributed to a lack of interaction between ARF6/GDP and CTA1, as previous studies have demonstrated ARF6 can bind to CTA1 at 25°C and 37°C in either the absence or presence of GTP (Pande et al., 2007).
Figure 1.
ARF6- or lipid raft-induced alterations to the structure of CTA1.
A. As indicated, equimolar amounts of ARF6/GDP or ARF6/GTP were added to either folded CTA1 before warming to 37°C or to CTA1 disordered by warming for 30 min at 37°C. Following a total of 1 h at 37°C, toxin samples were placed on ice and exposed to the protease thermolysin for 1 h at 4°C. Proteins were detected by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie staining. CTA1 was initially present in all samples. The addition of GTP induces a conformational change in ARF6 that renders it susceptible to nicking by thermolysin.
B. Lipid raft LUVs were added to either folded CTA1 before warming to 37°C or to CTA1 disordered by warming for 30 min at 37°C. Following a total of 1 h at 37°C, toxin samples were placed on ice and exposed to thermolysin for 1 h at 4°C. Proteins were detected by SDS-PAGE with Coomassie staining.
C. Lipid raft LUVs in the absence or presence of 1% Triton X-100 were added to either folded CTA1 before warming to 37°C or to CTA1 disordered by warming for 30 min at 37°C. Following a total of 1 h at 37°C, toxin samples were placed on ice and exposed to thermolysin for 1 h at 4°C. Proteins were detected by SDS-PAGE with Coomassie staining.
D. CTA1 was incubated at 25°C for 1 h in the presence or absence of 1% Triton X-100. Toxin samples were then placed on ice and exposed to thermolysin for 1 h at 4°C. Proteins were detected by SDS-PAGE with Coomassie staining.
E. Samples of α-casein were incubated for 1 h at 4°C with thermolysin in the absence or presence of lipid raft LUVs. Proteins were then detected by SDS-PAGE with Coomassie staining.
F. Non-raft LUVs mimicking the charge and fluidity of the plasma membrane were added to either folded CTA1 before warming to 37°C or to CTA1 disordered by warming for 30 min at 37°C. Additional CTA1 samples were incubated at 37°C or 25°C in the absence of LUVs. Following a total of 1 h at the indicated temperature, toxin samples were placed on ice and exposed to thermolysin for 1 h at 4°C. Proteins were detected by SDS-PAGE with Coomassie staining.
In contrast to ARF6, large unilamellar vesicles (LUVs) mimicking the composition of a lipid raft could protect CTA1 from proteolysis even when they were added after toxin unfolding had already occurred (Fig. 1B). This was consistent with previous biophysical measurements demonstrating the chaperone-like ability of lipid rafts to stabilize folded CTA1 and to induce both a gain-of-structure and gain-of-function in the disordered CTA1 polypeptide (Ray et al., 2012). ARF6 and lipid rafts thus exhibited overlapping yet distinct interactions with CTA1: both could prevent the thermal unfolding of CTA1 to a protease-sensitive conformation, but only lipid rafts could shift disordered CTA1 back to a protease-resistant structure.
Solubilization of the lipid raft LUVs with 1% Triton X-100 eliminated the protective lipochaperone effect against CTA1 proteolysis (Fig. 1C). Exposure of folded CTA1 to Triton X-100 at 25°C did not shift the toxin to a protease-sensitive conformation (Fig. 1D), so the proteolysis of CTA1 in the presence of LUVs and Triton X-100 most likely resulted from an effect of the detergent on the LUVs rather than the toxin. Lipid raft LUVs did not prevent the degradation of α-casein (Fig. 1E), a protein with an open conformation (Swaisgood, 1993) that is susceptible to proteolysis at all temperatures. Thus, the stabilizing and/or renaturation effect of LUVs were due to an effect on the toxin rather than the protease.
Non-raft LUVs mimicking the charge and fluidity of the plasma membrane neither stabilized nor refolded CTA1: the toxin was in a protease-sensitive conformation when exposed to these LUVs either before or after warming CTA1 to 37°C (Fig. 1F). This was comparable to the result obtained for a CTA1 sample incubated at 37°C in the absence of LUVs. In contrast, incubation of CTA1 alone at 25°C did not result in subsequent toxin degradation by thermolysin. These collective observations indicated lipid rafts represent the relevant structure for the lipochaperone effect which generates a gain-of-structure in disordered CTA1.
CTA1 is stabilized but not refolded by ARF6
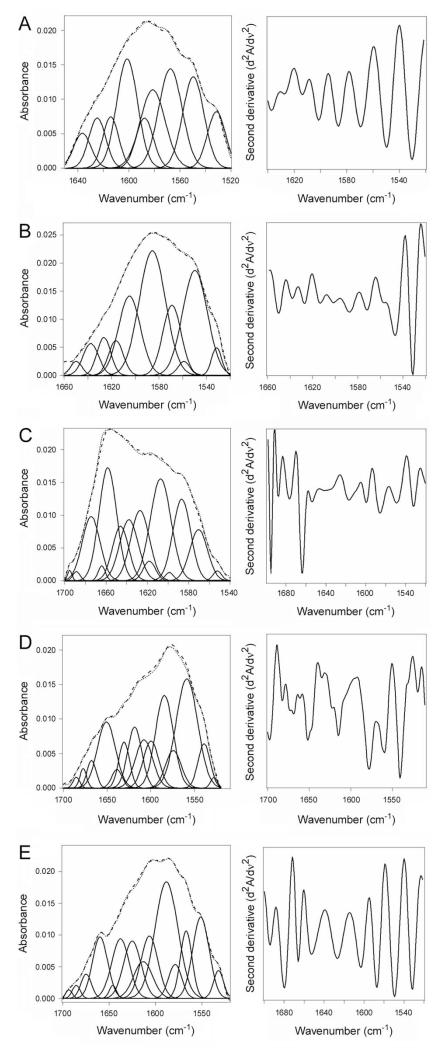
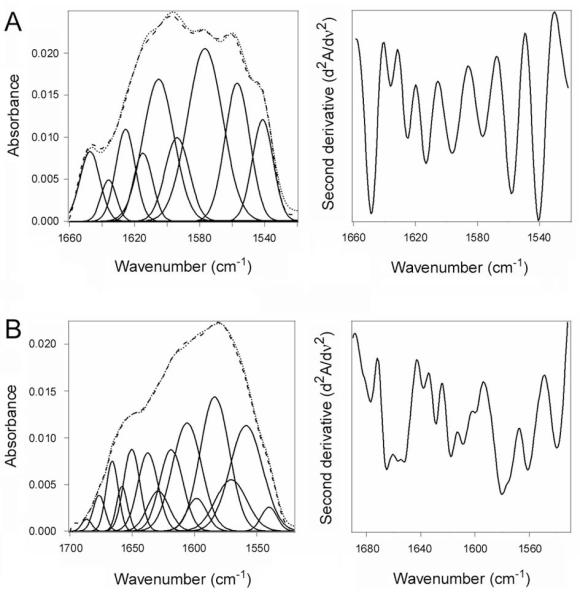
Isotope-edited Fourier transform infrared (FTIR) spectroscopy was used to directly examine the impact of ARF6 on the structure and stability of CTA1 (Fig. 2). FTIR absorption spectra of proteins contain a number of amide bands, among which the amide I mode in the 1700-1600 cm−1 region is of particular interest because of its sensitivity to the protein’s secondary structure (Tatulian, 2013). The amide I modes of α-helix, β-sheet, and unordered conformations occur in distinct spectral ranges of 1652 ± 4 cm−1, 1633 ± 5 cm−1, and 1644 ± 4 cm−1, respectively. Various types of turns absorb in the higher frequency range of 1700-1660 cm−1. The secondary structure content of a protein is evaluated by deconvolution of the amide I band, i.e. identification of the contributions of different structures in the total amide I band area, which can be facilitated by determination of the precise locations of the components from the second derivative spectra. Moreover, in cases when two proteins are combined in one sample, the structures of both can be determined by isotope-edited FTIR in which one of the proteins is uniformly labeled with 13C. 13C labeling is a common procedure that does not alter the conformation or function of a protein. However, the heavier nuclear mass of the stable 13C isotope generates a spectral downshift which allows the FTIR spectrum of a 13C-labeled protein to be resolved from the spectra of unlabeled proteins (Tatulian et al., 2002; Tatulian et al., 2005; Tatulian, 2010; Taylor et al., 2014). We have previously employed this approach and have reported the FTIR amide I band of 13C-labeled CTA1 exhibits a 45-50 cm−1 downshift in comparison to the spectrum of unlabeled CTA1. No structural differences between 13C-labeled CTA1 and unlabeled CTA1 were detected by far-UV circular dichroism (CD) over a range of temperatures (Taylor et al., 2011b).
Figure 2.
CTA1 structure in the absence or presence of ARF6.
Curve fitting (left panels) and second derivatives (right panels) for the FTIR spectrum of 13C-lableled CTA1 are shown. For curve fitting, the dotted line represents the sum of all deconvoluted components (solid lines) from the measured spectrum (dashed line). All spectra were recorded at 37°C with the exception of the 10°C spectrum presented in panel A.
A. CTA1 was incubated at 10°C for 30 min.
B. CTA1 was incubated at 37°C for 30 min.
C. CTA1 was mixed with ARF6/GTP at 4°C before warming to 37°C for 30 min.
D. CTA1 was mixed with ARF6/GDP at 4°C before warming to 37°C for 30 min.
E. CTA1 was incubated at 37°C for 30 min before ARF6/GTP was added for 1 h at 37°C.
CTA1 is in a folded conformation at low temperatures (Pande et al., 2007). We therefore used the FTIR spectrum of CTA1 at 10°C as a reference for the structure of folded CTA1 (Fig. 2A). Analysis of the 10°C FTIR spectrum from CTA1 indicated that the folded toxin contains 35 ± 4% α-helical and 49 ± 3% β-sheet content (Table 1). This result was consistent with our previous measurements (Taylor et al., 2011b) and the crystal structure of the folded, holotoxin-associated CTA1 subunit (PDB 1S5F) (O’Neal et al., 2004). Due to its intrinsic thermal instability, heating CTA1 to 37°C resulted in a loss of both α-helical and β-sheet structures (Fig. 2B). Concomitant with this loss of secondary structure, the percentage of CTA1 irregular structure shifted from 10 ± 3% at 10°C to 55 ± 2% at 37°C (Table 1). When ARF6/GTP was added to CTA1 before heating to 37°C, the toxin retained its full α-helical structure and a substantial amount of its β-sheet structure at 37°C (Fig. 2C, Table 1). There was some temperature-induced loss of CTA1 β-sheet structure in the presence of ARF6/GTP, but the toxin still retained more β-sheet structure in the presence of ARF6/GTP than in the absence of ARF6 (24% vs. 15%; Table 1). The impact of ARF6/GTP on CTA1 α-helical and β-sheet structure thus represents a partial stabilization of the folded toxin, and this is sufficient to maintain folded CTA1 in a protease-resistant conformation (Fig. 1A). No stabilizing effect on either CTA1 α-helical or β-sheet structure was seen with ARF6/GDP (Fig. 2D, Table 1), which demonstrated the specificity of the ARF6/GTP interaction and was consistent with the results of our protease sensitivity assay (Fig. 1A). Addition of ARF6/GTP to CTA1 after heating to 37°C did not induce a gain-of-structure in the disordered toxin (Fig. 2E). Under this condition, ARF-treated toxin resembled the untreated 37°C toxin and contained 54 ± 1% irregular structure with only 19 ± 1% α-helical and 17 ± 3% β-sheet content (Table 1). The same ARF6 protein thus produced different structural effects depending on which guanine nucleotide and which conformation of CTA1 was present. Additional experiments using a four-fold molar excess of ARF6 confirmed that ARF6/GTP does not lower the percentage of irregular structure in the disordered CTA1 subunit (data not shown). Collectively, these results demonstrated ARF6/GTP can partially stabilize the folded conformation of CTA1 but cannot promote the refolding of disordered CTA1.
Table 1.
Effect of ARF6 and/or lipid rafts on the structure of CTA1.
Deconvolution of the conformation-sensitive amide I bands from FTIR spectroscopy data presented in Figures 2 and 4 was used to calculate the percentages of CTA1 structure under the indicated conditions. The means ± standard deviations from three separate curve-fitting iterations are shown.
| % of CTA1 Structure |
||||
|---|---|---|---|---|
| Condition | α-Helix | β-Sheet | Irregular | Other |
| 10°C | 35 ± 4 | 49 ± 3 | 10 ± 3 | 7 ± 2 |
| 37°C | 26 ± 1 | 15 ± 2 | 55 ± 2 | 4 ± 1 |
| 37°C, ARF6/GTP added at 25°C | 35 ± 1 | 24 ± 3 | 27 ± 2 | 14 ± 2 |
| 37°C, ARF6/GDP added at 25°C | 21 ± 1 | 13 ± 4 | 55 ± 2 | 11 ± 1 |
| 37°C, ARF6/GTP added at 37°C | 19 ± 1 | 17 ± 3 | 54 ± 1 | 9 ± 4 |
| 37°C, lipid rafts added at 37°C | 33 ± 3 | 37 ± 2 | 18 ± 2 | 12 ± 1 |
| 37°C, lipid rafts and ARF6/GTP added at 37°C | 36 ± 4 | 48 ± 3 | 8 ± 1 | 8 ± 1 |
Folded but not disordered CTA1 is activated by ARF6
Our biochemical and biophysical data found that ARF6/GTP induces structural alterations in the folded but not disordered CTA1 subunit. This indicated ARF6/GTP would serve as an allosteric activator for folded but not disordered CTA1. An in vitro enzymatic assay was used to test this prediction (Fig. 3). At 25°C, the folded CTA1 subunit displayed a basal level of ADP-ribosyltransferase activity that was stimulated by ARF6/GTP but not ARF6/GDP (Fig. 3A). Our data recorded a 2-fold stimulation of CTA1 activity by ARF6/GTP, which was consistent with the 2-3 fold level of stimulation commonly reported for ARF/GTP-CTA1 interactions involving equimolar protein concentrations and no other additions (Bobak et al., 1990; Murayama et al., 1993; Price et al., 1992; Tsai et al., 1987; Tsai et al., 1988). ARF6/GTP could stimulate CTA1 activity at 37°C when mixed with the folded toxin before heating to 37°C. However, CTA1 remained inactive when ARF6/GTP was added to the unfolded toxin at 37°C (Fig. 3B). These results strongly suggested that, in vivo, ARF must be presented with a folded CTA1 polypeptide in order to act as an allosteric activator for the toxin.
Figure 3.
CTA1 activity in the absence or presence of ARF6.
For both panels, toxin activity against a diethylamino(benzylidine-amino)guanidine (DEA-BAG) substrate was detected from the increase in fluorescent units.
A. Two-fold dilutions of CTA1 were incubated at 25°C with ARF6/GTP (filled circles), ARF6/GDP (open circles), or in the absence of ARF6 (squares). Data are presented as the means ± standard errors of the means (SEMs) of 8 samples from two independent experiments.
B. Two-fold dilutions of CTA1 were incubated at 37°C in the presence of ARF6/GTP. Open squares represent toxin samples that were pre-incubated with ARF6/GTP at 25°C before warming to 37°C; filled squares represent toxin samples that were heated to 37°C for 30 min before the addition of ARF6/GTP. Data are presented as the means ± SEMs of 12 samples from three independent experiments.
Refolding of disordered CTA1 by lipid rafts is enhanced in the presence of ARF6
The lipochaperone function of lipid rafts (Ray et al., 2012) could act in conjunction with ARF to place disordered CTA1 in a folded, active conformation. This possibility was examined by FTIR spectroscopy (Fig. 4). CTA1 was heated to 37°C for 15 minutes before exposure to lipid raft-mimicking LUVs. Measurements of toxin structure were then recorded after an additional 30 minutes at 37°C (Fig. 4A). In the presence of lipid raft LUVs, the disordered conformation of CTA1 gained both α-helical and β-sheet content (Table 1). An additional gain of β-sheet structure and a further loss of irregular structure were detected when the disordered CTA1 subunit was incubated with both lipid rafts and ARF6/GTP (Fig. 4B). Under this condition, CTA1 exhibited an ordered conformation that matched the structural content of the folded, 10°C toxin (Table 1). The difference between raft-treated and ARF6/raft-treated CTA1 secondary structure content was rather modest because CTA1 gained a substantial amount of α-helix and β-sheet structure in the presence of lipid rafts alone; the additional gain-of-structure found in ARF6/raft-treated CTA1 simply returned the remaining minor percentage of disordered toxin structure to its native content. Thus, ARF6/GTP enhanced the raft-induced refolding of CTA1 and thereby allowed the disordered toxin to fully regain its native content of secondary structure.
Figure 4.
Lipid raft-induced alterations to the structure of disordered CTA1 in the absence or presence of ARF6.
Curve fitting (left panels) and second derivatives (right panels) for the FTIR spectrum of 13C-lableled CTA1 are shown. For curve fitting, the dotted line represents the sum of all deconvoluted components (solid lines) from the measured spectrum (dashed line).
A. CTA1 mixed with lipid raft LUVs after warming to 37°C.
B. CTA1 mixed with lipid raft LUVs and ARF6/GTP after warming to 37°C.
The raft-induced refolding of CTA1 facilitates further toxin activation by ARF6
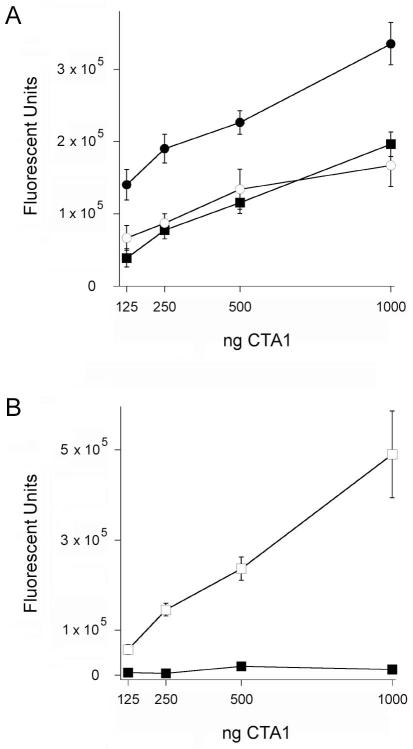
Lipid rafts might present ARF6 with a folded CTA1 polypeptide for allosteric activation at 37°C. Additional in vitro enzymatic assays were performed to examine this possibility (Fig. 5A). CTA1 was heated to 37°C before exposure to lipid raft LUVs or both ARF6/GTP and lipid raft LUVs. In contrast to ARF6/GTP (Fig. 3B), lipid rafts induced a gain-of-function in the disordered CTA1 polypeptide which produced a dose-dependent increase in CTA1 activity at 37°C. Higher levels of toxin activity were obtained when ARF6/GTP was allowed to bind and stabilize the folded, 25°C conformation of CTA1 before heating to 37°C. However, the greatest level of toxin activity was observed when a combination of ARF6/GTP and lipid rafts was added to the (initially) disordered CTA1 polypeptide at 37°C. These collective observations indicated that, at 37°C, lipid rafts place CTA1 in a folded conformation with a basal level of activity that can be enhanced through ARF-mediated allosteric activation. Lipid rafts alone do not appear to act through allostery, as addition of lipid rafts to the folded, 25°C conformation of CTA1 does not result in enhanced toxin activity (Ray et al., 2012). ARF proteins and lipid rafts thus play distinct but complementary roles in the activation of cytosolic CTA1.
Figure 5.
Raft-induced alterations to the activity of CTA1 in the absence or presence of ARF6.
A. Two-fold dilutions of CTA1 were incubated at 37°C with ARF6/GTP (squares), lipid raft LUVs (circles), or both lipid raft LUVs and ARF6/GTP (diamonds). Open symbols represent toxin samples that were pre-incubated with ARF6/GTP at 25°C before warming to 37°C; filled symbols represent toxin samples that were heated to 37°C for 30 min before the addition of LUVs or LUVs and ARF6/GTP. Toxin activity against a DEA-BAG substrate was detected from the increase in fluorescent units. Data are presented as the means ± SEMs of 8 samples from two independent experiments.
B. Two-fold dilutions of CTA1 were incubated at 37°C with ARF6/GTP (squares), non-raft LUVs (circles), or both non-raft LUVs and ARF6/GTP (diamonds). Open symbols represent toxin samples that were pre-incubated with ARF6/GTP at 25°C before warming to 37°C; filled symbols represent toxin samples that were heated to 37°C for 30 min before the addition of LUVs or LUVs and ARF6/GTP. Toxin activity against a DEA-BAG substrate was detected from the increase in fluorescent units. Four samples were used for each condition; data are presented as the means ± SEMs.
To ensure lipid rafts were the relevant structure for the gain-of-function in CTA1 activity at 37°C, we repeated the ADP-ribosylation assay with non-raft LUVs mimicking the charge and fluidity of the plasma membrane. As shown in Figure 5B, non-raft LUVs added to disordered CTA1 at 37°C did not produce a gain-of-function in either the absence or presence of ARF6/GTP. This was consistent with the results from our protease sensitivity assay (Fig. 1F) and previous biophysical studies which indicated plasma membrane LUVs do not refold CTA1 (Ray et al., 2012). Lipid rafts, where the G protein target of CTA1 is located (Allen et al., 2007; Kamata et al., 2008; Oh and Schnitzer, 2001), thus play a specific role in the refolding and activation of disordered CTA1.
CT Y149 point mutations block ARF-stimulated toxin activity and in vivo intoxication
The functional link between ARF-stimulated CTA1 activity and CT intoxication was examined with the use of two CT mutants that each contained a single amino acid substitution at residue Y149: CT Y149A and CT Y149S. The Y149 amino acid residue is not part of the CTA1 active site but instead represents a location for multiple contacts with ARF6 (O’Neal et al., 2005). Thus, point mutations in the Y149 residue were predicted to disrupt CTA1-ARF interactions without eliminating the basal enzymatic activity of the toxin. We chose this approach to study CTA1-ARF interactions because any ARF protein can activate CTA1 (Price et al., 1992; Welsh et al., 1994), and it is not practical to eliminate the in vivo expression or activity of all ARFs. The established role of ARF1 in CT trafficking to the ER (Morinaga et al., 2001; Richards et al., 2002) would further complicate studies of CTA1-ARF interactions in the cytosol, as the loss of ARF1 function would disrupt an event upstream of toxin activity in the cytosol.
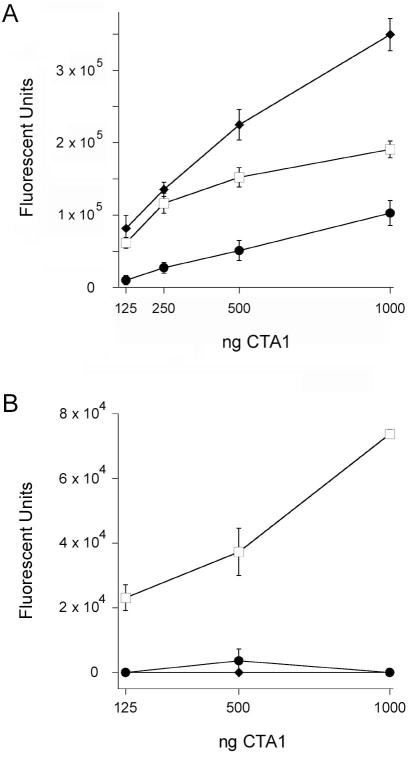
We found that ARF6 stimulated the in vitro activity of wild-type CTA1 but did not enhance the basal activities of CT Y149A or CT Y149S (Fig. 6A). Although both CT Y149A and CT Y149S exhibited in vitro enzymatic activity (albeit at less than wild-type levels), neither mutant toxin elicited a cAMP response from intoxicated CHO cells (Fig. 6B). A detection system based on surface plasmon resonance (SPR) demonstrated the A1 subunits from both CT mutants entered the host cytosol with approximately equal efficiency to the A1 subunit from wild-type toxin (Fig. 6C). Thus, the lack of in vivo toxicity for CT Y149A and CT Y149S could not be attributed to defects in toxin trafficking or translocation to the cytosol. These experiments strongly suggest the cytosolic activity of CTA1 requires allosteric activation by an ARF protein.
Figure 6.
CT Y149A and CT Y149S mutants are not activated by ARF6 and lack cytopathic activity.
A. Wild-type and mutant CT holotoxins were mixed with 2 mM DEA-BAG in the absence (grey bars) or presence (black bars) of ARF6/GTP for 2 h at 30°C. The ADP-ribosylation of DEA-BAG was then assessed by fluorometry; increasing fluorescent units correspond to increasing levels of ADP ribosylation. Background-subtracted data are presented as the averages ± ranges of two replicate samples per condition. One of three representative experiments is shown.
B. CHO cells were challenged with the stated concentrations of wild-type CT (filled circles), CT Y149A (open circles), or CT Y149S (open squares). Cell extracts generated after 2 h of continual toxin exposure were then screened for cAMP content. Data are presented as the means ± SEMs of 4 independent experiments with triplicate samples.
C. CHO cells pulse-labeled at 4°C with wild-type or mutant CT were subsequently chased at 37°C in toxin-free medium for 2 h. The cytosolic fractions extracted from intoxicated cells were perfused over a SPR sensor coated with an anti-CTA1 antibody. Unintoxicated cells and cells exposed to wild-type CT in the presence of brefeldin A (BfA), a drug that prevents toxin delivery to the cytosol, were used as negative controls. Known quantities of CTA (0.1 and 1 ng/mL) were used as positive controls. One of three representative experiments is shown.
Discussion
In order to reach the host cytosol, CTA1 undergoes what Ampapathi et. al. (2008) termed an order-disorder-order transition. The ordered CTA1 subunit travels from the cell surface to the ER as part of an intact AB toxin (Wernick et al., 2010). CTA1 then shifts to a disordered conformation after its chaperone-assisted displacement from the holotoxin (Pande et al., 2007; Taylor et al., 2011b; Taylor et al., 2014). Unfolding of the dissociated CTA1 polypeptide triggers its export to the cytosol through the quality control mechanism of ERAD (Banerjee et al., 2010; Massey et al., 2009; Taylor et al., 2011a). Because free CTA1 is an unstable protein, the translocated pool of toxin would remain in a disordered state at 37°C and must therefore engage components of the host cell to regain an ordered, active conformation. ARF proteins were originally thought to promote the ordering of cytosolic CTA1 (Ampapathi et al., 2008; Cho et al., 2012; Pande et al., 2007). Here, we instead demonstrated that the activation of CTA1 by ARF6 requires an interaction with the folded CTA1 polypeptide. The allosteric interaction between ARF6 and CTA1 thus appears to occur at the end of the order-disorder-order transition.
ARF6 can bind to both ordered and disordered conformations of CTA1 in either the absence or presence of GTP (Pande et al., 2007). However, the outcome of this interaction depends upon both the initial conformation of CTA1 and the state of the ARF-bound guanine nucleotide. The in vitro association between CTA1 and either ARF6/GTP or ARF6/GDP will protect the disordered toxin from ubiquitin-independent degradation by the 20S proteasome (Pande et al., 2007). In contrast, only ARF6/GTP will stimulate the ADP-ribosyltransferase activity of CTA1 (Bobak et al., 1990; Kahn and Gilman, 1984, 1986; Tsai et al., 1987). ARF6/GDP can bind to CTA1, but it does not induce appreciable changes in the secondary structure or thermal stability of CTA1 (Fig. 2D). GTP binding thus places ARF6 in a conformation that can both stabilize (Fig. 2C) and activate (Fig. 3A) CTA1. Yet ARF6/GTP can only impact the structure and function of folded CTA1: ARF6/GTP induces neither a gain-of-structure (Fig. 2E) nor a gain-of-function (Fig. 3B) in the disordered CTA1 polypeptide.
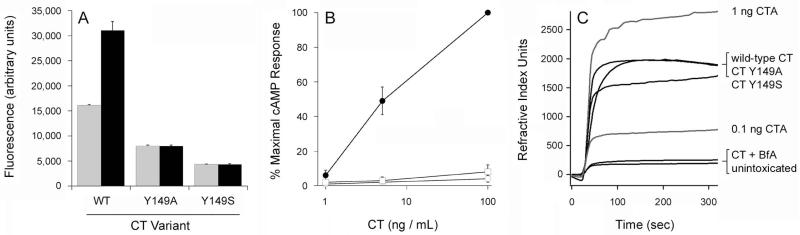
The inability of ARF6/GTP to stimulate the activity of disordered CTA1 cannot be attributed to the quality of our purified protein. Biophysical and biochemical studies confirmed the proper conformation of recombinant ARF6 (Fig. 7, Table 2), documented a structural difference between GDP- and GTP-bound ARF6 (Fig. 1A), and demonstrated the time- and concentration-dependent activation of CTA1 by ARF6/GTP but not ARF6/GDP (Figs. 3A, 8). Figures 1, 2, and 3 show the same ARF6/GTP protein yields a different result (for protease sensitivity, toxin structure, and toxin activity) based on the starting temperature of the experiment and, thus, the initial conformation of CTA1. Figures 1, 2, and 3 also show the same ARF6 protein yields a different result (for protease sensitivity, toxin structure, and toxin activity) based on the presence of either GDP or GTP. ARF6/GDP served as a negative control and allowed us to directly compare results obtained with ARF6 in the on (GTP) vs. off (GDP) state. Since ARF6 is a molecular switch, we have shown the specificity of the ARF6/GTP effects by demonstrating the same results were not obtained when ARF6 was in the inactive, GDP-bound state. The toxicity assays of Figures 3 and 5 further demonstrate ARF6/GTP can function at 37°C to enhance the activity of a previously stabilized CTA1 polypeptide. Our ARF6 protein thus produced the expected results at 25°C and at 37°C when mixed with the folded CTA1 subunit. Since ARF6/GTP functioned properly under multiple experimental conditions, its surprising inability to activate an unfolded CTA1 polypeptide at 37°C could not be attributed to problems with the ARF protein itself. It therefore appears that, contrary to the current model, ARF6/GTP requires additional host factors to stimulate CTA1 activity at the physiological temperature of 37°C.
Figure 7.
Structure of recombinant ARF6.
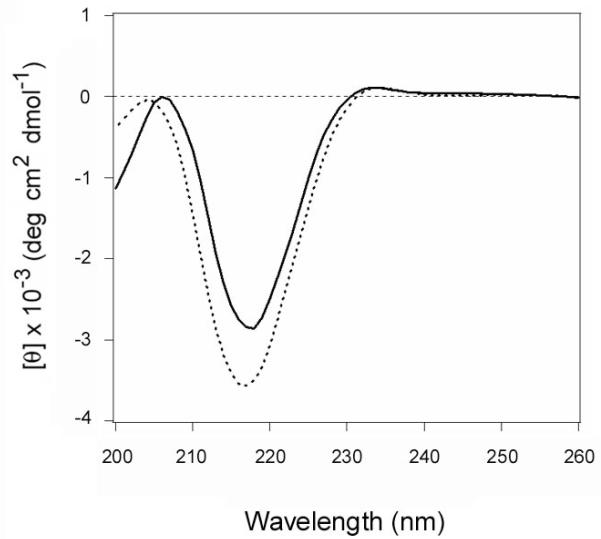
A. Far-UV CD was used to record the spectrum of ARF6 (50 μg) at 25°C in 220 μL of 10 mM sodium borate buffer (pH 7.0) containing 100 mM NaCl and excess GTP.
B. Curve fitting (left panel) and second derivatives (right panel) for the FTIR spectrum of ARF6/GTP are shown. For curve fitting, the dotted line represents the sum of all deconvoluted components (solid lines) from the measured spectrum (dashed line). The FTIR spectrum was recorded at 25°C in a D2O-based 10 mM sodium borate buffer (pH 7.0) containing 100 mM NaCl, 0.7 mM GTP, and 10 μM ARF6.
Table 2.
Measured and predicted secondary structure content of ARF6.
Deconvolution of the conformation-sensitive amide I bands from FTIR data presented in Figure 7 was used to calculate the percentages of ARF6 structure in the presence of GTP. The means ± standard deviations from three separate curve-fitting iterations are shown. The predicted secondary structure content of ARF6/GTP was derived from PDB 2A5G (O’Neal et al., 2005). n/a, not applicable: irregular structure would not be resolved by X-ray crystallography.
| % of ARF6 Structure |
||||
|---|---|---|---|---|
| α-Helix | β-Sheet | Irregular | Other | |
| ARF6/GTP Measurement | 58 ± 3 | 22 ± 1 | 10 ± 3 | 9 ± 4 |
| ARF6/GTP Prediction | 60 | 24 | n/a | 16 |
Figure 8.
Time- and concentration-dependent activation of CTA1 by ARF6/GTP.
A. 1 μg aliquots of CTA1 were incubated at 25°C in the presence of equimolar ARF6 and excess GTP. Toxin activity against the DEA-BAG substrate was detected from the increase in fluorescent units detected at the indicated time intervals. Four samples were used for each condition; data are presented as the means ± SEMs. One of two representative experiments is shown.
B. 1 μg aliquots of CTA1 were incubated at 25°C with the stated quantities of ARF6/GTP. Toxin activity against the DEA-BAG substrate was detected from the increase in fluorescent units after 2 h of incubation. Data are presented as the averages ± standard deviations from three replicate samples.
Allosteric activation of CTA1 by ARF6/GTP is thought to involve the ARF-induced stabilization of a CTA1 conformation in which the “activation loop” that occludes the active site in CT holotoxin is displaced to allow NAD and Gs to enter the active site (O’Neal et al., 2005). This mechanism and the findings reported in this study explain why ARF6/GTP alone cannot activate CTA1 at physiological temperature. Because CTA1 is disordered at 37°C and ARF6/GTP does not promote the refolding of disordered CTA1 (Fig. 2, Table 1), it is necessary for other host factors such as lipid rafts to participate in shifting CTA1 from a disordered conformation to a partially folded active state that is competent for functional interaction with ARFs. The gain-of-structure resulting from toxin-raft interactions allows ARF6/GTP to induce an additional conformational change (Fig. 4, Table 1) that presumably stabilizes displacement of the occluding CTA1 “activation loop” and consequently optimizes the enzymatic activity of CTA1 (Fig. 5A).
CTA1 mutants that cannot be activated by ARF6/GTP exhibit in vitro but not in vivo activity despite their delivery to the host cytosol (Fig. 6). Both CT Y149A and CT Y149S exhibited in vitro activity in the absence of ARF6, respectfully representing 50% and 27% of the basal enzymatic activity for wild-type CTA1. However, neither mutant was activated in vitro by ARF6/GTP: toxin activity against a DEA-BAG substrate was identical in both the absence and presence of ARF6/GTP. Furthermore, neither mutant toxin could elicit a cAMP response from intoxicated cells despite A1 subunit delivery to the cytosol. Allosteric activation of CTA1 by ARF6/GTP thus appears to be essential for in vivo toxin function, even when the toxin exhibits basal in vitro enzymatic activity in the absence of ARF.
ARF6/GTP can only enhance the activity of a folded CTA1 polypeptide, so the host factor or factors that present folded CTA1 to a GTP-bound ARF protein should therefore be essential for intoxication as well. Consistent with this model, we have reported that lipid rafts exhibit a chaperone-like interaction with CTA1 which is required for optimal in vivo toxin activity (Ray et al., 2012). Gs is located in lipid rafts at the cytoplasmic face of the plasma membrane (Allen et al., 2007; Kamata et al., 2008; Oh and Schnitzer, 2001), and, in vivo, GTP-bound ARF proteins associate with membranes by virtue of their N-terminal lipid modifications (Moss and Vaughan, 1995; Welsh et al., 1994). Thus, the host factors required for optimal CTA1 activity are in proximity to the Gs target. Our model affirms the key role of ARF proteins in CT intoxication, but it also proposes the CTA1-ARF interaction is more complex than originally thought: ARF does not refold the disordered CTA1 polypeptide, so the allosteric activation of CTA1 will only occur when additional host factors place CTA1 in a folded conformation that can be stimulated by ARF proteins. These novel observations provide additional insights about the molecular mechanisms required for allosteric activation of CTA1 by ARF6/GTP and identify lipid rafts as a host factor that can place CTA1 in the folded conformation required for ARF-stimulated toxin activity.
Experimental procedures
CTA1 proteins
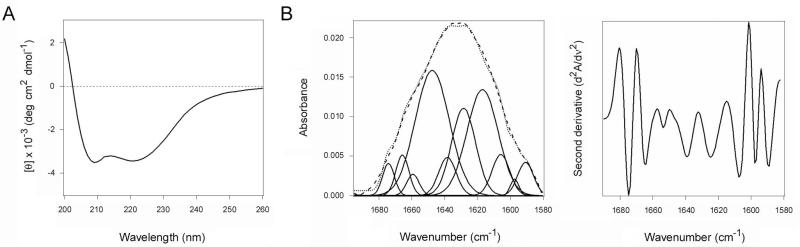
The CTA1/CTA2 heterodimer was purchased from Sigma-Aldrich (St. Louis, MO), while His-tagged CTA1 was expressed in Escherichia coli and purified in the lab (Banerjee et al., 2010; Massey et al., 2009; Teter et al., 2006). Both reduced CTA1 from the CTA1/CTA2 heterodimer and reduced, His-tagged CTA1 exhibit similar structures (Fig. 9) and thermal stabilities (Massey et al., 2009; Pande et al., 2007). 13C-labeling of CTA1-His6 was performed as previously described (Taylor et al., 2011b).
Figure 9.
Structures of reduced CTA1 from the CTA1/CTA2 heterodimer and the recombinant His-tagged protein.
A purified CTA1/CTA2 heterodimer was treated with 30 mM GSH and subjected to dialysis in 30 mM GSH buffer with a 3500 MWCO filter. Far-UV CD was then used to record the spectrum of the retained CTA1 subunit (solid line). A far-UV CD spectrum was also recorded for CTA1-His6 in the presence of 30 mM GSH (dashed line). Both measurements used 50 μg of toxin in 220 μL of 10 mM sodium borate buffer (pH 7.0) containing 100 mM NaCl. Measurements were recorded with a Jasco 810 spectrofluoropolarimeter at 18°C, and the spectra were averaged from three scans.
Purification of ARF6
E. coli strain BL21(pLysS) transformed with an inducible pT7ARF6 expression plasmid (generously provided by Dr. Joel Moss, National Heart, Lung, and Blood Institute, Bethesda, MD) was grown at room temperature for 4 h to a 0.6 OD600 in 500 mL Luria-Bertani broth containing 100 μg/mL of ampicillin. IPTG at a concentration of 1 mM was added to the growth medium for induction of ARF6 expression. At 4 h post-induction, the cells were pelleted and resuspended in lysis buffer (20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM MgCl2, 1 mM dithiothreitol, 2 mM EDTA, and 1 mg/mL lysozyme). After a 30 min incubation in lysis buffer, the cells were subjected to three freeze-thaw cycles. The lysate was clarified with a 45 min spin at 10,000 × g, and the soluble lysate was dialyzed against two changes of 4 L low-salt buffer (20 mM Tris (pH 7.6), 50 mM NaCl, 1 mM dithiothreitol, 1 mM MgCl2, and 2 mM EDTA). The sample was then run over a DEAE column, and the flow-through containing ARF6 was collected. The purity of eluted ARF6 was confirmed by SDS-PAGE and Coomassie staining.
CD and FTIR spectroscopy were used to examine the structure of the recombinant ARF6 protein (Fig. 7). The far-UV CD spectrum of ARF6 exhibited double minima centered at 208 and 222 nm that was indicative of a predominantly α-helical structure (Fig. 7A). Recombinant ARF1 produces a nearly identical far-UV CD spectrum (Honda et al., 2005). As assessed by FTIR spectroscopy, the secondary structure of ARF6/GTP contained 58% α-helix and 22% β-sheet content (Fig. 7B, Table 2). The measured secondary structure of purified ARF6 matched well with the secondary structure content predicted from the crystal structure of ARF6/GTP (PDB 2A5G) (O’Neal et al., 2005). As expected, titration experiments with ARF6/GTP documented its time- and dose-dependent stimulation of CTA1 enzymatic activity (Fig. 8). Our purified ARF6 thus exhibited the expected structural and biochemical properties.
The N-terminal myristoylation of ARF proteins is involved with their in vivo membrane association and the kinetics of GDP/GTP exchange (Franco et al., 1995; Haun et al., 1993; Welsh et al., 1994). Recombinant ARF proteins are not myristoylated, but they retain functional activity and are commonly used for structural studies and biochemical assays including CTA1 activation (Ampapathi et al., 2008; Haun et al., 1993; Menetrey et al., 2000; Moss and Vaughan, 1995; O’Neal et al., 2005; Pande et al., 2007; Pasqualato et al., 2001; Price et al., 1992; Teter et al., 2006; Weiss et al., 1989; Welsh et al., 1994). In addition, ARF6/GTP can serve as an allosteric activator of CTA1 in the absence of phospholipids (Price et al., 1992). The in vivo role of ARF myristoylation is therefore unlikely to impact the current, as well as previous, in vitro structure/function studies.
Saturating amounts (1 mM) of GDP or GTP were used to ensure ARF6 (≤ 10 μM for all experiments) was in an inactive (GDP-bound) or active (GTP-bound) conformation. Multiple, independent assays demonstrated this method was sufficient to convert ARF6 between GTP- and GDP-bound forms: in contrast to ARF6-GTP, ARF6-GDP did not (i) resolve as a protein doublet in SDS-PAGE after exposure to thermolysin (Fig. 1A); (ii) prevent the temperature-induced shift of CTA1 from a protease-resistant conformation to a protease-sensitive conformation (Fig. 1A); (iii) inhibit the thermal unfolding of CTA1 (Fig. 2C-D, Table 1); or (iv) enhance the enzymatic activity of CTA1 (Fig. 3A).
Generation of LUVs
The production of 100 nm LUVs was performed as previously described (Ray et al., 2012). LUVs mimicking the composition of lipid rafts contained 30% cholesterol, 30% palmitoyl sphingomyelin, 25% 1-Palmitoyl-2-Linoleoyl-sn-Glycero-3-Phosphoethanolamine, and 15% 1-Palmitoyl-2-Linoleoyl-sn-Glycero-3-Phospho-L-Serine, all from Avanti Polar Lipids (Alabaster, AL). Non-raft LUVs mimicking the charge and fluidity of the plasma membrane contained 15% 1-Palmitoyl-2-Linoleoyl-sn-Glycero-3-Phospho-L-Serine and 85% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids).
Construction and purification of CT mutants
A co-crystal of the CTA1 E110D/E112D mutant and ARF6/GTP indicated that the Y149 amino acid residue was an important site of contact for the ARF6 interface (O’Neal et al., 2005). We therefore constructed two CT mutants that each contained a single amino acid substitution at residue Y149: CT Y149A and CT Y149S. The CT holotoxin with a Y149A substitution was made by QuickChange mutagenesis of pARCT5 (Jobling and Holmes, 2000), a vector with an arabinose-inducible CT operon. The CT holotoxin with a Y149S substitution was generated by cloning an XbaI – ClaI fragment of a SOE-PCR (Horton et al., 1990) product encoding Y149S back into pARCT5. The following oligonucleotides (and their reverse complements, underlined bases changed) were used for these procedures: Y149S, GGGCTACAGAGATAGATCTTACAGTAACTTAG (adds BglII site); Y149A, GGGCTACAGAGATAGAGCTTACAGTAACTTAG (adds AluI site). Mutations were confirmed by restriction digest and DNA sequencing. Production and purification of recombinant holotoxin from arabinose-induced cultures was by Talon affinity chromatography essentially as described (Jobling and Holmes, 2001).
Protease sensitivity assay
The thermolysin protease sensitivity assay was performed as described in (Pande et al., 2007); 1 μg toxin samples, 1 μg ARF6, and 800 μM LUVs were used for the assay. 1 mM GTP or GDP was also present as indicated.
Isotope-edited FTIR spectroscopy
Experiments involving FTIR spectroscopy and isotope-edited FTIR spectroscopy were conducted and analyzed as described in (Taylor et al., 2011b). As detailed in (Tatulian, 2013), amide I components at the following wavenumbers were assigned to specific secondary structures for unlabeled ARF6 in a D2O-based buffer: 1652 ± 4 cm−1, α-helix; 1633 ± 7 cm−1, β-sheet; 1644 ± 4 cm−1, irregular. The higher frequency components between 1700-1660 cm−1 were assigned to turns and tabulated as “other” structures, while the components at the lower frequencies (<1620 cm−1) were assigned to side chains and excluded from the procedure of secondary structure evaluation (Tatulian, 2013; Taylor et al., 2011b). Isotope-edited FTIR spectroscopy used 10 μM 13C-labeled CTA1, 10 μM ARF6, and/or 800 μM lipid raft LUVs in 100 μL of a D20-based 10 mM sodium borate buffer (pD 7.0) containing 100 mM NaCl. 1 mM GTP or GDP was also present as indicated. Analysis of the FTIR spectra and deduction of the secondary structures of the unlabeled and 13C-labeled proteins has been described in great detail in the Supplemental Data of Taylor et al. (2011b) as well as elsewhere (Tatulian, 2013). Briefly, the spectra have been doubly differentiated to identify the locations (peak wavenumbers) of the amide I spectral components. For the unlabeled protein (i.e., ARF6), the components have been assigned to α-helix, β-sheet, irregular, or “other” structures according to the spectral ranges specified above. The content of each secondary structure in the protein was determined as the area of the corresponding component relative to the total amide I area, corrected for the respective extinction coefficient (Taylor et al., 2011b; Tatulian, 2013). Since uniform 13C-labeling of proteins results in a 45-50 cm−1 downshift of the amide I band (Taylor et al., 2011b; Tatulian 2010; Tatulian, 2013), the assignments of amide I components of the 13C-labeled CTA1 to secondary structure types were done in a similar manner, taking into account the spectral downshift. Finally, the secondary structures of combined unlabeled ARF6 and 13C-labeled CTA1 were estimated by dividing each amide I band fraction assigned to ARF6 or 13C-CTA1 by the fraction of the respective protein in the sample in terms of the number of amino acid residues, as described earlier in the Supplemental Data of Taylor et al. (2011b).
Measurement of in vitro toxin activity
DEA-BAG, a substrate for the ADP-ribosyltransferase activity of CTA1, was synthesized and used as previously described (Reddy et al., 2013). DEA-BAG loses its ability to bind AG-50W-X4 ion exchange resin after ADP-ribosylation and exhibits an intrinsic fluorescence (Soman et al., 1986), so the fluorescent signal remaining after pull-down with AG-50W-X4 resin represents the ADP-ribosylated pool of DEA-BAG. We have found different preparations of DEA-BAG yield different levels of background fluorescence, but the qualitative results are consistent between batches. Toxin was mixed with DEA-BAG for 2 h at 25°C or 37°C before measurements of fluorescent intensity were recorded with a Bio-TEK (Winooski, VT) plate reader. When indicated, CTA1 was heated to 37°C for 30 min before the addition of equimolar ARF6 and/or 800 μM lipid raft LUVs. 1 mM GTP or GDP was also present as indicated. Toxin samples were then incubated for another 60 min at 37°C before the 2 h exposure to DEA-BAG. As previously noted for CTA1 alone (Murayama et al., 1993; Ray et al., 2012), CTA1 disordered by warming to 37°C has little to no enzymatic activity upon subsequent exposure to ARF6/GTP: the signal recorded under this condition represented a background reading as it did not increase with increasing toxin concentrations and was no greater than the flat-line response obtained from heat-denatured CTA1 (Fig. 10). Background measurements obtained with heat-denatured CTA1 or CTA1 alone at 37°C were therefore subtracted from all experimental results before presentation of the data.
Figure 10.
ARF6/GTP enhances the activity of folded but not disordered CTA1.
Two-fold dilutions of CTA1 were incubated at 37°C in the presence of ARF6/GTP (squares). Open squares represent toxin samples that were pre-incubated with ARF6/GTP at 25°C before warming to 37°C; filled squares represent toxin samples that were heated to 37°C before the addition of ARF6/GTP. Circles represent two-fold dilutions of a CTA1 sample that was boiled for 10 min before placement at 37°C. Toxin activity against a DEA-BAG substrate at 37°C was then was detected from the increase in fluorescent units. Four samples were used for each condition; data are presented as the means ± SEMs.
Measurement of in vivo toxin activity
CHO cells grown to ~80% confluency in a 24-well plate were challenged for 2 h at 37°C with various concentrations of wild-type or mutant CT in serum-free medium. The cAMP content from intoxicated cells was then determined with a commercial kit according to the manufacturer’s instructions (GE Healthcare, Piscataway, NJ). The resting levels of cAMP from unintoxicated cells were background subtracted from the cAMP levels in toxin-challenged cells. The cAMP response elicited from 100 ng/mL of wild-type CT was then set at 100%, and all other cAMP data were expressed as percentages of this maximal value.
SPR
To detect the cytosolic pool of CTA1 from wild-type and mutant toxins, CHO cells grown to 80% confluency in 6-well plates were placed on ice for 30 min with 1 μg/mL of toxin. Unbound toxin was removed, and the cells were returned to 37°C in toxin-free medium for 2 h. Selective permeabilization of the plasma membrane with digitonin was followed by centrifugation to isolate the cytosolic fraction from CT-challenged cells. Perfusion of the cytosolic fraction over an SPR sensor coated with an anti-CTA1 antibody was subsequently used for toxin detection. Cells treated with BfA were used as a control to ensure CTA1 retained in the endomembrane system did not leak into the cytosolic fraction: BfA prevents the transport of endocytosed toxin to the ER (Lencer et al., 1993), so no CTA1 should be found in the cytosolic fraction of intoxicated, BfA-treated cells. A detailed protocol has been published in (Taylor et al., 2012).
Acknowledgement
This work was supported by NIH grants to K. Teter (R01 AI073783 and R01 AI099493) and R.K. Holmes (R01 AI031940).
References
- Allen JA, Halverson-Tamboli RA, Rasenick MM. Lipid raft microdomains and neurotransmitter signaling. Nat Rev Neurosci. 2007;8:128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- Ampapathi RS, Creath AL, Lou DI, Craft JW, Jr., Blanke SR, Legge GB. Order-disorder-order transitions mediate the activation of cholera toxin. J Mol Biol. 2008;377:748–760. doi: 10.1016/j.jmb.2007.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T, Pande A, Jobling MG, Taylor M, Massey S, Holmes RK, Tatulian SA, Teter K. Contribution of subdomain structure to the thermal stability of the cholera toxin A1 subunit. Biochemistry. 2010;49:8839–8846. doi: 10.1021/bi101201c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Forster ML, Lencer WI, Tsai B. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol Biol Cell. 2008;19:877–884. doi: 10.1091/mbc.E07-08-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Williams JM, Kikkert M, van Voorden S, Wiertz EJ, Ye Y, Tsai B. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol Biol Cell. 2010;21:140–151. doi: 10.1091/mbc.E09-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobak DA, Bliziotes MM, Noda M, Tsai SC, Adamik R, Moss J. Mechanism of activation of cholera toxin by ADP-ribosylation factor (ARF): both low- and high-affinity interactions of ARF with guanine nucleotides promote toxin activation. Biochemistry. 1990;29:855–861. doi: 10.1021/bi00456a600. [DOI] [PubMed] [Google Scholar]
- Cho JA, Chinnapen DJ, Aamar E, Welscher YM, Lencer WI, Massol R. Insights on the trafficking and retro-translocation of glycosphingolipid-binding bacterial toxins. Frontiers in Cellular and Infection Microbiology. 2012;2 doi: 10.3389/fcimb.2012.00051. doi: 10.3389/fcimb.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit G, Mikoryak C, Hayslett T, Bhat A, Draper RK. Cholera toxin up-regulates endoplasmic reticulum proteins that correlate with sensitivity to the toxin. Exp Biol Med (Maywood) 2008;233:163–175. doi: 10.3181/0705-RM-132. [DOI] [PubMed] [Google Scholar]
- Franco M, Chardin P, Chabre M, Paris S. Myristoylation of ADP-ribosylation factor 1 facilitates nucleotide exchange at physiological Mg2+ levels. J Biol Chem. 1995;270:1337–1341. doi: 10.1074/jbc.270.3.1337. [DOI] [PubMed] [Google Scholar]
- Haun RS, Tsai SC, Adamik R, Moss J, Vaughan M. Effect of myristoylation on GTP-dependent binding of ADP-ribosylation factor to Golgi. J Biol Chem. 1993;268:7064–7068. [PubMed] [Google Scholar]
- Hazes B, Read RJ. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry. 1997;36:11051–11054. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- Honda A, Al-Awar OS, Hay JC, Donaldson JG. Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J Cell Biol. 2005;168:1039–1051. doi: 10.1083/jcb.200409138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Jobling MG, Holmes RK. Identification of motifs in cholera toxin A1 polypeptide that are required for its interaction with human ADP-ribosylation factor 6 in a bacterial two-hybrid system. Proc Natl Acad Sci U S A. 2000;97:14662–14667. doi: 10.1073/pnas.011442598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MG, Holmes RK. Biological and biochemical characterization of variant A subunits of cholera toxin constructed by site-directed mutagenesis. J Bacteriol. 2001;183:4024–4032. doi: 10.1128/JB.183.13.4024-4032.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG. Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J Biol Chem. 1984;259:6228–6234. [PubMed] [Google Scholar]
- Kahn RA, Gilman AG. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J Biol Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- Kamata K, Manno S, Ozaki M, Takakuwa Y. Functional evidence for presence of lipid rafts in erythrocyte membranes: Gsalpha in rafts is essential for signal transduction. Am J Hematol. 2008;83:371–375. doi: 10.1002/ajh.21126. [DOI] [PubMed] [Google Scholar]
- Lai CY, Xia QC, Salotra PT. Location and amino acid sequence around the ADP-ribosylation site in the cholera toxin active subunit A1. Biochem Biophys Res Commun. 1983;116:341–348. doi: 10.1016/0006-291x(83)90420-5. [DOI] [PubMed] [Google Scholar]
- Lencer WI, de Almeida JB, Moe S, Stow JL, Ausiello DA, Madara JL. Entry of cholera toxin into polarized human intestinal epithelial cells. Identification of an early brefeldin A sensitive event required for A1-peptide generation. J Clin Invest. 1993;92:2941–2951. doi: 10.1172/JCI116917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey S, Banerjee T, Pande AH, Taylor M, Tatulian SA, Teter K. Stabilization of the tertiary structure of the cholera toxin A1 subunit inhibits toxin dislocation and cellular intoxication. J Mol Biol. 2009;393:1083–1096. doi: 10.1016/j.jmb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J, Macia E, Pasqualato S, Franco M, Cherfils J. Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nat Struct Biol. 2000;7:466–469. doi: 10.1038/75863. [DOI] [PubMed] [Google Scholar]
- Morinaga N, Kaihou Y, Vitale N, Moss J, Noda M. Involvement of ADP-ribosylation factor 1 in cholera toxin-induced morphological changes of Chinese hamster ovary cells. J Biol Chem. 2001;276:22838–22843. doi: 10.1074/jbc.M101184200. [DOI] [PubMed] [Google Scholar]
- Moss J, Vaughan M. Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- Murayama T, Tsai SC, Adamik R, Moss J, Vaughan M. Effects of temperature on ADP-ribosylation factor stimulation of cholera toxin activity. Biochemistry. 1993;32:561–566. doi: 10.1021/bi00053a022. [DOI] [PubMed] [Google Scholar]
- O’Neal CJ, Amaya EI, Jobling MG, Holmes RK, Hol WG. Crystal structures of an intrinsically active cholera toxin mutant yield insight into the toxin activation mechanism. Biochemistry. 2004;43:3772–3782. doi: 10.1021/bi0360152. [DOI] [PubMed] [Google Scholar]
- O’Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- Oh P, Schnitzer JE. Segregation of heterotrimeric G proteins in cell surface microdomains. G(q) binds caveolin to concentrate in caveolae, whereas G(i) and G(s) target lipid rafts by default. Mol Biol Cell. 2001;12:685–698. doi: 10.1091/mbc.12.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande AH, Scaglione P, Taylor M, Nemec KN, Tuthill S, Moe D, Holmes RK, Tatulian SA, Teter K. Conformational instability of the cholera toxin A1 polypeptide. J Mol Biol. 2007;374:1114–1128. doi: 10.1016/j.jmb.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato S, Menetrey J, Franco M, Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Welsh CF, Haun RS, Stanley SJ, Moss J, Vaughan M. Effects of phospholipid and GTP on recombinant ADP-ribosylation factors (ARFs). Molecular basis for differences in requirements for activity of mammalian ARFs. J Biol Chem. 1992;267:17766–17772. [PubMed] [Google Scholar]
- Ray S, Taylor M, Banerjee T, Tatulian SA, Teter K. Lipid rafts alter the stability and activity of the cholera toxin A1 subunit. J Biol Chem. 2012;287:30395–30405. doi: 10.1074/jbc.M112.385575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S, Taylor M, Zhao M, Cherubin P, Geden S, Ray S, Francis D, Teter K. Grape extracts inhibit multiple events in the cell biology of cholera intoxication. PLoS One. 2013;8:e73390. doi: 10.1371/journal.pone.0073390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AA, Stang E, Pepperkok R, Parton RG. Inhibitors of COP-mediated transport and cholera toxin action inhibit simian virus 40 infection. Mol Biol Cell. 2002;13:1750–1764. doi: 10.1091/mbc.01-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodighiero C, Tsai B, Rapoport TA, Lencer WI. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 2002;3:1222–1227. doi: 10.1093/embo-reports/kvf239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky DE, Cho JA, Chinnapen H, Massol RH, Chinnapen DJ, Wagner JS, De Luca HE, Kam W, Paw BH, Lencer WI. Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J Clin Invest. 2010;120:4399–4409. doi: 10.1172/JCI42958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A, Herrgen H, Winkeler A, Herzog V. Cholera toxin is exported from microsomes by the Sec61p complex. J Cell Biol. 2000;148:1203–1212. doi: 10.1083/jcb.148.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soman G, Narayanan J, Martin BL, Graves DJ. Use of substituted (benzylidineamino)guanidines in the study of guanidino group specific ADP-ribosyltransferase. Biochemistry. 1986;25:4113–4119. doi: 10.1021/bi00362a019. [DOI] [PubMed] [Google Scholar]
- Swaisgood HE. Review and update of casein chemistry. J Dairy Sci. 1993;76:3054–3061. doi: 10.3168/jds.S0022-0302(93)77645-6. [DOI] [PubMed] [Google Scholar]
- Tatulian SA, Chen B, Li J, Negash S, Middaugh CR, Bigelow DJ, Squier TC. The inhibitory action of phospholamban involves stabilization of alpha-helices within the Ca-ATPase. Biochemistry. 2002;41:741–751. doi: 10.1021/bi011148d. [DOI] [PubMed] [Google Scholar]
- Tatulian SA, Qin S, Pande AH, He X. Positioning membrane proteins by novel protein engineering and biophysical approaches. J Mol Biol. 2005;351:939–947. doi: 10.1016/j.jmb.2005.06.080. [DOI] [PubMed] [Google Scholar]
- Tatulian SA. Structural analysis of proteins by isotope-edited FTIR spectroscopy. Spectroscopy Int. J. 2010;24:37–43. [Google Scholar]
- Tatulian SA. Structural characterization of membrane proteins and peptides by FTIR and ATR-FTIR spectroscopy. In: Kleinschmidt JH, editor. Lipid-Protein Interactions: Methods and Protocols. Humana Press; New York: 2013. p. 177.p. 218. Chapter 9. [DOI] [PubMed] [Google Scholar]
- Taylor M, Banerjee T, Navarro-Garcia F, Huerta J, Massey S, Burlingame M, Pande AH, Tatulian SA, Teter K. A therapeutic chemical chaperone inhibits cholera intoxication and unfolding/translocation of the cholera toxin A1 subunit. PLoS ONE. 2011a;6:e18825. doi: 10.1371/journal.pone.0018825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Banerjee T, Ray S, Tatulian SA, Teter K. Protein disulfide isomerase displaces the cholera toxin A1 subunit from the holotoxin without unfolding the A1 subunit. J Biol Chem. 2011b;286:22090–22100. doi: 10.1074/jbc.M111.237966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Banerjee T, VanBennekom N, Teter K. Detection of toxin translocation into the host cytosol by surface plasmon resonance. Journal of Visualized Experiments. 2012;59 doi: 10.3791/3686. doi: 10.3791/3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Burress H, Banerjee T, Ray S, Curtis D, Tatulian SA, Teter K. Substrate-induced unfolding of protein disulfide isomerase displaces the cholera toxin A1 subunit from its holotoxin. PLoS Pathogens. 2014;10:e1003925. doi: 10.1371/journal.ppat.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K, Holmes RK. Inhibition of endoplasmic reticulum-associated degradation in CHO cells resistant to cholera toxin, Pseudomonas aeruginosa exotoxin A, and ricin. Infect Immun. 2002;70:6172–6179. doi: 10.1128/IAI.70.11.6172-6179.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K, Jobling MG, Holmes RK. A class of mutant CHO cells resistant to cholera toxin rapidly degrades the catalytic polypeptide of cholera toxin and exhibits increased endoplasmic reticulum-associated degradation. Traffic. 2003;4:232–242. doi: 10.1034/j.1600-0854.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- Teter K, Jobling MG, Sentz D, Holmes RK. The cholera toxin A13 subdomain is essential for interaction with ADP-ribosylation factor 6 and full toxic activity but is not required for translocation from the endoplasmic reticulum to the cytosol. Infect Immun. 2006;74:2259–2267. doi: 10.1128/IAI.74.4.2259-2267.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Tsai SC, Noda M, Adamik R, Moss J, Vaughan M. Enhancement of choleragen ADP-ribosyltransferase activities by guanyl nucleotides and a 19-kDa membrane protein. Proc Natl Acad Sci U S A. 1987;84:5139–5142. doi: 10.1073/pnas.84.15.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SC, Noda M, Adamik R, Chang PP, Chen HC, Moss J, Vaughan M. Stimulation of choleragen enzymatic activities by GTP and two soluble proteins purified from bovine brain. J Biol Chem. 1988;263:1768–1772. [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss O, Holden J, Rulka C, Kahn RA. Nucleotide binding and cofactor activities of purified bovine brain and bacterially expressed ADP-ribosylation factor. J Biol Chem. 1989;264:21066–21072. [PubMed] [Google Scholar]
- Welsh CF, Moss J, Vaughan M. ADP-ribosylation factors: a family of approximately 20-kDa guanine nucleotide-binding proteins that activate cholera toxin. Mol Cell Biochem. 1994;138:157–166. doi: 10.1007/BF00928458. [DOI] [PubMed] [Google Scholar]
- Wernick NLB, Chinnapen DJ-F, Cho JA, Lencer WI. Cholera toxin: an intracellular journey into the cytosol by way of the endoplasmic reticulum. Toxins. 2010;2:310–325. doi: 10.3390/toxins2030310. [DOI] [PMC free article] [PubMed] [Google Scholar]