Abstract
Dispersion enables the transition from the biofilm to the planktonic growth state in response to various cues. While several P. aeruginosa proteins, including BdlA and the c-di-GMP phosphodiesterases DipA, RbdA, and NbdA, have been shown to be required for dispersion to occur, little is known about dispersion cue sensing and the signaling translating these cues into the modulation c-di-GMP levels to enable dispersion. Using glutamate-induced dispersion as a model, we report that dispersion-inducing nutrient cues are sensed via an outside-in signaling mechanism by the diguanylate cyclase NicD belonging to a family of seven transmembrane (7TM) receptors. NicD directly interacts with BdlA and the phosphodiesterase DipA, with NicD, BdlA, and DipA being part of the same pathway required for dispersion. Glutamate-sensing by NicD results in NicD dephosphorylation and increased cyclase activity. Active NicD contributes to the non-processive proteolysis and activation of BdlA via phosphorylation and temporarily elevated c-di-GMP levels. BdlA, in turn, activates DipA, resulting in the overall reduction of c-di-GMP levels. Our results provide a basis for understanding the signaling mechanism based on NicD to induce biofilm dispersion that may be applicable to various biofilm-forming species and may have implications for the control of biofilm-related infections.
Keywords: dispersion, phosphorylation, pulldown, human pathogen, RbdA
INTRODUCTION
The formation of bacterial biofilms, or surface-associated communities is a developmental process that is initiated with surface attachment by planktonic cells. Biofilms return to the planktonic state upon dispersion, a process in which sessile, surface-attached organisms liberate themselves from the biofilm, apparent by biofilm microcolonies having central voids, and transition to the free-living state, enabling bacteria to spawn novel communities in new locales. It is thus not surprising that while biofilms are considered the root cause of chronic and persistent infections (Costerton et al., 1999, Kaplan, 2010), biofilm dispersion is believed to be part of an inherent strategy of biofilm-forming pathogens to initiate a disseminating phenotype, causing acute and periodic infections. The human pathogen Pseudomonas aeruginosa is the principal pathogen associated with cystic fibrosis (CF) pulmonary infection and ranks 2nd among the most common pathogens isolated from chronic and burn wounds (Emori & Gaynes, 1993). Dispersion by this pathogen has been shown to be induced by the self-synthesized signaling molecule cis-2-decenoic acid, changes in growth medium composition, oxygen limitation, exposure to nitric oxide (NO), and variation in the concentration of carbon and energy sources (Petrova & Sauer, 2012a, Basu Roy et al., 2012, Davies & Marques, 2009, Sauer et al., 2004, Li et al., 2013, Barraud et al., 2006, Barraud et al., 2009). Dispersion is not limited to P. aeruginosa biofilms, as environmental or physiological cues such as variation in iron, oxygen or carbon substrate concentration have also been described to induce dispersion by a large number of Gram-negative and –positive biofilm forming bacteria including Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Acinetobacter sp. str. GJ12 , P. putida and Schewanella oneidensis (reviewed in (McDougald et al., 2012)). Moreover, biofilm dispersion has been shown in various bacteria to be induced by exposure to matrix-degrading enzymes and surface protein releasing factors (Gjermansen et al., 2005, Kaplan et al., 2004, Lee et al., 1996). In P. putida and P. fluorescens, the large adhesive outer-membrane protein, LapA, mediates attachment to surfaces and to matrix components (Gjermansen et al., 2010, Hinsa et al., 2003, Monds et al., 2007). Gjermansen et al. (Gjermansen et al., 2010) demonstrated that in P. putida, release of LapA from the cell surface results in biofilm dispersal and is mediated through the activity of the periplasmic protease LapG. Additional mechanisms linked to dispersion include the production of the biosurfactant rhamnolipid (Schooling et al., 2004, Boles et al., 2005), and cell death, with filamentous phage Pf1-mediated cell death being an important mechanism of differentiation inside microcolonies that facilitates dispersal of a subpopulation of surviving cells (Webb et al., 2003). Oxidative or nitrosative stress, induced upon exposure to exogenous or endogenous nitric oxide (NO), has been linked to biofilm dispersion. A role of oxidative or nitrosative stress was further supported by a P. aeruginosa mutant lacking the only enzyme capable of generating metabolic NO through anaerobic respiration (nitrite reductase, ΔnirS) not dispersing (Barraud et al., 2006).
Dispersion in response to NO and changes in the carbon source concentration has been suggested to be a coordinated process coinciding with unique protein phosphorylation patterns and requiring specific regulatory events including phosphotransfers (Sauer et al., 2004, Barraud et al., 2009, Petrova & Sauer, 2012a). Moreover, biofilm dispersion has also been linked to the modulation of the intracellular signaling molecule cyclic di-GMP (c-di-GMP). C-di-GMP contributes to bacterial cells transitioning between a motile and a biofilm lifestyle by regulating gene expression profiles via interaction with transcriptional regulators such as FleQ and PelD. High levels of c-di-GMP promoting sessile growth, while low levels correlate with planktonic existence (Barraud et al., 2009, Basu Roy et al., 2012, Thormann et al., 2006, Gjermansen et al., 2005, Cotter & Stibitz, 2007). Levels of c-di-GMP are enzymatically modulated by diguanylate cyclases (DGC), proteins containing a GGDEF domain, and phosphodiesterases (PDE) harboring either an EAL or HD-GYP domain (Cotter & Stibitz, 2007, Schirmer & Jenal, 2009). In P. putida, two genes (PP0164, PP0165), encoding a putative periplasmic protein and a putative transmembrane protein involved in c-di-GMP modulation, were found to be required for biofilm formation and starvation-induced dispersion, with mutants in PP0164 being unable to disperse from biofilms in response to carbon starvation (Gjermansen et al., 2005). Likewise, inactivation of yhjH, encoding an enzyme having phosphodiesterase activity, impaired cellular detachment by S. oneidensis induced upon sudden depletion of molecular oxygen (Thormann et al., 2006). In P. aeruginosa, dispersion upon exposure to NO and elevated nutrient concentrations has been linked to the reduction of the cellular c-di-GMP levels, requiring the PDEs DipA, RbdA and NbdA (Basu Roy et al., 2012, Li et al., 2013, An et al., 2010, Morgan et al., 2006). In addition, the membrane-bound phosphodiesterase NbdA was found to be specific to the dispersion response following exposure to NO (Li et al., 2013). While lacking domains required for c-di-GMP modulation, the chemotaxis transducer protein BdlA has been described to contribute to intracellular sensing of dispersion-inducing conditions and together with DipA to form a regulatory network that modulates an intracellular c-di-GMP pool to enable dispersion. BdlA function requires an unusual, non-processive proteolytic cleavage found to be stimulated by increased c-di-GMP levels present in biofilms, and dependent on the protease ClpP, the chaperone ClpD, and BdlA phosphorylation (Petrova & Sauer, 2012b, Petrova & Sauer, 2012a).
While previous findings indicate biofilm dispersion to occur in response to various cues, to require BdlA and PDEs (for instance DipA and RbdA), and to coincide with increased PDE activity and a reduction of c-di-GMP levels (Basu Roy et al., 2012, Barraud et al., 2009), little is known about how these proteins interact to translate sensing of dispersion cues into the modulation of the intracellular c-di-GMP pool to enable dispersion. Moreover, no protein that either directly senses or responds to carbon sources, thereby controlling c-di-GMP turnover, has been identified. Using nutrient-induced dispersion as a model, we asked how P. aeruginosa senses or responds to carbon and energy sources acting as dispersion cues. Specifically, we wanted to test whether dispersion is triggered following intracellular sensing of cellular energy state changes or extracellular sensing via a membrane-localized sensor, and if so, how dispersion cue perception is translated across cellular compartments into the modulation of the intracellular c-di-GMP pool.
RESULTS
D-glutamate does not support growth but induces dispersion of P. aeruginosa biofilms
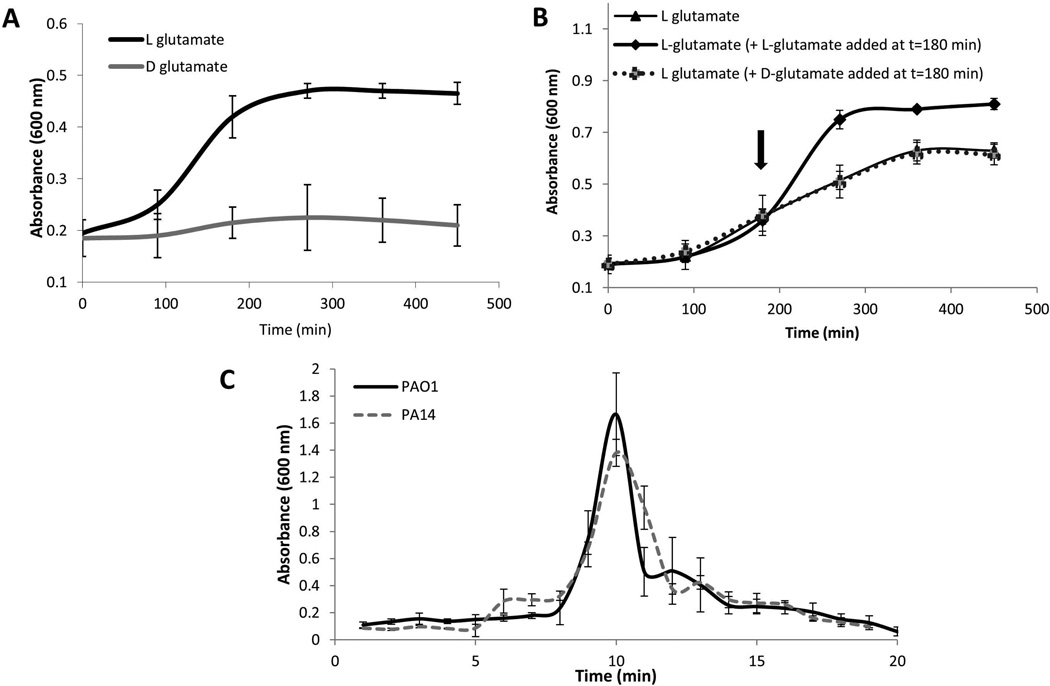
To address the question of how P. aeruginosa senses or responds to dispersion-inducing nutrient cues, we first asked whether the nutrient cues are required to be metabolized. To do so, we made use of glutamate, which was previously shown to induce dispersion. While L-glutamate as the sole carbon source supported growth of P. aeruginosa and further addition of L-glutamate to exponential phase cultures resulted in extended exponential growth, the isoform D-glutamate did not support growth or affect the onset of stationary phase (Fig. 1A–B). However, despite the isoform D–glutamate not supporting growth of P. aeruginosa, exposure of P. aeruginosa biofilms to D-glutamate induced dispersion in a manner similar to L-glutamate (Fig. 1C). The finding supported the notion that dispersion in response to nutrient cues, here glutamate, can be independent of the cues being used as carbon and energy sources. The finding suggested that dispersion in response to elevated levels of nutrients is either linked to osmotic stress or alternatively, relies on a mechanism involving a sensory protein capable of detecting these carbon sources and consequently relaying a dispersion-inducing signal. To elucidate by which mechanism nutrients induced dispersion of biofilms, we first asked whether a specific carbohydrate sensory protein contributes to nutrient-induced dispersion of P. aeruginosa biofilms, assuming that in the absence of such sensory protein, osmotic stress may be the likely cause.
Figure 1. Dispersion occurs in response to D-glutamate despite D-glutamate being a poor growth substrate for P. aeruginosa.
Growth of P. aeruginosa PAO1 in the presence of D-glutamate (130 mg/L) or L-glutamate (130 mg/L) as the sole carbon source (A) and (B) upon addition of extra D- or L-glutamate (130mg/L) at the 3 hr time point. P. aeruginosa was grown in minimal medium at 37°C with shaking at 220 rpm and absorbance was recorded at 600 nm every 90 min over a period of 8 hr. Experiments were repeated at least three times Error bars indicate standard deviation. (C) Dispersion by P. aeruginosa PA14 and PAO1 biofilms in response to L-glutamate, as indicated by measurements of effluent absorbance. P. aeruginosa strain PA14 harbors a mutation in ladS that renders the protein non-functional (Mikkelsen et al., 2011). Dispersion assays were carried out using biofilm tube reactors. Experiments were repeated at least three times Error bars indicate standard deviation.
Inactivation of nicD impairs the dispersion response by P. aeruginosa PA14 and PAO1 in response to glutamate
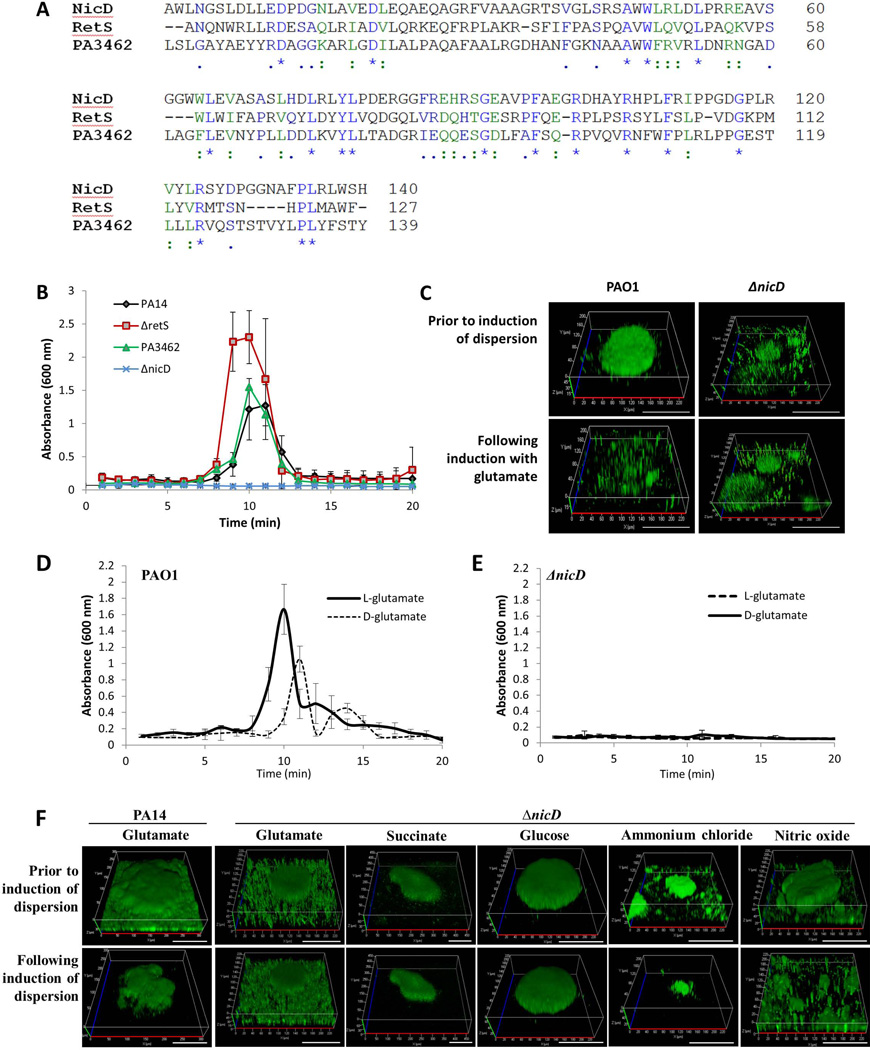
A sensory domain predicted to bind carbohydrates and their derivatives is the 7TMR-DISMED2 domain that is exposed to the periplasmic side of the cytoplasmic membrane, followed by the transmembrane 7TM-DISM domain characterized by the presence of 7 transmembrane helices (7TM-DISM) (Anantharaman & Aravind, 2003). A screen of the Pseudomonas genome for proteins harboring both 7TMR-DISMED2/DISM domains resulted in the identification of four proteins including the two-component hybrids LadS and RetS, the sensor/response regulator hybrid PA3462, and the hypothetical diguanylate cyclase PA4929 (Winsor et al., 2009) (Figs. 2A, S1) that we named nicD for nutrient-induced cyclase D as discussed below. We hypothesized that if the 7TMR-DISMED2-harboring proteins are involved in nutrient-sensing, inactivation of the respective genes would impair nutrient-induced dispersion. P. aeruginosa strain PA14 harbors a mutation in ladS that renders the protein non-functional (Mikkelsen et al., 2011). Despite lack of a functional LadS, biofilms formed by P. aeruginosa PA14 dispersed upon exposure to glutamate in a manner similar to biofilms formed by P. aeruginosa PAO1, which harbors an intact ladS gene (Fig. 1C). This finding indicated that LadS is not required for glutamate-induced dispersion. Therefore, a ladS mutant was not tested. However, using tube reactor-based dispersion assays as previously described (Petrova & Sauer, 2012a), we tested retS, nicD, and PA3462 mutants of P. aeruginosa PA14 which have been previously characterized in the literature (Kulasekara et al., 2005, Mikkelsen et al., 2011, Venkataraman et al., 2010) P. aeruginosa PA14 biofilms inactivated in retS and PA3462 dispersed in response to elevated concentrations of glutamate (Fig. 2B). However, P. aeruginosa PA14 biofilms inactivated in nicD were impaired with respect to the dispersion response to glutamate (Fig. 2B, Table S1). Impaired dispersion upon inactivation of nicD was confirmed by confocal microscopy (Fig. S2A, Table S1). Confocal microscopy furthermore indicated inactivation of nicD to correlate with reduced biofilm biomass accumulation as well as reduced average and maximum biofilm height (Table S1). Multicopy expression of nicD restored the biofilm architecture of nicD mutant biofilms to wild-type levels. Moreover, multicopy expression of nicD restored the dispersion-deficient phenotype of ΔnicD to glutamate to wild-type PA14 levels (Fig. S2B, Table S1).
Figure 2. Dispersion in response to nutrient cues requires NicD.
(A) Alignment of the NicD 7TMR-DISMED2 with the 7TMR-DISMED2 domains of RetS and PA3462. ":" denotes residues with strong conservation (score >0.5 in PAM250 matrix) and "." denotes residues with weak conservation (score ≤0.5); "*", denotes identical amino acids. Identical amino acid residues are shown in blue while similar amino acids are highlighted in green. (B) Inactivation of nicD but not retS or PA3462 renders P. aeruginosa dispersion-deficient in response to glutamate as indicated by measurements of effluent absorbance. Biofilms were grown for 5 days under flowing conditions in tube reactors prior to induction of dispersion inducing conditions. Experiments were repeated at least three times. Error bars indicate standard deviation. (C) Inactivation of nicD impairs dispersion of P. aeruginosa PAO1 biofilms to L-glutamate as indicated by confocal microscopy. Biofilms were grown for 5 days in flow cells under flowing conditions. Confocal images were acquired at the same position, prior to and post-induction of dispersion. Representative images are shown. Size bar, 100 µm. Dispersion by P. aeruginosa PAO1 biofilms (D) and ΔnicD biofilms (E) in response to D- and L-glutamate as indicated by measurements of effluent absorbance. Biofilms were grown for 5 days in tube reactors under flowing conditions prior to induction of dispersion inducing conditions. Experiments were repeated at least five times. Error bars indicate standard deviation. (F) Confocal images of P. aeruginosa PA14 ΔnicD biofilms prior to and following the addition of glutamate, succinate, glucose, ammonium chloride and nitric oxide to induce biofilm dispersion. Inactivation of nicD renders P. aeruginosa dispersion-deficient in response to glutamate, glucose and succinate but not ammonium chloride or nitric oxide. Shown confocal images were acquired at the same position, prior to and post-induction of dispersion. Size bar, 100 µm.
It is of interest to note that inactivation of nicD not only affected the dispersion response by P. aeruginosa PA14 biofilms but also dispersion by P. aeruginosa PAO1 biofilms. This was apparent as P. aeruginosa PAO1 biofilms inactivated in nicD were likewise impaired with respect to the dispersion response to L- and D-glutamate as determined by confocal microscopy and by using tube reactors (Fig. 2C–E, Table S1).
NicD is required for dispersion induced in response to carbon sources including glutamate, succinate and glucose but does not play a role in dispersion induced by NO or ammonium chloride
Impaired dispersion response by ΔnicD biofilms was not limited to glutamate. Inactivation of nicD likewise impaired dispersion by P. aeruginosa PA14 biofilms upon exposure to glucose and succinate (Fig. 2F, Table S1). In contrast, however, ΔnicD biofilms dispersed in response to nitric oxide and ammonium chloride (Fig.2F, Table S1). The findings indicated that lack of biofilm dispersion by ΔnicD biofilms was unrelated to the reduced biomass accumulation and reduced height of nicD mutant biofilms (Table S1). Instead, the findings indicated NicD to play a specific role in the dispersion response induced by carbon or nutrient sources, as inactivation of nicD did not impair dispersion in response to NO and ammonium chloride. The findings also demonstrated that the lack of nicD biofilm dispersion in response to glutamate was not the non-specific result of reduced biomass, as biofilms formed by this mutant could disperse in response to other cues.
NicD is an active diguanylate cyclase
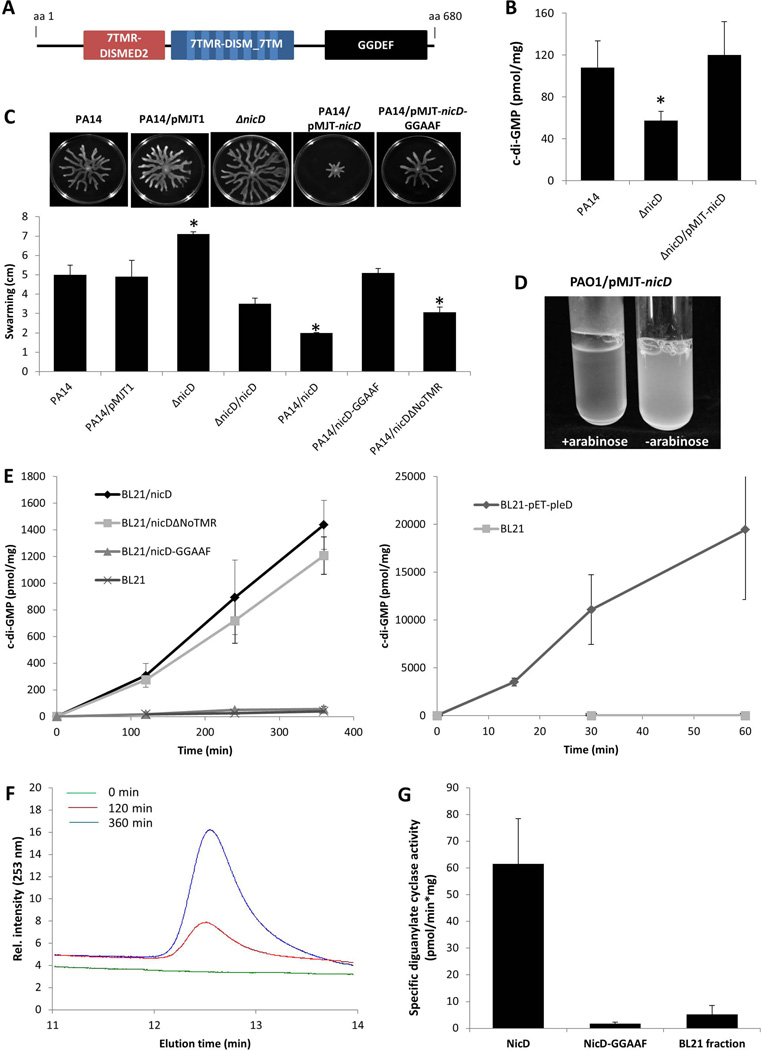
In addition to the periplasmic and membrane spanning 7TMR-DISMED2/DISM domains, NicD is predicted to harbor a diguanylate cyclase GGDEF domain (Fig. 3A). We therefore asked whether NicD exhibits diguanylate cyclase (DGC) activity and produces c-di-GMP. While not abolishing biofilm formation, inactivation of nicD correlated with reduced biofilm biomass accumulation by P. aeruginosa PA14 and PAO1 (Table S1), reduced P. aeruginosa PA14 and PAO1 biofilm c-di-GMP levels (Figs. 3B, S3), but enhanced swarming motility (Fig. 3C). In contrast, overexpression of nicD reduced swarming and rendered P. aeruginosa PAO1 hyperaggregative when grown in liquid (Fig. 3D). It is of interest to note that the hyperaggregative phenotype was specific to P. aeruginosa PAO1 /pMJT-nicD and not observed in P. aeruginosa PA14 background. The observed phenotypes are consistent with nicD encoding an active diguanylate cyclase.
Figure 3. nicD harbors in addition to 7TMR-DISMED2 and 7TMR-DISM_7TM domains a GGDEF domain, and encodes an active diguanylate cyclase.
(A) NicD (PA4929) is predicted to be a membrane-bound protein harboring three domains including a periplasmic 7TMR-DISMED2 sensory domain, a membrane-spanning 7TMR-DISM_7TM domain and a cytoplasmic GGDEF domain. Model of NicD domain organization and putative subcellular localization of NicD domains. NicD comprises of three domains: an extracellular 7TMR-DISMED2 (7 Trans-membrane Receptor with Diverse Intracellular Signaling Modules extracellular) domain, a membrane-spanning 7TMR-DISM_7TM (7TMR-DISM, 7 Trans-membranes) domain, and a cytoplasmic diguanylate cyclase (GGDEF) domain. Domain localization is based on TMHMM prediction. (B) C-di-GMP levels in P. aeruginosa PA14, ΔnicD and ΔnicD/pMJT-nicD biofilms. C-di-GMP was quantitated using an HLC-based method with commercially available c-di-GMP as standard. c-di-GMP (pmol/mg) refers to c-di-GMP levels (pmol) per total cell pellet protein (in mg). Experiments were repeated at least three times. Error bars indicate standard deviation. *, significantly different from wild type biofilms; p < 0.05 as determined by ANOVA and SigmaStat (C) Swarming motility of P. aeruginosa PA14, ΔnicD, and strains overexpressing nicD, nicD-GGAAF, and nicDΔNoTMR. The swarming diameter was recorded following 48 hr of growth on NA agar. Representative images of swarming motility P. aeruginosa PA14, ΔnicD, and strains overexpressing nicD, nicD-GGAAF, and nicDΔNoTMR are shown above the graph. P. aeruginosa PA14 harboring the empty vector pMJT1 was used as vector control. Experiments were repeated at least three times. Error bars indicate standard deviation. *, significantly different from wild type biofilms; p < 0.05 as determined by ANOVA and SigmaStat. (D) Image of P. aeruginosa PAO1/pMJT-nicD planktonic culture prior to and following addition of arabinose to induce nicD expression. –arabinose, no arabinose was added; +arabinose, 1% arabinose was added to the growth medium. (E) Formation of c-di-GMP by total cell extracts of E. coli BL21 overexpressing nicD, nicD variants and pleD. DGC assays were carried out using a total of 200 µg of E. coli BL21 cell extracts overexpressing nicD and nicDΔNoTMR lacking the N-terminal 7TMR-DISMED2 domain (NicDΔNoTMR) or nicD-GGAAF in which the GGDEF motif was substituted with GGAAF under the control of the IPTG-inducible promoter of the pET101D vector or a total of 20 µg of E. coli BL21 cell extracts overexpressing pleD cloned into pET11b (Merritt et al., 2007, Paul et al., 2004) were used. Cell extracts of E. coli BL21 not harboring any vector (200 and 20 µg) were used as controls while E. coli BL21 overexpressing pleD were used as positive control. Experiments were carried out in triplicate. Error bars indicate standard deviation. (F) Elution profiles of c-di-GMP produced by purified NicD. The reaction mixtures were analyzed by HPLC for the presence of c-di-GMP 0, 120, and 360 min post initiation of DGC assays. Representative peaks corresponding to c-di-GMP are shown. (G). Specific diguanylate cyclase activity of purified NicD and NicD-GGAAF. Diguanylate cyclase assays were performed using 25 µM GTP and 70 µg of Ni-affinity chromatography purified protein. BL21 control, protein purified from E. coli cell extracts not harboring any vector was used as negative control. Experiments were carried out in triplicate. Error bars indicate standard deviation.
In order to determine whether NicD is an active DGC, cyclase assays were performed using protein extracts of E. coli strains harboring pET-nicD or protein extracts of E. coli BL21 not harboring any vector. The formation of c-di-GMP was determined using HPLC analysis following incubation at 37° for 120, 240, and 360 min (Fig. 3E). While very little to no c-di-GMP was detected in the fractions obtained from extracts of the E. coli BL21 control, protein extracts of E. coli strains harboring pET-nicD were found to produce c-di-GMP in a linear manner (Fig. 3E). On average, more than 1400 pmol c-di-GMP per mg of total cell extract was produced over a period of 360 min. However, compared to protein extracts of E. coli BL21 harboring pET-pleD, the diguanylate cyclase activity detectable in protein extracts of BL21/pET-nicD was significantly reduced (Fig. 3E), indicating NicD to harbor reduced diguanylate cyclase activity compared to PleD. To further confirm NicD diguanylate cyclase activity, NicD was purified by Ni-affinity chromatography, and the purified protein used in diguanylate cyclase assays. Similar to protein extracts, purified NicD protein was capable of converting GTP to c-di-GMP as indicated by the HPLC elution profiles of c-di-GMP produced by purified NicD following 0, 120, and 360 min post initiation of DGC assays (Fig. 3F) indicating purified NIcD to catalyze the formation of c-di-GMP in vitro. The specific activity of purified NicD was found to be 61±16.6 pmol/mg*min (Fig. 3G). In contrast, no c-di-GMP was detected in the Ni-affinity chromatography fractions obtained using the E. coli BL21 control (Fig. 3G). The overall specific activity of NicD is comparatively low compared to PleD or WspR (Paul et al., 2007, Paul et al., 2004, Hickman et al., 2005). It is likely that the reduced NicD activity is related to its phosphorylation state given that PleD activity has been shown to be activated by phosphorylation and more specifically, by phosphorylation-mediated dimerization, with PleD phosphorylation correlating with significantly increased formation of c-di-GMP (Paul et al., 2007, Paul et al., 2004). Likewise, while purified WspR catalyzed the formation of c-di-GMP, phosphorylation of WspR stimulated this activity both in vitro and in vivo (Hickman et al., 2005, Huangyutitham et al., 2013). Moreover, The DGC TbpB was found to be phosphorylated at tyrosine located at amino acid positions 48 and 62, with phosphorylated TbpB correlating with increased cellular c-di-GMP levels (Ueda & Wood, 2009, Pu & Wood, 2010).
To determine whether the extracellular domain of NicD was required for diguanylate cyclase activity, we also generated a NicD variant lacking the periplasmic 7TMR-DISMED2 domain, NicDΔNoTMR. Both NicD and NicDΔNoTMR were produced in a soluble manner (Fig. S4A). Moreover, similarly to NicD, NicDΔNoTMR was found to be membrane-associated as determined using ultracentrifugation and subsequent immunoblot analysis (Fig. S4B) indicating that deletion of the periplasmic domain did not affect cellular localization of NicDΔNoTMR compared to NicD. Diguanylate cyclase assays were performed using protein extracts of E. coli strains harboring pET-nicDΔNoTMR and found to produce c-di-GMP in a manner similar to protein extracts of E. coli strains harboring pET-nicD (Fig. 3E). Moreover, overexpression of nicDΔNoTMR resulted in significantly reduced swarming motility compared to the wild type (Fig. 3C).
Several studies have demonstrated that the amino acids present in the GGDEF motif of the GGDEF domain are essential for cyclase activity with alanine substitution of aspartic acid (D) and/or glutamic acid (E) residues resulting in diguanylate cyclases being rendered inactive (Aldridge et al., 2003, Ferreira et al., 2008, De et al., 2008). To confirm NicD to be an active diguanylate cyclase, site-directed mutagenesis was carried out using alanine substitutions to generate a NicD variant harboring a GGAAF motif instead of the active site GGDEF motif. Similar to intact and truncated NicD, resulting variant protein NicD-GGAAF was found to be produced in a soluble manner (Fig.S4A). Under the conditions tested, however, very little to no c-di-GMP was detected in the fractions obtained from extracts of E. coli strains harboring pET-nicD-GGAAF (Fig. 3E). Likewise, very little to no c-di-GMP was detected when purified NicD-GGAAF was used (Fig. 3G). In agreement with NicD-GGAAF not harboring diguanylate cyclase activity was the finding of overexpression of nicD-GGAAF not affecting swarming motility relative to the wild type (Fig. 3A). Taken together, our findings suggested NicD to be an active diguanylate cyclase, with activity being independent of the presence of the periplasmic 7TMR-DISMED2 sensory domain but dependent on the presence of the GGDEF motif, as substituting the GGDEF motif present in the GGDEF domain to GGAAF rendered NicD inactive (Fig. 3C, E).
ΔnicD biofilms expressing nicD-GGAAF or nicDΔNoTMR are impaired in dispersion in response to L-glutamate
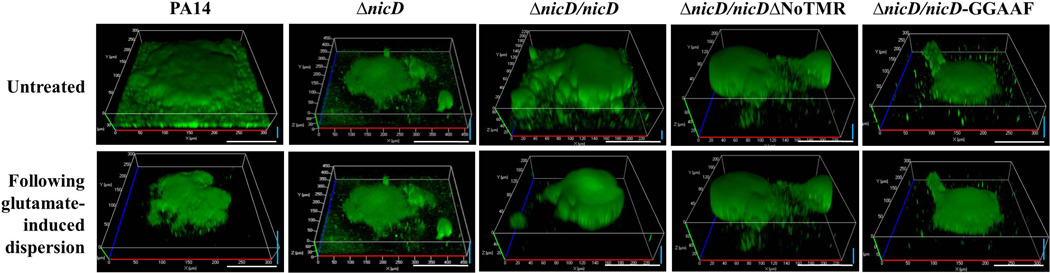
Having confirmed NicD to be an active cyclase that contributes to the biofilm architecture and intracellular c-di-GMP levels present in biofilm cells, we next asked about the role of the diguanylate cyclase NicD in dispersion. In particular, we were interested in determining whether diguanylate activity played a role in the dispersion response. To address this question, we made use of the NicD-GGAAF variant that was demonstrated via cyclase and swarming assays to be inactive. To do so, confocal microscopy-based dispersion assays were carried out as previously described (Sauer et al., 2004, Petrova & Sauer, 2012a). While wild-type P. aeruginosa PA14 and ΔnicD/pMJT-nicD biofilms dispersed in response to glutamate, ΔnicD biofilms expressing nicD-GGAAF did not (Fig. 4, Table S1).
Figure 4. Dispersion in response to glutamate requires NicD DGC activity and the presence of the periplasmic 7TMR-DISMED2 domain.
Biofilms formed by P. aeruginosa PA14, ΔnicD, and ΔnicD mutant strains overexpressing nicD, nicD-GGAAF and nicDΔNoTMR under the control of the arabinose-inducible promoter of the pMJT1 vector were grown for 5 days in flow cells under flowing conditions. Multicopy expression of nicD but not nicD-GGAAF or nicDΔNoTMR restores biofilm dispersion upon glutamate exposure to wild type levels. Confocal images were acquired at the same position, prior to and following induction of dispersion by L-glutamate. Representative images are shown. White size bar, 100 µm. Blue vertical size bar (indicating height), 40 µm.
We furthermore asked whether NicD plays a direct role in sensing dispersion cues such as glutamate. Based on available crystal structures and protein folding prediction models, the 7TMR-DISMED2 domain is believed to the located on the periplasmic side of the cytoplasmic membrane, and to be the domain involved in binding of carbohydrates and their derivatives. Considering that nicD inactivation only impaired dispersion in response to nutrients such as glutamate, succinate and glucose but not NO or ammonium chloride, we reasoned that NicD may play a direct role in sensing nutrient cues. To explore the possibility of the periplasmic domain of NicD playing a sensory role in the dispersion response, we made use of the NicD variant lacking the periplasmic 7TMR-DISMED2 domain, NicDΔNoTMR. While the biofilm architecture of ΔnicD/nicDΔNoTMR biofilms was more similar to that of wild-type PA14 biofilms (Table S1), ΔnicD biofilms producing the truncated NicD variant NicDΔNoTMR did not disperse upon exposure to glutamate (Fig. 4, Table S1). Our finding suggests that NicD cyclase activity is required for dispersion to occur. However, our findings also suggest that active NicD diguanylate cyclase activity alone is not sufficient to restore the dispersion response. Instead, the data indicated NicD to specifically play a role in nutrient-induced dispersion by likely perceiving nutrient cues via the periplasmic 7TMR-DISMED2.
Exposure to glutamate enhances NicD diguanylate cyclase activity and results in increased cellular c-di-GMP levels
The requirement for diguanylate cyclase activity and the periplasmic sensory domain of NicD in the dispersion response upon exposure to glutamate hinted at an integration of nutrient dispersion cues sensed via the sensory 7TMR-DISMED2 domain into intracellular regulation of c-di-GMP levels via the diguanylate cyclase activity of NicD. However, the finding was surprising considering that dispersion has been correlated with a reduction rather than an increase in c-di-GMP levels (Basu Roy et al., 2012, Li et al., 2013). This unusual finding led to the question of whether NicD responds to dispersion-inducing cues by becoming inactive or, in contrast, elevating its DGC activity. To determine how NicD integrates sensing of environmental cues into the regulation of c-di-GMP levels, we made use of the finding that overexpression of DGCs, including nicD, results in an aggregative phenotype in liquid, characterized by clearing of the culture medium upon settling of the cell aggregates (Fig. 3D). We reasoned that if NicD cyclase activity is induced upon sensing dispersion cues such as glutamate, aggregation will be observed as a decrease in turbidity of the medium supernatant. If NicD cyclase activity instead is reduced or abrogated, no aggregation or no change in medium turbidity compared to control would be noted. As the hyperaggregative phenotype due to nicD overexpression was more pronounced in P. aeruginosa PAO1 than PA14, all subsequent experiments were carried out using P. aeruginosa PAO1 strains.
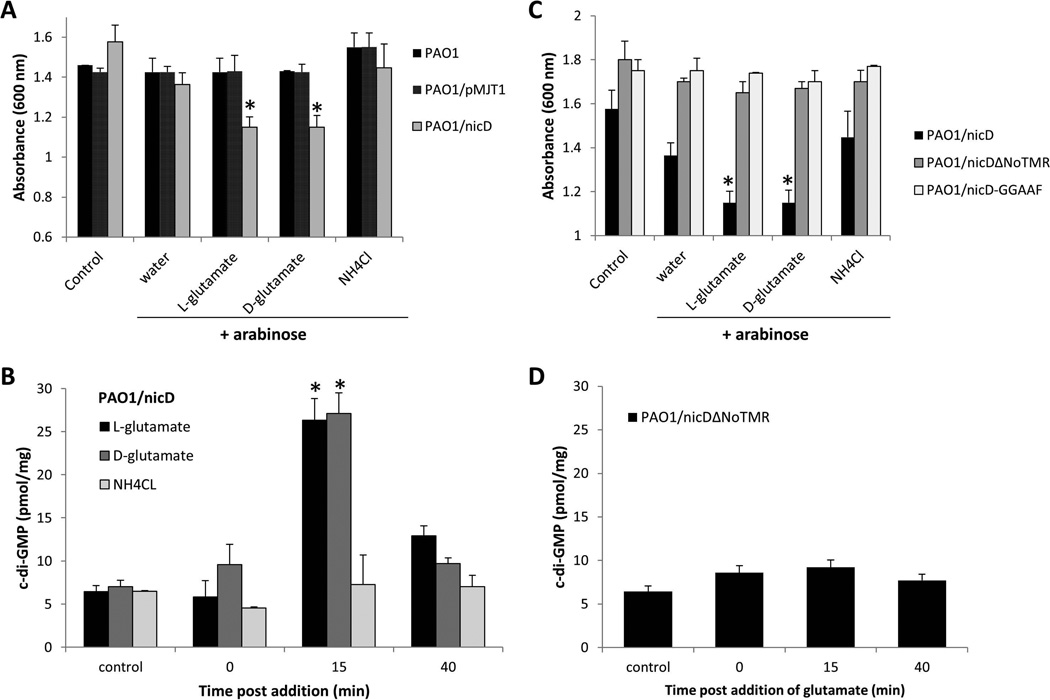
To determine the effect of glutamate on medium turbidity, P. aeruginosa PAO1 was grown to early exponential phase, at which point nicD expression was induced by the addition of arabinose. Following 3 hrs of continued growth in the presence of arabinose, glutamate was added and the medium turbidity determined post 30 min of incubation. Addition of L-glutamate to a P. aeruginosa PAO1 culture overexpressing nicD resulted in significantly increased aggregation in liquid and correlated with a decrease in the culture turbidity compared to controls including cultures prior to the addition of L-glutamate or cultures only exposed to water (Fig. 5A). Similar results were obtained when D-glutamate was used (Fig. 5A). In contrast, addition of water or ammonium chloride did not affect the aggregative phenotype of PAO1/pMJT-nicD (Fig. 5A). Likewise, no significant change in the culture turbidity was observed for the wild type or vector control regardless of the dispersion cues used, indicating that the observed aggregative behavior of strain of PAO1/pMJT-nicD upon exposure to L- and D-glutamate required the presence of NicD (Fig. 5A). The enhanced aggregative phenotype of PAO1/pMJT-nicD upon exposure to glutamate correlated with a 3-fold increase in c-di-GMP levels, from an average of 8 pmol/mg to 28 pmol/mg within 15 min post glutamate addition (Fig. 5B). Interestingly, the increase in c-di-GMP levels was temporary and returned to pre-glutamate levels within 40 min of incubation. In contrast, no effect on the cellular c-di-GMP levels was noted upon addition of ammonium chloride (Fig. 5B), indicating increased aggregation and increased c-di-GMP levels to be specifically associated with glutamate. The findings furthermore suggested NicD cyclase activity to be activated upon sensing glutamate (a nutrient cue) but not ammonium chloride (a non-nutrient cue).
Figure 5. NicD DGC activity is induced upon exposure to glutamate, resulting in increased cellular c-di-GMP levels.
(A) Absorbance of culture medium of exponential phase P. aeruginosa PAO1/pMJT-nicD, prior to addition of arabinose (control) and following addition of arabinose (3 hr) and subsequent addition of water, D- and L-glutamate and ammonium chloride (NH4Cl). PAO1 and PAO1/pMJT1 were used as vector control. Experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from “water” control to which only arabinose (3 hr) plus water was added. (B) Cellular c-di-GMP levels in PAO1/nicD following addition of arabinose (3 hr, control), and 0, 15 and 40 min post-addition of D- and L-glutamate and ammonium chloride (NH4Cl). Experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from 0 min time point. (C) Absorbance of culture medium of exponential phase P. aeruginosa PAO1 overexpressing nicD, nicDΔNoTMR, or nicD-GGAAF, prior to addition of arabinose (control) and following addition of arabinose (3 hr) and subsequent addition of water, D- and L-glutamate and ammonium chloride (NH4Cl). Experiments were carried out at least in triplicate. Error bars indicate standard deviation. (D) Cellular c-di-GMP levels in PAO1/nicDΔNoTMR mutant following addition of arabinose (3 hr), and 0, 15 and 40 min post-addition of glutamate. Experiments were carried out at least in triplicate. Error bars indicate standard deviation.
To further determine how NicD integrates sensing of environmental cues into the regulation of c-di-GMP levels, we made use of strains overexpressing nicD-GGAAF and nicDΔNoTMR. Similar to the vector control (Fig. 5A), no change in the culture turbidity was observed for strains overexpressing nicD-GGAAF regardless of the dispersion cues used, indicating that the observed aggregative behavior by PAO1/pMJT-nicD upon exposure to L- and D-glutamate required NicD diguanylate cyclase activity (Fig. 5C). While the NicD variant NicDΔNoTMR harbors diguanylate cyclase activity, exposure of PAO1/nicDΔNoTMR to L- and D-glutamate had no effect on the culture turbidity (Fig. 5C). Likewise, no change in the culture turbidity was noted upon exposure to ammonium chloride. While the cellular c-di-GMP levels present in PAO1/nicDΔNoTMR cells were comparable to PAO1/nicD prior to addition of glutamate, addition of glutamate did not result in increased c-di-GMP levels (Fig. 5B, D).
NicD is dephosphorylated upon addition of glutamate
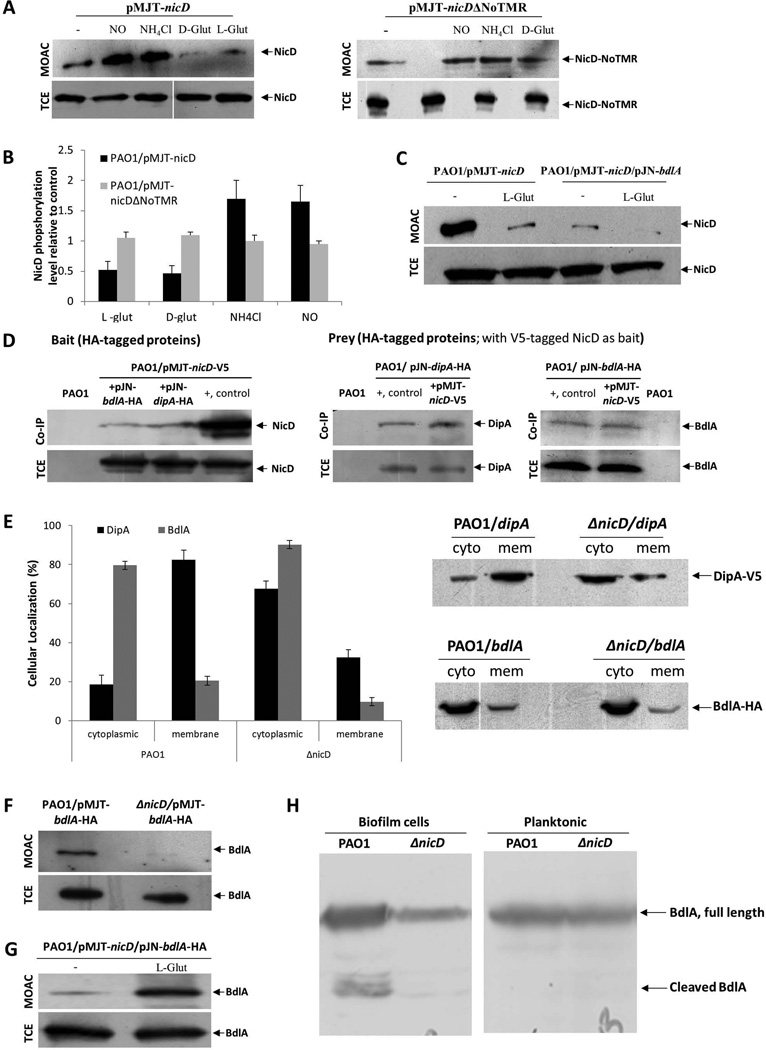
Our findings indicated NicD cyclase activity to be stimulated by glutamate and pointed at NicD activation being specific to glutamate with the compound likely being sensed via the periplasmic sensory domain of NicD. To explore the mechanism by which NicD relays extracellular dispersion cues to affect NicD cyclase activity and thus, modulation of the intracellular c-di-GMP pool, we made use of the finding that information relay from one cellular location to another has been linked to posttranslational modifications, in particular phosphorylation-dephosphorylation events of intracellular regulatory domains or proteins (Scott & Pawson, 2009). To determine whether NicD is phosphorylated and whether the level of NicD phosphorylation varies upon exposure to glutamate and other dispersion-inducing cues, the phosphorproteome, enriched via metaloxide affinity chromatography (MOAC), of P. aeruginosa PAO1 planktonic cells was analyzed by immunoblot analysis for the presence of V5-tagged NicD. Samples tested corresponded to the untreated controls and the time point (15 min) of highest NicD induction as revealed by the turbidity and c-di-GMP determination assays (Fig. 5A–B). Under the conditions tested, NicD was found to be phosphorylated (Fig. 6A). Addition of L- and D-glutamate correlated with a 2-2.5-fold decrease in NicD phosphorylation while exposure to ammonium chloride and NO resulted in increased phosphorylation (Fig. 6A–C). No difference in phosphorylation was noted when the cells expressing nicDΔNoTMR instead of nicD were used, regardless of the absence or presence of D-glutamate (Fig. 6A–B). Overall, the phosphorylation status of NicD was inversely correlated with the aggregative phenotype and the c-di-GMP levels, with increased aggregation correlating with decreased phosphorylation, suggesting NicD dephosphorylation upon activation.
Figure 6. NicD forms a membrane associated complex with DipA and BdlA and contributes to BdlA activation.
(A) Detection of V5-tagged NicD and NicDΔNoTMR in total cell extracts (TCE) or metaloxide affinity chromatography-enriched phosphoproteomes (MOAC) of planktonic P. aeruginosa PAO1/pMJT-nicD and PAO1/pMJT-nicDΔNoTMR cells prior to (−) and following addition of nitric oxide (NO), ammonium chloride (NH4Cl), and D- and L-glutamate (D-glut, L-glut, respectively) by immunoblot analysis using anti-V5 antibodies. A total of 10 µg per TCE was loaded prior to MOAC purification and used as loading control. For MOAC samples, the entire MOAC eluate was loaded. (B) Band intensity was used to determine the relative levels of NicD and NicDΔNoTMR phopshorylation following addition of nitric oxide (NO), ammonium chloride (NH4Cl), and D- and L-glutamate (D-glut, L-glut, respectively) relative to untreated controls (−). (C) Detection of V5-tagged NicD in MOAC-enriched fractions is dependent on the addition of glutamate and the presence of BdlA, as determined by immunoblot analysis using anti-V5 antibodies. NicD phopshorylation levels were determined using of PAO1/pMJT-nicD and PAO1/pMJT-nicD/pJN-bdlA. A total of 10 µg per TCE, obtained prior to MOAC purification, was used as loading control. For MOAC samples, the entire MOAC eluate was loaded. (−) no L-glutamate was added to cells grown planktonically to exponential phase prior to cell lysis. L-glut, L-glutamate was added to cells grown planktonically to exponential phase prior to cell lysis. (D) Immunoblot analysis of in vivo pull-down assays (Co-IP) demonstrating complex formation between NicD, BdlA and DipA using BdlA-HA or DipA-HA either as prey or bait. Total cell extracts obtained from P. aeruginosa PAO1 were used as negative control. Cell extracts containing the protein of interest were used as positive control (+, control). Total cell extracts (TCE) obtained prior to Co-IP were used as loading control. (E) Inactivation of nicD affects the subcellular localization of BdlA and DipA. Detection of subcellular localization was achieved by ultracentrifugation and subsequent analysis by SDS/PAGE and immunoblot analysis using anti-V5 and anti-HA antibodies. A total of 15 µg per cytoplasmic (cyto) and membrane (mem) fraction was loaded. Band intensity was determined using ImageJ to determine the ratio of cellular localization (%) of BdlA and DipA in the absence and presence of NicD. (F) NicD contributes to BdlA phosphorylation. Detection of BdlA-V5 in total cell extracts (TCE) or MOAC-enriched phosphoproteomes (MOAC) of P. aeruginosa PAO1 and ΔnicD biofilms as determined by immunoblot analysis. A total of 10 µg per TCE, obtained prior to MOAC purification, was used as loading control. For MOAC samples, the entire MOAC eluate was loaded. (G) Exposure to L-glutamate enhances BdlA phosphorylation in a NicD-dependent manner. (−) no L-glutamate was added to cells grown planktonically to exponential phase prior to cell lysis. L-glut, L-glutamate was added to cells grown planktonically to exponential phase prior to cell lysis. (H) Non-processive cleavage of BdlA is dependent on the mode of growth and the presence of NicD. Inactivation of nicD impairs BdlA processing under biofilm growth conditions. No BdlA processing is observed under planktonic growth conditions. BdlA was detected by immunoblot analysis. All experiments were carried out at least in triplicate and only representative images are shown. Error bars indicate standard deviation.
NicD contributes to subcellular localization and activation of BdlA
Considering that the increase in c-di-GMP levels upon glutamate addition was found to be temporary (Fig. 5B), the finding suggested the subsequent induction of mechanisms resulting in the degradation of c-di-GMP. We therefore asked whether NicD activity is regulated via interaction with known proteins involved in biofilm dispersion in response to glutamate, in particular BdlA. BdlA is central to glutamate-induced dispersion and has been previously demonstrated to interact with DipA and RbdA, two PDEs playing a role in glutamate-induced dispersion (Basu Roy et al., 2012, Petrova & Sauer, 2012b). Protein interaction studies were carried out in P. aeruginosa PAO1 using pulldown assays with HA-tagged BdlA and DipA serving as bait, and V5-tagged NicD serving as prey. Subsequent immunoblot analysis confirmed NicD to interact with BdlA and DipA in vivo (Fig. 6D). Similar results were obtained when HA-tagged BdlA and DipA were used as prey (Fig. 6D). Moreover, while in accord with previous observations, BdlA interacted with DipA, the membrane bound two-component sensory protein BfiS did not co-purify with HA-tagged BdlA (Fig. S5). The findings suggested that NicD interacts, at least indirectly, with BdlA and DipA.
The interaction with NicD furthermore affected the cellular localization of BdlA. While predicted to be located in the cytoplasm, BdlA was consistently found in both the cytoplasmic and membrane fractions as determined using ultracentrifugation and subsequent immunoblot analysis. Inactivation of nicD enhanced the cytoplasmic localization of BdlA by 10%, while the fraction of DipA present in the cytoplasm increased from less than 20% to more than 70% (Fig. 6E).
BdlA has been shown to be activated in a biofilm-specific manner, via non-processive cleavage at high c-di-GMP levels upon phosphorylation at tyrosine-238 (Petrova & Sauer, 2012a). In addition to NicD affecting BdlA localization, NicD was also found to contribute to BdlA activation. The phosphorylation level of BdlA was analyzed by comparing the detectable levels of BdlA in total cell extracts and the phosphoproteomes, enriched via metaloxide affinity chromatography (MOAC), of wild-type P. aeruginosa PAO1 and ΔnicD biofilm cells. While V5/His-tagged BdlA was present in total cell extracts of both wild-type and ΔnicD biofilm cells, the construct was only detectable in the MOAC-enriched phosphoproteomes of wild-type biofilms, but not ΔnicD biofilms (Fig. 6F). Additionally, while full length and cleaved BdlA were detectable in wild type biofilms, only full-length BdlA was detected in ΔnicD biofilms (Fig. 6H). The findings suggested NicD to likely contribute to both BdlA phoshorylation and non-processive cleavage and thus, BdlA activation. The finding of NicD playing a role in BdlA activation was further supported by the finding of BdlA phopshorylation inversely correlating with NicD dephosphorylation, with BdlA phopshorylation being further enhanced by glutamate addition (Fig. 6G). BdlA in turn, was found to exert an effect on NicD, as co-expression of nicD and bdlA under planktonic growth conditions resulted in a significant decrease in NicD phosphorylation compared to multicopy expression of nicD alone, with NicD phosphorylation being further reduced upon glutamate addition (Fig. 6C).
BdlA contributes to the reduction of c-di-GMP levels upon glutamate addition by stimulating DipA PDE activity
The findings indicated a link between BdlA activation and NicD DGC activity. We reasoned that if BdlA activated or enhanced NicD function, an additional increase in the culture turbidity or c-di-GMP levels upon glutamate addition would be observed. Compared to strains overexpressing only nicD, coexpresion of bdlA and nicD resulted in an overall similar increase in aggregation and thus, similarly decreased medium turbidity prior to and following addition of L- and D- glutamate (Fig. 7A). Given the lack of significant difference in the medium turbidity, the finding suggested BdlA to have no effect on NicD function, However, previous findings demonstrated BdlA to be inactive under planktonic growth conditions (Petrova & Sauer, 2012a). To better mimic the presence of active BdlA, we therefore made use of the hyperactive/constant-on BdlA variant, BdlA-G31A, which was demonstrated to result in a hyperdispersive phenotype (Petrova & Sauer, 2012b). While coexpression of bdlA-G31A and nicD did not impair increased aggregation of PAO1/pMJT-nicD/pJN-bdlA-G31A upon glutamate addition, we noticed that coexpression of bdlA-G31A and nicD resulted in an overall reduced aggregative phenotype, as indicated by an overall significantly higher medium turbidity compared to multicopy expression of nicD and bdlA or nicD alone (Fig. 7A). The finding suggested inactive BdlA to enhance the hyperaggregative phenotype associated with overexpression of NicD, while active constant-on BdlA reduced the hyperaggregative phenotype.
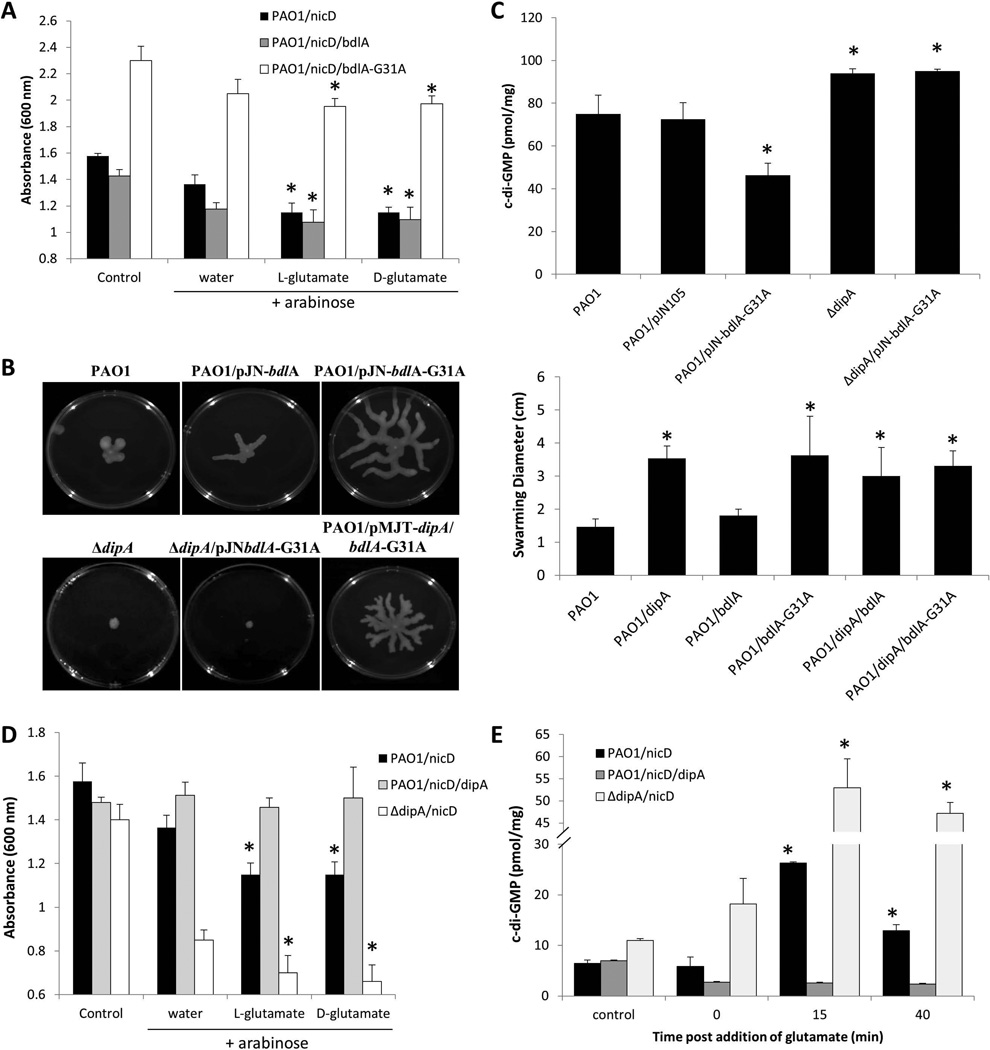
Figure 7. Multicopy expression of constant-on bdlA-G31 enhances DipA phosphodiesterase activity.
(A) Absorbance of culture medium of exponential phase P. aeruginosa PAO1/pMJT-nicD, PAO1/pMJT-nicD/pJN-bdlA, and PAO1/pMJT-nicD/pJN-bdlA-G31A, prior to addition of arabinose (control) and 3 hr post addition of arabinose plus subsequent addition of water, and D- and L-glutamate. Experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from “water” sample to which only arabinose and water was added. (B) Representative images of swarming motility of P. aeruginosa PAO1 and strains overexpressing dipA, bdlA, and bdlA-G31. Swarming motility of P. aeruginosa PAO1 and strains overexpressing dipA, bdlA, and bdlA-G31 as well as strains co-expressing dipA and bdlA or dipA and bdlA-G31A. The swarming diameter was recorded following 48 hr of growth on M8 agar. Experiments were repeated at least three times. Error bars indicate standard deviation.*, significantly different from wild type biofilms; p < 0.05 as determined by ANOVA and SigmaStat. (C) Overexpression of active bdlA-G31A affects the cellular c-di-GMP level present in P. aeruginosa PAO1 biofilm cells in a DipA-dependent manner. C-di-GMP levels present in P. aeruginosa PAO1, PAO1/pJN-bdlA-G31A, ΔdipA and ΔdipA/pJN-bdlA-G31A were quantitated using an HPLC-based method with commercially available c-di-GMP as standard. c-di-GMP (pmol/mg) refers to c-di-GMP levels (pmol) per total cell pellet protein (in mg). Experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from PAO1 and PAO1 biofilms harboring the empty vector pJN105. (D) Absorbance of culture medium of exponential phase P. aeruginosa PAO1/pMJT-nicD, PAO1/pMJT-nicD/pJN-dipA, and ΔnicD/pJN-dipA, prior to (control) and following addition of arabinose (3 hr) and subsequent addition of water, and D- and L-glutamate. Experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from water only sample. (E) Cellular c-di-GMP levels in PAO1/pMJT-nicD, PAO1/pMJT-nicD/pJN-dipA, and ΔdipA/pMJT-nicD following addition of arabinose (3 hr, control), and 0, 15 and 40 min post-addition of L-glutamate. Experiments were carried out at least in triplicate. Error bars indicate standard deviation *, significantly different from 0 min time point.
Considering that BdlA does not harbor any c-di-GMP modulating domains, the observation pointed at active BdlA-G31 indirectly modulating c-di-GMP levels. Previous reports indicated BdlA-G31 to have increased interaction with RbdA and DipA, correlating with increased overall PDE activity in biofilms (Petrova & Sauer, 2012b). BdlA enhancing PDE activity was further supported by overexpression of bdlA-G31A, but not of wild-type bdlA, significantly enhancing swarming motility by P. aeruginosa (Fig. 7B). Moreover, overexpression of bdlA-G31A correlated with a significant reduction in the cellular levels of c-di-GMP present in biofilms cells (Fig. 7C). We therefore asked which phoshodiesterase is stimulated by active BdlA by screening phosphodiesterase mutants for their ability to swarm. A total of six phosphodiesterase mutants were screened, namely PA3825, ΔrbdA (PA0861), Δarr (PA2188), PA5295, PA2133, and ΔdipA (PA5017). Compared to P. aeruginosa wild type, the mutants with the exception of Δarr demonstrated reduced swarming motility (Fig. S6). Overexpression of bdlA-G31A enhanced the swarming motility of PA3825, ΔrbdA, Δarr, PA5295, and PA2133, indicating that these PDEs are not affected by BdlA (Fig. S6). However, swarming motility was unaltered in a ΔdipA mutant (Figs. 7B, S6). As no difference in dipA transcript abundance was noted in strains overexpressing bdlA and bdlA-G31A compared to the vector control (1.1±0.04 fold change), the findings strongly suggested active BdlA to enhance or stimulate DipA PDE activity.
The phopshodiestearse DipA counters NicD function
The finding suggested DipA activity to be modulated by active BdlA. The notion was further supported by the finding of BdlA contributing to an overall reduction in cellular c-di-GMP levels of biofilms, however, BdlA affecting c-di-GMP levels was found to be DipA dependent, as no difference in c-di-GMP levels were noted in dipA mutant biofilms and dipA mutant biofilms overexpressing bdlA-G31A (Fig. 7C). To determine whether DipA is capable of countering the increased NicD activity noted upon glutamate exposure, turbidity assays were carried out using strains expressing nicD and/or dipA. Compared to strain PAO1/pMJT-nicD, expression of nicD in a ΔdipA mutant resulted in a significant decrease in the medium turbidity noted upon induction of nicD gene expression prior to and following arabinose addition (Fig. 7D). Moreover, an additional decrease was observed post-addition of D- and L-glutamate (Fig. 7D). In contrast, however, no change in the medium turbidity was noted for strain PAO1/nicD/dipA upon addition of arabinose or arabinose plus glutamate compared to the control (Fig. 7D).
The effect of coexpression of dipA and nicD on the hyperagregative phenotype of P. aeruginosa PAO1 was also reflected in the c-di-GMP levels. No difference in cellular c-di-GMP levels was noted between PAO1/pMJT-nicD and PAO1/dipA/nicD controls, prior to the addition of arabinose (Fig. 7E). However, c-di-GMP levels present in PAO1/dipA/nicD cells were reduced compared to PAO1/nicD alone following the addition of arabinose. Moreover, while the enhanced aggregative phenotype of PAO1/pMJT-nicD upon exposure to glutamate correlated with a 3-fold increase in c-di-GMP levels compared to untreated cells, no increase in the c-di-GMP levels present in PAO1/dipA/nicD was noted upon addition of glutamte (Fig. 7E). The finding suggested DipA to counter the effect of NicD diguanylate cyclase activity. In support of DipA countering the effect of NicD was the finding that c-di-GMP levels were found to be significantly increased in ΔdipA/pMJT-nicD compared to levels present in PAO1/pMJT-nicD 15 min post-addition of glutamate (Fig. 7E). Moreover, inactivation of dipA eliminated the decrease in c-di-GMP levels noted within 40 min of incubation post-glutamate addition for PAO1/pMJT-nicD (Fig. 7E), indicating DipA to not only be responsible for the degradation of c-di-GMP but also to be responsible for the return to pre-glutamate levels.
NicD, BdlA, and DipA are part of the same pathway required for glutamate-induced disperison
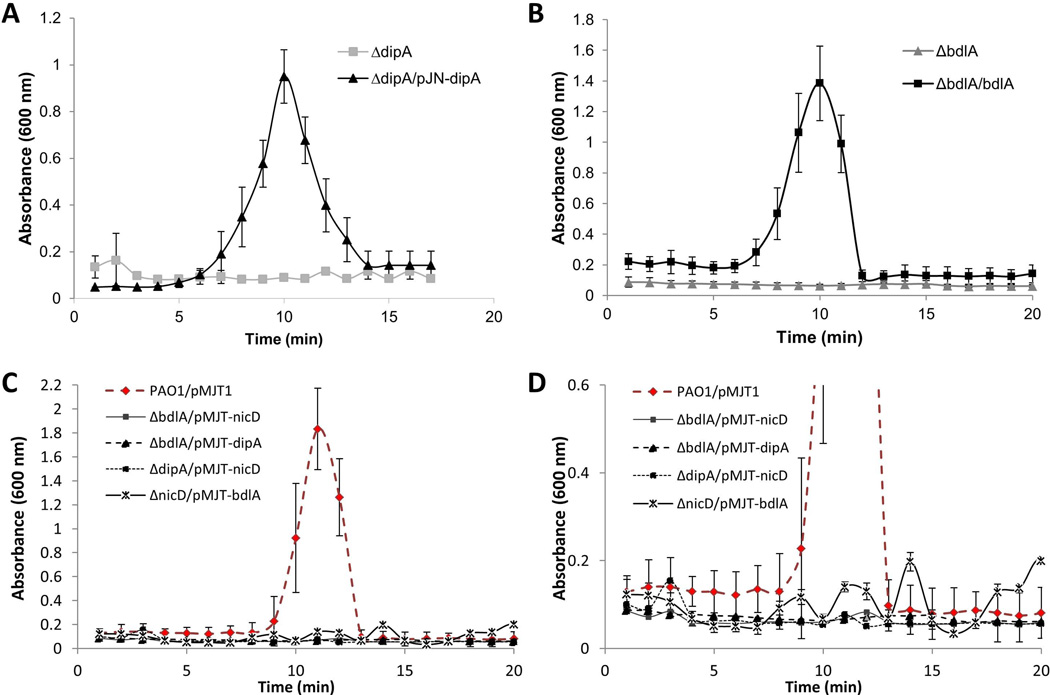
The findings indicated NicD to likely transduce an extracellular cue via phosphorylation to first raise intracellular c-di-GMP through its own DGC activity and, subsequently, via phosphotransfer and/or elevated c-di-GMP levels contributing to BdlA activation. Active BdlA, in turn, activates DipA phosphodiesterase activity, thus, reducing the intracellular c-di-GMP pool and enabling biofilm dispersion. The finding suggested BdlA, DipA and NicD to work in concert to enable dispersion. To confirm this interaction and its contribution to dispersion, biofilm tube reactor-based dispersion assays were carried out. We reasoned that if the proteins indeed work in concert, expression of, for instance, dipA in a ΔnicD mutant background or expression of nicD in a ΔbdlA mutant would not restore dispersion to wild type P. aeruginosa PAO1 levels. However, if the proteins contribute independently to the dispersion response, overproduction of any downstream acting proteins was anticipated to restore dispersion to wild-type levels. While ΔdipA/dipA, ΔnicD/nicD, and ΔbdlA/bdlA exhibited wild-type biofilm dispersion responses (Figs. 4, 8A–B), overexpression of nicD in ΔbdlA or ΔdipA mutants did not enable biofilm dispersion (Fig. 8C–D). Likewise, overexpression of dipA in a ΔbdlA mutant did not restore biofilm dispersion to wild-type levels (Fig. 8C–D). Our findings strongly suggested the three proteins to form a complex that is part of a signaling cascade required for dispersion.
Figure 8. Dispersion of mutants inactivated in nicD, bdlA and dipA in response to glutamate cannot be restored by cross-complementation.
Biofilms were grown for 5 days under flowing conditions in tube reactors prior to induction of dispersion inducing conditions, via exposure to L-glutamate. (A) Multicopy expression dipA in ΔdipA biofilms, and (B) multicopy expression of bdlA in ΔbdlA biofilms restores biofilm dispersion response to glutamate to wild-type levels. (C) Multicopy expression nicD and dipA in ΔbdlA biofilms, nicD in ΔdipA biofilms, or bdlA in ΔnicD biofilms does not restore biofilm dispersion response to glutamate to wild-type levels, as indicated by measurements of effluent absorbance. (D) Expanded view of data shown in Figure 8C. All experiments were repeated at least three times. Error bars indicate standard deviation.
DISCUSSION
The process of biofilm dispersion has been described to occur in response to various endogenous and exogenous cues, with dispersed cells differing physically and physiologically from biofilm and planktonic cells with respect to levels of motility, protein and gene expression, and even susceptibility. Numerous adhesins, factors and enzymes have been described to play a role in dispersion of various bacterial biofilms, with the next challenge being to determine how these pieces work in concert to induce dispersion. Here, we demonstrate that nutrient-induced dispersion, specifically dispersion in response to glutamate, occurs via a signal transduction mechanism, requiring the membrane-localized NicD. Several lines of evidence support the notion of NicD being an active c-di-GMP synthase. Consistent with the presence of a GGDEF domain, overexpression of nicD significantly increased the cellular c-di-GMP levels present in biofilm cells compared to cells in which the gene was not overexpressed or inactivated. Moreover, we show that both inactivation and overexpression of nicD alter several phenotypes previously associated with the modulation of c-di-GMP levels including biofilm biomass accumulation, aggregative phenotype in liquid and swarming motility. While protein extracts of E. coli overexpressing pleD produced significantly larger amounts of c-di-GMP than extracts overexpressing nicD, increased levels of c-di-GMP were nevertheless detected compared to E. coli control extracts or extracts overexpressing nicD-GGAAF, in which the GGDEF motif was substituted by GGAAF. Moreover, purified NicD but not NicD-GGAAF was found to produce c-di-GMP, although with low specific activity. The low activity of NicD is supported by a recent report by Gupta et al. demonstrating that cell extracts obtained from planktonic P. aeruginosa overexpressing nicD only contained 1.5-times higher c-di-GMP than cell extracts in which the gene was not overexpressed (Gupta et al., 2014). The activity of several DGCs and PDEs has been suggested to be modulated in response to environmental or intracellular stimuli as a manner to fine-tune the c-di-GMP signaling network (Romling et al., 2005). For instance, the activities of PleD and of the Rrp1 protein from Borrelia burgdorferi are strongly dependent on the phosphorylation status of the CheY domains (Chan et al., 2004, Ryjenkov et al., 2005), thus allowing the enzymes to only produce or degrade c-di-GMP only when a specific signal has been received. While we have not identified the amino acids at which NicD is phosphorylated or the respective kinase, our findings indicate that NicD activity is likely regulated in a manner similar to PleD or Rrp1, as NicD enzyme activity was found to be modulated in response to environmental stimuli. Moreover, we demonstrate that upon extracellular glutamate sensing, NicD is less phosphorylated, with dephosphorylated NicD correlating with increased diguanylate cyclase activity, as indicated by the increase in the intracellular c-di-GMP pool. The role of phosphorylation in regulating NicD activity will be the focus of future research. While our finding of an active DGC playing a role in dispersion by temporarily increasing the c-di-GMP levels was surprising, our results indicate that active NicD likely contributes to BdlA activation in two different ways, likely via phosphorylation and by increasing c-di-GMP levels, both of which are required for BdlA activation. BdlA, in turn, then activates the phosphodiesterase DipA, with active BdlA furthermore recruiting a second phosphodiesterase, RbdA, to the protein complex (Petrova & Sauer, 2012b), with the result being an overall reduction in the intracellular c-di-GMP levels, enabling biofilm dispersion. A model summarizing our findings is shown in Figure 9. Several DGCs and PDEs have been shown to work in concert with other proteins to enable c-di-GMP signaling and to integrate components of the c-di-GMP signaling pathway on several levels. Numerous DGCs and PDEs are intimately linked to two-component signaling systems. In some bacteria, genes encoding EAL domain proteins are coexpressed with sensor kinase and response regulator genes (Kulasekara et al., 2005, Tischler & Camilli, 2004). Many functionally characterized GGDEF/EAL domain proteins have an N-terminal receiver domain (Drenkard & Ausubel, 2002, Gronewold & Kaiser, 2001), that is phosphorylated by cognate sensor kinases, or for which c-di-GMP modulating activity has been shown to be modulated by phosphorylation (Aldridge et al., 2003, Kulasekara et al., 2005, De et al., 2009, Hickman et al., 2005, Paul et al., 2007). In Salmonella enterica, production of the GGDEF domain protein AdrA is transcriptionally activated by the response regulator CsgD (Römling et al., 2000). Ryan et al. (Ryan et al., 2010) reported the dynamic interaction between a two-component system comprising HD-GYP domain harboring RpfG and the sensor kinase RpfC and two proteins with diguanylate cyclase (GGDEF) domains, with the interaction controlling a subset of RpfG-regulated virulence factors such as extracellular enzymes, biofilm structure, and motility. In E. coli, several DGCs (YegE, YdaM) and PDEs (YhjH, YciR) and the MerR-like transcription factor MlrA regulate the transcription of csgD, which encodes a biofilm regulator essential for producing amyloid curli fibres of the biofilm matrix. Lindenberg et al. (Lindenberg et al., 2013) demonstrated that this system operates as a signaling cascade, in which c-di-GMP controlled by the DGC/PDE pair YegE/YhjH (module I) regulates the activity of the YdaM/YciR pair (module II). Via multiple direct interactions, the two module II proteins form a signaling complex with MlrA and YciR acting as a connector between modules I and II (Lindenberg et al., 2013).
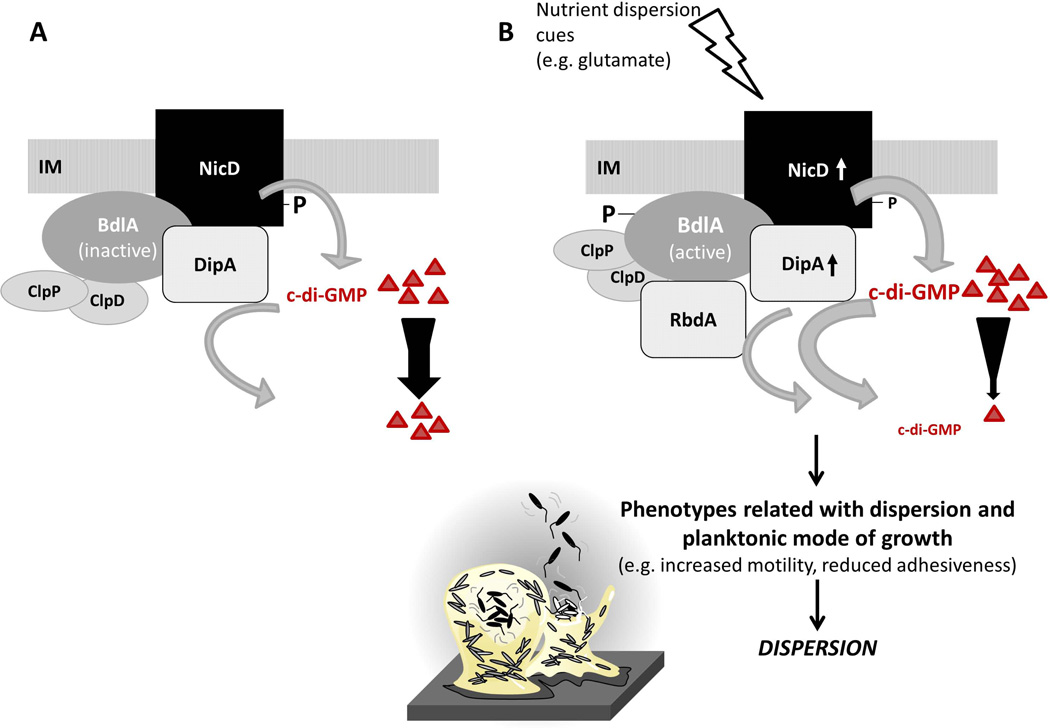
Figure 9. Model of signal transduction upon sensing the dispersion-inducing cue glutamate.
(A) The diguanylate cyclase NicD is membrane-bound and phosphorylated. NicD forms a multiprotein complex with the phosphodiesterase DipA and the sensory protein BdlA. BdlA is intact but inactive under planktonic growth conditions (Petrova & Sauer, 2012a). BdlA interacts with the chaperone ClpD and the protease ClpP (Petrova & Sauer, 2012a). (B) Upon perceiving a dispersion-inducing nutrient cue, e.g. glutamate, NicD is dephosphorylated and its diguanylate cyclase activity increases, resulting in elevated levels of c-di-GMP. BdlA is phosphorylated. Both phosphorylation and elevated levels of c-di-GMP contribute to BdlA being cleaved in a non-processive manner, a process requiring the chaperone ClpD, the protease ClpP, and BdlA phosphorylation at Y238 (Petrova & Sauer, 2012a). Cleaved BdlA is active with respect to enabling P. aeruginosa biofilms to respond to dispersion inducing conditions (Petrova & Sauer, 2012a). Active BdlA in turn enhances the activity of the phosphodiesterase DipA, resulting in decreased c-di-GMP levels. It has been well demonstrated that increased DipA activity results in decreased biofilm c-di-GMP levels (Basu Roy et al., 2012). Moreover, active BdlA was recently demonstrated to interact with the phosphodiesterase RbdA in vivo (Petrova & Sauer, 2012b). The recruitment of RbdA to the multiprotein complex likely further contributes to the observed reduction of c-di-GMP in dispersed cells compared to biofilm cells (Basu Roy et al., 2012, An et al., 2010, Barraud et al., 2009). Dispersion has been described to require or coincide with the breakdown of extracellular polymeric matrix surrounding the biofilms, induction of flagellar gene expression, increased motility, and reduced adhesiveness (Morgan et al., 2006, Sauer et al., 2002, Sauer et al., 2004, Basu Roy et al., 2012). IM, inner membrane; triangles and number of triangles represent c-di-GMP and cellular c-di-GMP levels. P, phosphorylation. Size of P correlates with level of phosphorylation. Arrows associated with NicD and DipA indicate increased enzyme activity. Width of grey arrows indicates level of activity.
Our findings suggest that the membrane-bound DGC NicD with its periplasmic/7TM sensory and cytoplasmic GGDEF domains appears to relay dispersion cues across the bacterial membrane, with signal relay resulting in NicD dephosphorylation, activation of DGC activity, and temporarily increased levels of the intracellular c-di-GMP pool. NicD function thus spatially restricts dispersion-signaling events to extracellular dispersion cues that are sensed via the periplasmic 7TMR-DISMED domain. A similar relay was recently described for NO-induced dispersion requiring the membrane bound PDE NbdA harboring the periplasmic NO-sensing MHYT domain (Li et al., 2013). While the finding hinted at NO being sensed outside the cell and relayed via NbdA PDE activity into c-di-GMP modulation, the mechanism of signal transduction is unknown. NO sensing in other bacterial species has been shown to require HNOX-harboring proteins (Plate & Marletta, 2012, Price et al., 2007, Wang et al., 2010). While the manner by which P. aeruginosa senses or responds to NO remains to be elucidated, our findings indicate glutamate-induced dispersion to be likely governed by a signal transduction mechanism, via a multiprotein complex composed of the DGC NicD, PDE DipA, and BdlA, based on phosphorylation events and modulation of the overall cellular c-di-GMP pool. Protein complex formation was found to be NicD-dependent, with NicD enabling the generation of membrane-associated pockets of concentrated enzymes/proteins, with each interacting partner contributing, directly or indirectly, to signal processing and thus, dispersion. Such enzyme configuration is believed in eukaryotic signal transduction pathways to not only enhance the precision of information flow but also to improve the fidelity of signaling events by clustering successive enzymes into a transduction pathway, with intermediate enzymes exhibiting restricted substrate specificities and limited spheres of action (Scott & Pawson, 2009). Here, we demonstrate that sensing glutamate dispersion cues not only activates NicD activity but triggers a sequence of events resulting in BdlA activation via phosphorylation, with active BdlA in turn activating the PDE DipA. While it is unclear how DipA activity is modulated by BdlA, the enhanced PDE activity was not due to increased dipA transcript abundance. It is likewise unclear whether the composition of the multiprotein complex is stable prior to and post addition of dispersion cues. Our findings, however, indicate the protein complex to be modular due to posttranslational modifications, with BdlA activation likely resulting in the recruitment of the PDE RbdA to the complex (Petrova & Sauer, 2012b) (Fig. 9).
Eukaryotic cells possess sophisticated regulatory mechanisms for signal transduction, ensuring that signaling enzymes encounter their substrates in the right place and at the right time (Scott & Pawson, 2009). A variety of signal transduction mechanisms have evolved including, (i) signal-dependent formation of protein complexes; (ii) processing of signals through preassembled multiprotein complexes; (iii) enzyme regulation by subcellular localization; and (iv) temporal control of signaling pathways, with many pathways requiring posttranslational modifications (Scott & Pawson, 2009). The NicD-dependent signal transduction pathway is somewhat similar to eukaryotic signal transduction mechanisms, by incorporating signal processing across compartments via multiprotein complexes and temporal posttranslational modifications. An additional similarity to eukaryotic signal transduction pathways across membranes lies in the 7TMR DISMED2/DISM domains. An enormous family of over 800 genes encodes receptor proteins that are characterized by a signature seven-transmembrane (7TM) configuration (reviewed in (Pierce et al., 2002)). Members of this family regulate virtually all known physiological processes in humans and include receptors for many hormones, neurotransmitters, chemokines and calcium ions, as well as sensory receptors for various odorants, bitter and sweet taste, and even photons of light (Pierce et al., 2002). 7TM receptors are commonly referred to as G-protein-coupled receptors, because most of them signal by activating heterotrimeric G-proteins. For instance, visual signaling in the retina and hormonal signaling in other tissues are both processes involving a ‘receptor’ or input receiver, a transducer ‘G protein’ (transducin in the retina, or Gs in the case of adenylyl cyclase), and an effector enzyme (for example, cyclic GMP phosphodiesterase in the retina, or adenylyl cyclase) (Pierce et al., 2002). While the presence of 7TM homologs was believed to be restricted to the well-characterized sensory rhodopsins of various phototropic prokaryotes, a comparative genomic approach by Anantharaman and Vivek (Anantharaman & Aravind, 2003) demonstrated the presence of two widespread families of 7TM receptors in bacteria that are distantly related to the eukaryotic 7 TM receptors and prokaryotic rhodopsins. The members of the first family of these receptors possess an α-helical extracellular domain, and are predicted to transduce signals via an intracellular HD hydrolase domains. Based on comparative analysis of gene neighborhoods, these receptors are predicted to function as regulators of the diacylglycerol-kinase-dependent glycerolipid pathway. The second family of bacterial 7TM receptors contain one of two distinct N-terminal extracellular globular domains, which are predicted to bind ligands such as carbohydrates. In their intracellular portions they contain fusions to a variety of signaling domains, which suggest that they are likely to transduce signals via cyclic AMP, cyclic diguanylate, histidine phosphorylation, dephosphorylation, and through direct interactions with DNA (Anantharaman & Aravind, 2003). The latter respresents the family to which NicD belongs to.
The signaling mechanism described here to enable dispersion and thus, the transition from the surface associated to planktonic state may be applicable to a large number of pathogenic species, as BLAST and BLINK analysis revealed the presence of NicD orthologs in the genomes of numerous pathogenic and non-pathogenic, Gram-negative bacteria including Burkholderia sp., Pseudoalteromonas sp., Shewanella sp., Methylobacter sp., Vibrio sp., Azotobacter sp., and Alteromonas sp. These pathogens are capable of forming biofilms with the motile/sessile transitions recently implicated as switches in pathogenicity phenotypes. Thus, sensing of exogenous dispersion cues and subsequent signaling to trigger biofilm dispersion may be a contributor to the migration of bacteria to locations remote from an initial site of infection, with cells dispersed from biofilms causing acute and periodic infections.
MATERIAL AND METHODS
Bacterial strains, plasmids, media, and culture conditions
All bacterial strains and plasmids used in this study are listed in Table S2. P. aeruginosa strains PA14 and PAO1 were used as parental strains. All planktonic strains were grown in Lennox Broth (LB, BD Biosciences) or Vogel-Bonner Minimal Medium (VBMM) (Schweizer, 1991) in shake flasks at 220 rpm in the absence or presence of 0.1–1.0% arabinose. Escherichia coli cultures were grown in LB in the absence or presence of 1mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Antibiotics were used at the following concentrations: 50–75 µg/ml gentamicin and 200–250 µg/ml carbenicillin for P. aeruginosa; and 20 µg/ml gentamicin, 25 µg/ml kanamycin, and 50 µg/ml ampicillin for E. coli.
Strain Construction
Complementation and overexpression of nicD (PA4929) was accomplished by placing the respective genes under the control of an arabinose-inducible promoter in the pMJT1 vector. C-terminal V5/6xHis tagging of PA4929 was accomplished by subcloning into pET101D (Invitrogen). Site-directed mutagenesis of indicated nicD sequences was accomplished by using the GeneArt Site-Directed Mutagenesis Kit (Invitrogen) according to the manufacturer’s protocol. The identity of vector inserts was confirmed by sequencing. Plasmids were introduced into P. aeruginosa via conjugation or electroporation. Primers used for strain construction are listed in Table S3.
Growth on D- and L-glutamate
Two isoforms of glutamate are known, D- and L-glutamate. To determine whether P. aeruginosa is capable of using both L- and D-glutamate as a carbon source, growth curves were carried out. Briefly, a total of 5ml of PAO1 grown over-night in LB was split into 5 tubes, spun down (4500xg, 5 min, 4°C) and the resulting cell pellet subsequently washed twice with 1ml minimal medium MMA (Sauer et al., 2002) lacking any carbon source. The pellets were pooled and resuspended in a total of 500µl MMA minimal medium (Sauer et al., 2002) lacking carbon sources. This suspension was used to inoculate 50 ml of minimal medium containing either D- or L- glutamate (130 mg/L) as a carbon source. Growth was carried out at 37°C with shaking at 220 rpm and absorbance was recorded at 600 nm every 90 min over a period of 8 hr. In addition, following growth in L-glutamate supplemented minimal medium, additional L- or D- glutamate (130 mg/L) was added to the cultures at 180 min and absorbance recorded every 90 min for an additional 5 hr to observe growth stimulating effects.
Biofilm formation
Biofilms were grown in a continuous flow tube reactor system (1 m long size 14 silicone tubing, Masterflex, Cole Parmer, Inc.) in VBMM or 20-fold diluted LB medium in presence of 0.1% arabinose at 22°C for up to 6 days at a flow rate of 0.2 ml/min. Biofilms were likewise grown in flow cells to view the biofilm architecture by confocal scanning laser microscopy (CSLM) using a LSM510 Meta confocal microscope (Zeiss, Germany) as previously described (Allegrucci et al., 2006, Allegrucci & Sauer, 2007, Petrova & Sauer, 2009, Sauer et al., 2002, Southey-Pillig et al., 2005). Quantitative analysis of CSLM images was performed using COMSTAT (Heydorn et al., 2000).
Biofilm dispersion assays
For biofilm dispersion assays, biofilms were cultivated in once-through continuous flow tube reactor system at 22°C for 5 days. After 5 days of biofilm growth, biofilm dispersion was induced by the sudden addition of D- or L-glutamate (18 mM), succinate (20 mM), glucose (20 mM), or ammonium chloride (100 mM), to the growth medium as previously described (Morgan et al., 2006). In addition, 500 µM sodium nitroprusside (SNP) was used as a source of nitric oxide (Barraud et al., 2006). Dispersion was indicated by an increase in turbidity at 600 nm in the effluent from the silicone tubing. Dispersion of flow cell grown biofilms was induced in a similar manner. Confocal images prior to and following induction of dispersion were analyzed using COMSTAT as previously described (Morgan et al., 2006).
Diguanylate cyclase assays
Diguanylate cyclase assays were performed essentially as previously described (Paul et al., 2004, Monds et al., 2007). Briefly, diguanylate cyclase assays were performed using total cell extracts obtained from E. coli overexpressing nicD, nicDΔNoTMR, nicD-GGAAF, and pleD under the control of an IPTG-inducible promoter (pET101D, pET11b), or purified protein. Total cell extracts from E. coli BL21 not harboring an plasmid were used as negative control while E. coli extracts overexpressing the diguanylate cyclase pleD were used as positive control. Total extracts were obtained by sonication followed by centrifugation to remove cell debris as previously described (Southey-Pillig et al., 2005) or detailed below. Protein determination was carried out using a modified Lowry as previously described (Southey-Pillig et al., 2005). Diguanylate cyclase assays were performed essentially as previously described (Paul et al., 2004, Monds et al., 2007) with some modifications. Briefly, the reaction mixtures contained 75 mM Tris-HCl at pH 7.8, 250 mM NaCl, 25 mM KCl, 10 mM MgSO4 in 200 µL volume and were started by the addition of a mixture of 25 µM GTP. A total of 200 µg of total extract was used per assay. The reaction mixtures were incubated for up to 360 min at 37°C, terminated by heating to 95°C for 5 min, and subsequently analyzed by HPLC as described below.
Purification of His-tagged proteins
V5/6xHis-tagged proteins were purified from E. coli supernatants following sonication of LB-grown planktonic cells, and centrifugation at 21,200 × g. The supernatant was loaded onto nickel-nitrilotriacetic acid (Ni-NTA) affinity resin (Qiagen), washed with buffer, and eluted with an imidazole gradient according to the manufacturer’s instructions for native protein purification. Since NicD was found to be membrane-associated, the purification was carried out with the detergent tween-20 (0.1 % final concentration) present in all buffers. Protein preparations were examined for purity by SDS-PAGE, and fractions containing pure protein were pooled and desalted using VivaSpin centrifugal concentrator columns (10 kDa cut-off, Sartorius) (Fig. S2). For di-guanylate cyclase assays, protein fractions obtained following Ni-NTA affinity chromatography from E. coli BL21 were used as negative controls.
In vivo quantification of c-di-GMP from P. aeruginosa
Cyclic di-GMP (c-di-GMP) was extracted in triplicate from wild type and mutant strains using heat and ethanol precipitation (Morgan et al., 2006) and quantitated essentially as previously described (Petrova & Sauer, 2011). Briefly, c-di-GMP was extracted in triplicate from wild-type and mutant strains grown planktonically to exponential phase or as biofilms for 6 days using heat and ethanol precipitation followed by centrifugation. Supernatants were combined, dried using a Speed-Vac and resuspended in water. Samples (20 µl) were analyzed using an Agilent 1100 HPLC equipped with an autosampler, degasser, and detector set to 253 nm, and separated using a reverse-phase C18 Targa column (2.1 × 40 mm; 5 µm) at a flow rate of 0.2 ml/min with the following gradient: 0 to 9 min, 1% B; 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 26 min, 90% B; 26 to 40 min, 1% B (buffer A, 10 mM ammonium acetate; buffer B, methanol plus 10 mM ammonium acetate) (Basu Roy et al., 2013, Basu Roy et al., 2012). Commercially available cyclic di-GMP was used as a reference for the identification and quantification of cyclic di-GMP in cell extracts.
Swarming assays
P. aeruginosa PAO1 swarming motility was determined using M8 medium in the presence of 0.5% agar and 1% arabinose. P. aeruginosa PA14 swarming motility was determined using 0.8% nutrient broth (Difco, BD Company) supplemented with 0.5% glucose in the presence of 0.5% agar and 1% arabinose. The swarming diameter was determined following 48 hr of incubation at 37°C.
Turbidity Assays
To determine the effect of dispersion inducing cues on NicD activity, turbidity assays were used. The assays are based on the finding that overexpression of cyclases including nicD, has been shown to induce auto-aggregation of cells growing planktonically, with aggregates settling to the bottom of the growth tube under static conditions, resulting in a decrease in the absorbance of the growth medium. Briefly, PAO1 strains over-expressing nicD (PA4929) under the control of the PBAD promoter were grown as two successive over-nights, first in 5ml and then in 50 ml LB. On the third day, 4ml of the over-night grown culture was used to inoculate 50 ml fresh LB medium and allowed to grow for 3 hr at 37°C with shaking at 220 rpm. The bacterial culture was then split into 5ml/tube and arabinose (1% final concentration) was added to induce gene expression. Following 3.5 hr of continued growth at 37°C, dispersion cues (18 mM L-glutamate, 18 mM D-glutamate or 100 mM ammonium chloride final concentration) or equal volume of sterile water was added, and the bacterial cultures were incubated for an additional 30 min at 37°C. The absorbance of the supernatant was recorded at 600 nm the after cell aggregates were allowed to settle at room temperature for 10 min. In addition, the cultures were used for c-di-GMP extraction.
Quantitative real-time reverse-transcription PCR (qRT-PCR)
qRT-PCR was used to determine the expression levels of dipA (PA5017) using 1 µg of total RNA isolated from wild type P. aeruginosa PAO1, PAO1/pJN-bdlA, and PAO1/pJN-bdlA-G31A grown planktonically. Isolation of mRNA and cDNA synthesis was carried out as previously described (Petrova & Sauer, 2010, Petrova & Sauer, 2011). qRT-PCR was performed using the CFX Connect realtime PCR cycler (Biorad) and the SsoAdvanced SYBR Green supermix (Biorad), with oligonucleotides listed in Table S3. mreB was used as control. The stability of mreB levels were verified by 16S RNA abundance using primers HDA1/HDA2 (McBain et al., 2003). Relative transcript quantitation was accomplished using the CFX Connect software (Biorad) by first normalizing transcript abundance (based on Ct value) to mreB followed by determining transcript abundance ratios. Melting curve analyses were employed to verify specific single product amplification.
Protein localization
To determine cellular localization of NicD, DipA, and BdlA, and to determine whether presence or absence of NicD affects the cellular localization of DipA and BdlA, P. aeruginosa PAO1 and ΔnicD expressing V5/His-tagged DipA, and BdlA were grown planktonically to exponential phase in VBMM medium containing 0.1% arabinose. The cultures were centrifuged for 5 min at 10,000 g and the resulting pellet resuspended in 1 ml TE buffer (10 mM Tris/HCl pH 8.0, 1 mM EDTA, plus 0.3 mg/ml PMSF) and lysed by sonication. The samples were centrifuged for 6 min at 21,200 × g to pellet unbroken cells and the resulting supernatant spun again at 30,000 × g to remove any debris. The resulting supernatant was spun at 100,000 × g for 90 min at 4°C. The supernatant containing cytoplasmic proteins was retained and the pellet was washed with cold TE buffer and centrifuged for another 90 min at 4°C at 100,000 g. The final pellet containing membrane proteins was resuspended in 1 ml TE buffer containing 1% CHAPS. SDS loading buffer was mixed with the cytoplasmic and membrane samples (having equal protein concentrations), followed by heat denaturation at 100°C for 10 min. The samples were resolved on an 11% polyacrylamide gel and subsequently transferred onto PVDF membrane using a TransTurbo blot apparatus (Biorad). Western blots were probed with anti-V5 antibodies and developed with ImmunStar WesternC detection reagents (Biorad). Considering that catalase has been demonstrated to be located in the cytoplasm but absent from the membrane (Katsuwon & Anderson, 1990, Brown et al., 1995), proper fractionation was determined via the absence of catalase activity in the membrane fraction but presence in the cytoplasmic fraction using 1 mM hydrogen peroxide as substrate. No catalase activity was detected in thus prepared membrane fractions. Commercially available catalase (purified from Aspergillus sp.) was used as control.
Immunoblot analysis and pulldowns
Pulldown assays were used to assess the in vivo interactions between various NicD, BdlA, and DipA, using total protein cell extracts from P. aeruginosa PAO1/pMJT-nicD-V5/pJN-bdlA-HA and PAO1/pMJT-nicD-V5/pJN-dipA-HA. Pulldown assays using the membrane-bound two-component sensory BfiS (PAO1/pMJT-bdlA-HA/pJN-bfiS-V5) were used as control. Briefly, HA-tagged proteins were immunoprecipitated using immobilized anti-HA antibodies. Immunoprecipitation eluates were resolved on an 11% polyacrylamide gel and subsequently transferred onto PVDF membrane using a TurboTransblot apparatus (Biorad). Western blots were probed with anti-V5 antibodies (Invitrogen). The blots were subsequently developed using ImmunStar WesternC chemiluminescent reagents (Biorad). The pulldowns were also reversed using V5-tagged proteins as bait and probing immunoblots for HA-tagged proteins using anti-HA antibodies (Covance). Aliquots obtained prior to pulldowns were used as loading controls to ensure equal loading. The indicated antibodies (Covance, Invitrogen Corp) were used for immunoblotting at 0.2 µg/mL while 2 µl each were used for immunoprecipitations.
Phosphoprotein enrichment, detection by immunoblot analysis
Qualitative analysis of phosphorylated NicD and BdlA levels in protein extracts obtained from P. aeruginosa biofilm cells was accomplished using phosphoprotein purification via metal oxide affinity chromatography (MOAC) essentially as described by Wolschin and colleagues (Wolschin et al., 2005). MOAC has been demonstrated by Krüger et al. to result in up to 20-fold enrichment of phosphoproteins and to approach 100% specificity (Krüger et al., 2007). Briefly, 750 µg of cell extract were diluted with MOAC incubation buffer (30 mM MES, 0.2 M potassium glutamate, 0.2 M sodium aspartate, 0.25 % Chaps, and 8 M urea) to a final volume of 1.5 mL, and subsequently incubated for 30 min at 4°C in the presence of 80 mg of aluminum hydroxide. Unbound phosphoproteins were removed by washing the aluminum hydroxide slurr with incubation buffer. Then, phosphoproteins were eluted from the slurr using 100 mM potassium pyrophosphate and 8M urea, desalted by methanol-chloroform precipitation, and subsequently vacuum-dried. Samples were then analyzed by SDS/PAGE, followed by the detection of V5 tagged proteins by immunoblotting with anti-V5 antibodies. Aliquots obtained prior to MOAC (15 µg) were used as loading controls.
Statistical analysis
A Student’s t-test was performed for pair-wise comparisons of groups, and multivariant analyses were performed using a 1-Way ANOVA followed by a posteriori test using Sigma Stat software. All experiments were performed at least in triplicate.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by a grant from NIH (1RO1 A107525701A2).
REFERNCES
- Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus . Mol. Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, Post JC, Sauer K. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 2006;188:2325–2335. doi: 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci M, Sauer K. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 2007;189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Wu Je, Zhang L-H. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing fomain. Appl. Environ. Microbiol. 2010;76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics. 2003;4:34. doi: 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa . J. Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy A, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 2012;194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy A, Petrova OE, Sauer K. Extraction and quantification of cyclic di-GMP from Pseudomonas aeruginosa . bio-protocol. 2013 doi: 10.21769/bioprotoc.828. http://www.bio-protocol.org/wenzhang.aspx?id=828. [DOI] [PMC free article] [PubMed]
- Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- Brown SM, Howell ML, Vasil ML, Anderson AJ, Hassett DJ. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. National Acad. Sci. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Davies DG, Marques CNH. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De N, Navarro MVAS, Raghavan RV, Sondermann H. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 2009;393:619–633. doi: 10.1016/j.jmb.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008;6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- Emori TG, Gaynes RP. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RBR, Antunes LCM, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 2008;190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 2010;75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 2005;7:894–904. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- Gronewold TMA, Kaiser D. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 2001;40:744–756. doi: 10.1046/j.1365-2958.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa . Mol. Microbiol. 2014;92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Huangyutitham V, Güvener ZT, Harwood CS. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio. 2013;4 doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB. Biofilm dispersal: mechanisms, clinical Implications, and potential therapeutic uses. J. Dent. Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2004;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuwon J, Anderson AJ. Catalase and superoxide dismutase of root-colonizing saprophytic fluorescent Pseudomonads. Appl. Environ. Microbiol. 1990;56:3576–3582. doi: 10.1128/aem.56.11.3576-3582.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger R, Wolschin F, Weckwerth W, Bettmer J, Lehmann WD. Plant protein phosphorylation monitored by capillary liquid chromatography--element mass spectrometry. Biochem. Biophys. Res. Commun. 2007;355:89–96. doi: 10.1016/j.bbrc.2007.01.108. [DOI] [PubMed] [Google Scholar]
- Kulasekara H, Ventre I, Kulasekara B, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- Lee SF, Li YH, Bowden GH. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infection and Immunity. 1996;64:1035–1038. doi: 10.1128/iai.64.3.1035-1038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J. Bacteriol. 2013;195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013;32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain AJ, Bartolo RG, Catrenich CE, Charbonneau D, Ledder RG, Rickard AH, Symmons SA, Gilbert P. Microbial characterization of biofilms in domestic drains and the establishment of stable biofilm microcosms. Appl. Environ. Microbiol. 2003;69:177–185. doi: 10.1128/AEM.69.1.177-185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Micro. 2012;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- Merritt JH, Brothers KM, Kuchma SL, O'Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 2007:585–507. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, McMullan R, Filloux A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS . PLoS ONE. 2011;6:e29113. doi: 10.1371/journal.pone.0029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, Newell PD, Gross RH, O'Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 2007;63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa . J. Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 2007;282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc. National Acad. Sci. 2012a;109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J. Bacteriol. 2012b;194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Plate L, Michael A, Marletta Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Molecular cell. 2012;46:449–460. doi: 10.1016/j.molcel.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu M, Wood TK. Tyrosine phosphatase TpbA controls rugose colony formation in Pseudomonas aeruginosa by dephosphorylating diguanylate cyclase TpbB. Biochem. Biophys. Res. Commun. 2010;402:351–355. doi: 10.1016/j.bbrc.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Rohde M, Olsén A, Normark S, Reinköster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris . Proc. National Acad. Sci. 2010;107:5989–5994. doi: 10.1073/pnas.0912839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Micro. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- Schooling SR, Charaf UK, Allison DG, Gilbert P. A role for rhamnolipid in biofilm dispersion. Biofilms. 2004;1:91–99. [Google Scholar]
- Schweizer HP. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa . J. Bacteriol. 1991;173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they're apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey-Pillig CJ, Davies DG, Sauer K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2005;187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Wood TK. Connecting Quorum Sensing, c-di-GMP, Pel Polysaccharide, and Biofilm Formation in Pseudomonas aeruginosa through Tyrosine Phosphatase TpbA (PA3885) PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A, Rosenbaum M, Arends JBA, Halitschke R, Angenent LT. Quorum sensing regulates electric current generation of Pseudomonas aeruginosa PA14 in bioelectrochemical systems. Electrochemistry Communications. 2010;12:459–462. [Google Scholar]
- Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri . Proc. National Acad. Sci. 2010;107:8375–8380. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock REW, Brinkman FSL. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucl. Acids Res. 2009;37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschin F, Wienkoop S, Weckwerth W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC) PROTEOMICS. 2005;5:4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.