Abstract
The transcriptional architecture of intracellular circadian clocks is similar across phyla, but in mammals interneuronal mechanisms confer a higher level of circadian integration. The suprachiasmatic nucleus (SCN) is a unique model to study these mechanisms, as it operates as a ∼24 h clock not only in the living animal, but also when isolated in culture. This “clock in a dish” can be used to address fundamental questions, such as how intraneuronal mechanisms are translated by SCN neurons into circuit-level emergent properties and how the circuit decodes, and responds to, light input. This review addresses recent developments in understanding the relationship between electrical activity, [Ca2+]i, and intracellular clocks. Furthermore, optogenetic and chemogenetic approaches to investigate the distinct roles of neurons and glial cells in circuit encoding of circadian time will be discussed, as well as the epigenetic and circuit-level mechanisms that enable the SCN to translate light input into coherent daily rhythms.
Introduction
Understanding the link between genetic programs, neuronal activity, and circuit processing that determine animal behavior is a long-standing goal of neuroscience. Connecting the several steps necessary to encode behaviors, from genes to neurons to circuits, has proven a formidable challenge, especially because of the difficulty in finding experimental approaches to observe the brain across the several levels of organization involved in this integration. Such a task is particularly daunting in mammals, whose exceptional richness of neuronal cell types and inherent connectivity make it extremely difficult to map both the anatomical and functional interactions responsible for different neural behaviors. In recent years, however, neuroscientists have developed an arsenal of “circuit-hacking” molecular tools, such as intersectional genetics, live imaging, viral transduction (Huang and Zeng, 2013), and optogenetics and chemogenetics (Rogan and Roth, 2011) to target and manipulate specific neuronal types and circuits. These developments have started to facilitate the deconstruction of the neural pathways responsible for complex, enduring behaviors, such as memory (Garner et al., 2012), sleep (Jego et al., 2013), and feeding (Aponte et al., 2011).
Researchers investigating the brain circuits responsible for circadian control of daily rhythms in mammalian physiology enjoy an advantageous position in this endeavor because, in contrast to other neural pathways, the principal brain area responsible for this behavior is highly localized and its neuronal type composition well known (Welsh et al., 2010; Hastings et al., 2014). Indeed, pioneer lesion experiments in rodents soon established the suprachiasmatic nuclei (SCN) of the anterior ventral hypothalamus as a necessary component of the circadian system, governing daily rhythms of behavior and hormone release in mammals (Moore and Eichler, 1972; Stephan and Zucker, 1972). Moreover, subsequent experiments showed that fetal and adult SCN grafts were sufficient to (partially) restore circadian rhythms in SCN-ablated recipient animals and impose on them the genetically specified periodicity of the donor tissue. Thus, the role of the SCN as the master circadian clock in mammals was established (Ralph et al., 1990; Sujino et al., 2003).
The discovery of an underlying intracellular transcription–translation feedback loop (TTFL), oscillating with a ∼24 h periodicity in SCN neurons, thus provided an elegant molecular counterpart to such highly localized neuronal function (Fig. 1A). The TTFL (for review, see Takahashi et al., 2008), in its simplest description, revolves around the timed transcription of period (per) and cryptochrome (cry) genes, activated by BMAL1/CLOCK dimers, and the subsequent accumulation of Per and Cry proteins that progressively inhibit BMAL1/CLOCK activity. Timely degradation of Per and Cry proteins subsequently removes the inhibition from BMAL1/CLOCK and licenses a new ∼24 h cycle. The prominent role of such a cell-autonomous TTFL in SCN timekeeping was, however, questioned when it appeared clear that almost all cells and tissues in mammals shared a similar clockwork (Balsalobre et al., 1998; Welsh et al., 2004). Yet the SCN alone is capable of both orchestrating daily rhythms across other tissues in vivo, as well as sustaining persistent and synchronous circadian oscillations when isolated in culture. Moreover, genetic deletions of several clock genes were shown to have limited effects in the SCN, in contrast to their debilitating consequences for peripheral tissue oscillators (Liu et al., 2007; Ko et al., 2010).
Figure 1.
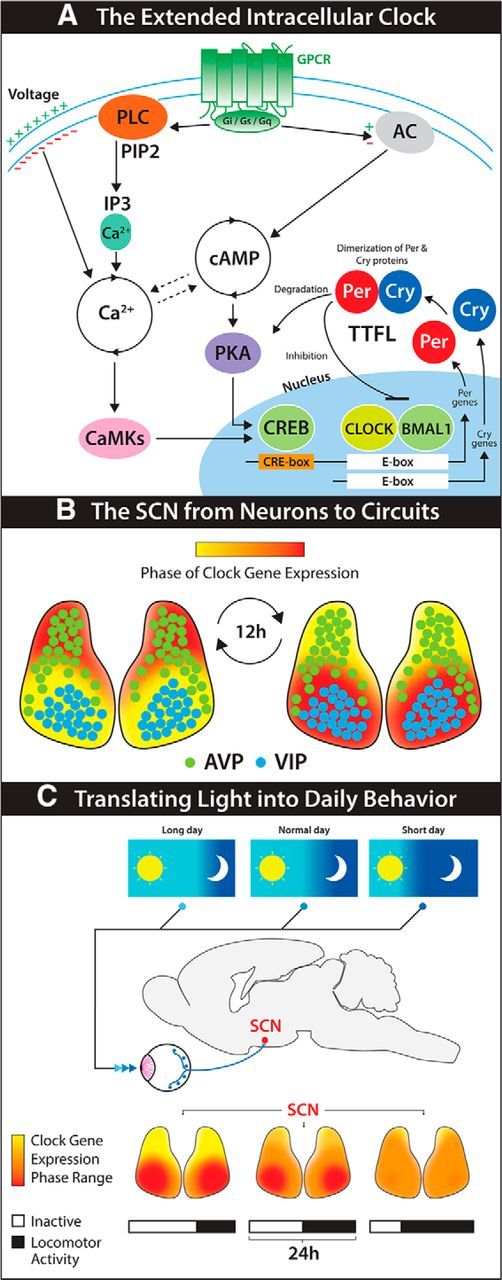
Encoding circadian time in mammals from genes to behavior. A, A simplified model of the intracellular molecular clockwork sustaining ∼24 h oscillations in SCN neurons. TTFLs, based on E-boxes, are synchronized to circadian oscillations of cytosolic Ca2+ and cAMP, via CRE boxes. GPCR-mediated peptidergic signaling and synaptic connectivity provide circuit-derived reinforcement cues to the intracellular clocks, by impinging on the Ca2+ and cAMP oscillations and CREs. B, Representation of the spatiotemporal wave of clock gene expression traveling across SCN tissue (frontal view). The anatomical and cellular architecture of the SCN, as well as the spatiotemporal wave, are preserved in isolated SCN cultures, thus showing them as intrinsic features of the SCN circuit. C, The intrinsically generated spatiotemporal wave is modified in vivo by subjective experience of light. Under short day lengths, SCN neurons display a similar timing of rhythms in protein expression. This phase clustering produces an overall waveform with short duration of peak expression. Phase differences among SCN neurons increase with day length, and this temporal segregation increases peak width at population level. Changes in the spatiotemporal wave are then translated into adaptive behavioral responses, such as variations in locomotor activity.
In sharp contrast with the shared underlying TTFL molecular clockwork, both the cytoarchitecture and the intrinsic and diffused connectivity of the SCN are very varied. Numerous neuronal subpopulations secreting various neurotransmitters can be distinguished within the SCN, such as vasoactive intestinal polypeptide (VIP), arginine-vasopressin (AVP), and gastrin-releasing peptide (GRP) (Abrahamson and Moore, 2001). The spatial distribution of these subpopulations is remarkably specific and highly conserved across species, thus suggesting that they may play differential roles in processing information flow within the nucleus or in relation to its input/output structure (Leak and Moore, 2001). In particular, GRP and VIPergic neurons populate the ventrolateral SCN (core region), which directly receives retinal input, and relay it throughout the entire SCN (through both ipsilateral and contralateral projections), as well as to other hypothalamic and thalamic areas. In contrast, the AVP subpopulation delineates the shell region, which receives less retinal innervation than the core (Abrahamson and Moore, 2001) but may receive inputs from other sources, such as limbic structures (Moga and Moore, 1997). Intranetwork connectivity patterns are less well understood, but it is known that the SCN shell receives dense innervation from VIP neurons and that reciprocal connections from shell to core are less pronounced (Daikoku et al., 1992; Romijn et al., 1997). Output from the shell influences visceral function (Ueyama et al., 1999), REM sleep (Lee et al., 2009), and timing of the LH surge (Williams et al., 2011).
These considerations have led scientists of the field to focus on intercellular and circuit properties of SCN timekeeping, armed with a new arsenal of molecular tools to finally unravel the mechanisms that confer its unique role as the master circadian clock.
In this review, we present current work aimed at expanding our knowledge of the mechanisms that, from intracellular TTFL clocks to neural connectivity, shape the emergent circuit logic of the SCN. In particular, the following two sections will highlight investigations aimed at understanding how SCN circadian pacemaking is intrinsically generated across genes, neural connectivity, and network control. The final two sections will deal with the epigenetic and circuit-mediated plasticity of the SCN, responsible for translating its principal input (light) into coherent adaptive circadian outputs. Particular attention will be given to current research involving live imaging, as well as intersectional and optogenetic and chemogenetic approaches to reveal causal relationships within SCN circadian pacemaking. These topics will feature in a related mini-symposium at the Society for Neuroscience Annual Meeting in Washington, DC in November 2014.
From neuronal clocks to SCN circuit encoding of circadian time
The SCN has three main functions: (1) to define stable circadian oscillations in the absence of external entraining agents (i.e., pacemaking); (2) to synchronize this representation of time to the solar cycle of light and darkness; and (3) to convey temporal cues to the rest of the body to sustain appropriately adaptive daily rhythms of metabolism and behavior. Although these functions coexist in vivo, the second and third require retinohypothalamic input to the SCN and output from it, respectively. In contrast, pacemaking is a remarkably autonomous SCN property. When SCN tissue is explanted from rodents in organotypic culture, it can retain structural coherence for months. More importantly, in these conditions, circadian oscillations in gene expression and neural activity are also preserved. To follow these oscillations in living tissue, several mouse models have been created in which clock genes, such as Per1, Per2, and Cry1, have been fused to bioluminescent and fluorescent reporters, such as luciferase or GFP (Kuhlman et al., 2000; Sujino et al., 2003; Yoo et al., 2004; Maywood et al., 2013). Real-time, long-term imaging of gene expression of this “circuit in a dish” has greatly expanded our knowledge of circadian pacemaking in the SCN. In particular, it has revealed that, even though SCN oscillators are synchronized, they do not share a common phase: the dorsomedial cells peaking on average 2–3 h before ventrolateral ones. As a consequence, a spatiotemporal wave of TTFL gene expression travels across the tissue, from the dorsomedial aspect toward the ventrolateral region of the SCN (Fig. 1B) (Yamaguchi et al., 2003; Evans et al., 2011).
The generation of such a recursive spatiotemporal wave is an emergent property, specific to the SCN and dependent on synaptic integration. Thus, application of TTX to SCN slices to inhibit voltage-gated sodium channels reduces the amplitude of Per1 oscillations and alters the phase relationships among individual cells, thereby impairing the coherence of the wave (Yamaguchi et al., 2003). Similarly, acutely hyperpolarizing SCN neurons with low extracellular K+ dampens the molecular clock by abolishing the rhythmic expression of Per1 and PER2 (Lundkvist et al., 2005). Unraveling this link between synaptic activity and the TTFL is therefore of paramount importance to understand the intrinsic organization of SCN circuits. Firing rates are synchronized in the SCN, so that neurons fire quickly during the day (6–10 Hz) and slowly at night (<1 Hz) (Atkinson et al., 2011; Colwell, 2011), and animal models have provided convincing evidence linking the intracellular molecular clock to these firing rhythms. As an example, the Tau mutation in the clock gene casein kinase 1ε causes a drastic 4 h reduction of the normal 24 h period of SCN electrical activity because of an acceleration of the molecular feedback loop (Liu et al., 1997; Meng et al., 2008). Similarly, SCN neurons from mice heterozygous for a mutation in Clock have a lengthened period of electrical activity rhythms, whereas behaviorally arrhythmic Cry1/2 double knock-out mice also exhibit a complete lack of firing rate rhythms (Albus et al., 2002). Although these results clearly indicate a necessary connection between the molecular clock and electrical activity, they do not, however, show how this interaction would take place, or the directional causality of it. Indeed, there may be a reciprocal dependence between these two processes.
Per genes have recently been suggested as a possible entry point to connect synaptic activity to the intracellular TTFL. In mice expressing GFP driven by Per1 promoter sequences, the degree of promoter activity positively correlates with spike frequency in individual SCN neurons, both after a nocturnal light pulse and also during the middle of circadian day, suggesting that a fixed phase relationship may exist between the molecular clock and electrical activity (Kuhlman et al., 2000; Quintero et al., 2003). Moreover, other GPCR pathways involved in SCN timekeeping, such as GRP or VIP, depend on activation of the endogenous Per1 gene to reset SCN spike frequency (Gamble et al., 2007; Kudo et al., 2013).
A specific feature of per genes compared with cry genes (the second component of negative feedback) is the presence of cAMP/Ca2+ Responsive Elements (CRE) in their promoter regions. Intracellular calcium ([Ca2+]i) and cAMP, which can both activate CRE sequences, show robust circadian oscillations in SCN slices (O'Neill et al., 2008; Brancaccio et al., 2013). These second messengers seem, therefore, ideal mediators to integrate fast membrane-bound cues, such as neuronal firing and GPCR signaling to the very slow temporal structure of circadian oscillations in gene expression, via CREs (Fig. 1A). In particular, the role of [Ca2+]i as an input to the TTFL has been recently reconsidered by using live imaging in SCN slices. Virally delivered, genetically encoded calcium indicators, either FRET based (Yellow Camaleon 3.60) (Enoki et al., 2012a, 2012b) or single wavelength (GCaMP3) (Brancaccio et al., 2013), have been used in combination with clock gene bioluminescent reporters to show that circadian [Ca2+]i oscillations follow a spatiotemporal wave similar to that of the TTFL oscillations. Critically, however, the phase of [Ca2+]i oscillations precedes the phase of per- and cry-mediated oscillations, which follow in series according to the presence and activity of CREs in their promoters (Brancaccio et al., 2013). Such a temporal structure is consistent with a scenario in which [Ca2+]i conveys synaptic and/or paracrine interneuronal cues to CRE sequences present in per genes, thereby integrating events happening in the short timescale to the very slow temporal structure of circadian gene expression. Accordingly, inhibition of synaptic activity by TTX (Enoki et al., 2012a), as well as chemogenetically mediated activation of Gq- and Gi-coupled receptors, are both capable of disrupting the spatiotemporal organization of [Ca2+]i in the SCN, eliciting the previously mentioned reorganization of the TTFL spatiotemporal wave (Brancaccio et al., 2013). Remarkably, although the effects of TTX or Gi are reversible, Gq activation permanently and yet specifically modifies the spatiotemporal wave of TTFL activation, thus suggesting a more profound role for this pathway in determining the internal circuit logic of SCN pacemaking (Brancaccio et al., 2013).
In work presented at the 2014 Society for Neuroscience Annual Meeting, the link between electrical, cytosolic, and molecular circadian rhythms will be further investigated by using simultaneous real-time imaging of gene expression, intracellular calcium, and membrane potential. Moreover, various optogenetic and chemogenetic approaches aimed at revealing the causal relationships between these different aspects of the “extended clock” will be presented.
The role of glial cells in SCN circadian pacemaking
A much debated and yet largely unanswered question in circadian neurobiology is whether or not glial cells play a role in circadian timekeeping (Jackson, 2011). Early studies showed high expression of the astrocytic marker GFAP in the SCN, compared with surrounding hypothalamic areas. Furthermore, GFAP expression undergoes high-amplitude daily oscillations in the hamster SCN, which persist in constant darkness (Lavialle and Servière, 1993). This suggested a possible role for glial cells in intrinsic SCN circadian pacemaking. Although subsequent studies in rats confirmed these oscillations and localized them to the VIP-ergic core region, they failed to reproduce the persistence of such rhythms in constant darkness. It was therefore proposed that such variations could instead be linked to photic entrainment of the SCN via retinal afferents (Becquet et al., 2008; Girardet et al., 2010).
Understanding whether glial cells do indeed play a role in SCN intrinsic pacemaking could be of paramount importance to unravel its intrinsic network logic. It is indeed increasingly evident that the robustness of circadian timekeeping in the SCN cannot be reduced uniquely to the TTFL but is rather a consequence of the concerted progression of multiple interlocked cycles of cytosolic, energetic, and electrical activity. It is commonly thought that such cycles are an exclusive property of SCN neurons, when embedded in the SCN neuronal circuitry. It cannot be excluded, however, that the numerous glial cells present in this tissue may also sustain some of these functions. Glial cells modulate both the energetic and electrical homeostasis of the brain and therefore seem ideally suited to sense misalignment between the electrical neuronal activity and the metabolic state of each SCN neuron and the surrounding ones. The advantage of such a network, when extended to (glial) nonexcitable cells, could be to protect the SCN from the naturally occurring electrical noise due to its neuronal activity (Freeman et al., 2013) that could otherwise desynchronize its oscillators. Feedback from glia could therefore provide a counteracting homeostatic drive to balance the intrinsic neuronal plasticity of the SCN circuit, which would be necessary to align body clocks to environmental cues (see below). Such a role for glial cells in the SCN can, however, only be postulated at the moment. Indeed, whereas a direct role for glial cells in intrinsic pacemaking has been shown in flies (Suh and Jackson, 2007), the evidence in mammals is only indirect: treating rats with fluorocitrate, a drug that poisons the Krebs cycle in astrocytes, makes them arrhythmic (Prosser et al., 1994), and administering antimitotic drugs to SCN slices alters the phase relationship of VIP and AVP release (Shinohara et al., 1995), the inference being that this drug compromises astrocytic proliferation in culture.
Cultured glial cells from cerebral cortices of Per2:luc mice show circadian oscillations that can be entrained by SCN-derived cues and by drugs acting on CRE sequences via calcium or cAMP signaling (Prolo et al., 2005). VIP may be responsible for this effect by CRE activation in astrocytes (Marpegan et al., 2009). As for the other arm of this interaction (glio-transmission to SCN neurons), many well-established glio-transmitters (ATP, serine, glutamate) have been proposed to play a role, but the evidence remains incomplete (Jackson, 2011). This may be partly due to the technical difficulties associated with the recording and manipulation of SCN glial cells in their natural SCN network environment, and the potential artifacts associated with pure astrocytic cultures, in the absence of neuronal feedback.
In the upcoming meeting, new evidence will be presented showing that glia are indeed required for sustaining network encoding of circadian time in the SCN. To achieve this, simultaneous imaging of circadian oscillations in neurons and glial cells has been performed, coupled with intersectional and chemogenetic manipulations of glial and/or neuronal clocks. Moreover, the mechanisms of glio-transmission in the SCN, as well as the role of glia in shaping the spatiotemporal wave of circadian gene expression, will be addressed.
From SCN circuits to behavior
As previously mentioned, the SCN anatomy and cellular structure are not homogeneous. Although the vast majority (∼95%) of SCN neurons are GABAergic (Abrahamson and Moore, 2001), they can be subclassified according to the differential expression of neuropeptides. Neurons in the ventrolateral aspect of the SCN (core) express VIP and GRP, whereas neurons in the dorsomedial region (shell) mainly express AVP (Buijs et al., 1995). VIP and AVP, secreted by ∼10% and 20% of SCN neurons, respectively (Abrahamson and Moore, 2001), act primarily on VPAC2 and V1a/V1b receptors throughout the SCN (Li et al., 2009; An et al., 2012).
This organization is conserved across mammals and has led to the hypothesis that different peptidergic subpopulations may play different roles, not only in circadian pacemaking, but also in relationship to SCN input/output organization. In particular, VIPergic neurons would be mainly involved in integrating light input through the retinohypothalamic tract and to confer intrinsic synchronization of SCN neurons, whereas AVP would amplify the endogenous SCN rhythms into coherent behavioral outputs. Although much is known about retinal signaling to the SCN (Baver et al., 2008), little is known about how VIP neurons process light information and relay it to the SCN circuit. Functionally, VIP signaling has a well-defined role in synchronizing and phase-shifting rhythms within the SCN. In vitro, a single pulse of VIP phase shifts Per2 gene rhythms and a daily VIP pulse entrains SCN rhythms. In addition, VIP- or VPAC2R-deficient mice start their daily activity about 8 h early in a light cycle and lose rhythmicity in constant darkness because of weakened synchrony among SCN neurons (Harmar et al., 2002; Colwell et al., 2003; Cutler et al., 2003; Aton et al., 2005; Maywood et al., 2006).
In addition to integrating light input, the SCN must drive robust rhythmic daily behaviors. Despite its relatively high expression within the SCN, AVP addition to the SCN in vivo (Albers et al., 1984) and ex vivo (Watanabe et al., 2000) yields no obvious effects on circadian behavior or gene expression. Unlike other SCN cell types, AVP neurons exhibit high-amplitude fluctuations in circadian firing, neuropeptide expression, and release. Furthermore, a decrease in this high-amplitude rhythm through aging or individual variability is correlated with decline of rhythmic behavior (Kalsbeek et al., 2006). Loss of V1a/V1b receptor signaling within the SCN enables faster entrainment to a shifted light schedule (Yamaguchi et al., 2013), consistent with a role for AVP signaling in influencing rhythmic behavior.
Although previous research has provided a solid basis for the roles of the neuropeptides VIP and AVP, it should be noted that exogenous application of VIP and AVP might not reflect the roles of these neuronal populations in an intact circuit. The use of optogenetics allows precise, direct control of neuronal firing both in vitro and in vivo. By targeting Channelrhodopsin2 to VIPergic or AVPergic neurons, respectively, and optogenetically tuning the frequency, duration, and circadian time of the stimulation, it is possible finally to dissect their specific roles in controlling circadian behavior.
A plastic clock: circuit-mediated and epigenetically mediated decoding of light/dark input in the SCN
The SCN acts not only as a daily clock, but also as a calendar (Hastings, 2001). Indeed, the seasonal changes in day length (photoperiod) are translated by the SCN into coherent adaptive behavioral and physiological responses. The SCN displays photoperiodic changes in the overall duration of circadian gene expression, where the duration of daily electrical firing and TTFL expression positively correlate with day length. This modulation of the waveform of SCN rhythms reflects changes in the phase relationships of its constituent neurons (Messager et al., 1999; Schaap et al., 2003; Hazlerigg et al., 2005; Inagaki et al., 2007; Naito et al., 2008; Evans et al., 2013). Thus, the SCN circuit is capable of encoding individual experience to ensure that circadian rhythms are adjusted to suit the local environment. It does so through plastic adaptation of its previously described spatiotemporal wave (Fig. 1C).
Recent research that exploits this adaptive plasticity has provided new insights into the signaling mechanisms that SCN neurons use to communicate with one another. Evans et al. (2013) showed that SCN neurons dissociated by long day lengths are able to resynchronize once released from these lighting conditions. This demonstrated that SCN neurons interact with one another during network resynchronization to shift their clocks earlier or later depending on their relative phase within the network. Using this analytical assay to quantify the strength of SCN coupling, it was revealed that this resynchronization is dependent on the integration of multiple, nonredundant signaling pathways (Evans et al., 2013). VIP signaling was found to be important for both maintaining and reestablishing synchrony in the SCN network, which confirms and extends previous work in VIP and VPAC2R receptor knock-out models and chemogenetic activation of VIP neurons (Aton et al., 2005; Maywood et al., 2006; Brancaccio et al., 2013). In the absence of VIP signaling, the ability of SCN neurons to resynchronize is attenuated, but not completely abolished, suggesting that other intercellular signaling factors also contribute to this coupling process, consistent with results from SCN grafting studies (Maywood et al., 2011).
Although GABA signaling synchronizes neurons in cortical circuits (Buzsáki and Chrobak, 1995; Cobb et al., 1995), its role in SCN coupling has been difficult to ascertain. Although GABA can synchronize dissociated SCN neurons (Liu and Reppert, 2000) and is involved in network communication (Albus et al., 2005; Han et al., 2012), GABA signaling is not necessary for maintaining synchronization in SCN slices (Aton et al., 2006). A solution to this puzzling contradiction was provided by showing that GABA signaling does promote synchrony, but only when the network is resynchronizing from the highly polarized state (Evans et al., 2013). Interestingly, GABA signaling interacts with VIP signaling in either a cooperative or antagonistic manner depending on the state of the network. These results indicate that environmental lighting conditions not only influence the spatiotemporal organization of the SCN network but also modulate the signaling mechanisms that SCN neurons use to communicate with one another. In the upcoming meeting, new data will be presented that indicate that plasticity in the coupling role of GABA signaling is associated with changes in the strength and polarity of GABA responses in the SCN.
The effects of day length on SCN circuit-level properties pose interesting questions in terms of the adaptive mechanisms that, at a cellular level, enable SCN neurons to change their reciprocal phase relationships in response to the variations in light input. These mechanisms likely involve epigenetic processes directly controlling gene expression. Epigenetic mechanisms are based upon chemical modifications of the chromatin and include histone post-translational modifications as well as DNA methylation (Strahl and Allis, 2000). Extensive studies have shown that transient or daily changes in histone post-translational modification play a role in facilitating or repressing circadian gene activity (Crosio et al., 2000; Naruse et al., 2004; Ripperger and Schibler, 2006).
Exposing mice to non-24 h light/dark cycles (T-cycles) results in long-lasting changes of their genetically determined circadian period, which persist for several weeks in constant conditions, a phenomenon known as “aftereffect” (Daan and Pittendrigh, 1976; Aton et al., 2004; Molyneux et al., 2008). This model has recently been used to investigate the involvement of a specific epigenetic modification, DNA methylation, in mediating such prolonged circadian plasticity (Azzi et al., 2014). In contrast to histone modifications, which are transient, DNA methylation leads to long-lasting changes in gene transcription. This epigenetic modification is catalyzed by DNA methyltransferases, which add a methyl group to the C5 position of cytosine, whereas DNA demethylation mediates its removal (Kohli and Zhang, 2013; Wu and Zhang, 2014). The effect of DNA methylation on gene activity is context-dependent: for example, methylation of promoter regions represses transcription, whereas gene body methylation enhances it (Jones, 2012). Analysis of the methylomes of model organisms has demonstrated that DNA methylation is very dynamic in response to environmental changes, particularly in the adult brain. For example, brain DNA methylation occurs dynamically upon neuronal activation (Guo et al., 2011), early life stress (Murgatroyd et al., 2009), or in response to external stimuli (Miller and Sweatt, 2007).
Exposing mice to <24 h light/dark cycles (short T-cycles) leads to alteration in the global SCN transcriptome. Interestingly, genome-wide methylation profiling reveals that short T-cycles also elicit global alterations in DNA methylation in the SCN, which correlate with the observed transcriptional changes. These changes are especially observed in genes involved in various aspects of neuronal plasticity, such as synaptogenesis and axonal guidance. Importantly, infusing methyltransferase inhibitors during entrainment to short T-cycles suppresses the aftereffect. Furthermore, circadian behavior and DNA methylation dynamics are reversible upon re-entrainment to the original normal (24 h) T-cycle (Azzi et al., 2014). This observation is consistent with previous findings that reported dynamic seasonal changes in DNA methylation at the level of the deiodinase Type III promoter region in the hamster hypothalamus, which establishes seasonal reproductive phenotypes (Stevenson and Prendergast, 2013).
Experimental evidence has shown that aberrant lighting can lead to pathological disorders, including altered mood, cognitive function deficits, and depression (Salgado-Delgado et al., 2011; LeGates et al., 2012; Leach et al., 2013; Fonken and Nelson, 2014). Even though the molecular mechanisms involved are not fully understood, studies that focused on peripheral clocks in humans reported that changes in methylation of clock genes occur in response to various conditions, including shiftwork, sleep deprivation, and aging (Zhu et al., 2011; Zhang et al., 2013; Massart et al., 2014). A better understanding of the DNA methylation dynamics in the SCN and its control of peripheral clocks could therefore provide important new insights into human health and the treatment of such conditions.
In conclusion, it seems difficult to believe that the activity of a few thousand neurons in the mouse or human SCN could direct behavior as complex as that of daily rhythms. Stress to this system is constant in modern society, and dysfunctions can challenge it at any level, with exacting consequences for human health (Toh et al., 2001; Nicholas et al., 2007; McCarthy and Welsh, 2012; Musiek et al., 2013). We are now unraveling how information flows in the SCN, through the multiple levels of abstraction (genes, neural activity, cell types, circuits, behavior) that finally define our daily life (O'Neill et al., 2013). What we learn in the process will not only satisfy mere scientific curiosity but will also inform the future therapeutic paths for human health and well-being.
Footnotes
M.B. is supported by Medical Research Council United Kingdom. R.E. is supported by PRESTO, Japan Science and Technology Agency, Tokyo, Japan. C.N.M. is supported by National Institutes of Health T32 Graduate Trainee on Systems and Molecular Neurobiology Sciences Training Grant. J.J. is supported by National Institutes of Health T32 MH064931 and F31 NS082213. A.A. is supported by the Velux Foundation and the Swiss National Science Foundation. We thank Dr. M. H. Hastings for comments and Leo Hillier for figure contribution.
The authors declare no competing financial interests.
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/S0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Albers HE, Ferris CF, Leeman SE, Goldman BD. Avian pancreatic polypeptide phase shifts hamster circadian rhythms when microinjected into the suprachiasmatic region. Science. 1984;223:833–835. doi: 10.1126/science.6546454. [DOI] [PubMed] [Google Scholar]
- Albus H, Bonnefont X, Chaves I, Yasui A, Doczy J, van der Horst GT, Meijer JH. Cryptochrome-deficient mice lack circadian electrical activity in the suprachiasmatic nuclei. Curr Biol. 2002;12:1130–1133. doi: 10.1016/S0960-9822(02)00923-5. [DOI] [PubMed] [Google Scholar]
- Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr Biol. 2005;15:886–893. doi: 10.1016/j.cub.2005.03.051. [DOI] [PubMed] [Google Scholar]
- An S, Tsai C, Ronecker J, Bayly A, Herzog ED. Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmatic nucleus. J Comp Neurol. 2012;520:2730–2741. doi: 10.1002/cne.23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SE, Maywood ES, Chesham JE, Wozny C, Colwell CS, Hastings MH, Williams SR. Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J Biol Rhythms. 2011;26:210–220. doi: 10.1177/0748730411402810. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Block GD, Tei H, Yamazaki S, Herzog ED. Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non-24-hour light cycles. J Biol Rhythms. 2004;19:198–207. doi: 10.1177/0748730404264156. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci U S A. 2006;103:19188–19193. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A, Dallmann R, Casserly A, Rehrauer H, Patrignani A, Maier B, Kramer A, Brown SA. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17:377–382. doi: 10.1038/nn.3651. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/S0092-8674(00)81199-X. [DOI] [PubMed] [Google Scholar]
- Baver SB, Pickard GE, Sollars PJ, Pickard GE. Two types of melanopsin retinal ganglion cell differentially innervate the hypothalamic suprachiasmatic nucleus and the olivary pretectal nucleus. Eur J Neurosci. 2008;27:1763–1770. doi: 10.1111/j.1460-9568.2008.06149.x. [DOI] [PubMed] [Google Scholar]
- Becquet D, Girardet C, Guillaumond F, François-Bellan AM, Bosler O. Ultrastructural plasticity in the rat suprachiasmatic nucleus: possible involvement in clock entrainment. Glia. 2008;56:294–305. doi: 10.1002/glia.20613. [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78:714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs RM, Wortel J, Hou YX. Colocalization of gamma-aminobutyric acid with vasopressin, vasoactive intestinal peptide, and somatostatin in the rat suprachiasmatic nucleus. J Comp Neurol. 1995;358:343–352. doi: 10.1002/cne.903580304. [DOI] [PubMed] [Google Scholar]
- Buzsáki G, Chrobak JJ. Temporal structure in spatially organized neuronal ensembles: a role for interneuronal networks. Curr Opin Neurobiol. 1995;5:504–510. doi: 10.1016/0959-4388(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Cobb SR, Buhl EH, Halasy K, Paulsen O, Somogyi P. Synchronization of neuronal activity in hippocampus by individual GABAergic interneurons. Nature. 1995;378:75–78. doi: 10.1038/378075a0. [DOI] [PubMed] [Google Scholar]
- Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci. 2011;12:553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R939–R949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Cutler DJ, Haraura M, Reed HE, Shen S, Sheward WJ, Morrison CF, Marston HM, Harmar AJ, Piggins HD. The mouse VPAC2 receptor confers suprachiasmatic nuclei cellular rhythmicity and responsiveness to vasoactive intestinal polypeptide in vitro. Eur J Neurosci. 2003;17:197–204. doi: 10.1046/j.1460-9568.2003.02425.x. [DOI] [PubMed] [Google Scholar]
- Daan S, Pittendrigh CS. A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol. 1976;106:253–266. doi: 10.1007/BF01417857. [DOI] [Google Scholar]
- Daikoku S, Hisano S, Kagotani Y. Neuronal associations in the rat suprachiasmatic nucleus demonstrated by immunoelectron microscopy. J Comp Neurol. 1992;325:559–571. doi: 10.1002/cne.903250408. [DOI] [PubMed] [Google Scholar]
- Enoki R, Kuroda S, Ono D, Hasan MT, Ueda T, Honma S, Honma K. Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 2012a;109:21498–21503. doi: 10.1073/pnas.1214415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki R, Ono D, Hasan MT, Honma S, Honma K. Single-cell resolution fluorescence imaging of circadian rhythms detected with a Nipkow spinning disk confocal system. J Neurosci Methods. 2012b;207:72–79. doi: 10.1016/j.jneumeth.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS One. 2011;6:e15869. doi: 10.1371/journal.pone.0015869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80:973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonken LK, Nelson RJ. The effects of light at night on circadian clocks and metabolism. Endocr Rev. 2014;35:648–670. doi: 10.1210/er.2013-1051. [DOI] [PubMed] [Google Scholar]
- Freeman GM, Jr, Krock RM, Aton SJ, Thaben P, Herzog ED. GABA networks destabilize genetic oscillations in the circadian pacemaker. Neuron. 2013;78:799–806. doi: 10.1016/j.neuron.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble KL, Allen GC, Zhou T, McMahon DG. Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J Neurosci. 2007;27:12078–12087. doi: 10.1523/JNEUROSCI.1109-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, Mayford M. Generation of a synthetic memory trace. Science. 2012;335:1513–1516. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardet C, Blanchard MP, Ferracci G, Lévêque C, Moreno M, François-Bellan AM, Becquet D, Bosler O. Daily changes in synaptic innervation of VIP neurons in the rat suprachiasmatic nucleus: contribution of glutamatergic afferents. Eur J Neurosci. 2010;31:359–370. doi: 10.1111/j.1460-9568.2009.07071.x. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Yu FH, Schwartz MD, Linton JD, Bosma MM, Hurley JB, Catterall WA, de la Iglesia HO. Na(V)1.1 channels are critical for intercellular communication in the suprachiasmatic nucleus and for normal circadian rhythms. Proc Natl Acad Sci U S A. 2012;109:E368–E377. doi: 10.1073/pnas.1115729109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/S0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Hastings M. Modeling the molecular calendar. J Biol Rhythms. 2001;16:117–123. doi: 10.1177/074873001129001818. discussion 124. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Brancaccio M, Maywood ES. Circadian pacemaking in cells and circuits of the suprachiasmatic nucleus. J Neuroendocrinol. 2014;26:2–10. doi: 10.1111/jne.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlerigg DG, Ebling FJ, Johnston JD. Photoperiod differentially regulates gene expression rhythms in the rostral and caudal SCN. Curr Biol. 2005;15:R449–R450. doi: 10.1016/j.cub.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Huang JZ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR. Glial cell modulation of circadian rhythms. Glia. 2011;59:1341–1350. doi: 10.1002/glia.21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jego S, Glasgow SD, Herrera CG, Ekstrand M, Reed SJ, Boyce R, Friedman J, Burdakov D, Adamantidis AR. Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat Neurosci. 2013;16:1637–1643. doi: 10.1038/nn.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010;8:e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Tahara Y, Gamble KL, McMahon DG, Block GD, Colwell CS. Vasoactive intestinal peptide produces long-lasting changes in neural activity in the suprachiasmatic nucleus. J Neurophysiol. 2013;110:1097–1106. doi: 10.1152/jn.00114.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Quintero JE, McMahon DG. GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. Neuroreport. 2000;11:1479–1482. doi: 10.1097/00001756-200005150-00023. [DOI] [PubMed] [Google Scholar]
- Lavialle M, Servière J. Circadian fluctuations in GFAP distribution in the Syrian hamster suprachiasmatic nucleus. Neuroreport. 1993;4:1243–1246. doi: 10.1097/00001756-199309000-00008. [DOI] [PubMed] [Google Scholar]
- Leach G, Adidharma W, Yan L. Depression-like responses induced by daytime light deficiency in the diurnal grass rat (Arvicanthis niloticus) PLoS One. 2013;8:e57115. doi: 10.1371/journal.pone.0057115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2001;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- Lee ML, Swanson BE, de la Iglesia HO. Circadian timing of REM sleep is coupled to an oscillator within the dorsomedial suprachiasmatic nucleus. Curr Biol. 2009;19:848–852. doi: 10.1016/j.cub.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGates TA, Altimus CM, Wang H, Lee HK, Yang S, Zhao H, Kirkwood A, Weber ET, Hattar S. Aberrant light directly impairs mood and learning through melanopsin-expressing neurons. Nature. 2012;491:594–598. doi: 10.1038/nature11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Burton KJ, Zhang C, Hu SB, Zhou QY. Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am J Physiol Regul Integr Comp Physiol. 2009;296:R824–R830. doi: 10.1152/ajpregu.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25:123–128. doi: 10.1016/S0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/S0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- Lundkvist GB, Kwak Y, Davis EK, Tei H, Block GD. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci. 2005;25:7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marpegan L, Krall TJ, Herzog ED. Vasoactive intestinal polypeptide entrains circadian rhythms in astrocytes. J Biol Rhythms. 2009;24:135–143. doi: 10.1177/0748730409332042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart R, Freyburger M, Suderman M, Paquet J, El Helou J, Belanger-Nelson E, Rachalski A, Koumar OC, Carrier J, Szyf M, Mongrain V. The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl Psychiatry. 2014;4:e347. doi: 10.1038/tp.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O'Neill JS, O'Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O'Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A. 2011;108:14306–14311. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Drynan L, Chesham JE, Edwards MD, Dardente H, Fustin JM, Hazlerigg DG, O'Neill JS, Codner GF, Smyllie NJ, Brancaccio M, Hastings MH. Analysis of core circadian feedback loop in suprachiasmatic nucleus of mCry1-luc transgenic reporter mouse. Proc Natl Acad Sci U S A. 2013;110:9547–9552. doi: 10.1073/pnas.1220894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MJ, Welsh DK. Cellular circadian clocks in mood disorders. J Biol Rhythms. 2012;27:339–352. doi: 10.1177/0748730412456367. [DOI] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sládek M, Semikhodskii AS, Glossop NR, Piggins HD, Chesham JE, Bechtold DA, Yoo SH, Takahashi JS, Virshup DM, Boot-Handford RP, Hastings MH, Loudon AS. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messager S, Ross AW, Barrett P, Morgan PJ. Decoding photoperiodic time through Per1 and ICER gene amplitude. Proc Natl Acad Sci U S A. 1999;96:9938–9943. doi: 10.1073/pnas.96.17.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Moga MM, Moore RY. Organization of neural inputs to the suprachiasmatic nucleus in the rat. J Comp Neurol. 1997;389:508–534. doi: 10.1002/(SICI)1096-9861(19971222)389:3<508::AID-CNE11>3.0.CO%3B2-H. [DOI] [PubMed] [Google Scholar]
- Molyneux PC, Dahlgren MK, Harrington ME. Circadian entrainment aftereffects in suprachiasmatic nuclei and peripheral tissues in vitro. Brain Res. 2008;1228:127–134. doi: 10.1016/j.brainres.2008.05.091. [DOI] [PubMed] [Google Scholar]
- Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, Roh JH, Ortiz-Gonzalez X, Dearborn JT, Culver JP, Herzog ED, Hogenesch JB, Wozniak DF, Dikranian K, Giasson BI, Weaver DR, Holtzman DM, Fitzgerald GA. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013;123:5389–5400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito E, Watanabe T, Tei H, Yoshimura T, Ebihara S. Reorganization of the suprachiasmatic nucleus coding for day length. J Biol Rhythms. 2008;23:140–149. doi: 10.1177/0748730408314572. [DOI] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol Cell Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas B, Rudrasingham V, Nash S, Kirov G, Owen MJ, Wimpory DC. Association of Per1 and Npas2 with autistic disorder: support for the clock genes/social timing hypothesis. Mol Psychiatry. 2007;12:581–592. doi: 10.1038/sj.mp.4001953. [DOI] [PubMed] [Google Scholar]
- O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill JS, Maywood ES, Hastings MH. Cellular mechanisms of circadian pacemaking: beyond transcriptional loops. Handb Exp Pharmacol. 2013:67–103. doi: 10.1007/978-3-642-25950-0_4. [DOI] [PubMed] [Google Scholar]
- Prolo LM, Takahashi JS, Herzog ED. Circadian rhythm generation and entrainment in astrocytes. J Neurosci. 2005;25:404–408. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser RA, Edgar DM, Craig Heller HC, Miller JD. A possible glial role in the mammalian circadian clock. Brain Res. 1994;643:296–301. doi: 10.1016/0006-8993(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacol Rev. 2011;63:291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romijn HJ, Sluiter AA, Pool CW, Wortel J, Buijs RM. Evidence from confocal fluorescence microscopy for a dense, reciprocal innervation between AVP-, somatostatin-, VIP/PHI-, GRP-, and VIP/PHI/GRP-immunoreactive neurons in the rat suprachiasmatic nucleus. Eur J Neurosci. 1997;9:2613–2623. doi: 10.1111/j.1460-9568.1997.tb01691.x. [DOI] [PubMed] [Google Scholar]
- Salgado-Delgado R, Tapia Osorio A, Saderi N, Escobar C. Disruption of circadian rhythms: a crucial factor in the etiology of depression. Depress Res Treat. 2011;2011:839743. doi: 10.1155/2011/839743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap J, Albus H, VanderLeest HT, Eilers PH, Détári L, Meijer JH. Heterogeneity of rhythmic suprachiasmatic nucleus neurons: implications for circadian waveform and photoperiodic encoding. Proc Natl Acad Sci U S A. 2003;100:15994–15999. doi: 10.1073/pnas.2436298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K, Honma S, Katsuno Y, Abe H, Honma K. Two distinct oscillators in the rat suprachiasmatic nucleus in vitro. Proc Natl Acad Sci U S A. 1995;92:7396–7400. doi: 10.1073/pnas.92.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Prendergast BJ. Reversible DNA methylation regulates seasonal photoperiodic time measurement. Proc Natl Acad Sci U S A. 2013;110:16651–16656. doi: 10.1073/pnas.1310643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino M, Masumoto KH, Yamaguchi S, van der Horst GT, Okamura H, Inouye ST. Suprachiasmatic nucleus grafts restore circadian behavioral rhythms of genetically arrhythmic mice. Curr Biol. 2003;13:664–668. doi: 10.1016/S0960-9822(03)00222-7. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptácek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Ueyama T, Krout KE, Nguyen XV, Karpitskiy V, Kollert A, Mettenleiter TC, Loewy AD. Suprachiasmatic nucleus: a central autonomic clock. Nat Neurosci. 1999;2:1051–1053. doi: 10.1038/15973. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Vanecek J, Yamaoka S. In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res. 2000;877:361–366. doi: 10.1016/S0006-8993(00)02724-4. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WP, 3rd, Jarjisian SG, Mikkelsen JD, Kriegsfeld LJ. Circadian control of kisspeptin and a gated GnRH response mediate the preovulatory luteinizing hormone surge. Endocrinology. 2011;152:595–606. doi: 10.1210/en.2010-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, Dong X, Tsujimoto G, Okuno Y, Doi M, Okamura H. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science. 2013;342:85–90. doi: 10.1126/science.1238599. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin QL, Lu L, Yang CC, Li YL, Sun FL, Wang DH, Cai YN, Li L. Tissue-specific modification of clock methylation in aging mice. Eur Rev Med Pharmacol Sci. 2013;17:1874–1880. [PubMed] [Google Scholar]
- Zhu Y, Stevens RG, Hoffman AE, Tjonneland A, Vogel UB, Zheng T, Hansen J. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol Int. 2011;28:852–861. doi: 10.3109/07420528.2011.618896. [DOI] [PMC free article] [PubMed] [Google Scholar]


