Summary
NK cells are innate lymphoid cells that are critical for host defense against infection, and mediate anti-tumor responses. MicroRNAs (miRNAs) are a large family of small non-coding RNAs that target the 3′UTR of mRNAs, thereby attenuating protein translation. The expression of miRNAs within human peripheral blood and mouse splenic NK cells has been cataloged, with the majority of the miRNA sequence pool represented in the top 60 most abundantly expressed miRNAs. Global miRNA deficiency within NK cells has confirmed their critical role in NK cell biology, including defects in NK cell development and altered functionality. Studies using gain- and loss-of-function of individual miRNAs in NK cells have demonstrated the role of specific miRNAs in regulating NK cell development, maturation, and activation. miRNAs also regulate fundamental NK cell processes including cytokine production, cytotoxicity, and proliferation. This review provides an update on the intrinsic miRNA regulation of NK cells, including miRNA expression profiles, as well as their impact on NK cell biology. Additional profiling is needed to better understand miRNA expression within NK cell developmental intermediates, subsets, tissues, and in the setting of disease. Furthermore, key open questions in the field as well as technical challenges in the study of miRNAs in NK cells are highlighted.
Keywords: NK cell, microRNA, development, activation, lymphocyte
Introduction
NK cells are lymphocytes that play an important role in host defense against pathogens, mediate anti-tumor effects (reviewed in [1],[2]), and were one of the first members characterized in a growing list of innate lymphoid cells (ILCs) (reviewed in [3]). NK cell progenitors arise in the bone marrow and undergo additional differentiation and maturation in peripheral lymphoid organs, during which they undergo a complex educational process to maintain self-tolerance, which is mediated in mature NK cells via activating, inhibitory, and cytokine receptors (reviewed in [4],[5]). Thus, mature NK cells may be activated in different situations, for example, when activating receptor signals dominate inhibitory signals upon interaction with a target cell, or by pro-inflammatory cytokines produced by dendritic cells (DCs) or macrophages (reviewed in [6]). Activation may trigger NK cell killing via granule exocytosis or death receptors, result in cytokine and chemokine production, and induce proliferation and changes in cell surface receptors, including the upregulation of the activating receptors NKp46 and KIR expression, and costimulatory molecules such as 2B4 [2]. NK cells traffic to most tissues and organs in the body, and the existence of tissue-specific functions of NK cells is being increasingly appreciated (reviewed in [7],[8]). More recently, adaptive or ‘memory’ properties have been identified in the NK cell lineage, suggesting that unlike other innate immune cells, recall of prior activation events may influence subsequent NK cell responses (reviewed in [9],[10]). In addition, it has become increasingly apparent that NK cells have a complex and dynamic interaction with many other immune cells, with the potential for both positive and negative outcomes for the NK cell and/or accessory cell, which include DCs, macrophages, neutrophils, and various T cell lineages. From a translational medicine perspective, NK cells are being manipulated by drugs or administered as adoptive immunotherapy to benefit cancer patients (reviewed in [11]).
Most studies of the molecular events that regulate NK cell development and function have focused on transcription factors and key signaling pathways (and their regulators) (reviewed in [12],[13]). Post-transcriptional control of effector molecules important for the NK cell response has been shown [14],[15], but the precise mechanisms responsible have received only minimal attention. microRNAs (miRNAs) are a large family of small non-coding RNAs that primarily mediate their function by repressing mRNA target translation, either through translational blockade or degradation [16],[17]. miRNAs are grouped into families based on the similarity of the mature miRNA sequence, especially nucleotides 2–7 which comprise the “seed” sequence and which play a major role in mRNA target specificity. There are over one thousand miRNAs identified in the mouse and human genomes, with many miRNA genes having multiple copies likely arising from gene duplication events [18]. Additionally, miRNA biogenesis is imperfect, and yield isomiRs that typically vary at the 3′ end of the mature miRNA sequence. These relatively unique aspects of miRNA expression and biogenesis mandate that all variations of a miRNA family be taken into account when evaluating their potential regulatory role. Mature miRNA sequences are bioinformatically predicted to target hundreds of mRNA sequences, most commonly in the 3′UTR, but more recently non-canonical miRNA targeting and repression has been identified [19]. For example, miR-155 binds to a number of target genes in their coding region, but in contrast to 3′ UTR regulation, contain seed mismatches and do not lead to visible mRNA downregulation [20]. Moreover, a complex interaction occurs between a miRNA and its cell context-specific set of mRNA targets [21]. Since a stoichiometric relationship between any single miRNA sequence and its mRNA targets exists, having a distinct complement of targeted mRNAs (and competitive endogenous RNAs that regulate the expression of other mRNAs through competition for shared miRNA regulators (reviewed in [22]) can markedly impact the relevant binding of miRNA to an established target. Therefore, it is imperative to i) define the miRNA expression profile, ii) define the expression of relevant targets and ceRNAs, and iii) experimentally test the impact of a miRNA : mRNA interaction within the cell type of interest.
Several initial reports identified the general importance of miRNAs to NK cell development and function through the deletion of key enzymes in the miRNA biogenesis pathway, such as Dicer and Dgcr8, or enzymatic regulators of miRNA abundance, such as Eri1 [23]–[25]. In these examples, the overall loss of miRNA generation resulted in a failure to generate mature NK cells, and alterations in NK cell function. Thus, miRNAs are critical to normal NK cell homeostasis and activation. While it is important to establish that mature miRNAs are critical for the normal development of NK cells, the impact of individual or families of miRNAs on NK cell development and function require in-depth experimental investigation. This review provides an update on miRNA expression and the intrinsic impact of miRNAs on NK cells. miRNAs may also extrinsically influence NK cell responses, exemplified by reports of human cytomegalovirus virus, Kaposi’s sarcoma associated herpesvirus, and Epstein-Barr virus derived miRNA targeting of NKG2D ligands [26]; however, this is not the focus of this review. We also highlight gaps in our understanding of miRNA expression and open questions about the role of key miRNAs in regulating NK cell biology. Finally, miRNA research has some technical challenges that require careful experimental design and control, which are also discussed.
1. What we know (and don’t know) about assessing miRNA expression in NK cells and how to properly measure this)
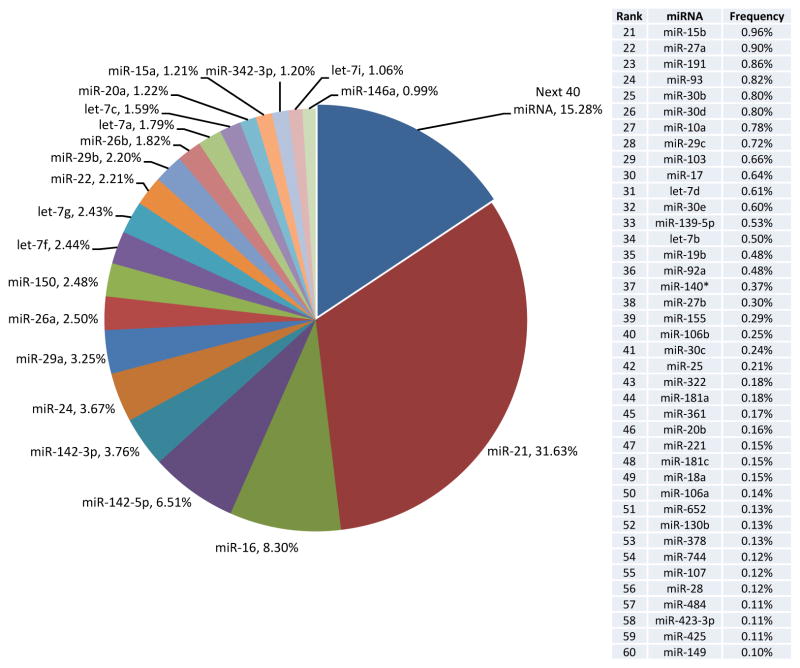
With the advent of massively parallel DNA sequencing techniques, several groups, albeit working with distinct methodologies, sample preparation, and bioinformatic analysis pipelines, have now defined the miRNA transcriptomes of mouse and human NK cells using next generation sequencing (NGS, small RNA seq) [27]–[29]. The ranked expression of the top expressed miRNAs by NGS has been comparable among different groups examining resting NK cells, and there are some differences depending on the species profiled. It is unclear at this point whether this reflects true species differences, nuances of NK cell subsets in mice or humans, or simply a limited number of profiled individuals. Experiments performed on mouse NK cells, including profiling on two distinct NGS platforms with orthogonal validation, suggest that approximately 60 mature miRNA sequences comprise the vast majority (>97%) of miRNA sequences in a resting splenic NK cell population (Figure 1). In order to understand the full complement of mature miRNA expression relevant for NK cell biology, additional profiling needs to be performed, including miRNA expression in maturation subsets, tissue-specific NK cells, as well as NK progenitors, precursors, and immature NK cells. Furthermore, comparison of miRNA profiles of NK cells to those of other innate and adaptive lymphoid cells will allow identification of NK cell-restricted miRNA expression, as well as NK cell-selective lack of expression, which may play a critical role in facilitating the NK cell genetic program. Another priority in the field would be to compare the miRNA profiles associated with specific NK cell functional attributes, such as licensed vs. unlicensed, naïve vs. memory, and tumor-infiltrating vs. peripheral NK cells, where the molecular mechanisms regulating their biology remain unclear. Given the objectives outlined above, much work needs to be done to further characterize the miRNA transcriptome in the context of dynamic NK cell biology.
Figure 1. An abundance of resting splenic mouse NK cell miRNAs defined by small RNA sequencing.
The relative expression of the top 60 expressed miRNAs from Fehniger et. al. [27], are shown with the 20 most abundant featured in the circle chart (left) and the remaining 40 rank ordered in the table (right). Relative expression was determined by enumerating the number of unique sequence reads that corresponded to a miRNA mature sequence, as a percentage of all mature miRNA sequences identified.
The miRNA profiling studies carried out to date in mouse and human NK cells have provided a critical starting point to study miRNA regulation during NK cell activation. However, the small RNA seq quantification of miRNA changes during cytokine activation have only been described for mouse NK cells stimulated for 24 hours with IL-15 [27], or human NK cells stimulated with combined cytokine activation (IL-2, IL-15, and IL-21) or IFN-α stimulation [28],[29]. Therefore, defining miRNA alterations under specific stimuli or with physiologic stimuli, such as viral infection [30] or the response to tumor targets, would provide mechanistic links between extracellular stimuli and alterations in NK cell function. Ideally, NK cell miRNA profiling would occur concomitantly with mRNA and proteomic profiling to provide a complete set of interaction data. Whether mammalian miRNA-mediated targeting is is mechanistically due to mRNA degradation or translational repression is controversial [31],[32], and thus concurrent profiling of miRNA and mRNA alone may be inadequate to identify all relevant miRNA : mRNA interactions, but may be useful in identifying select targets. To provide clear evidence of NK cell-intrinsic miRNA : mRNA targets, there remains a strong need for more studies using profiling approaches such as CLIP-Seq [33] or RISC-Seq [34],[35] both of which are techniques that identify miRNAs and mRNAs bound in the RNA-induced silencing complex (RISC), the site of miRNA regulatory action. These approaches allow direct evidence of miRNA:mRNA interaction within the cell of interest. Additional considerations for accurately identifying relevant targets include performing such experiments in primary NK cells whenever possible, since the competing mRNA or ceRNA targets of a given miRNA will influence the mRNAs targeted.
All miRNA profiling approaches do not yield the same results and there are discrepancies between profiling using microarrays and small RNA sequencing. While both the microarray and small RNA sequencing techniques quantify relative miRNA abundance, microarrays in general have lower sensitivities and dynamic range compared with those of small RNA sequencing, which can sequence down to a single miRNA sequence read. In contrast, small RNA sequencing depends on a robust, well-designed bioinformatics pipeline for analysis, which correctly maps high quality small sequence reads within the genome. When miRNA genes and sequences were being rapidly discovered primarily using miRNA-sequencing and early inaccurate bioinformatic approaches, novel miRNA genes were identified that were later found to likely comprise other small RNAs [36]. This issue has been largely resolved with improved small read alignment algorithms and ongoing experimental evaluation of putative miRNA genes, but initial annotation errors have filtered through to other profiling techniques, creating false positive results. For example, microarrays use a single probe for a mature miRNA sequence, potentially allowing the simultaneous detection of primary, precursor, and mature sequences, and the possibility of “off target” hybridization signals. In NK cells, this is exemplified by mmu-mir-720, which was minimally detected in NK-cell small RNA sequencing experiments (rank order 208 in resting NK cells, [27]), but consistently was identified on microarrays in the top 20 expressed miRNAs in NK cells [23],[37]. The corresponding mature sequence is no longer identified as a miRNA within miRBase (www.mirbase.org), and rather a tRNA fragment resulting from normal processing [36]. Since small RNA sequencing includes mapping individual sequence reads to the best fit in the genome, this processed tRNA sequence is mapped to the tRNA gene, thereby excluding it as a “false positive”. Such an example suggests that hybridization-based techniques may not be the optimal method for initial miRNA profiling, and that confirmation is necessary with an orthogonal platform to support clear quantification of mature miRNAs via microarrays. One alternative method, although yet to be widely used for NK cell miRNA expression studies is that of NanoString, which allows absolute quantification without the bias of prior amplification. Thorough evaluation of this technology, especially in comparison to currently used methods, is still required, but use of this platform still restricts analysis to known miRNAs and does not allow assessment of other small RNA species or novel miRNAs. Currently, our preferred method is small RNA sequencing, which accurately defines the relative abundance of miRNAs by direct sequencing within the small RNA pool. In concert, we utilize real-time RT-qPCR, which when appropriately normalized [38], allows for sensitive detection of relative differences between cell populations, including limiting numbers of sorted cells.
2. miRNA regulation of the effector molecules IFN-γ, granzyme B, and perforin
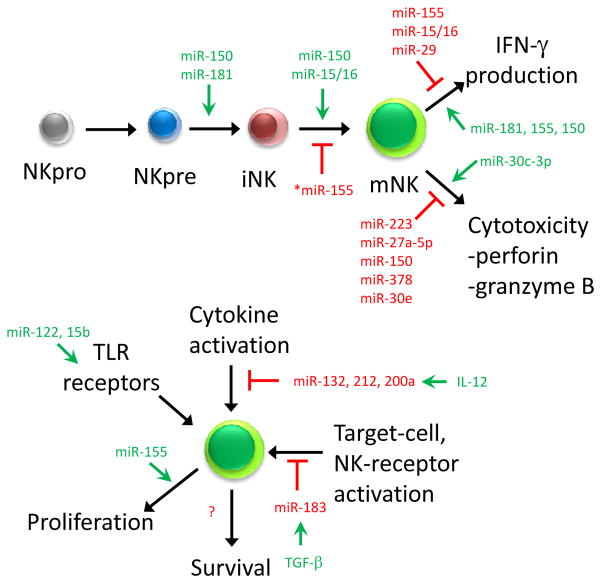
The post-transcriptional regulation of IFN-γ [14] and the cytotoxic effector proteins perforin and granzyme B in NK cells [15] have been documented in murine models (Figure 2). IFN-γ production in human and mouse NK cells is now known to be regulated by several miRNAs, including miR-155, miR-15/16, miR-150, miR-181, and miR-29 [24],[35],[39],[40]. In these reports, IFN-γ translation was found to be regulated either through direct repression of the IFN-γ 3′ UTR (miR-15/16 [41] and miR-29 [42] in mouse NK cells ) or through indirect repression of upstream targets (miR-181 in human NK cells [43] or miR-155 in both human and mouse NK cells [44],[45]). Further studies will be necessary to understand the complex interplay between multiple IFN-γ-regulating miRNAs and other post-transcriptional regulatory mechanisms, such as 5′ UTR pseudoknot regulatory elements [46]. A number of groups have reported miRNAs that potentially regulate granzyme B and/or perforin production, including luciferase reporter assays of miR-223 against the murine granzyme B 3′ UTR [27], miR-27a-5p against the human granzyme B and perforin 3′ UTR [47], miR-150 against the human perforin 3′ UTR [48], and miR-378 and miR-30e against the human granzyme B and perforin 3′ UTR [29]. Several important considerations exist for studies of miRNAs to date. First, the majority of these studies have largely relied on their manipulation in NK cell lines, which only partially retain the characteristics of primary cells. Additional techniques, including the creation of NK cell-specific miRNA knockout models and effective transfection methods for primary human cells, will be useful for definitively assessing the contributions of miRNAs to NK cell function. Secondly, miRNA target 3′ UTR conservation must be considered, especially in the application of murine mouse models to human miRNA regulation. For example, miR-223 was found to regulate the mouse granzyme B 3′ UTR [27], but the site is not conserved in the human granzyme B 3′ UTR and there is no evidence for alternate binding sites by bioinformatics predictions. Finally, it remains unknown how the multitude of miRNAs that regulate the same 3′ UTR contribute to gene regulation. Are specific miRNAs responsible for the basal regulation of gene expression and others for the inducible changes during cell activation? The assessment of miRNA expression, together with the context in which miRNA alterations occur and their intracellular targets, will be helpful in evaluating the physiologic importance of specific miRNAs.
Figure 2. Schematic summary of how miRNAs regulate NK cell development and function.
The green arrows represent a positive influence of the miRNA on the indicated developmental stage or NK cell function, while red bars indicate inhibition. This summary integrates findings within both mouse and human NK cells. miR-155 (denoted by an *) has unclear roles in maturation, displaying either no effect on maturation or a repression.
3. miRNA regulation of NK cell activation: miR-155 as a case in point
Two studies have reported miRNA regulation events following IL-12 or TGF-β stimulation of human NK cells, while miRNA regulation of other typical NK cell activation pathways, including IL-15 stimulation or triggering of NK cell activating receptors, has not yet been extensively evaluated. miRs-132, 212, and 200a were shown to be induced in human NK cells by IL-12 stimulation, which in turn target STAT4 mRNA, thereby providing a negative regulatory pathway for IL-12 signaling [49]. In another study, miR-183 was shown to be induced by TGF-β, a hallmark cytokine of tumor-derived immunosuppression [50], and TGF-β in turn regulates the expression of the signaling adapter DAP12, a critical ITAM-containing adapter for transducing activating signals [51]. Recently, several miRNAs, including miR-122 and miR-15b, have been shown to stimulate NK cells extrinsically, probably through binding and activation of TLR1 [42]. This led to the upregulation of CD69, increased degranulation, greater IFN-γ production, and provided a potential physiologic context for the presence of circulating small RNAs in the plasma [52]. By far, the most extensively studied individual miRNA in NK cells is miR-155, and the multiple studies defining miR-155 regulation of NK cell activation are discussed in depth in the next section.
miR-155 is a miRNA processed from the Bic non-coding RNA, and was originally shown to have numerous roles in B cells and T cells [53]–[56]. miR-155 is modestly expressed in mouse [27] and human [35],[39] NK cells, but is upregulated after activation with cytokines (such as IL-12, IL-15, and IL-18) or activating receptor ligation (such as NK1.1), as well as during MCMV infection [30],[35],[39]. Trotta et. al. showed, using a Bic−/− mouse as well as lentiviral-mediated overexpression and knockdown in human NK cell lines, that miR-155 regulates murine and human IFN-γ production through modulation of the phosphatase SHIP-1 [39]. The proposed model includes miR-155 augmenting NK cell activation by repressing SHIP-1, and thus increasing activation-induced signaling via the PI3K pathway. This group extended these findings using a mouse model in which miR-155 overexpression is driven by a Lck transgene [45], and reported that constitutive miR-155 overexpression led to baseline NK cell activation and enhanced target cell conjugation. This resulted in increased anti-tumor activity in vitro, and improved survival of lymphoma-bearing mice in vivo [45]. In this miR-155-overexpressing model, NK cells were also hyperproliferative, and were developmentally skewed towards immaturity.
Zawislak et. al. [30] used a global miR-155−/− mouse model to investigate the ramifications of miR-155 loss on naive NK cells and NK cells during MCMV infection. The authors reported that miR-155 was largely dispensable for cytotoxicity and cytokine production in response to activating receptors and cytokine stimulation [30]. The miR-155−/− NK cells also showed a skewing toward a more mature phenotype. However, this group found that miR-155−/− mice had severely impaired responses to MCMV infection, featuring impaired NK cell expansion. The authors suggest that this effect was mediated through Noxa and SOCS1, based upon CLIP-Seq of T cells and validated in NK cells. The authors further showed that the transplantation of transduced hematopoietic stem cells, which resulted in pan-hematopoietic Noxa or SOCS1 overexpression, led to defective NK cell expansion after MCMV infection [30].
Sullivan et. al. [35], utilizing mice with global miR-155 ablation but NK cell-specific miR-155 overexpression, and lentiviral overexpression of miR-155 in mature NK cells, investigated the role of miR-155 in NK cell activation. No differences between wild-type and miR-155-overexpressing mice or cells were observed in NK cell maturation in either model. In addition, both miR-155-overexpression and miR-155−/− models had NK cells with increased capacity for IFN-γ production. Distinct cellular mechanisms were shown to be responsible, with increased per-cell IFN-γ production driven by miR-155 overexpression, and an increased percentage of IFN-γ-producing NK cells in the miR-155−/− mice. Moreover, RISC-seq of activated wild-type and miR-155−/− murine NK cells biochemically identified miR-155 targets, which included a large number of predicted and non-canonical miR-155 targets. Many of the identified targets were members of activating intracellular signaling pathways, such as IKBKE, SLP76, PKCθ. Through the use of pathway-specific inhibitors, a role for miR-155 in tuning the threshold of NK cell activation was defined when miR-155 was absent. The targets identified in NK cells were distinct from those identified in a study performed looking at miR-155 targets in T cells [57], highlighting the importance of cell context.
Common phenotypes among all miR-155 studies to date include augmented IFN-γ production when miR-155 is overexpressed in mature NK cells, at least partially via the repression of SHIP-1. Numerous differences exist between the above-described studies, especially regarding maturation, IFN-γ production in the absence of miR-155, and miR-155 targets. Differences between these studies which may explain the phenotype differences include i) the mouse source and strain background, ii) the cell-extrinsic effects of global expression alterations and/or integration events, iii) the age of the mice used in each study, and iv) NK and non-NK cell sources of cells for RISC-Seq and CLIP-Seq. Some of these issues will be resolved by the generation and analysis of a B6 conditional miR-155 floxed knockout allele, combined with an NK cell specific Cre, as well as a direct comparison between the models.
4. microRNA regulation of NK cell development and maturation
Three miRNAs have been shown to regulate NK cell development: miR-181[43], miR-150 [40], and miR-15/16 [41]. Using an in vitro human NK cell differentiation system, miR-181 was shown to correlate with NK cell development and promotes NK cell generation through repression of nemo-like kinase. miR-150 and miR-15/16, both highly expressed miRNAs which belong to distinct miRNA families, appear to exert their effects by targeting the transcription factor c-Myb. Bezman et. al. [40] showed that the genetic loss of miR-150 led to a deficiency of mature NK cells, and found that c-Myb expression was increased in the absence of miR-150. The authors also showed that a Myb+/− mouse had NK cells with increased maturity, indirectly supporting this hypothesis. Similarly, Sullivan et al. [41] presented that c-Myb levels are increased with a deficiency of miR-15/16, and NK cells from NK-specific mir-15a/16-1 knockout mice have a similar defect in maturation, which can be restored by shRNA directed against c-Myb [41]. The confluence of these two highly expressed miRNAs on the repression of a single transcription factor strongly suggests that c-Myb is critical for proper NK cell maturation. As deletion of c-Myb is embryonic lethal, there have been no studies to date investigating the effect of genetic modification of c-Myb on NK cell maturation. The development of an NK cell-specific c-Myb deletion and overexpression will be crucial for directly addressing the role of this important transcription factor in NK cells, which is targeted by two distinct miRNA families. It is also likely that additional miRNAs regulate NK cell development and maturation, the study of which will be facilitated by miRNA profiling of NK progenitors, precursors, and immature NK cells.
Conclusions and Perspective
The unraveling of the complex miRNA regulation of NK cell development and function is in its infancy. While selected miRNAs, such as miR-155, have been studied and demonstrated to play a role in NK cell biology, with more than 60 miRNA sequences expressed in abundance in resting mature NK cells alone, this mode of post-transcriptional regulation will likely be involved at many levels. Furthermore, profiling the miRNA expression of key developmental intermediates, functionally distinct as well as tissue-specific subsets, remains to be performed. Studies of miRNA expression in NK cells should derive from validated platforms of miRNA expression, such as small RNA sequencing and orthogonal validation by quantitative PCR and demonstrate physiologically relevant abundance of differentially expressed miRNAs by direct manipulation in primary NK cells. For human NK cells, the relevance of miRNA manipulation in primary NK cells is paramount, and further investigation of miRNAs in NK cells of patients with infections, malignancies, autoimmunity, and other diseases are warranted.
Acknowledgments
This was supported by T32 HL708836 from the NIH (RPS), K08HL093299 (TAF) and R01AI102924 (TAF) from the NIH.
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 3.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–7. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 4.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142:847–56. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonsson HA, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol. 2009;101:27–79. doi: 10.1016/S0065-2776(08)01002-X. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sojka DK, Tian Z, Yokoyama WM. Tissue-resident natural killer cells and their potential diversity. Semin Immunol. 2014:1–5. doi: 10.1016/j.smim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Santo JP. Natural killer cells: diversity in search of a niche. Nat Immunol. 2008;9:473–5. doi: 10.1038/ni.f.201. [DOI] [PubMed] [Google Scholar]
- 9.Cooper MA, Colonna M, Yokoyama WM. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep. 2009;10:1103–10. doi: 10.1038/embor.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min-Oo G, Kamimura Y, Hendricks DW, Nabekura T, Lanier LL. Natural killer cells: walking three paths down memory lane. Trends Immunol. 2013 doi: 10.1016/j.it.2013.02.005. Epub Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy WJ, Parham P, Miller JS. NK cells--from bench to clinic. Biol Blood Marrow Transpl. 2012;18:S2–7. doi: 10.1016/j.bbmt.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hesslein DGT, Lanier LL. Transcriptional control of natural killer cell development and function. Adv Immunol. 2011;109:45–85. doi: 10.1016/B978-0-12-387664-5.00002-9. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez K, Kee BL. Transcriptional regulation of natural killer cell development. Curr Opin Immunol. 2010;22:193–8. doi: 10.1016/j.coi.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang Z-E, Gapin L, Kronenberg M, et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med. 2003;198:1069–76. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, Ley TJ. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 16.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 17.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 18.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–60. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 19.Khorshid M, Hausser J, Zavolan M, van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods. 2013;10:253–5. doi: 10.1038/nmeth.2341. [DOI] [PubMed] [Google Scholar]
- 20.Loeb GB, Khan Aa, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–70. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll AP, Tooney Pa, Cairns MJ. Context-specific microRNA function in developmental complexity. J Mol Cell Biol. 2013;5:73–84. doi: 10.1093/jmcb/mjt004. [DOI] [PubMed] [Google Scholar]
- 22.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DGT, Lanier LL. Distinct Requirements of MicroRNAs in NK Cell Activation, Survival, and Function. J Immunol. 2010;185:3835–46. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan RP, Leong JW, Schneider SE, Keppel CR, Germino E, French AR, Fehniger TA. MicroRNA Deficient NK Cells Exhibit Decreased Survival but Enhanced Function. J Immunol. 2012 doi: 10.4049/jimmunol.1102294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas MF, Abdul-Wajid S, Panduro M, Babiarz JE, Rajaram M, Woodruff P, Lanier LL, et al. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and anti-viral function. Blood. 2012 doi: 10.1182/blood-2011-11-394072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elias S, Mandelboim O. Innate microRNA networking Battle of the midgets. RNA Biol. 2012;9:792–798. doi: 10.4161/rna.19717. [DOI] [PubMed] [Google Scholar]
- 27.Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, Keppel CR, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Wang Y, Sun Q, Yan J, Huang J, Zhu S, Yu J. Identification of microRNA transcriptome involved in human natural killer cell activation. Immunol Lett. 2012;143:208–217. doi: 10.1016/j.imlet.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, Gu Y, Zhang Q, Han Y, Hou J, Lin L, Wu C, et al. Identification of Resting and Type I IFN-Activated Human NK Cell miRNomes Reveals MicroRNA-378 and MicroRNA-30e as Negative Regulators of NK Cell Cytotoxicity. J Immunol. 2012 doi: 10.4049/jimmunol.1200609. jimmunol.1200609–. [DOI] [PubMed] [Google Scholar]
- 30.Zawislak CL, Beaulieu AM, Loeb GB, Karo J, Canner D, Bezman Na, Lanier LL, et al. Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc Natl Acad Sci USA. 2013;110:6967–72. doi: 10.1073/pnas.1304410110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baek D, Villén J, Shin C, Camargo F. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW. RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan RP, Fogel LA, Leong JW, Schneider SE, Wong R, Romee R, Thai T-H, et al. miR-155 tunes both the threshold and extent of NK cell activation via targeting of multiple signaling pathways. J Immunol. 2013 doi: 10.4049/jimmunol.1301950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schopman NCT, Heynen S, Haasnoot J, Berkhout B. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 2010;7:573–576. doi: 10.4161/rna.7.5.13141. [DOI] [PubMed] [Google Scholar]
- 37.Beaulieu AM, Bezman Na, Lee JE, Matloubian M, Sun JC, Lanier LL. MicroRNA function in NK-cell biology. Immunol Rev. 2013;253:40–52. doi: 10.1111/imr.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X. A PCR-based platform for microRNA expression profiling studies. RNA. 2009;15:716–23. doi: 10.1261/rna.1460509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trotta R, Chen L, Ciarlariello D, Josyula S, Mao C, Costinean S, Yu L, et al. MiR-155 regulates IFN-γ production in natural killer cells. Blood. 2012:3478–3485. doi: 10.1182/blood-2011-12-398099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Exp Med. 2011:1–15. doi: 10.1084/jem.20111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan R, Leong J, Schneider S, Romee R, Sexl V, Dalla-Faver R, Fehniger T. Mir-15/16 Antagonizes Myb To Control Natural Killer Cell Differentiation and Maturation. Blood. 2013;122:A17. [Google Scholar]
- 42.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 43.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting Edge: MicroRNA-181 Promotes Human NK Cell Development by Regulating Notch Signaling. J Immunol. 2011 doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan RP, Fogel LA, Leong JW, Schneider SE, Wong R, Romee R, Thai T-H, et al. miR-155 tunes both the threshold and extent of NK cell activation via targeting of multiple signaling pathways. J Immunol. 2013;191:5904–13. doi: 10.4049/jimmunol.1301950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trotta R, Chen L, Costinean S, Josyula S, Mundy-Bosse BL, Ciarlariello D, Mao C, et al. Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells. Blood. 2013 doi: 10.1182/blood-2012-12-467597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–32. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 47.Kim T-D, Lee SUH, Yun S, Sun H, Kim JW, Kim HM, Park S-K, et al. Human microRNA-27a * targets Prf1 and GzmB expression to regulate NK cell cytotoxicity. Blood. 2011;118:5476–5486. doi: 10.1182/blood-2011-04-347526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim N, Kim M, Yun S, Doh J, Greenberg PD, Kim T-D, Choi I. MicroRNA-150 regulates the cytotoxicity of natural killers by targeting perforin-1(*) J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Lei Y, Zhang H, Hou L, Zhang M, Dayton AI. MicroRNA regulation of STAT4 protein expression: rapid and sensitive modulation of interleukin-12 signaling in human natural killer cells. Blood. 2011;118:6793–6802. doi: 10.1182/blood-2011-05-356162. [DOI] [PubMed] [Google Scholar]
- 50.Donatelli SS, Zhou J-M, Gilvary DL, Eksioglu Ea, Chen X, Cress WD, Haura EB, et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bryceson YT, Chiang SCC, Darmanin S, Fauriat C, Schlums H, Theorell J, Wood SM. Molecular mechanisms of natural killer cell activation. J Innate Immun. 2011;3:216–26. doi: 10.1159/000325265. [DOI] [PubMed] [Google Scholar]
- 52.He S, Chu J, Wu L-C, Mao H, Peng Y, Alvarez-Breckenridge Ca, Hughes T, et al. MicroRNAs activate natural killer cells through toll-like receptor signaling. Blood. 2013 Apr 11; doi: 10.1182/blood-2012-07-441360. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lind EF, Ohashi PS. Mir-155, a central modulator of T-cell responses. Eur J Immunol. 2014;44:11–15. doi: 10.1002/eji.201343962. [DOI] [PubMed] [Google Scholar]
- 54.Thai T-H, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, et al. Regulation of the germinal center response by microRNA-155. Science (80- ) 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, et al. Requirement of bic/microRNA-155 for normal immune function. Science (80- ) 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Connell RM, Rao DS, Chaudhuri Aa, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 57.Loeb GB, Khan Aa, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–70. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]