Abstract
A key obstacle to creating sophisticated genetic circuits has been the lack of scalable device libraries. Here we present a modular transcriptional repression architecture based on clustered regularly interspaced palindromic repeats (CRISPR) system and examine approaches for regulated expression of guide RNAs in human cells. Subsequently we demonstrate that CRISPR regulatory devices can be layered to create functional cascaded circuits, which provide a valuable toolbox for engineering purposes.
Engineered biological circuits provide insights into the underlying biology of living cells and offer potential solutions to a range of medical and industrial challenges1, 2. A prerequisite for efficient engineering of such sophisticated circuits is the availability of a library of regulatory devices that can be connected in various contexts to create new and predictable behaviors. In synthetic biology, a regulatory device is loosely defined as a set of biochemical regulatory interactions that implement a basic information-processing relationship between inputs and outputs1, 3. Here we investigate transcriptional devices where the input is expression of a gene product that regulates production of output from a corresponding promoter. In particular, we focus on repressor devices, as these can in principle be used to build any computational circuit, whereas activators cannot4, 5. To date, however, an impediment to engineering larger and more complex circuits in any living organism is the lack of an efficient framework for generating a sufficient number of composable regulatory devices (i.e., having matching input and output types and expression levels) that can interconnect to form functional circuits3.
Recent efforts toward developing a transcriptional framework for a large library of composable devices include the creation of synthetic transcriptional modifiers by fusing effector domains to zinc-finger proteins or transcription activator–like effector (TALE) proteins, albeit with limitations such as extensive DNA assembly protocols6–8 or slow temporal responses because of the epigenetic modifications caused by the effector domains at target promoters9. The Cas9 protein from the Streptococcus pyogenes CRISPR-Cas immune system has recently been adapted for both RNA-guided genome editing and gene regulation in a variety of organisms8, 10–13. This mechanism is attractive for engineering a large library of devices in mammalian cells because Cas9 can be targeted to virtually any DNA sequence by means of a small guide RNA (gRNA) and thus can be easily programmed for the generation of a diverse device library6. In addition, catalytically inactive Cas9 protein (Cas9m), not fused to any effector domains, has been shown to repress both synthetic and endogenous genes through steric blocking of transcription initiation and elongation8–11. Therefore, we decided to focus on the CRISPR system to generate synthetic gene regulatory devices and circuits. Specifically, we devised strategies for regulated expression of gRNAs in human cells using both RNA polymerase type II (RNA Pol II) and RNA Pol III promoters, and demonstrated that CRISPR repressor devices can be layered to create functional circuits with high on/off ratios.
We designed two CRISPR families of promoters regulated by Cas9m-mediated steric blocking of transcription: CRISPR-responsive RNA Pol II promoters (CRP; Supplementary Fig. 1) and CRISPR-responsive RNA Pol III U6 promoters (CR-U6; Supplementary Fig. 2a). Both families are modular and extensible because orthogonal and highly specific repressor-promoter pairs can be created by altering the Cas9 target sequence and corresponding gRNA. Initially, we tested the ability of CRPs to regulate expression of enhanced YFP (EYFP) based on the presence or absence of gRNA constitutively expressed from a standard U6 promoter. Flow cytometry analysis 48 h after transfection of regulatory circuitry into human embryonic kidney 293 (HEK293) cells showed ~100-fold repression for two different gRNA and CRP pairs (gRNA-a and gRNA-b; Supplementary Fig. 3), and minimal cross-talk between the two devices, which demonstrated the desired orthogonality (Supplementary Fig. 4 and Supplementary Discussion). For all circuits discussed in this paper, one of these two CRPs regulates expression of output reporter EYFP.
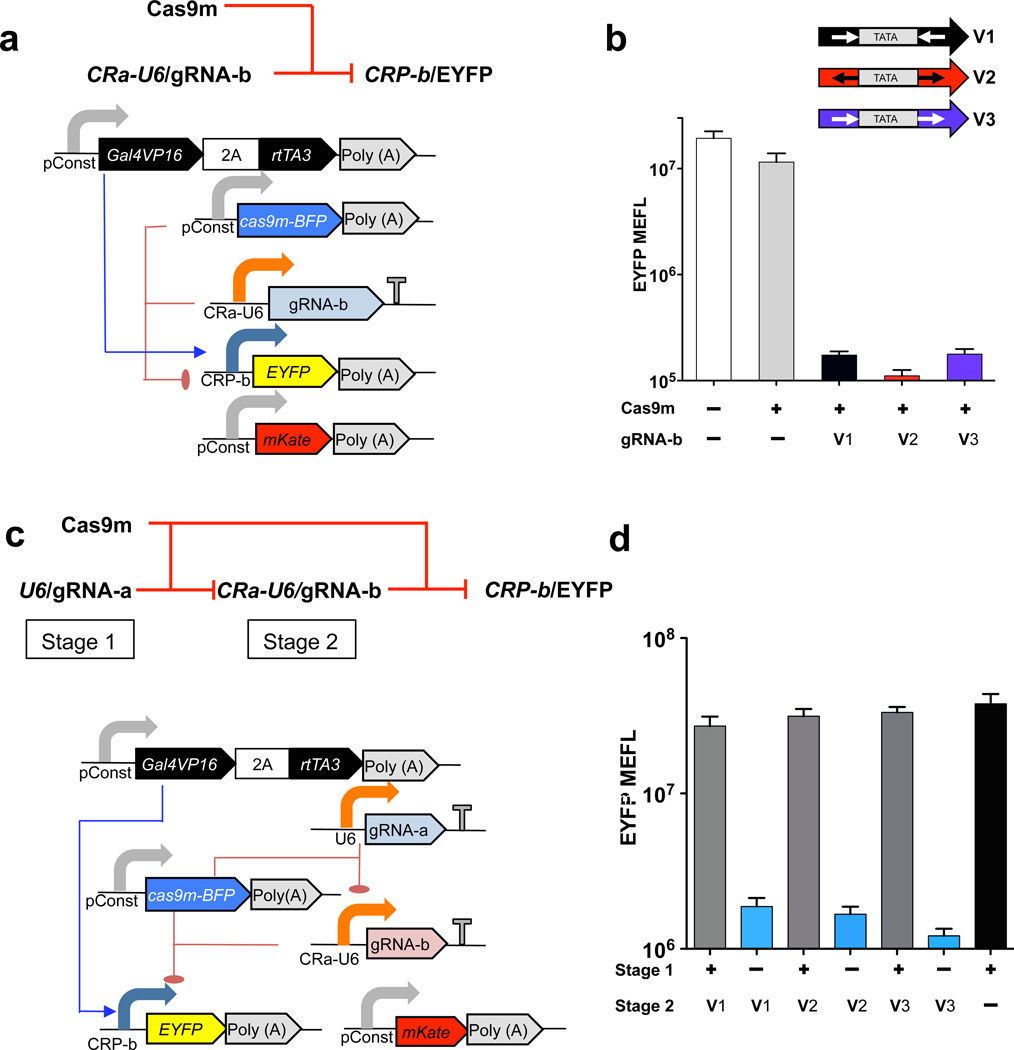
An important objective for us was to investigate the potential of using CRISPR devices to create layered circuitry, defined as the ability to compose complex multilevel control or regulatory operations by interconnecting CRISPR-based devices14. We designed the CR-U6 architecture to both express and be regulated by gRNA, hence creating composable devices. We accomplished this by inserting one Cas9m target site upstream and another downstream of the U6 promoter TATA box, similar to the case with tetracycline-responsive U6 promoter variants15. We created three versions of the CRa-U6 promoter that differ in the directionality of gRNA-a target sites flanking the TATA box (Supplementary Fig. 2a), and our experiments indicate that these regulate CRP-b with comparable efficiency to the unmodified U6 promoter (Fig. 1a,b). We then tested composability of these three variants in a cascade circuit where U6-driven gRNA-a regulates CRa-U6–driven expression of gRNA-b, which in turn regulates CRP-b expression of EYFP output (Fig. 1c). Transfection into HEK293 cells demonstrated highly functional layered CRISPR circuits that exhibited up to 27-fold derepression. (Fig. 1d and Supplementary Fig. 5).
Figure 1.
Design and experimental analysis in human cells of CRISPR repression devices and circuits based on the RNA Pol III U6 promoter. (a) Schematic of CRP repression device. CRa-U6 drives expression of gRNA-b, which in turn regulates EYFP output. (b) Flow cytometry–based analysis of three repression devices based on CRa-U6–driven gRNA-b expression, in HEK293 cells transfected with the indicated CRa-U6 promoter variants (V1–V3). Shown is geometric mean and s.d. of means of molecules of equivalent fluorescein (MEFL) of EYFP for cells expressing >3 × 106 MEFL of transfection marker mKate. n = 4 independent technical replicates, combined from two experiments. (c) Schematic of CRISPR transcriptional repression device cascade. U6-driven gRNA-a regulates CRa-U6–driven expression of gRNA-b, which in turn regulates CRP-b expression of output EYFP. (d) EYFP fluorescence for samples transfected either with all transcriptional units (+ for stage 1; V1 or V2 or V3 for stage 2), with all units but without U6-driven gRNA-a (– for stage 1) and with all units but without CRa-U6–driven gRNA-b (+ for stage 1; – for stage 2). Data represent geometric mean and s.d. of means of EYFP MEFL for cells expressing >1 × 107 MEFL of transfection marker mKate for n = 4 biological replicates pooled from two representative experiments.
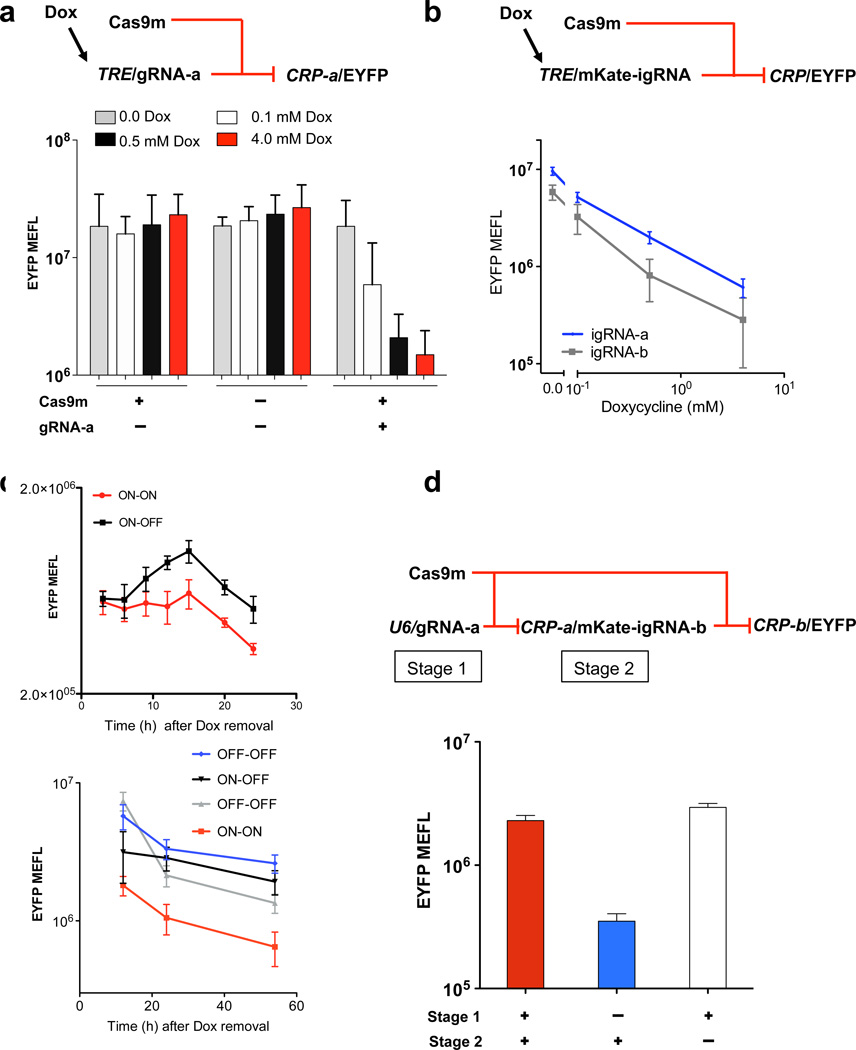
We next developed strategies for expressing gRNA from RNA Pol II promoters, specifically the well-studied mini-cytomegalovirus (mini-CMV) promoter, so that CRP devices can be likewise composed (Fig. 2). This will allow CRISPR devices to be regulated and tuned by commonly used modulators such as Gal4VP16 or rtTA3 (ref. 16), and to participate in the same framework as other protein-based regulators, sensors, actuators and reporters. We first inserted a gRNA sequence directly downstream of a tetracycline response element (TRE) promoter (Supplementary Fig. 6), a design inspired by RNA Pol II–mediated expression of short hairpin RNA17, 18 used for cell context–dependent expression and in vivo16, 17. Flow cytometry analysis 48 h after transfection of a characterization circuit with gRNA-a transcribed from TRE and controlling CRP-a showed substantial dose-dependent repression upon induction with doxycycline (Fig. 2a and Supplementary Fig. 6). Analysis of the reversibility of the circuit revealed an increase in EYFP expression after we removed doxycycline (Fig. 2c).
Figure 2.
Design and experimental analysis in human cells of CRISPR repression devices and circuits based on RNA Pol II promoters. (a) Schematic of a gRNA-a repression device regulated by TRE promoter and inducible by doxycycline (Dox) (top). EYFP output fluorescence was measured for samples transfected with or without Cas9m vector and TRE-driven gRNA-a with indicated amounts of Dox (top). Shown are geometric mean and s.d. of means of molecules of equivalent fluorescein (MEFL) of EYFP for cells expressing >106 MEFL of transfection marker mKate. n = 3 independent technical replicates combined from three experiments. (b) Schematic of igRNA repression devices expressing mKate and igRNA regulated by TRE promoter. EYFP fluorescence was measured for samples transfected with circuits containing igRNA-a or igRNA-b devices under indicated amounts of Dox. Geometric mean and s.d. of means of EYFP MEFL for cells expressing >106 MEFL of transfection marker mKate. n=2 (gRNA-a) and 3 (gRNA-b) independent technical replicates combined from two and three experiments, respectively. (c) Analysis of reversible repression: TRE–gRNA-a circuit from a (top) and TRE/mKate-igRNA-b circuit from b (bottom). Two phases, where in each phase “ON” indicates induction with 4 mM Dox, and “OFF” indicates zero Dox. Initial phase is 24 h (top) or 12 h (bottom) after transfection. Experimental results are reported for the second phase that includes indicated modulations of Dox. Reversibility is shown by comparison between the different groups. Geometric mean and s.d. of EYFP MEFL means for cells expressing >106 MEFL of Cas9m-BFP. n = 2 independent technical replicates combined from two experiments. (d) Schematic of cascades with igRNA (top). EYFP output fluorescence for samples transfected with or without U6–gRNA-a (stage 1) and CRP-a–igRNA-b (stage 2) (bottom). Geometric mean and s.d. of EYFP means for cells expressing >3 × 106 MEFL. n = 4 independent technical replicates combined from two experiments.
We also encoded gRNA as an intron with flanking splicing sequences using a strategy similar to that employed for intronic microRNAs12, 19(Supplementary Fig. 7). Intronic gRNA (igRNA) co-expressed with a protein allows additional capabilities, such as monitoring device regulation by observing a coexpressed fluorescent reporter. Expression of igRNA also potentially allows multiple gRNAs to be expressed from separate introns inserted in a single coding gene. Flow cytometry analysis 48 h after transfection of characterization circuits for both igRNA-a and igRNA-b showed substantial dose-dependent repression upon induction with doxycycline (Fig. 2b), as well as reversibility (Fig. 2c, Supplementary Figs. 8–10 and Supplementary Discussion).
To test the extensibility of CRISPR regulatory devices, we designed a small library of igRNA and CRP pairs that differ only in the nucleotide sequence of the designated target sites in the CRPs and corresponding igRNA sequences (Supplementary Fig. 11a). Characterization of this library showed a range of repression efficiencies from 2-fold to 30-fold (Supplementary Fig. 11b), which suggested that we can achieve devices with varying regulatory properties.
We next tested whether igRNA from a CRP can regulate another CRP, forming a layered CRISPR cascade with connected RNA Pol-II promoters (Supplementary Fig. 12). We replaced the CRa-U6 device shown in Figure 1c with CRP-a that drives expression of an igRNA-b as an intron of near-infrared fluorescent protein (iRFP) (Supplementary Fig. 12), which yielded a cascade with repression of about six fold (Fig. 2xd). A similar configuration of cascaded promoters (U6 followed by two CRPs) but with exchanged gRNA-a and gRNA-b resulted in a moderately functional cascade (Supplementary Fig. 13). Circuits comprising only RNA Pol II promoters did not yield substantial regulation, and additional testing is required to address possible compositional or host-context issues related to these circuits (Supplementary Fig. 14)20–23.
Further analysis should directly capture the input-output behavior of our devices and quantify any apparent cooperativity, helping determine whether additional optimization is needed (e.g., more operators) for improved signal restoration to create more complex layered circuitry. Compared to other approaches for generating synthetic transcriptional regulators (e.g., TALE or zinc-finger proteins), CRISPR-based devices are likely to be better suited for scaling to larger and more sophisticated circuits, as the DNA required for each additional device is much smaller. This means that more complex circuits can be encoded when there are size limits on the DNA to be delivered, and that any given circuit topology can be encoded in a much smaller amount of DNA. Regulation by RNA interference is expected to integrate well with CRISPR-based devices, providing a useful means of sensing and effecting cell state, though at present RNA interference cannot be used on its own to build layered circuits. Additional scaling of the CRISPR technology and the creation of more complex logic may also benefit from using multiple orthogonal Cas9 proteins24. Finally, given the simplicity of creating additional gRNAs and corresponding promoters, and the high performance of the devices presented here, it should be possible to rapidly generate, characterize and optimize a large library of effective regulatory devices. Taken together, the scalability and ability to rapidly design these devices should allow CRISPR-based circuits to facilitate a wide range of applications in human cells.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Cell culture and transfection
HEK293FT cells were obtained from Invitrogen and maintained in DMEM (CellGro) supplemented with 10% FBS (PAA Laboratories), 1% L-glutamine–streptomycin–penicillin mix (CellGro) and 1% nonesential amino acids (NEAA; HyClone) at 37 °C and 5% CO2. rtTA3 stable cell lines (HEK293-rtTA3) were created by lentiviral transduction of HEK293 cells with rtTA3 coding sequence under a constitutive promoter and antibiotic selection with hygromycin for 2 weeks. All experiments were done in HEK293-rtTA3 cell lines. Transfections were performed using Attractene reagent (QIAGEN). Cells were seeded the day before at 2 × 105 cells per well in a 24-well plate. Dosages of plasmids used for the transfections were identified after optimization experiments for each component of the devices and circuits (data not shown). For transfections involving the repression devices, 500 ng of input gRNA plasmid was mixed with a cocktail of other plasmids (ratio of 1x:4x:14x:4x for Gal4VP16-2A-rtTa3 plasmid, EYFP (output) expression plasmid, Cas9m-BFP expression plasmid and mKate expression plasmid, respectively, where x = 5 ng) in 70 µl of DMEM (without supplements). For transfection of the cascade circuits of two U6-based devices, 500 ng of the stage 1 gRNA–encoding plasmid was mixed with a cocktail of other plasmids (ratio of 2x:x:14x:10x:5x for Gal4VP16-2A-rtta3 plasmid, EYFP (output) expression plasmid, Cas9m-BFP expression plasmid, stage 2 gRNA plasmid and mKate expression plasmid, respectively, where x = 5 ng). For transfections of the cascade circuits of other devices the concentration of the stage 2 gRNA encoding plasmid was twice the value of stage 2 gRNA in U6-only cascades. In control experiments, we replaced the DNA plasmid under study with an equivalent amount of empty DNA plasmid to maintain the total amount of transfected DNA constant among the groups. 1.5 µl of Attractene was added to DNA mixtures, and the tube was gently mixed and kept at room temperature for 20 min to form the DNA-liposome complex. Fresh medium was added to the cells directly before transfection (500 µl of DMEM with supplements). The DNA-Attractene solution was then added drop-wise to the wells. Induction of the circuit was performed at this time as well by addition of doxycycline. In experiments involving the cascade, the ratio of stage 1 gRNA encoding plasmid to the stage 2 gRNA encoding plasmid was 5:1, except for U6-only cascades in which the ratio was 10:1.
Plasmids
Plasmids used for this project were constructed using the Gateway system (Invitrogen). A plasmid encoding catalytically mutant Cas9 fused to BFP was obtained from Addgene (plasmid 46910). The expression vectors were made by Gateway cloning. The U6-driven gRNA expression cassettes were ordered as gblocks from IDT and cloned into a pCR2.1-TOPO TA vector by Topo TA cloning. The library of CRPs were ordered as gene fragments from IDT and assembled into an appropriate promoter entry vector. igRNA library elements were also ordered as gblocks from IDT and assembled into the mKate entry vector by appropriate restriction digest. Sequences are available in the Supplementary Note.
Flow cytometry
Flow cytometry data were collected 48 h after transfection. Cells were trypsinized and centrifuged at 453 g for 5 min at 4 °C. The supernatant was then removed, and the cells were resuspended in Hank’s Balanced Salt Solution without calcium or magnesium supplemented with 2.5% FBS. BD LSRII was used to obtain flow cytometry measurements with the following settings: EBFP, measured with a 405 nm laser and a 450/50 filter; EYFP, measured with a 488 nm laser and a 530/30 filter; mKate, measured with a 561 nm laser and a 695/40 filter. At least 100,000 events were gathered from each sample, ensuring that any 1/10 decade interval with more than 5% of the mean density of events would contain at least 100 expected events.
Statistical analysis
Flow cytometry data were converted from arbitrary units to compensated molecules of equivalent fluorescein (MEFL) using the Tool-Chain to Accelerate Synthetic Biological Engineering (TASBE) characterization method (MIT CSAIL Tech. Report 2012–008 (2012). An affine compensation matrix is computed from single positive and blank controls. FITC measurements are calibrated to MEFL using SpheroTech RCP-30-5-A beads, and mappings from other channels to equivalent FITC are computed from DNA co-transfection of constitutive EBFP, EYFP and mKate (plus iRFP, for four-color experiments) each controlled by the Hef1a promoter on its own otherwise identical plasmid. nontransfected controls were included in each experiment. MEFL data are segmented by constitutive fluorescent protein expression into logarithmic bins at 10 bins per decade, and geometric mean and variance are computed for those data points in each bin. Based on the observed constitutive fluorescence distributions (Supplementary Fig. 15), a threshold was selected as a cutoff for each data set, below which data were excluded as being too close to the non-transfected population. Data shown in the figures are geometric mean and s.d. of means for cells expressing the transfection marker mKate based on the MEFL threshold set. High outliers were removed by excluding all bins without at least 100 data points. Both population and per-bin geometric statistics were computed over this filtered set of data. Sample sizes were pre-determined for each experiment. During analysis of flow cytometry data, samples were excluded by the following predetermined criteria: if they contained less than 10% of the number of events or less than 10% of the fraction of successful transfections of the mode for the batch in which they were collected.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health grants 5R01CA155320-04 and P50 GM098792. We thank L. Wrobleska and P. Guye (Massachusetts Institute of Technology) for providing the initial intronic miRNA–based plasmid and the primary Cas9 construct, and for helpful discussions, and M. Graziano (Massachusetts Institute of Technology) for providing us the HEK293 cell lines that constitutively express rtTA3. J.H. was partially supported by the Intelligent Synthetic Biology Center of Global Frontier Project (2013M3A6A8073557) funded by the Ministry of Science, Information and Communication Technology (ICT) and Future Planning of Korea.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
R.W. and S.K. conceived the idea. S.K., R.W., M.R.E. and J.B. designed experiments. S.K. performed the majority of experiments. M.R.E. and R.N.H. helped with flow cytometry and transfections. Y.L. and Z.X. built initial versions of CRP-a and CRP-b promoters. J.H. helped with DNA constructions. J.B. developed and applied computational analysis techniques. J.B. and S.K. performed the flow cytometry and statistical analysis. S.K. wrote the manuscript. R.W., J.B. and M.R.E. edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Andrianantoandro E, Basu S, Karig DK, Weiss R. Mol. Systems Biology. 2006;2:0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruder WC, Lu T, Collins JJ. Science. 2011;333:1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 3.Slusarczyk AL, Lin A, Weiss R. Nat. Rev. Genet. 2012;13:406–420. doi: 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- 4.Bird J. Engineering Mathematics. Vol. 532. Elsevier Science; 2007. [Google Scholar]
- 5.Peirce CS. Collected Papers of Charles Sanders Peirce. Vol. 4. Harvard University Press; 1933. pp. 12–20. [Google Scholar]
- 6.Farzadfard F, Perli SD, Lu TK. ACS Synthetic Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil AS, et al. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garg A, et al. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer BP, Fischer C, Fussenegger M. Biotechnol. Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 10.Stanton BC, et al. Nat. Chem. Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, et al. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P, et al. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi LS, et al. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen AA, Segall-Shapiro TH, Voigt CA. Curr. Opin. Chem. Biol. 2013;17:878–892. doi: 10.1016/j.cbpa.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Henriksen JR, et al. Nucleic Acids Res. 2007;35:e67. doi: 10.1093/nar/gkm193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko JK, Choi KH, Zhao X, et al. FASEB J. 2011;25:2638–2649. doi: 10.1096/fj.11-181560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia H, et al. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 18.Giering JC, Grimm D, Storm TA, Kay MA. Molecular therapy. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 19.Lin SL, et al. Biochem. Biophys. Res. Commun. 2003;310:754–760. doi: 10.1016/j.bbrc.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 20.Cardinale S, Arkin AP. Biotechnol. J. 2012;7:856–866. doi: 10.1002/biot.201200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyorgy A, Del Vecchio D. PLoS Comp. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis PP, Ehrenberg M, Bremer H. Microbiol. Mol. Biol. Rev. 2004;68:639–668. doi: 10.1128/MMBR.68.4.639-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klumpp S, Hwa T. Proc. Natl. Acad. Sci. USA. 2008;105:20245–20250. doi: 10.1073/pnas.0804953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esvelt KM, et al. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.