Abstract
Glycosylation is an abundant post-translational modification that alters the fate and function of its substrate proteins. To aid in understanding the significance of protein glycosylation, identification of target proteins is key. As with all proteomics experiments, mass spectrometry has been established as the desired method for substrate identification. However, these approaches require selective enrichment and purification of modified proteins. Chemical reporters in combination with bioorthogonal reactions have emerged as robust tools for identifying post-translational modifications including glycosylation. We provide here a method for the use of bioorthogonal chemical reporters for isolation and identification of glycosylated proteins. More specifically, this protocol is a representative procedure from our own work using an alkyne-bearing O-GlcNAc chemical reporter (GlcNAlk) and a chemically cleavable azido-azo-biotin probe for the identification of O-GlcNAc-modified proteins.
Keywords: Proteomics, Glycosylation, Bioorthogonal chemical reporter, Click chemistry, Azide, Mass spectrometry, O-GlcNAc
1. Introduction
The identification of glycosylated proteins via mass spectrometry (MS) has proven invaluable in elucidating the function of glycosylation events as well as the relationship between mono- and polysaccharides and their substrates. Several technologies have been developed to facilitate this investigation, which typically involves affinity enrichment for target proteins followed by mass spectrometry analysis. Lectins (1) are helpful in isolating and identifying N-linked glycoproteins, but lectins are not generally applicable to other forms of glycosylation. Several antibodies have been raised for the recognition of O-GlcNAc-modified proteins (2); however, they all display some sequence requirement for the underlying peptide. Chemical reporters are an alternative to these methods. Originally developed in the Bertozzi laboratory, metabolic bioorthogonal chemical reporters deliver unique reactivity to glycosylated proteins (3). There are currently several azide- or alkyne-bearing monosaccharide analogs that can be used as chemical reporters of glycosylation (Fig. 1). This metabolic labeling approach has been effective for the visualization and identification of cell-surface glycoproteins (4-8) and O-GlcNAc-modified proteins (9-12). Post-lysis enzymatic transfer of an azide-modified chemical reporter to O-GlcNAcylated proteins is an alternative delivery method developed by the Hseih-Wilson laboratory (13, 14). A mutant galacytosyltranferase transfers an N-azidoacetylgalactosamine (GalNAz) residue onto O-GlcNAc-modified proteins. Regardless of the method of incorporation, these bioorthogonally functionalized cell lysates can then be subjected to Staudinger ligation, Cu-catalyzed Azide-Alkyne Cycloaddition (CuAAC), or Strain-promoted Azide-Alkyne Cycloaddition with a corresponding phosphine-, azide-, or alkyne-modified affinity probe (Fig. 2).
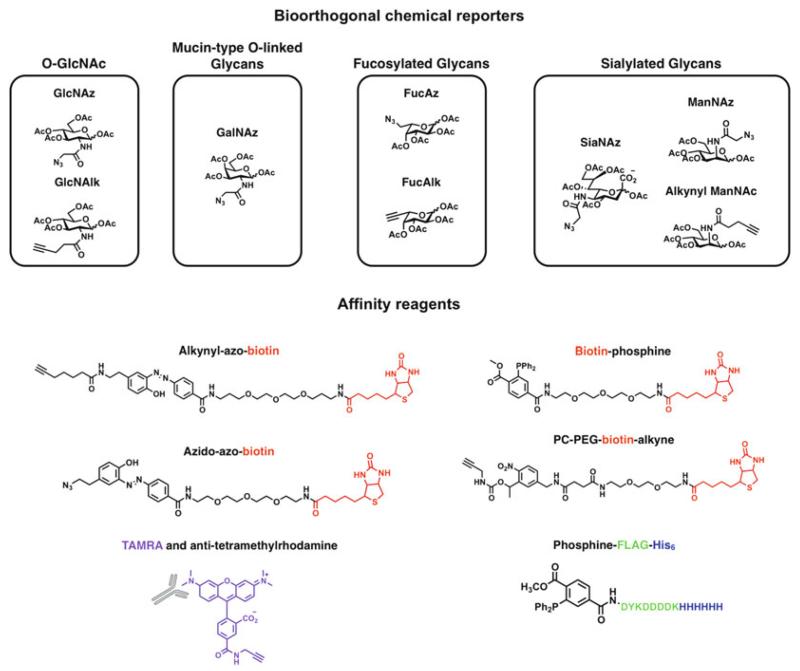
Fig. 1.
Bioorthogonal chemical reporters for glycosylation: GlcNAz (21), GlcNAlk (20), GalNAz (4), FucAz (22), FucAlk (23), ManNAz (21), alkynyl ManNAc (23) and SiaNAz (24). Affinity probes: azido-azo-biotin (16), alkynyl-azo-biotin (16), PC-PEG-biotin-alkyne (14), phosphine-FLAG-His6 (9) and alkynyl tetramethyl-6-carboxyrhodamine (TAMRA) and antibody.
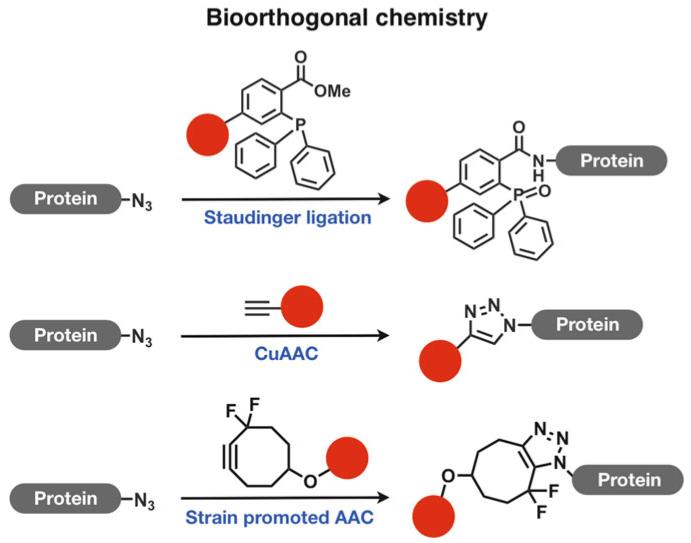
Fig. 2.
Bioorthogonal chemistry: Staudinger Ligation, Cu-catalyzed Azide-Alkyne Cycloaddition and Strain Promoted Azide-Alkyne Cycloaddtion.
There are several approaches to affinity purification. The Hsieh-Wilson laboratory developed an alkyne-modified fluorescent rhodamine derivative, tetramethyl-6-carboxyrhodamine alkyne (TAMRA), and a corresponding antibody is commercially available that allows for the selective enrichment of rhodamine-modified proteins (13) (Fig. 1). Cleavable azido- and alkynyl-biotin probes facilitate the enrichment and subsequent elution of target proteins for mass spectrometry analysis (15, 16). Notably, the Hart laboratory developed a photo-cleavable probe termed PC-PEG-biotin-alkyne (Fig. 1). Additionally, chemically cleavable biotin reagents that contain an azo moiety that is readily reduced to liberate proteins from streptavidin beads for downstream MS analysis can be used (17-19).
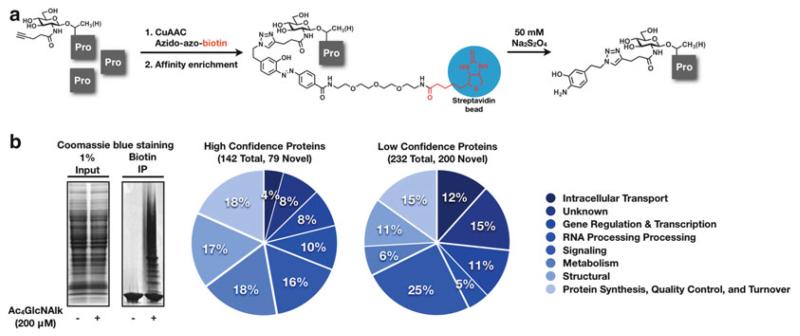
This protocol is representative of the identification of O-GlcNAc-modified proteins (Fig. 3a) but should be readily applicable to other types of glycosylation including mucin-type O-linked glycans and N-linked glycans. Specifically, we describe the metabolic incorporation of the alkyne-bearing N-acetylglucosamine analog GlcNAlk onto O-GlcNAcylated proteins in NIH-3T3 cells. Following lysis in 4% SDS, soluble proteins were subjected to CuAAC with azido-azo-biotin. Subsequent affinity enrichment with streptavidin beads was used to isolated GlcNAlk-modified proteins. The biotinylated proteins were then liberated from the beads using sodium dithionite (50 mM), separated by SDS-PAGE and subjected to trypsinolysis. Finally, LC-MS was conducted to identify 374 GlcNAlk-modified proteins (Fig. 3b).
Fig. 3.
(a) GlcNAlk was metabolically incorporated into NIH-3T3 cells. Following lysis in 4% SDS, soluble proteins were subjected to CuAAC with azido-azo-biotin. Affinity enrichment using streptavidin beads isolated GlcNAlk-modified proteins. The proteins were then liberated from the beads using sodium dithionite (50 mM), separated by SDS-PAGE and subjected to proteolysis. (b) Inputs and biotin enriched proteins stained with Coomassie blue. *Large smear at bottom of the gel is streptavidin. LC-MS proteomics were conducted to identify 374 GlcNAlk-modified proteins of high and medium confidence (10).
2. Materials
All solutions and buffers should be prepared with 18 MΩ H2O at 25°C. Reagents should be stored at room temperature unless otherwise noted. Dispose of hazardous waste appropriately.
2.1. Materials for Metabolic Incorporation of Chemical Reporters and Preparation of Cell Lysates
1,3,4,6-tetra-O-acetyl-N-4-pentynylglucosamine (Ac4GlcNAlk): 200 mM stock solution in DMSO. Ac4GlcNAlk was synthesized according to literature procedure (20) . To 84.5 mg of Ac4GlcNAlk add 1 mL DMSO in a microcentrifuge tube. Vortex until homogeneous. Store at −20°C.
HyClone Dulbecco’s Phosphate Buffered Saline (DPBS) (Thermo Scientific, Rockford, IL, USA): Combine 9.6 g DPBS powder with 1 L H2O and autoclave.
Phenylmethanesulfonylfluoride (PMSF) (Sigma Chemical Company, St. Louis, MO, USA) in H2O: 250 mM stock solution. Add 43.5 mg PMSF to a microcentrifuge tube and add 1 mL H2O. Vortex. Store at −20°C.
0.05% SDS Buffer: 0.05% SDS, 10 mM triethanolamine (TEA) pH 7.4, 150 mM NaCl with Complete Mini protease inhibitor cocktail (Roche Biosciences, Indianapolis, IN, USA). Add 25 mg SDS, 746 mg TEA, 4.38 g NaCl to a 1 L graduated cylinder, and add 450 mL H2O. Mix and adjust pH to 7.4. Add additional H2O to final volume of 500 mL.
Benzonase nuclease (Sigma Chemical Company, St. Louis, MO, USA). Store at −20°C.
4% SDS buffer: 4% SDS, 150 mM NaCl, 50 mM TEA pH 7.4. Combine 40 g SDS, 1.49 g NaCl, 7.46 g TEA, and 950 mL H2O. Mix and adjust pH to 7.4. Add additional H2O to final volume of 1 L.
Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA).
Bovine serum albumin standard (2 mg/mL): 5.52 mg bovine serum albumin (Sigma Chemical Company, St. Louis, MO, USA), 260 μL H2O, 500 μL 0.05% SDS buffer, 2,000 μL 4% SDS buffer. Aliquot into 500 μL stocks and store at −20°C.
1% Nonidet P-40 (NP-40) buffer: 1% NP-40, 150 mM NaCl, 50 mM TEA pH 7.4, Complete Mini protease inhibitor cocktail (Roche Biosciences, Indianapolis, IN, USA). In a 1 L glass bottle combine 10 g NP-40, 1.49 g NaCl, 7.46 g TEA, and 950 mL H2O and stir until all reagents go into solution. Adjust pH 7.4. Add additional H2O to a final volume of 1 L.
2.2. Materials for Click Chemistry, Biotin Enrichment, and Preparation of Samples for LC-MS Analysis
Azido-azo-biotin: 10 mM stock solution in DMSO. Azido-azo-biotin was synthesized according to literature procedure (16) . To 7.4 mg azido-azo-biotin was added 1 mL DMSO in a microcentrifuge tube. Vortex until solubilized. Pipette out into 100 μL aliquots. Store at −20°C.
Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (Calbiochem, San Diego, CA, USA): 50 mM freshly prepared stock solution in water (see Note 1). Combine 5 mg TCEP and 349 μL H2O in a microcentrifuge tube. Vortex until TCEP goes into solution. Store on ice during use.
Tris[(1-benzyl-1-H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (Anaspec, Fremont, CA, USA): 10 mM stock solution in DMSO. Combine 5 mg of TBTA and 942 μL DMSO in a microcentrifuge tube. Vortex until TBTA goes into solution. Divide into ~250 μL aliquots. Store at −20°C solution for short-term or −80°C for 6+ months. Store dry compound at −20°C.
CuSO4·5H2O (Sigma Chemical Company, St. Louis, MO, USA): 50 mM freshly prepared stock solution in water. Combine 5 mg CuSO4·5H2O with 400 μL H2O in a microcentrifuge tube. Vortex until solubilized. Store on ice during use.
HEPES buffer: 6 M urea, 2 M thiourea, 10 mM HEPES pH 8.0 (see Note 2). Combine 36 mg urea, 15.2 mg thiourea, 238 mg, and 90 mL H2O. Adjust pH to 8.0 and add H2O to a final volume of 100 mL.
Dithiothreitol (Sigma Chemical Company, St. Louis, MO, USA): 100 mM stock solution in H2O. Add 15.4 mg dithiothreitol and 1 mL H2O to a microcentrifuge tube. Vortex until dissolved. Store at −20°C.
Iodoacetamide (Sigma Chemical Company, St. Louis, MO, USA): 550 mM freshly prepared stock solution in H2O. Add 102 mg of iodoacetamide and 1 mL H2O to a microcentrifuge tube. Vortex until dissolved.
Streptavidin agarose resin (Thermo Scientific, Rockford, IL, USA). Store at 4°C.
Sodium dithionite solution: freshly prepared 25 mM sodium dithionite, 1% SDS in DPBS (see Note 3). To a 15 mL centrifuge tube add 22 mg sodium dithionite and 5 mL 1% SDS in DPBS. Invert gently until sodium dithionite goes into solution. Sonicate if necessary.
YM-10 Centricon 3,000 MWCO filters (Millipore, Billerca, MA, USA).
1× SDS-free loading buffer: 10% glycerol, 0.1% bromophenol blue, 0.7% β-mercaptoethanol. Combine 50 g glycerol, 500 mg bromophenol blue and 500 mL H2O. Store at room temperature. As needed, add 7 μL β-mercaptoethanol to a 1 mL aliquot of 1× loading buffer. Store the loading buffer containing β-mercaptoethanol at −20°C.
Criterion Tris–HCl 4–20% polyacrylamide gel (Bio-Rad, Hercules, CA, USA). Store at 4°C.
50 mM ammonium bicarbonate (ABC) solution in H2O: In a 250 mL bottle dissolve 395 mg ABC in 100 mL H2O.
Acetonitrile, anhydrous (EMD Chemicals, Gibbstown, NJ, USA).
Trifluoroacetic acid (TFA) (Sigma Chemical Company, St. Louis, MO, USA).
Trypsin solution: Combine 1 mg trypsin in 15 mL of 50 mM ABC solution in a 15 mL falcon tube. Vortex until solubilized. Aliquot into 1 mL portions. For long-term storage, place at 20°C. Once aliquot is thawed, store at 4°C.
3. Methods
3.1. Metabolic Incorporation of Chemical Reporters and Preparation of Cell Lysates
Replace media on cells (twenty 150 mm plates) at 80–85% confluency for 20 mL low-glucose media ( see Note 4 ) containing 200 μM Ac4 GlcNAlk or DMSO vehicle.
After 16 h, aspirate off media, and wash cells with 10 mL PBS per plate. Add 2 mL trypsin to each plate and return in incubator for ~3 min or until cells come off the plate.
Resuspend cells in 5 mL PBS per plate and combine into two 50 mL centrifuge tubes.
Centrifuge at 4°C for 4 min at 3,000 ×g. Aspirate off supernatant, resuspend both pellets in a total of 30 mL PBS and combine into 1 falcon tube. Centrifuge at 4°C for 4 min at 3,000 ×g. Aspirate off supernatant. Repeat wash and centrifugation one time.
Resuspend washed cell pellet in 200 μL H2O, 60 μL PMSF, and 500 μL 0.05% SDS buffer. Add 8 μL Benzonase and incubate cells on ice for 30 min.
Add 2,000 μL 4% SDS buffer, sonicate the cells in a bath sonicator for 5 min and collect by centrifugation at 20,000 ×g for 10 min at 15°C. Transfer the soluble fractions to a new 15 mL centrifuge tube.
Normalize protein concentration by BCA assay (Pierce, ThermoScientific). Combine 50 parts Reagent A to 1 part Reagent B in 15 mL falcon tube and vortex until green color is homogeneous. Aliquot out 1 mL of working reagent (WR) into a microcentrifuge tube for each sample and an additional four tubes for the standard curve. Pipette 1 μL of soluble lysate into the corresponding centrifuge tube filled with 1 mL WR. For the standard curve add 0, 1, 2, or 4 μL (0, 2, 4, or 8 μg, respectively) of albumin standard to the WR. Place in heat block at 60°C for 30 min. Upon completion, remove all samples from heat block. Transfer the samples to 1 cm plastic cuvettes. With the UV spectrophotometer set to 562 nm, blank the instrument using the standard sample not containing albumin. Take absorbance readings of each sample.
3.2. Click Chemistry, Biotin Enrichment, and Preparation of Samples for LC-MS Analysis
In a spreadsheet program, graph the absorbance vs. concentration of the BCA assay albumin standards. Generate a linear best-fit line and determine the concentration of each of the samples using this equation. Dilute the samples with 1% NP-40 buffer to a final concentration of 1 mg/mL (10 mg of total lysate per sample) and transfer to a 50 mL centrifuge tube.
Prepare click chemistry cocktail (1,200 μL per 10 mg sample). Combine azido-azo-biotin tag (200 μL, 100 μM, 10 mM stock solution in DMSO), TCEP (400 μL, 1 mM, 50 mM freshly prepared stock solution in water), tris[(1-benzyl-1-H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (200 μL, 100 μM, 10 mM stock solution in DMSO), and CuSO4·5H2O (400 μL, 1 mM, 50 mM freshly prepared stock solution in water). Vortex gently. Add appropriate amount of click chemistry cocktail to each sample. Vortex gently.
Place the samples in the dark and allow the reaction to proceed for 1 h. To quench the reaction and precipitate proteins, add ~10 volumes (12 mL) of ice-cold methanol and place at −80°C overnight.
Centrifuge precipitated proteins at 6,000 ×g for 30 min at 0°C. Wash 3× with 40 mL ice-cold MeOH, taking care to resuspend the pellet each time.
Allow the protein pellet to air-dry for 1 h and resuspend in 4 mL of HEPES buffer by bath sonication. Transfer to a new 15 mL centrifuge tube.
Incubate captured proteins in freshly made 1 mM dithiothreitol for 40 min to reduce cysteines. Cap cysteines by further incubation with freshly prepared 5.5 mM iodoacetamide for 30 min in the dark.
Wash 250 μL streptavidin beads with an equal volume PBS two times and with an equal volume HEPES buffer one time. Resuspend beads in an equal volume HEPES buffer; add beads to captured proteins. Incubate on a rotator for 2 h.
Collect beads by centrifugation (2,000 ×g for 2 min). Wash with HEPES buffer two times, PBS two times, and 1% SDS in PBS two times (10 mL per wash, 2,000 × g, 2 min). After the final wash, resuspend beads in 250 μL 1% SDS in PBS and transfer samples to 2 mL dolphin-nosed tubes. Collect beads by centrifugation (2,000 ×g for 2 min) and carefully pipette away supernatant.
Pipette 250 μL of sodium dithionite solution into each sample and incubate for 30 min at room temperature to elute captured proteins. Collect the beads by centrifugation for 2 min at 2,000 ×g and collect eluent. Repeat elution step with an additional 250 μL of sodium dithionite solution. Combine eluents from both steps.
Transfer eluent to a YM-10 Centricon 3,000 MWCO filter and centrifuge at 10,000 × g for 30 min. Add an additional 300 μL of PBS into the filter and centrifuge again at 10,000 ×g for 30 min at room temperature. Transfer the concentrated eluent to a microcentrifuge tube (see Note 5) and dry by SpeedVac overnight (see Note 6).
Resuspend the dried pellets in 40 μL 1× SDS-free loading buffer and boil at 98°C for 5 min (see Note 7). Load 36 μL of the sample onto a Criterion Tris–HCl 4–20% polyacrylamide gel for subsequent in-gel trypsin digestion. Load the remaining sample onto another Criterion Tris–HCl 4–20% polyacrylamide gel for validation of protein candidates by Western blot.
Remove each lane of the Criterion Tris–HCl 4–20% gel using a razor blade (see Note 8). Divide each lane evenly into 10 sections. Dice each section into ~0.5 cm squares (see Note 9) and transfer the pieces to a microcentrifuge tube. Add 300 μL of 50 mM ABC and incubate for 15 min. Carefully aspirate away the ABC solution. Repeat 2×.
Add 300 μL of a 1:1 solution of 50 mM ABC/acetonitrile and incubate for 30 min. Carefully aspirate away the solution and repeat. Add 300 μL 100% acetonitrile and SpeedVac until dry.
Rehydrate gel slices by adding 30 μL trypsin solution and incubate at 37°C in a water bath for 18 h.
Add 200 μL 50% acetonitrile in H2O with 0.1% TFA to elute peptides. Collect eluent and repeat elution. Dry combined eluents by SpeedVac.
3.3. LC-MS Analysis
Samples are now ready to be subjected to standard nano-HPLC-MS/MS analysis. Under our conditions, peptides were pressure-loaded onto a 75-μm (inner diameter), 15-cm C18 reverse-phase column, and separated with a gradient running from 95% buffer A (HPLC water with 0.1% (v/v) formic acid) and 5% buffer B (HPLC-grade CH3CN with 0.1% (v/v) formic acid) to 55% B over 30 min, increased to 95% B over 10 min and held at 95% (v/v) B for 10 min.
After one complete MS scan (300–2,000 MW), conduct three data-dependent scans of the nth most intense ions with dynamic exclusion enabled. For peptide identification, use SEQUEST version 28 (ThermoFisher Scientific) and search against the appropriate International Protein Index protein sequence database v3.45.v. Compile data using scaffold software (Proteome Software).
Acknowledgments
We thank Leslie Bateman for careful reading of the manuscript. This work was supported by the University of Southern California and the American Cancer Society Grant IRG-58-007-51 (to M.R.P.) and the Ellison Medical Foundation and the National Institutes of Health and General Medical Sciences Grant 1RO1GM087544 O1A2 (to H.C.H.).
Footnotes
TCEP degrades over time. Therefore, we recommend storing the reagent at 4°C and purchasing fresh TCEP every 6 months.
HEPES buffer should be made fresh for every use. The thiourea can be tricky to get into solution. Do not warm up the solution to solubilize. Sonicate instead.
The sodium dithionite solution should be made fresh for each use.
Metabolic incorporation conditions should be optimized for the type of glycosylation targeted. We have shown previously that treatment with Ac4GlcNAlk under low-glucose conditions optimizes the labeling of O-GlcNAc-modified proteins (10).
The easiest way to transfer the concentrated eluent is to take a clean microcentrifuge tube, place it over the top of the filter and gently invert the tube right side up. Centrifuge at 500 × g for 5 min. Eluent will collect in bottom of new microcentrifuge tube.
Set the temperature to 25°C on the SpeedVac and do not allow it to rise above 30°C.
Due to the large amount of SDS in the sample, resuspension of the dried pellet can be difficult. The best way to resuspend is to add the 40 μL 1× SDS-free loading buffer and then take a pipette tip, dip it in the loading buffer in the bottom of the tube and then wet the sides of the tube with the buffer to resuspend the sample on the sides. Boil the sample for 5 min at 98°C, take the sample pipette tip, wet the sides again, and then centrifuge at 13,000 × g for 1 min. Do not use gel-loading tips to load the sample. Instead use a standard 10–200 μL pipette tip.
In order to prevent contamination, use a different razor blade for each lane.
Take care in dividing each section into uniform squares as this will improve in-gel trypsin digestion.
References
- 1.Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141(5):897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Teo CF, et al. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 2010;6(5):338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287(5460):2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 4.Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci USA. 2003;100(25):14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehnert KW, et al. Metabolic labeling of fucosylated glycans in developing zebrafish. ACS Chem Biol. 2011;6(6):547–552. doi: 10.1021/cb100284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Probing mucin-type O-linked glycosylation in living animals. Proc Natl Acad Sci USA. 2006;103(13):4819–4824. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaro BW, Bateman LA, Pratt MR. Robust in-gel fluorescence detection of mucin-type O-linked glycosylation. Bioorg Med Chem Lett. 2011;21(17):5062–5066. doi: 10.1016/j.bmcl.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 8.Hanson SR, et al. Tailored glycoproteomics and glycan site mapping using saccharide-selective bioorthogonal probes. J Am Chem Soc. 2007;129(23):7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyce M, et al. Metabolic cross-talk allows labeling of O-linked {beta}-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci USA. 2011;108(8):3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaro BW, Yang Y-Y, Hang HC, Pratt MR. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc Natl Acad Sci USA. 2011;108(20):8146–8151. doi: 10.1073/pnas.1102458108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sprung R, et al. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J Proteome Res. 2005;4(3):950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 12.Nandi A, et al. Global identification of O-GlcNAc-modified proteins. Anal Chem. 2006;78(2):452–458. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 13.Clark PM, et al. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc. 2008;130(35):11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2010;9(1):153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Science STKE. 2010;3(104):ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y-Y, Ascano JM, Hang HC. Bioorthogonal chemical reporters for monitoring protein acetylation. J Am Chem Soc. 2010;132(11):3640–3641. doi: 10.1021/ja908871t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonovic M, Verhelst SHL, Sorum MT, Bogyo M. Proteomics evaluation of chemically cleavable activity-based probes. Mol Cell Proteomics. 2007;6(10):1761–1770. doi: 10.1074/mcp.M700124-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Verhelst SHL, Fonovic M, Bogyo M. A mild chemically cleavable linker system for functional proteomic applications. Angew Chem Int Ed. 2007;46(8):1284–1286. doi: 10.1002/anie.200603811. [DOI] [PubMed] [Google Scholar]
- 19.Denny JB, Blobel G. 125I-labeled crosslinking reagent that is hydrophilic, photo-activatable, and cleavable through an azo linkage. Proc Natl Acad Sci USA. 1984;81(17):5286–5290. doi: 10.1073/pnas.81.17.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurcel C, et al. Identification of new O-GlcNAc modified proteins using a click-chemistry-based tagging. Anal Bioanal Chem. 2008;390(8):2089–2097. doi: 10.1007/s00216-008-1950-y. [DOI] [PubMed] [Google Scholar]
- 21.Saxon E, et al. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J Am Chem Soc. 2002;124(50):14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 22.Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR. A chemical reporter strategy to probe glycoprotein fucosylation. J Am Chem Soc. 2006;128(37):12078–12079. doi: 10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu T-L, et al. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci USA. 2007;104(8):2614–2619. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goon S, Schilling B, Tullius MV, Gibson BW, Bertozzi CR. Metabolic incorporation of unnatural sialic acids into Haemophilus ducreyi lipooligosaccharides. Proc Natl Acad Sci USA. 2003;100(6):3089–3094. doi: 10.1073/pnas.0437851100. [DOI] [PMC free article] [PubMed] [Google Scholar]