Abstract
A reduction in the number of digits has evolved multiple times in tetrapods, particularly in cursorial mammals that travel over deserts and plains, yet the underlying developmental mechanisms have remained elusive. Here we show that digit loss can occur both during early limb patterning and at later post-patterning stages of chondrogenesis. In the “odd-toed” jerboa and horse and the “even-toed” camel, expansive cell death sculpts the tissue around the remaining toes. In contrast, digit loss in the pig is orchestrated by earlier limb patterning mechanisms including down regulation of Ptch1 expression but no increase in cell death. Together these data demonstrate remarkable plasticity in the mechanisms of vertebrate limb evolution and shed light on the complexity of morphological convergence, particularly within the artiodactyl lineage.
Introduction
Tetrapod limbs evolved adaptations for running, swimming, flying, and a myriad of other tasks, each reflected in functional modifications to their morphology. Digit reduction, a decrease in the number of digits from the basal pentadactyl, or five-digit, morphology, arose repeatedly in tetrapod evolution1. In broad strokes, there are two plausible developmental mechanisms by which this could take place. The first would be to specify fewer digit primordia during the time when developmental fates are patterned in the early limb bud. The second would be to initially organize the limb bud in a normal pentadactyl pattern but then fail to elaborate the full set of digits by resculpting the nascent limb through differential proliferation or cell death.
To date, the molecular developmental mechanism of evolutionary digit reduction has been explored in only one tetrapod group - the skinks of the genus Hemiergis. Distinct species of Hemiergis range in digit number from two to five2,3 with evolutionary progression to fewer digits correlating with increasingly early termination of Sonic hedgehog(Shh) expression in the posterior limb bud4. Shh serves a dual purpose in limb development, both to pattern the digits and to expand the hand/foot plate to allow for the formation of a full complement of digits5-7. Experimental truncation of the developmental timing of Shh expression removes digits in the reverse order of their formation7 thus providing a convenient way to evolutionarily tweak digit number without disturbing the overall structure of the limb, a mechanism first suggested by Alberch and Gale8. However, this mechanism would not, in a simple manner, generate the symmetrical reduction of anterior (pre-axial) and posterior (post-axial) digits seen, for example, in the evolution of the horse lineage.
To investigate how digit reduction evolved in other adaptive contexts we examined the mode of digit loss in a bipedal three-toed rodent and in three ungulates: the single-toed horse, an odd-toed ungulate or perissodactyl, and the pig with four toes and the camel with two, each representing the even-toed ungulates or artiodactyls (Fig. 1a, b).
Figure 1. Convergent evolution of the embryonic limb skeleton in multiple mammal species.
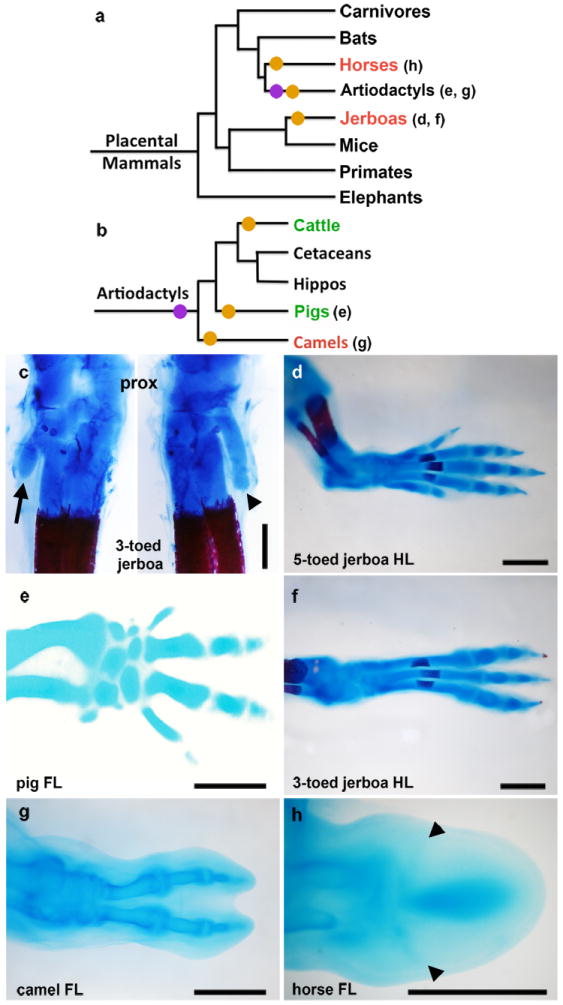
a-b, Phylogeny of (a) mammals and of (b) artiodactyls representing the major groups that have independently lost digits, based on Meredith et al37. Parenthetical lettering references skeletons in accompanying panels. Orange circles indicate an evolutionary incidence of digit loss. Purple circles represent the shift from mesaxonic to paraxonic limbs in basal artiodactyls. Species that sculpt the limb by cell death are highlighted in red, and those that show a restriction of Ptch1 expression are highlighted in green. C Alcian blue and alizarin red stained skeleton of postnatal day 0 three-toed jerboa, D sagitta with the ankle (proximal) at the top. Posterior view (left) highlights the fifth metatarsal (arrow). Anterior view (right) highlights the first metatarsal (arrow head). d, Alcian blue stained skeletons of the approximately 16 dpc five-toed jerboa, A elater, hind foot; e, 30 dpc pig fore foot; f, approximately 16 dpc D sagitta hind foot; g, 50 dpc camel hind foot; h, 34 dpc horse fore foot; c, d, f, scalebar = 0.5 mm. e, g, h, scalebar = 1 mm.
Mechanisms of digit loss in the three-toed jerboa
We first focused on the three-toed jerboa, Dipus sagitta (Fig. 1f). This species has several advantages in identifying meaningful alterations to ancestral developmental mechanisms. First, it has a close evolutionary relationship to the laboratory mouse and to a five-toed species of jerboa, Allactaga elater9 (Fig. 1d). Moreover, digit loss in D sagitta is limited to the hind limb while fore limbs maintained five fully formed fingers10,11. This provides a unique opportunity to identify differences specific to morphological divergence of the hind limb among a potential plethora of species-specific modifications shared in the development of both sets of paired appendages.
In the adult D sagitta, the three central metatarsals are fused into a single element that trifurcates distally and articulates with each of the three digits10. However, in the neonate, alcian blue and alizarin red staining of the chondrogenic skeleton reveals that the three complete digits and their associated metatarsals are flanked by small, truncated cartilage remnants of the first and fifth metatarsals (Fig. 1c; Extended Data Fig. 1). This suggests that at least the proximal-most portion of each of the five digit rays is patterned early in development and that digits I and V are either not fully patterned distally or are truncated at a subsequent differentiation stage.
To gain a better sense of when the patterning and/or morphogenesis of the lateral digits begins to diverge in the three-toed jerboa hind limb, we compared the contours of various staged limb buds between mice and D sagitta. We found that when scaled for size, the fore limbs of mice and three-toed jerboas are consistently identical in morphology. In contrast, the D sagitta hind limb starts to be noticeably narrower as early as E11.5 (Extended Data Fig. 2).
Accordingly, we conducted an expression screen of a series of genes known to be involved in limb patterning just prior to and at the time of morphological divergence in hind limb bud shape. None of the patterning genes we examined showed a significant difference in expression in the D sagitta hind limb, including Shh, Ptch1, Gli1, and HoxD13 (Fig. 2a, b).
Figure 2. Expression of early patterning genes: Shh, Ptch1, Gli1, and HoxD13.
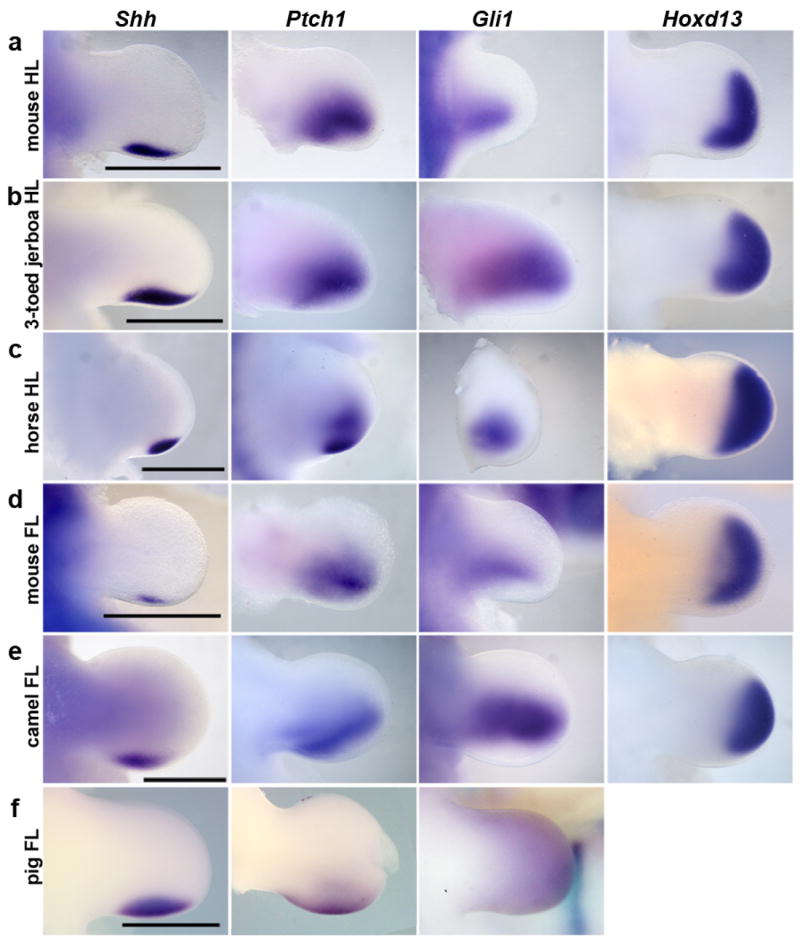
a, mouse hind limb (HL). b, three-toed jerboa, D sagitta, hind limb. c, horse hind limb. d, mouse fore limb (FL). e, camel fore limb. f, pig fore limb. Scalebars = 1 mm.
Turning to post-patterning stages, cell proliferation was assessed by phospho-histone H3 antigen detection. However, we did not see a decrease in proliferation in the hind limb of the three-toed jerboa, either at early stages of autopod expansion or later during digit out growth in any domain of the developing limbs (Extended Data Fig. 3). In contrast, we saw derived expanded domains of TUNEL positive nuclei, a marker for programmed cell death, specific to the jerboa hind limb as early as 12.5 days post conception (dpc) (Extended Data Fig. 6). These domains further expand by 13.5 dpc to encompass all of the tissue distal to what would become the truncated cartilage condensations (Fig. 3b). Thus digit loss in this species appears to result from the sculpting of anterior (pre-axial) and posterior (post-axial) tissues at the distal ends of properly patterned nascent digits.
Figure 3. Patterns of cell death.
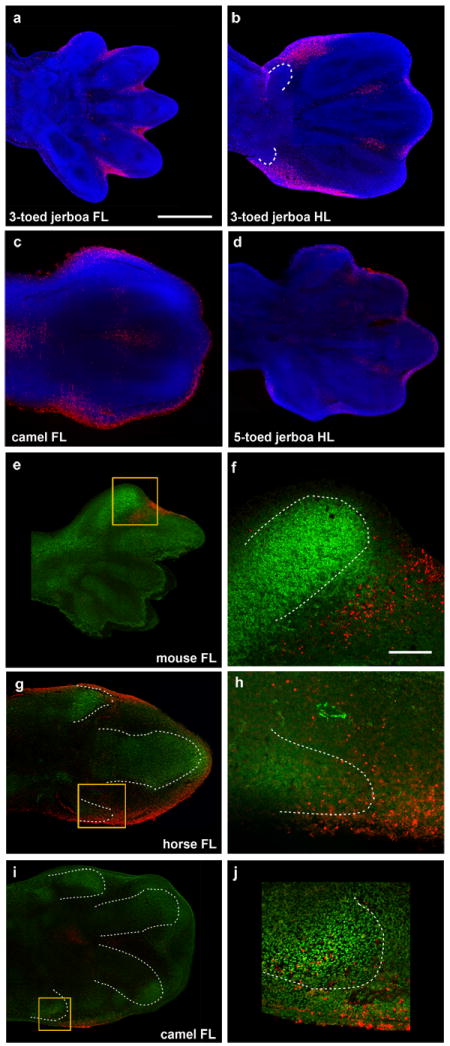
DAPI (blue), Sox9 IHC (green), TUNEL (red). a, approximately 13.5 dpc three-toed jerboa, D sagitta, fore limb; b, approximately 13.5 dpc D sagitta hind limb (white dashed line indicates truncated metatarsals I and V); c, 45 dpc camel fore limb; d, approximately 13.5 dpc five-toed jerboa, A elater, hind limb e, mouse E13.5 with Sox9 and TUNEL; f, magnification of boxed region in (e); g, 34 dpc horse fore limb; h, magnification of boxed region in (g); i, 42 dpc camel fore limb; j, magnification of boxed region in (i). Scalebar in (a) = 0.5 mm for a-d, e, g, and i. Scalebar in (f) = 0.1 mm for f, h, and j.
Apoptosis is used in basal tetrapods to sculpt the digits, removing interdigital tissue late in limb development12. This suggests that a potential evolutionary route for achieving cell death in the D sagitta hind limb digit I and V primordia might be through cooption of the apoptotic pathways normally used to direct interdigital cell death. The transcription factor Msx2 is strongly expressed in the interdigital tissue of the embryonic mouse and chicken13, and retroviral misexpression in chicken embryos induces a dramatic increase in cell death and loss of cartilaginous digit condensations14,15. We found that Msx2 is strongly expressed in the D sagitta hind limb in tissue surrounding and distal to the truncated first and fifth metatarsals and completely overlaps with domains of TUNEL-positive nuclei (Fig. 4a-c). In different contexts within the limb bud, the secreted protein Bmp4 can act both upstream and downstream of Msx215,16. We observe a transient spatial increase of Bmp4 expression specific to the D sagitta hind limb autopod starting at 12 dpc that resolves at 12.5 dpc into two strong and discrete domains of expression precisely prefiguring the proximal positions of the first and fifth digits (Extended Data Fig. 4). However, Msx2 is expanded in the D sagitta hind limb prior to expanded Bmp4 expression, as early as 11 dpc (Extended Data Fig. 5). This is when the D sagitta hind limb first shows signs of narrowing relative to limbs that will develop five digits (Extended Data Fig. 2), consistent with altered Msx2 expression potentially being the primary causal mechanism of digit loss in this species.
Figure 4. Expression of Msx2 at the start of digit chondrogenesis.
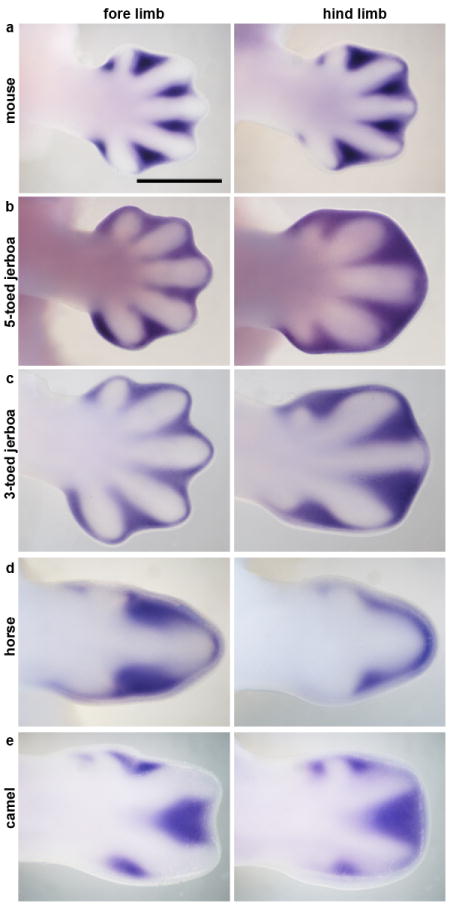
Fore limb and hind limb of a, 13 dpcmouse; b, approximately 13 dpc five-toed jerboa, A elater; c, approximately 13 dpc three-toed jerboa, D sagitta; d, 34 dpc horse; e, 42 dpc camel. Scalebar = 1 mm.
As the interdigital tissue begins to undergo apoptosis during mouse limb development, Fgf8 expression is lost in the overlying apical ectodermal ridge (AER), while Fgf8 expression is maintained above the growing digits (Fig. 5a). Fgf8 is both necessary and sufficient for digit outgrowth in mouse and chicken embryos17-20. From about 12.75 dpc in the D sagitta hind limb, Fgf8 expression regresses away from the posterior and then anterior AER as well as the interdigital domains, persisting only over the digits that continue to develop to completion (Fig 5a, b; Extended Data Fig. 6).
Figure 5. Fgf8 expression is restricted to the AER overlying nascent digits.
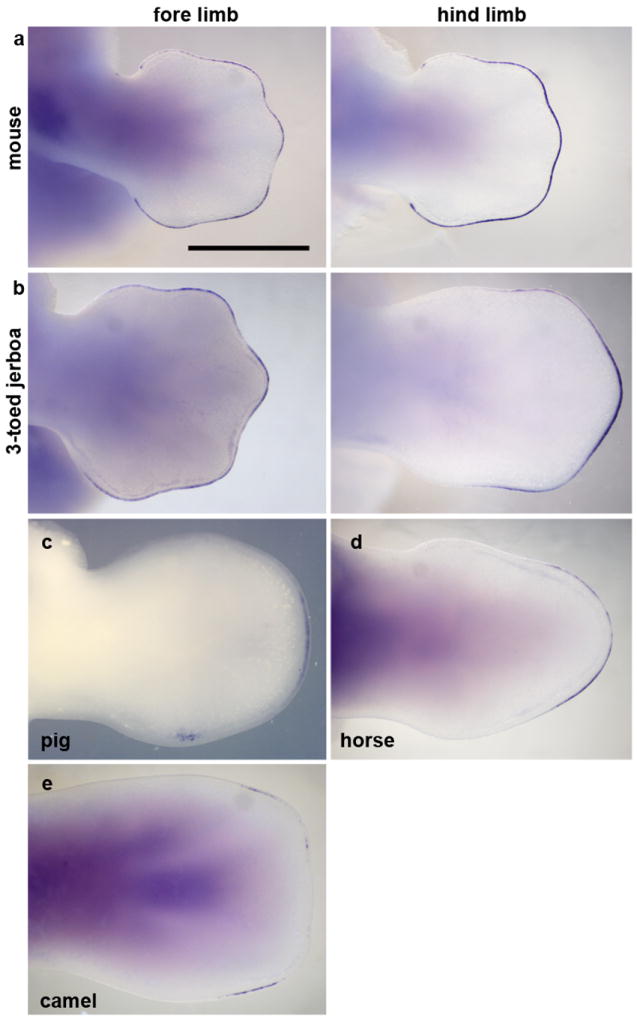
Fore limb and hind limb of a, 13 dpc mouse; b, approximately 13 dpc three-toed jerboa, D sagitta; c, 25 dpc pig; d, 34 dpc horse hind limb; e, 42 dpc camel fore limb. Scalebar = 1 mm.
Convergence of post-pattern sculpting in the horse
The three-toed jerboa hind limb remarkably resembles the limb structure of some of the early ancestral equine species with three toes21. To test possible mechanisms for digit reduction in the horse, we once again started by examining the expression of genes known to be involved in patterning the early limb bud. We observed no obvious differences in expression of Shh, Ptch1, Gli1, or HoxD13 relative to those previously described in mice (Fig. 2c). In contrast, we did observe TUNEL-positive nuclei entirely surrounding the central toe and within the distal ends of nascent Sox9+ truncated condensations of metacarpals 2 and 4 (Fig. 3g, h), a condition not observed in mouse (Fig. 3 e, f). Moreover, we found expanded Msx2 expression in domains correlating with those regions of anterior and posterior cell death (Fig. 4d). We also observed increased posterior expression of Msx2 earlier in development (Extended Data Fig. 5) and distal expansion of Bmp4 in both fore and hind limbs (Extended Data Fig. 4) similar to D sagitta hind limbs. Fgf8 expression is also maintained in the horse AER only over the nascent central digit III (Fig. 5d). Thus, in the horse as in the three-toed jerboa, digit reduction appears to have a post-patterning contribution involving expanded domains of lateral apoptosis, possibly in part through similar molecular mechanisms. It is likely that mechanisms yet to be identified eliminate the first and fifth digits while a jerboa-like carving away of digits II and IV occurs by transforming cells from a chondrogenic to an apopototic fate. A more extensive investigation of early patterning may be worthwhile with additional precisely staged early horse embryos.
Plasticity of digit loss mechanisms in the artiodactyls
The even-toed ungulates present yet another opportunity to explore the possible convergence of digit reduction mechanisms in the context of additional skeletal remodeling. The distal artiodactyl limb has shifted the central axis of symmetry from digit III in the ancestral mesaxonic limb to a derived paraxonic limb where the axis of symmetry runs through the interdigital space between digits III and IV22. To explore whether digit loss in these species occurs via patterning and/or post-patterning changes, we obtained embryos from two species of artiodactyls, the pig and camel. While this work was in progress, we learned of similar studies by Lopez-Rios et al23 in a third artiodactyl species with convergent digit loss to two toes, the cow. The accompanying paper identifies a gene regulatory control region for Ptch1 expression in the limb that is altered in the cow. The resulting expression of Ptch1 is reduced and more posteriorly restricted than in non-artiodactyl species. One role thatPtch1 expression serves is to restrict the movement of the morphogen Shh across the limb bud24,25. As a consequence of the change in Ptch1 expression in the cow, Shh targets, including Gli1 and the Hoxd genes, are expressed more uniformly across the limb bud23. Mice in which Ptch1 expression is reduced in the limb display similar changes in downstream genes and a concomitant shift in the central axis of the limb to the space between digits III and IV and loss of the first digit26. Importantly, after learning of our results with the three-toed jerboa and horse, Lopez-Rios and colleagues looked closely and saw no evidence of expanded apoptosis in the developing cow limb23. Together these results suggest that, as in Hemiergis, the even-toed ungulates might have lost digits through a Shh-dependent patterning mechanism, albeit by a different genetic alteration, allowing the digits to be lost in an a symmetrical manner in the artiodactyls.
As would be expected if mutations affecting Ptch1 regulation play a prominent role in artiodactyl limb evolution, we find Ptch1 expression in the pig is also posteriorly restricted and down-regulated concomitant with an up-regulation of Gli1 (Fig. 2f). Further, as in the cow, there is no evidence of increased cell death in developing pig limbs27. Surprisingly, however, Ptch1 expression is not down-regulated and restricted in the camel and is instead expressed much like non-artiodactyls (Fig. 2e). Additionally, Shh, Gli1, and HoxD13 exhibit ancestral patterns of expression indicating early patterning of the digit field by this subset of molecules is conserved in the camel (Fig. 2e). In contrast, when we examined patterns of cell death in the camel, we found expansive apoptosis throughout outgrowths of tissue flanking digits III and IV at 45 dpc (Fig. 3c) as well as at 42 dpc within small Sox9+ pre-cartilaginous nodules in the positions of missing digits II and V (Fig. 3i, j). As in the three-toed jerboa and horse, this correlates with domains of Msx2 expression in the anterior and posterior limb bud at the time of digit condensation (Fig. 4e), though earlier expression of Bmp4 and Msx2does not correlate suggesting a distinct initiating mechanism for camel (Extended Data Fig. 4, 5).
Regardless of the mechanism by which digit loss occurs, at patterning or post-patterning stages, Fgf8 expression is lost from the AER anterior and posterior to the digits that continue to develop in the pig, camel (Fig. 5c, e), and cow23 as seen with the three-toed jerboa and horse. Regression of Fgf8 in the pig and cow, two species that lack expanded domains of cell death, uncouples this expression change from the direct cause of apoptosis and may rather reflect an independent requirement for its elimination to allow for digit termination in all species.
Discussion
These data indicate that at least two mechanisms of digit reduction are employed in the even-toed ungulates, one (exemplified by the pig and cow) involving changes in early patterning by Shh and not involving apoptosis, and a distinct mechanism (seen in camels) involving changes in domains of apoptosis that resculpt the limb after the patterning phase. These data do, however, present a paradox in the context of the well-established artiodactyl phylogeny and fossil record (Fig. 1b). Although the morphology of the cow and pig is remarkably similar to the mouse phenotype when Ptch1 is lost from the limbs, both in the reduction of digits and shift in the symmetry of toes to the interdigit of III-IV23, it can not have been responsible for both phenotypes in the artiodactyls as they occurred at different stages evolutionarily. The fact that a change in Ptch1 regulation is seen in both pigs and cattle indicates that it was likely present in their last common ancestor. As such, it cannot have been solely responsible for the loss of digit 1, as this occurred convergently in these two lineages. Indeed, digit reduction occurred at multiple independent times within the artiodactyl clade (Fig. 1b, orange circles), as the stem group of each major lineage was pentadactyl at least in the forelimb28. The common ancestor of pigs and cattle would also have been ancestral to the hippos and their Cetacean relatives, the dolphins and whales (Fig. 1b). Like extinct basal artiodactyls, hippos and basal Cetaceans have a relatively small first digit22,27,29. Thus, a restriction of Ptch1 in a basal member of the group including pigs, hippos, cetaceans, and cattle may have served to reduce the size of the first digit and predispose the limb to further digit loss.
Perhaps even more striking is the absence of altered Ptch1 regulation in the camel. Without this information, one might have speculated that the Ptch1 mutation was responsible for the reorientation of the axis of symmetry in artiodactyls similar to the mutant mouse. However, the shift in the position of digits from mesaxonic to paraxonicis believed to be ancestral to the split of modern artiodactyl suborders (and indeed is a defining characteristic trait for this clade30-32) (Fig. 1b, purple circle). Given the camel evidence, one has to either conclude that the shift actually arose independently in the ancestors of the camels and those of other artiodactyl lineages, or alternatively, any role Ptch1 may have in the establishment of digit position in the pig and cow arose secondary to a separate mechanism established prior to the split of camels and their relatives.
The identification of several distinct molecular and cellular mechanisms of digit loss with recurring motifs suggests the developmental program of the tetrapod limb is fairly plastic. This would have provided some flexibility to allow adaptation in different circumstances and ultimately contributed to the diversity of limbs seen today.
Full Methods
Skeletal Preparation
For mouse and jerboa
Embryos were eviscerated and fixed in 95% ethanol overnight and then in acetone overnight at room temperature. Embryos were subsequently stained overnight at room temperature in alcian/alizarin staining solution – 5 ml each alcian blue and alizarin red S stock, 5 ml glacial acetic acid, 85 ml 70% ethanol. [Alcian stock: 0.3 g Alcian Blue 8GX (Sigma-Aldrich A5268) in 70% ethanol; Alizarin stock: 0.1 g Alizarin Red S (Sigma-Aldrich A5533), 5 ml H2O, 95 ml absolute ethanol]. Embryos were then cleared in 1% KOH that was replaced with fresh when it turned purple. When skeletal features were visible and soft tissues only slightly blue, embryos were carried through a graded series of 25%, 50%, 75% glycerol in 1% KOH and then into 100% glycerol for imaging and storage.
For horse and camel
Embryos were fixed in 4% paraformaldehyde in 1X PBS and dehydrated through a graded methanol series into 100% methanol for storage at -20°C. Embryos were then rehydrated to PBS and transferred to 0.1% ammonium hydroxide in 70% ethanol overnight to bleach. Embryos were then washed twice for an hour in acid alcohol (5% acetic acid, 70% ethanol). Staining was carried out for three hours at room temperature in 0.05% alcian blue, 5% acetic acid, 70% ethanol. Embryos were washed two times for an hour in acid alcohol, cleared in 100% methanol (two times for an hour) and then transferred to BABB (1:2 benzyl alcohol: benzyl benzoate) for further clearing, imaging, and storage.
For pig
Limbs were removed from pig embryos and stored in 100% MeOH. Skin was dissected off limbs and limbs were briefly rehydrated through a methanol series into 100% PBS. Limbs were then placed in Alcian blue overnight (0.02% Alcian blue in ethanol and 30% glacial acetic acid). The next day limbs were washed for an hour each in 100%, 95% and 70% EtOH, then washed with deionized water for 1 hour. Limbs were stained with Alizarin red (0.1% in 1% KOH) overnight. The next day the limbs were cleared in 1% KOH and solution changed every day until skeletal features were visible. Limbs were moved to glycerol for extended preservation.
Whole-mount in situ Hybridization
Probe templates for all species were generated by PCR from first strand cDNA synthesis (primers and accession numbers provided in Extended Data Table 1). PCR products were ligated into pGEM-TEasy (Promega), transformed into DH5alpha competent cells, and plated for blue white selection on IPTG/XGal/Amp plates. White colonies were selected for sequence verification and then plasmid prepped (Qiagen). Plasmids were linearized with the appropriate restriction enzyme and then a transcription reaction was carried out using the appropriate anti-sense transcription enzyme (SP6 or T7) with digoxygenin RNA labeling mix (Roche). RNA probes were precipitated with LiCl2 and ethanol and resuspended in 50 ul nuclease free water plus 1 ul RNAse Inhibitor. One microliter of probe was run on an agarose gel to confirm probe synthesis.
All embryos for whole-mount in situ hybridization (WISH) were fixed with 4% paraformaldehyde in 1X PBS and dehydrated through a graded methanol series (25%, 50%, 75% MeOH in PBS, 100% MeOH) and stored at -20° C until use. Before WISH, embryos were treated with 6% H2O2 in methanol for 1 hour, and rehydrated through a methanol series to PBST (PBS + 0.1% Tween-20).
After three 5-minute washes in PBST, proteinase K (10μg/mL in PBST) was added and embryos were incubated at room temperature for 25 (embryonic day [E] 21.5 to 22.5 pigs), 35 (E22.5 to 23.5 pigs), or 45 (E25.5 to 26.5 pigs) minutes. Mouse and jerboa embryos were permeabilized in 10 ug/ml proteinase K as follows: E11 for 20 min, E11.5 for 22 min, E12 for 25 min, E12.5 for 27 min, E13 for 30 min. For fgf8 WISH, embryos of all species were incubated in Proteinase K for 10 minutes regardless of age. Camel and horse embryos in Figure 2 were incubated for 30 minutes in proteinase K, camel and horse embryos in Figure 4 for 45 minutes, and those in Extended Data Figure 4 and 5 for 40 minutes.
After permeablization, embryos were washed in PBST and then fixed for 20 minutes in 4% PFA/0.2% glutaraldehyde in PBST. After several washes with PBST, embryos were added to pre-warmed prehybridization solution (Pig: 50% formamide, 5X SSC pH 4.5, 2% SDS, 2% Roche blocking reagent, 250 μg/mL tRNA, 100 μg/mL heparin sodium salt; Other species: 50% formamide, 5X SSC pH 4.5, 1% SDS, 50 ug/ml yeast tRNA, 50 ug/ml heparin) and incubated for at least 1 hour at 70°C (pig) or 65°C (other species). After pig embryo incubation, the prehybridization solution was changed to fresh with 1μL probe added followed by overnight incubation at 70°C. For other species, prehybridization solution was replaced with fresh solution containing 1:200 dilution of the appropriate probe followed by overnight incubation at 65°C.
For pig
On day two, four 30-minute washes in solution I (50% formamide, 2X SSC pH 4.5, 1% SDS) were performed at 70°C. Embryos were then briefly washed at room temperature in a 50/50 mix of Solution I and MABT (100mM maleic acid, 150mM NaCl, 0.1% Tween-20, pH 7.5). Next, two 30 minute washes were done in MABT. Embryos were then blocked in 2% Blocking reagent/MABT for one hour. A final blocking step was done for at least 1 hour in 20% Heat inactivated goat serum/2% Blocking reagent/MABT. Anti-DIG antibody (Roche) was added at 1:2000 to fresh blocking solution and embryos incubated overnight at 4°C. The next day embryos were washed all day (changing solution five times) and overnight in MABT. The following day embryos were washed in NTMT (100mM NaCl, 100mM Tris pH 9.5, 50mM MgCl2, 0.1% Tween-20) for four 10-minute washes and then placed in a BM Purple solution (Roche). During their time in BM purple, samples were wrapped in foil and monitored for the appearance of staining. After the color reaction had reached an appropriate level, embryos were rinsed several times in NTMT, then PBST, and then fixed with 4% PFA overnight. Embryos were transferred to 1% PFA for long-term storage.
For other species
On day two, embryos were washed three times for 30 minutes in solution I (50% formamide, 5X SSC pH 4.5, 1% SDS) at 65°C followed by three times for 30 minutes in solution III (50% formamide, 2X SSC pH 4.5) at 65°C. Embryos were then washed three times for 5 minutes in TBST (1X TBS + 1% Tween 20) and blocked for one hour at room temperature in block solution (10% heat inactivated sheep serum and 0.1% Roche blocking reagent in TBST). Embryos were then incubated overnight at 4°C in block solution plus 1:2500 anti-digoxygenin AP antibody (Roche). On day three, embryos were washed three times for 5 minutes in TBST and then five times for at least an hour each in TBST followed by overnight at 4°C in TBST. On day four, embryos were washed three times 10 minutes in NTMT (100mM NaCl, 100mM Tris pH 9.5, 50mM MgCl2, 1% Tween-20) before coloration in AP reaction mix (125 ug/ml BCIP and 250 ug/ml NBT in NTMT). Coloration was carried to completion in the dark. Embryos were then washed 10 minutes in NTMT followed by three times 10 minutes in TBST and finally overnight in TBST to reduce background and increase signal. Embryos were postfixed in 4% paraformaldehyde for 30 minutes at room temperature and imaged and stored in 1% paraformaldehyde in 1X PBS.
Immunohistochemistry and TUNEL
Paraformaldehyde fixed embryos for paraffin sectioning were dehydrated through an ethanol series, cleared in xylenes, and infiltrated with paraffin for embedding and sectioning. Embryos for frozen sections were paraformaldehyde fixed, dehydrated through a graded series to 100% methanol for storage and subsequently rehydrated into PBST before transfer to 15% sucrose in 1X PBS. Limbs were then dissected from the embryos and embedded in gelatin solution (7.5% gelatin and 15% sucrose in PBS) in disposable cryomolds. Blocks were frozen and stored at -80 until cryosectioned.
Slides for immunohistochemistry were rinsed in xylenes to dewax and rehydrated through an ethanol series to PBS (for paraffin sections) or thawed (for frozen sections) and washed twice in PBS. All immunohistochemistry was carried out after antigen retrieval in 1:100 dilution citric acid antigen unmasking solution (Vector labs) by boiling for 2 minutes in the microwave followed by incubation at room temperature wrapped tightly in foil and then cooling to room temperature for 20 minutes at 4°C. Slides were washed in PBS (or TBS for phosphoantibodies) and then blocked in a solution of 5% heat inactivated goat serum, 0.1% TritonX-100, 0.02% SDS in PBS (or TBS). Primary antibody was added at a concentration of 1:500 for Sox 9 (Millipore AB5535) or 1:200 for phospho histone H3 (Cell Signal #9701) and slides incubated overnight in a humidified chamber at 4°C. On day 2, slides were washed three times in PBST (or TBST) and incubated at room temperature in secondary antibody (goat anti- rabbit Alexa 594, Life Technologies) diluted 1:250 in block plus 0.1 ug/ml DAPI. Slides were then washed three times 5 minutes in PBS (or TBS) and mounted in Fluoromount-G (Southern Biotech).
For TUNEL, slides were rehydrated or thawed, washed in PBS, and then permeabilized for 10 minutes in 5 ug/ml proteinase K in PBS followed by 5 minutes postfix in 4% paraformaldehyde in PBS and 3 washes in PBS. Slides were then incubated in TUNEL reaction mixture (Roche In Situ Cell Death Detection Kit, TM-Red) for 60 minutes at 37°C, rinsed three times in PBS, and mounted in Prolong Gold. Slides that had been previously processed for Sox9 IHC were placed immediately into the TUNEL reaction mixture.
Extended Data
Extended Data Figure 1. The proximal remnants of truncated skeletal elements in D sagitta are correctly patterned.
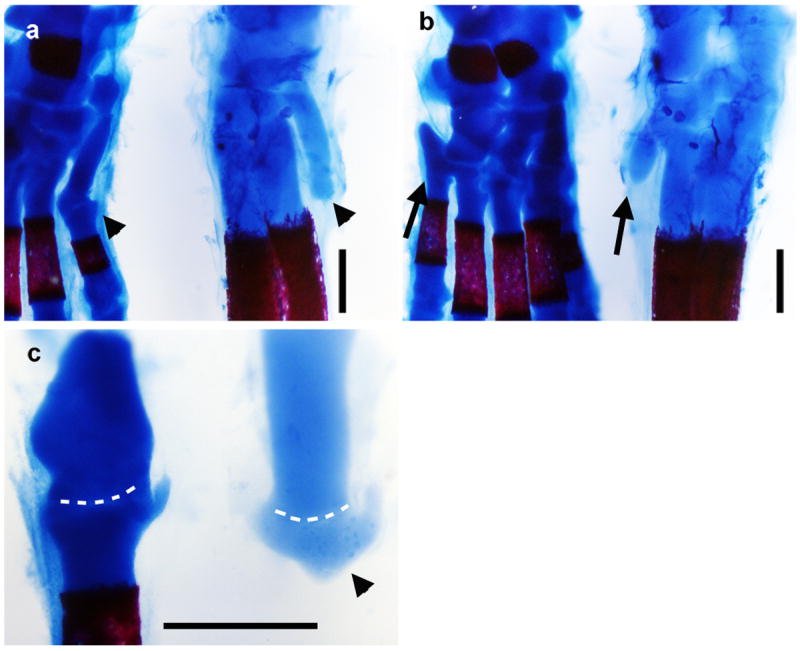
Alcian blue and alizarin red stained skeletons of postnatal day 0 mouse (left) and three-toed jerboa, D sagitta (right) with proximal (ankle) at the top. a, Anterior view highlights the first metatarsal (arrow head). b, Posterior view highlights the fifth metatarsal (arrow). c, Dissected first tarsal-metatarsal elements demonstrate the morphology of the truncated first metatarsal of D sagitta (right, arrow head) compared to mouse (left). Joint interzone indicated by white dashed line. Scalebars = 0.5 mm.
Extended Data Figure 2. The shape of the three-toed jerboa hind limb differs from the mouse as early as 11.5 dpc.
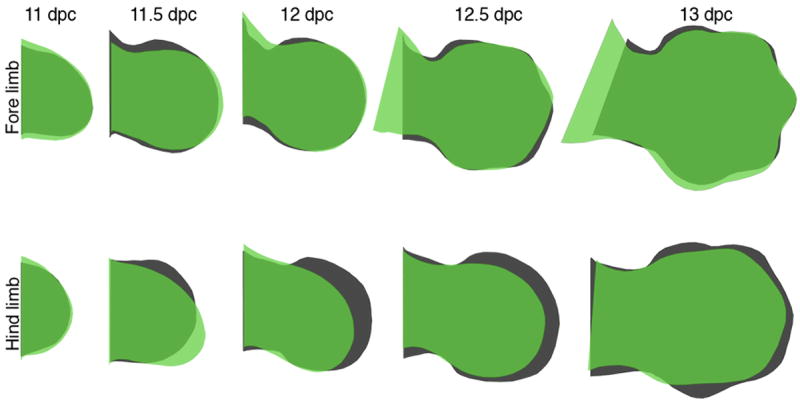
Trace outlines of limb buds of the mouse (black) and three-toed jerboa, D sagitta (green) over a developmental time series.
Extended Data Figure 3. Proliferation is unchanged in the D sagitta hind limb bud.
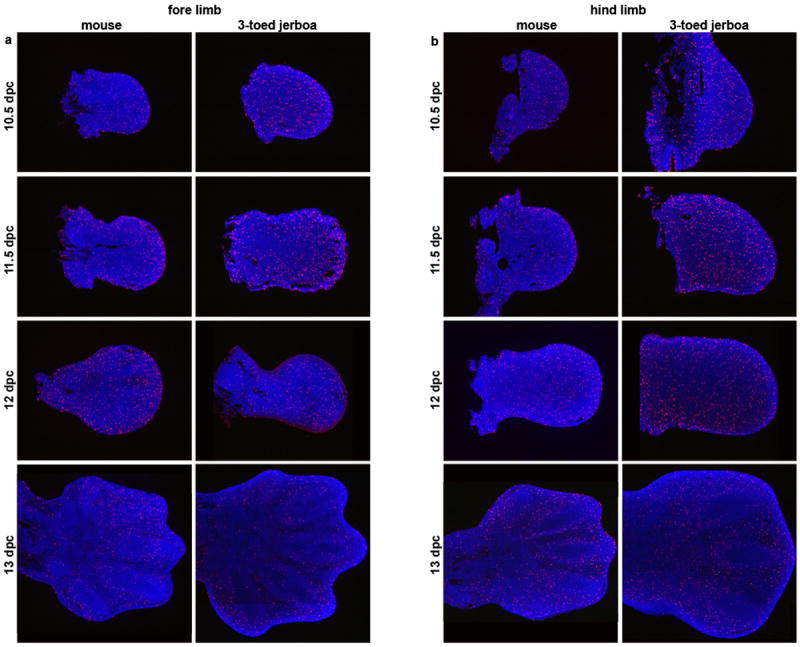
Phospho-histone H3 detection in sections of mouse and three-toed jerboa, D sagitta, limb buds. a, fore limbs; b, hind limbs.
Extended Data Figure 4. Developmental time course and species comparisons of Bmp4 expression.
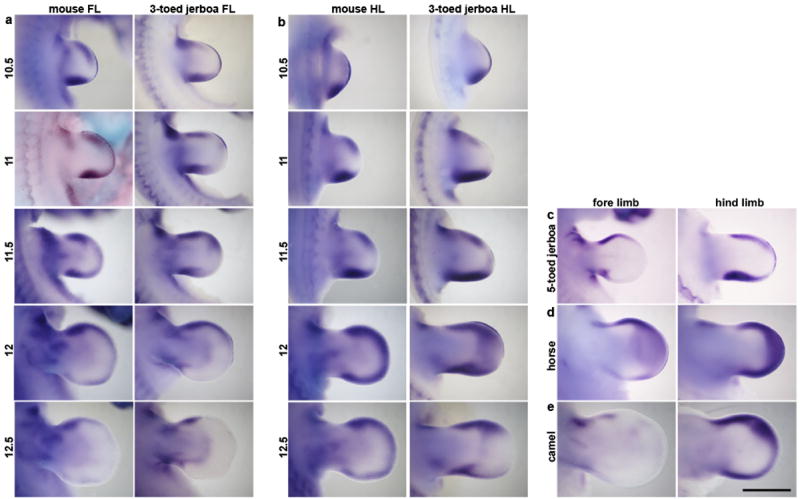
a, Fore limb buds (FL) and b, hind limb buds (HL) of mouse and the three-toed jerboa, Dipus sagitta, at 10.5, 11, 11.5, 12, and 12.5 dpc. c, Fore limb and hind limb of the five-toed jerboa, A elater, at approximately 12.25 dpc. d, Fore limb and hind limb of the horse at 30 dpc (approximately equivalent to mouse 12 dpc). e, Fore limb and hind limb of the camel at 38 dpc (approximately equivalent to mouse 12.5 dpc). Scalebar = 1 mm for D sagitta, A elater, horse, and camel and 0.8 mm for mouse limbs.
Extended Data Figure 5. a, Developmental time course and species comparisons of Msx2 expression.
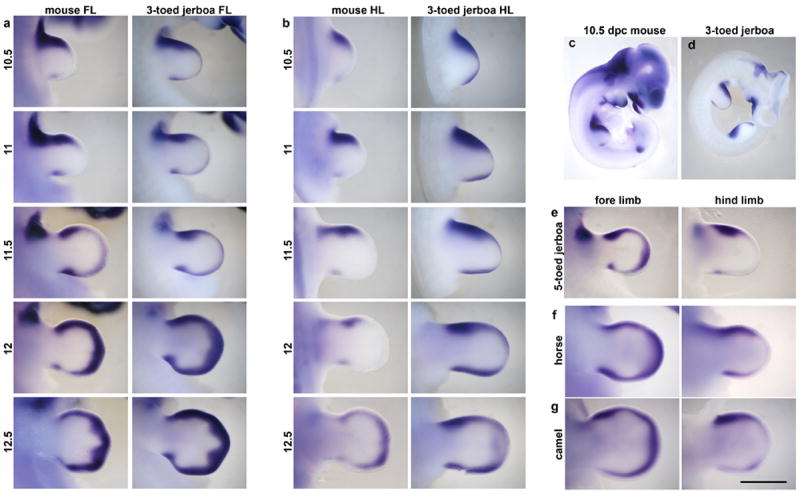
a, Fore limb buds (FL) and b, hind limb buds (HL) of mouse and the three-toed jerboa, Dipus sagitta, at 10.5, 11, 11.5, 12, and 12.5 dpc. c, d, Msx2 expression in the (c) mouse and (d) D sagitta embryo at 10.5 dpc. e, Fore limb and hind limb of the five-toed jerboa, A elater, at approximately 12.25 dpc. f, Fore limb and hind limb of the horse at 30 dpc (approximately equivalent to mouse 12 dpc). g, Fore limb and hind limb of the camel at 38 dpc (approximately equivalent to mouse 12.5 dpc). Scalebar = 1 mm for D sagitta, A elater, horse, and camel and 0.8 mm for mouse limbs.
Extended Data Figure 6. Developmental time course of Fgf8 expression and early TUNEL in the jerboa hind limb.
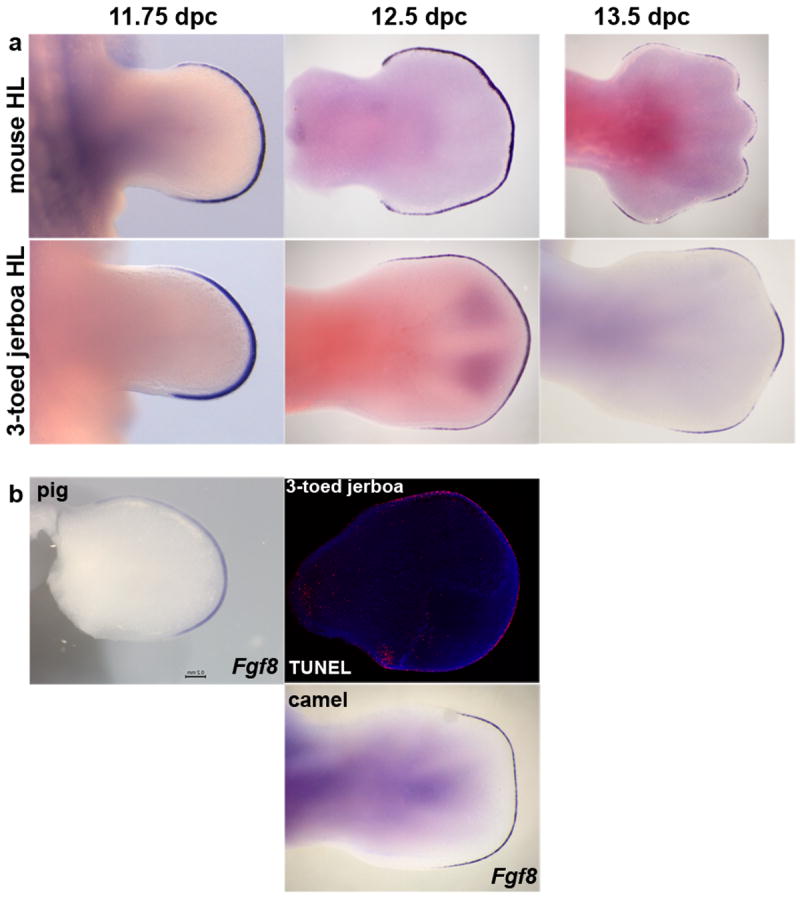
a, Time series of Fgf8 expression in the mouse and three-toed jerboa, D sagitta, hind limb. b, Fgf8 expression in the pig (25 dpc) and camel (42 dpc) hind limbs of embryos in Figure 7. TUNEL labeling of cell death in the 12.5 dpc D sagitta hind limb. Limbs in (b) are aligned with the closest stage matched embryos in (a).
Extended Data Table 1. Whole mount in situ hybridization probe information.
Primer sequences used for amplification and accession numbers for probe sequences are provided for each species and gene.
| Shh | Forward | Reverse | NCBI Accession # |
|---|---|---|---|
| mouse | GACCCCTTTAGCCTACAAGCAGTTT | GCGTCTCGATCACGTAGAAGACCT | Pr032067205 |
| jerboa | GACCCCTTTAGCCTACAAGCAGTTT | GCGTCTCGATCACGTAGAAGACCT | Pr032067198 |
| horse | CTGGTGGTTCTGGTCTCCTC | CCCTCGTCCGATCACGTA | Pr032067191 |
| camel | used horse probe | ||
| pig | CCGGCTGATGACTCAGAGAT | GCAGGTCCTTCACCAGCTT | Pr032067208 |
| Ptch1 | |||
| mouse | CTTCGCTCTGGAGCAGATTT | GCATGGTTAAACAGGCATAGG | Pr032067204 |
| jerboa | CTTCGCTCTGGAGCAGATTT | GCATGGTTAAACAGGCATAGG | Pr032067197 |
| horse | CGCCAGAAGATTGGAGAAGA | CCTGAGTTGTTGCAGCGTTA | Pr032067190 |
| camel | CGCCAGAAGATTGGAGAAGA | CCTGAGTTGTTGCAGCGTTA | Pr032067184 |
| pig | GGAGCAGATTTCCAAGGGGA | CGGAGAGCTTCTGTGGTCAG | Pr032067207 |
| Gli1 | |||
| mouse | TACATGCTGGTGGTGCACAT | GGCTGTGGCGAATAGACAGA | Pr032067201 |
| jerboa | TACATGCTGGTGGTGCACAT | GGCTGTGGCGAATAGACAGA | Pr032067194 |
| horse | GTGACCACTCCCCAGCAG | GATTCAGACCACTGCCCATC | Pr032067187 |
| camel | TACATGCTGGTGGTGCACAT | GGCGGTCAAGAGAAACTGG | Pr032067182 |
| Hoxd13 | |||
| mouse | CTATGGCTACCATTTCGGCAAC | ACTGGTAGCCCTCCATGGAAAT | Pr032067202 |
| jerboa | CTATGGCTACCATTTCGGCAAC | ACTGGTAGCCCTCCATGGAAAT | Pr032067195 |
| horse | TTCCCGGTGGAGAAGTACA | TTGAGCTTGGAGACGATTTTC | Pr032067188 |
| camel | TTCCCGGTGGAGAAGTACA | TTGAGCTTGGAGACGATTTTC | Pr032067183 |
| Msx2 | |||
| mouse | CTCTCGTCAAGCCCTTCGAGAC | AGCCATTTTCAGCTTTTCCAGTT | Pr032067203 |
| jerboa | CTCTCGTCAAGCCCTTCGAGAC | AGCCATTTTCAGCTTTTCCAGTT | Pr032067196 |
| horse | TCGCTTAGGGTGGTGTAAGC | TTGCTAATTCACCCCTCTCTG | Pr032067189 |
| camel | used horse probe | ||
| Bmp4 | |||
| mouse | AGTGAGAGCTCTGCTTTTCGTTTC | GGCAGTAGAAGGCCTGGTAGCC | Pr032067199 |
| jerboa | AGTGAGAGCTCTGCTTTTCGTTTC | GGCAGTAGAAGGCCTGGTAGCC | Pr032067192 |
| horse | CCAGCGAAAACTCTGCTTTT | GATCAATATGGTCAAAACATTTGC | Pr032067185 |
| camel | CCAGCGAAAACTCTGCTTTT | GATCAATATGGTCAAAACATTTGC | Pr032067180 |
| Fgf8 | |||
| mouse | TGCTGTGCCTGCAGGCNCARGARGG | CAGCTTGCCCTTCTTGTTCATRCADAT | Pr032067200 |
| jerboa | TGCTGTGCCTGCAGGCNCARGARGG | CAGCTTGCCCTTCTTGTTCATRCADAT | Pr032067193 |
| horse | CCTAATTTTACACAGCATGTGAGG | GGCGGGTAGTTGAGGAACTC | Pr032067186 |
| camel | CCTAATTTTACACAGCATGTGAGG | GGCGGGTAGTTGAGGAACTC | Pr032067181 |
| pig | CAGGGTGTTTCCCAACAGGT | GGCAATCAGCTTCCCCTTCT | Pr032067206 |
Acknowledgments
We thank Javier Lopez-Rios and Rolf Zeller (University of Basel, Switzerland) for generously providing data and discussion prior to publication. We also thank Juan Carlos Izpisua Belmonte and Aitor Aguirre for sharing space and materials to complete experiments subsequent to review. Jerboa embryos were harvested with the assistance of Shaoyuan Wu and colleagues in Xinjiang, China. Pig embryos were harvested with the assistance of Daniel Urban. Additional horse embryos were provided by Regina Turner and Hannah Galatino-Homer (University of Pennsylvania) and by Reto Fritsche and Sara Lyle (Louisiana State University). Mouse Gli1 probe plasmid, used in the pig in situ, was provided by Alexandra Joyner. This work was supported by NIH grant R37HD032443 to C.J.T., and NSF IOS grant 1257873 to K.E.S.
Footnotes
Author Contributions
K.L.C., K.E.S., and C.J.T. conceived of and initiated the project. K.L.C. and C.J.T. wrote the manuscript. K.L.C performed the mouse, three- and five-toed jerboa, horse, and camel in situ hybridizations, PH3 IHC, and skeletal stains. A.U. performed TUNEL and Sox9 IHC. J.M. and K.E.S. performed the pig in situ hybridizations. K.S.-B. cloned the pig probes. M.B. and D.A. provided most of the horse embryos and material for cloning the horse probes. J.S. provided the camel embryos and material for cloning the camel probes.
References
- 1.Clack JA. The Fish–Tetrapod Transition: New Fossils and Interpretations. Evol Educ Outreach. 2009;2:213–223. [Google Scholar]
- 2.Skinner A, Lee MS, Hutchinson MN. Rapid and repeated limb loss in a clade of scincid lizards. BMC Evol Biol. 2008;8:310. doi: 10.1186/1471-2148-8-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro M. Developmental morphology of limb reduction inhemiergis (squamata: scincidae): chondrogenesis, osteogenesis, and heterochrony. J Morphol. 2002;254:211–231. doi: 10.1002/jmor.10027. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro MD, Hanken J, Rosenthal N. Developmental basis of evolutionary digit loss in the Australian lizard Hemiergis. J Exp Zool B Mol Dev Evol. 2003;297:48–56. doi: 10.1002/jez.b.19. [DOI] [PubMed] [Google Scholar]
- 5.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–28. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Towers M, Mahood R, Yin Y, Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452:882–886. doi: 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, et al. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14:624–32. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberch P, Gale EA. Size dependence during the development of the amphibian foot. Colchicine-induced digital loss and reduction. J Embryol Exp Morphol. 1983;76:177–97. [PubMed] [Google Scholar]
- 9.Walker EP. Mammals of the world. John Hopkins Press; 1964. [Google Scholar]
- 10.Shenbrot GI, Sokolov VE, Heptner VG. Jerboas: Mammals of Russia and Adjacent Regions. Science Publishers; 2008. [Google Scholar]
- 11.Cooper KL. The lesser Egyptian jerboa, Jaculus jaculus: a unique rodent model for evolution and development. Cold Spring Harb Protoc. 2011;2011:1451–1456. doi: 10.1101/pdb.emo066704. [DOI] [PubMed] [Google Scholar]
- 12.Zuzarte-Luis V, Hurle JM. Programmed cell death in the embryonic vertebrate limb. Semin Cell Dev Biol. 2005;16:261–9. doi: 10.1016/j.semcdb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Fernández-Terán Ma, Hinchliffe Jr, Ros Ma. Birth and death of cells in limb development: A mapping study. Dev Dyn. 2006;235:2521–2537. doi: 10.1002/dvdy.20916. [DOI] [PubMed] [Google Scholar]
- 14.Marazzi G, Wang Y, Sassoon D. Msx2 Is a Transcriptional Regulator in the BMP4-Mediated Programmed Cell Death Pathway. Dev Biol. 1997;186:127–138. doi: 10.1006/dbio.1997.8576. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari D, et al. Ectopic expression of Msx-2 in posterior limb bud mesoderm impairs limb morphogenesis while inducing BMP-4 expression, inhibiting cell proliferation, and promoting apoptosis. Dev Biol. 1998;197:12–24. doi: 10.1006/dbio.1998.8880. [DOI] [PubMed] [Google Scholar]
- 16.Pizette S, Abate-Shen C, Niswander L. BMP controls proximodistal outgrowth, via induction of the apical ectodermal ridge, and dorsoventral patterning in the vertebrate limb. Dev Camb Engl. 2001;128:4463–4474. doi: 10.1242/dev.128.22.4463. [DOI] [PubMed] [Google Scholar]
- 17.Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nat Genet. 2000;26:460–3. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- 18.Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–5. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–8. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- 20.Sanz-Ezquerro JJ, Tickle C. Fgf signaling controls the number of phalanges and tip formation in developing digits. Curr Biol. 2003;13:1830–6. doi: 10.1016/j.cub.2003.09.040. [DOI] [PubMed] [Google Scholar]
- 21.Romer AS. Vertebrate Paleontology. University of Chicago Press; 1936. [Google Scholar]
- 22.Prothero DR, Foss SE. The Evolution of Artiodactyls. JHU Press; 2007. [Google Scholar]
- 23.Lopez-Rios J, et al. Attenuated sensing of SHH by Ptch1 underlies adaptive evolution of bovine limbs. doi: 10.1038/nature13289. Submitted. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Zhang H, Litingtung Y, Chiang C. Cholesterol modification restricts the spread of Shh gradient in the limb bud. Proc Natl Acad Sci U S A. 2006;103:6548–6553. doi: 10.1073/pnas.0600124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butterfield NC, et al. Patched 1 is a crucial determinant of asymmetry and digit number in the vertebrate limb. Dev Camb Engl. 2009;136:3515–3524. doi: 10.1242/dev.037507. [DOI] [PubMed] [Google Scholar]
- 27.Sears KE, et al. Developmental basis of mammalian digit reduction: a case study in pigs. Evol Dev. 2011;13:533–541. doi: 10.1111/j.1525-142X.2011.00509.x. [DOI] [PubMed] [Google Scholar]
- 28.Clifford AB. The Evolution of the Unguligrade Manus in Artiodactyls. J Vertebr Paleontol. 2010;30:1827–1839. [Google Scholar]
- 29.Cooper LN, Berta A, Dawson SD, Reidenberg JS. Evolution of hyperphalangy and digit reduction in the cetacean manus. Anat Rec Adv Integr Anat Evol Biol. 2007;290:654–672. doi: 10.1002/ar.20532. [DOI] [PubMed] [Google Scholar]
- 30.Rose KD. Skeleton of Diacodexis, Oldest Known Artiodactyl. Science. 1982;216:621–623. doi: 10.1126/science.216.4546.621. [DOI] [PubMed] [Google Scholar]
- 31.Rose KD. On the origin of the order Artiodactyla. Proc Natl Acad Sci. 1996;93:1705–1709. doi: 10.1073/pnas.93.4.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theodor J, Erfurt J, Metais G. Evol Artiodactyls. JHU Press; 2007. [Google Scholar]
- 33.Rasweiler JJ, Cretekos CJ, Behringer RR. Alcian Blue Staining of Cartilage of Short-Tailed Fruit Bat (Carollia perspicillata) Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5165. pdb.prot5165. [DOI] [PubMed] [Google Scholar]
- 34.Ovchinnikov D. Alcian Blue/Alizarin Red Staining of Cartilage and Bone in Mouse. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5170. pdb.prot5170. [DOI] [PubMed] [Google Scholar]
- 35.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–16. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 36.Rasweiler JJ, Cretekos CJ, Behringer RR. Whole-Mount In Situ Hybridization of Short-Tailed Fruit Bat (Carollia perspicillata) Embryos with RNA Probes. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5164. pdb.prot5164. [DOI] [PubMed] [Google Scholar]
- 37.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]


