Abstract
Systemic delivery of therapeutic nucleic acids to target cells and tissues outside of the liver remains a major challenge. We synthesized a biomimetic high density lipoprotein nanoparticle (HDL NP) for delivery of a cholesteryl modified therapeutic nucleic acid (RNAi) to vascular endothelial cells, a cell type naturally targeted by HDL. HDL NPs adsorb cholesteryl modified oligonucleotides and protect them from nuclease degradation. As proof of principle, we delivered RNAi targeting vascular endothelial growth factor receptor 2 (VEGFR2) to endothelial cells to effectively silence target mRNA and protein expression in vitro. In addition, data show that treatment strongly attenuated in vivo neovascularization measured using a standard angiogenesis assay and in hypervascular tumor allografts where a striking reduction in tumor growth was observed. For effective delivery, HDL NPs required the expression of the cell surface protein scavenger receptor type-B1 (SR-B1). No toxicity of HDL NPs was measured in vitro or after in vivo administration. Thus, by using a biomimetic approach to nucleic acid delivery, data demonstrate that systemically administered RNAi-HDL NPs target SR-B1 expressing endothelial cells to deliver functional anti-angiogenic RNAi as a potential treatment of cancer and other neo-vascular diseases.
Keywords: Tumor, VEGFR2, HDL, Angiogenesis, RNAi
1. Introduction
Since the discovery of RNA interference (RNAi), RNAi molecules have emerged as alluring candidates for gene therapy of countless diseases whereby they act via regulation of target mRNA and/or protein expression.[1] However, despite significant advances,[2] the ultimate therapeutic goal of efficacious, targeted systemic RNAi delivery has not been fully realized. The ideal RNAi-based therapy requires an optimal therapeutic RNAi molecule with minimal off-target effects, and a delivery vehicle that stabilizes the RNAi cargo during transport and specifically delivers the cargo to targeted cells while avoiding off-target toxicity. Additionally, RNAi delivery vehicles should display high bioavailability at both the organismal and cellular levels. While significant progress has been made towards developing delivery vehicles to address each of these issues, more work remains.
Previous studies have revealed that high-density lipoproteins (HDL), naturally occurring nanoparticles involved in cholesterol transport, are capable of binding, stabilizing, and delivering systemically administered lipidated oligonucleotides to cells that express the high affinity HDL receptor, scavenger receptor B-1 (SR-B1).[3] Further, HDLs naturally adsorb endogenous single-stranded RNA (ssRNA), such as microRNAs, and deliver them to cells that express SR-B1.[4] For therapeutic nucleic acid delivery, our and other groups have shown that synthetic HDLs can be formulated with oligonucleotides for targeting specific cell populations and regulating target gene expression.[5-10] Together, synthetic HDL nanoparticles are an intriguing and promising platform for targeted delivery of systemically administered nucleic acids.[4-7, 9, 10]
Endothelial cells (ECs) lining blood vessels naturally express SR-B1 through which they bind mature, spherical HDLs and transport them across the endothelium.[11] These cells also play a vital role in angiogenesis - the formation of new blood vessels - whereby they proliferate and migrate in response to gradients of pro-angiogenic factors creating new vascular channels. Vascular endothelial growth factors (VEGF) are well characterized and highly potent inducers of angiogenesis, and VEGF signaling pathways are crucial in the regulation of developmental, physiological, and pathological angiogenesis.[12-14] A multitude of studies have validated the importance of VEGF driven neo-angiogenesis, especially VEGF-A, as a critical step in tumor progression.[12-15] VEGF-A binds to cell-surface VEGFR2 and causes receptor dimerization followed by auto-phosphorylation of cytosolic domain tyrosine residues. This initiates signaling events that promote endothelial cell proliferation, chemotaxis, and survival resulting in neovascularization.[16] Disrupting VEGF/VEGFR2 signaling arrests the angiogenic cascade at the point of signal initiation and can potently inhibit angiogenesis.[17] Two FDA-approved humanized VEGF antibodies (Avastin, Lucentis) and the soluble VEGF receptor (Eyelea), are now in clinical use for the treatment of cancer and age-related macular degeneration.[18-21] Also some receptor tyrosine kinase (RTK) inhibitors are reported to inhibit VEGF/VEGFR2 signaling.[22-24] However, systemic administration of VEGF targeted antibodies and signaling inhibitors is plagued by toxicities that stem from the dependence of non-endothelial cells, such as neural crest cells, on VEGF for survival and/or function.[25-28] Most of these inhibitors act by sequestering VEGF-A and only a few specifically disrupt VEGF/VEGFR2 signaling. RTK inhibitors, like Sunitinib, inhibit VEGFR2 phosphorylation, but with moderate target specificity.[29, 30] To date, there are no therapies that modulate VEGFR2 protein levels. Thus, therapies based on VEGF/VEGFR2 blockade could benefit from a targeted delivery strategy that relies on inhibition of VEGFR2 expression in endothelial cells.
We report the synthesis of biomimetic HDL nanoparticles (HDL NPs) that adsorb, stabilize, and efficiently deliver RNAi molecules to endothelial cells resulting in the knockdown of VEGFR2 expression (both mRNA and protein). We further demonstrate that VEGFR2 knockdown results in modulation of all critical VEGF responses in vitro and inhibits angiogenesis in vivo after systemic administration. Thus, we demonstrate that HDL NPs are an effective vehicle for delivering RNAi to cell types naturally targeted by HDLs, including endothelial cells, and they overcome many of the hurdles inherent to the systemic delivery of nucleic acids.
2. Results and Discussion
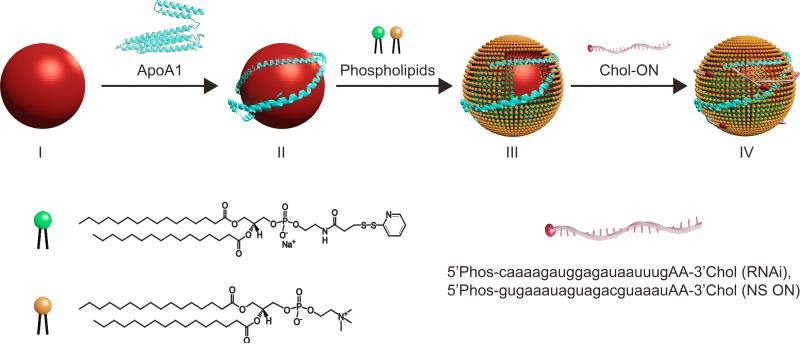
Biomimetic high-density lipoprotein nanoparticles (HDL NPs) were synthesized following the outline depicted in Figure 1 (see Experimental Section). Briefly, citrate-stabilized 5 nm diameter colloidal gold nanoparticles (gold NP) were utilized to control conjugate size, shape, and surface chemistry. Gold NPs were first surface-functionalized with purified human apolipoprotein A-I (ApoA1), the major protein constituent of natural HDLs. Then, the particles were loaded with the following phospholipids: 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio) propionate] (Di-S). Following surface assembly and HDL NP formation, unbound components were purified away via repeated centrifugation and re-suspension in nuclease free water. 3’ cholesterylated antisense VEGFR2 (RNAi) and control non-silencing (NS) oligonucleotides (ON), were added to the suspension of HDL NPs to adsorb onto their surface. RNAi-HDL NPs or control NS-HDL NPs were purified by centrifugation and re-suspension in nuclease free water. At each step, the size, surface chemical composition, zeta potential, and stability of the nanostructures were measured (Table S1). The hydrodynamic diameter of the conjugates exhibited a step-wise increase with the addition of surface components from 6.15±1.34 nm to a final 15.09±3.64 nm (HDL NPs). The HDL NPs had 2-3 ApoA1 molecules per gold NP. Data show that conjugates with ONs retained Apo A1 and the RNAi-HDL NPs had 15.52±2.91 RNAi molecules and NS-HDL NPs had 17.01±3.19 NS ON molecules per particle. The nanoparticle conjugates demonstrated an increase in negative zeta potential at each step of the synthetic process. To visualize the biomolecule corona surrounding the gold NP core, conjugates were stained with uranyl acetate for transmission electron microscopy (TEM) imaging (Table S1). The conjugates were stable to aggregation under physiological conditions as evidenced by the stable UV-Vis absorbance peak at 527 nm in PBS, consistent with monodisperse gold NPs.
Figure 1.
Scheme for synthesis of ON-HDL NPs. Gold NPs with a diameter of 5 nm (red spheres) were surface-functionalized with apolipoprotein A1 (Apo A1, blue) and two phospholipids [DPPC (yellow) and Di-S (green)] to form HDL NPs. Next, cholesterylated ONs (red) are adsorbed to HDL NPs to form ON-HDL NPs.
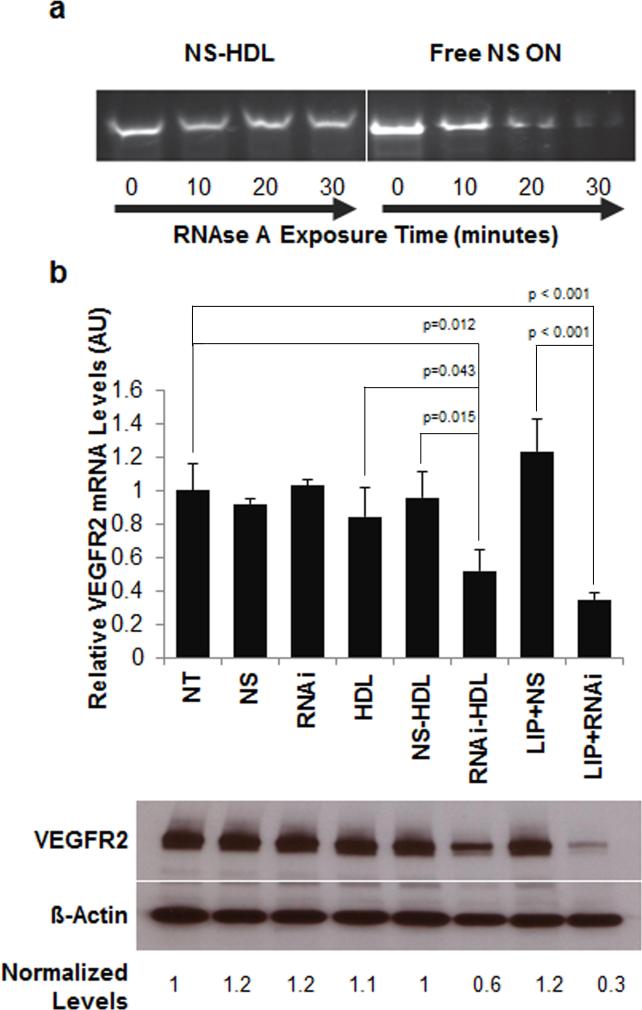
Natural HDLs are known to stabilize RNA against nuclease digestion[4] and our group has previously demonstrated the enhanced stability of cholesterylated DNA molecules bound to HDL NPs.[5] Here, the ability of synthetic HDL NPs to protect their ON cargo was assayed by comparing nuclease resistance of the free, cholesterylated NS ONs and the same sequence adsorbed to HDL NPs. Data demonstrate an increase in stability of the cholesterylated ONs when bound to the HDL NPs (Figure 2a). These data suggest that the nucleic acid is at least partially buried among the surface constituents of the ON-HDL NP, which is consistent with previous reports demonstrating similar localization of microRNAs on the surface of natural and reconstituted HDLs.[4]
Figure 2.
HDL NPs stabilize conjugated ONs to nuclease degradation and RNAi-HDL NPs regulate in vitro VEGFR2 expression. (a) NS ONs mixed with HDL NPs or free in solution were exposed to RNAse A for different time intervals (0, 10, 20, and 30 mins). Reactions were stopped and reaction contents were subjected to polyacrylamide gel electrophoresis and stained with ethidium bromide. (b) RT-qPCR (plot) and Western blotting demonstrate knockdown of VEGFR2 mRNA and protein, relative to ß-actin mRNA and protein levels, respectively, after treatment with RNAi-HDL NP (RNAi-HDL) or positive control Lipofectamine/RNAi (LIP+RNAi).
Endothelial cells (EC) have inherent capacity for HDL uptake via SR-B1 for the maintenance of cholesterol homeostasis.[31] Therefore, we hypothesized that ECs would efficiently internalize RNAi-HDL NPs via SR-B1 without inherent toxicity. To test this hypothesis, we first measured cytotoxicity as the amount of lactate dehydrogenase (LDH) released from cultured human umbilical vein endothelial cells (HUVECs) exposed to HDL NPs and NS-HDL NPs. As expected, neither HDL NPs nor NS-HDL NPs caused a significant increase in LDH release (Figure S1). In addition, there were no significant increase in apoptosis after 48 hours of exposure to the HDL NPs as measured via flow cytometric analysis of the cells stained for Annexin V and propidium iodide (AV/PI, Figure S2) even at doses well above those used for nucleic acid delivery (vide infra). Next, we compared the ability of HDL NPs carrying RNAi against VEGFR2 to regulate target gene expression in HUVECs with comparison made to RNAi delivered via the conventional transfection reagent, Lipofectamine RNAiMax (Invitrogen) (Figure 2b). The RNAi-HDL NPs inhibited VEGFR2 expression at the protein and mRNA levels with potency comparable to that of RNAi delivered with Lipofectamine RNAiMax. Importantly, HDL NPs alone and NS-HDL NPs were ineffective (Figure 2b). Finally, an equivalent amount of free RNAi targeting VEGFR2 directly added to HUVECs caused no measurable knockdown, demonstrating that the delivery vehicle is crucial (Figure 2b). This requirement should be even more pressing in vivo, where nuclease exposure and rapid clearance are imminent.
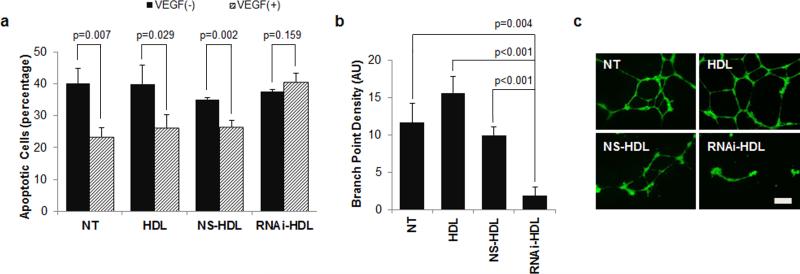
In HUVECs, VEGFR2 activation causes increased cell proliferation, chemotaxis, and survival, which collectively culminate in neovascularization.[16] We assessed each of these responses in vitro after treatment with RNAi-HDL NPs. First, we performed a tritiated thymidine incorporation assay to measure cell proliferation after treatment with RNAi-HDL NPs. Data show that RNAi-HDL NPs significantly attenuated VEGF-induced proliferation after 48 hours of treatment while control NS-HDL NPs had no effect (Figure S3). Next we utilized an AV/PI assay to measure apoptosis following a similar treatment. Apoptotic response of HUVECs to serum deprivation was significantly reduced in the presence of VEGF-A (Figure 3a). Treatment with RNAi-HDL NPs abolished the rescue by addition of VEGF-A (Figure 3a). Next VEGF-A promoted HUVEC migration in a wound closure assay was suppressed by RNAi-HDL NP but not by control HDL NPs or NS-HDL NPs (Figure S4). Finally, we measured the effects of RNAi-HDL NPs on VEGF-A induced morphogenesis in a Matrigel tube formation assay. In this assay, HUVECs form branched tubular networks in response to VEGF-A. Data show that RNAibut not control NS-HDL NPs dramatically reduced overall network complexity and significantly reduced branching (Figure 3b, c).
Figure 3.
RNAi-HDL NPs reduce VEGF-A responsive endothelial cell survival and morphogenesis. (a) Annexin V/PI flow-cytometric assay conducted on VEGF-A induced (grey, 20 ng/ml) or un-induced (black) HUVECs revealed that treatment with RNAi-HDL NPs (RNAi-HDL) leads to a significant reduction in rescue from apoptosis as compared to controls. (b) RNAi-HDL NP (RNAi-HDL) treated HUVECs display reduced branch point density and overall network complexity in a Matrigel tube formation assay, where the HUVECs were induced to form branched tubular networks by stimulation with VEGF-A. (c) Fluorescence images obtained after Calcein AM staining of treated endothelial cells embedded within the Matrigel layer (scale bar 200 μm).
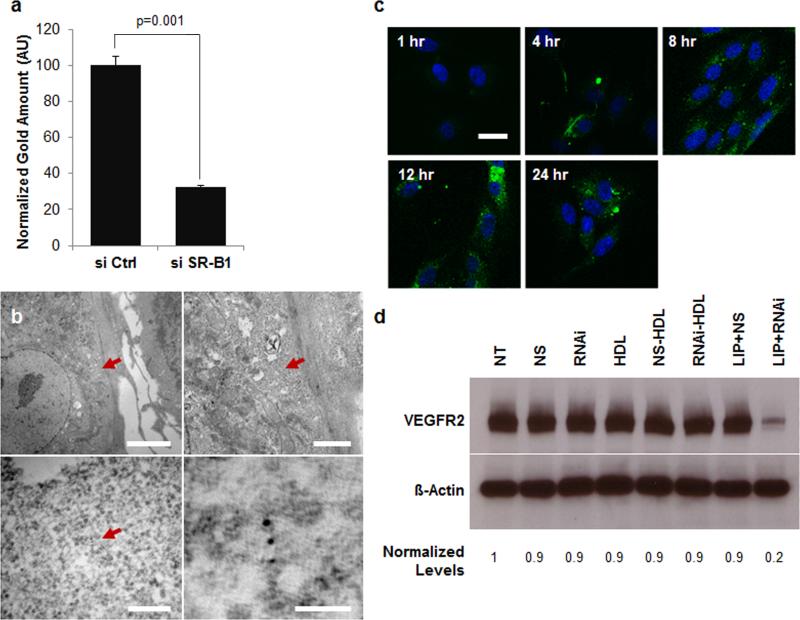
Since the ECs internalize natural HDLs,[11] we hypothesized that HDL NPs and ON-HDL NPs would be internalized. We explored the uptake of NS-HDL NPs in HUVECs, by measuring intracellular gold content using inductively coupled plasma mass spectrometry (ICP-MS), by TEM (Figure 4b), and by confocal fluorescence microscopy (Figure 4c). All methods confirmed cellular uptake of the conjugates. TEM images demonstrated the presence of gold NPs in the cytosol (Figure 4b) and in vesicular structures (Figure S5). To explore the uptake of the nucleic acid component of the conjugates, we treated HUVECs with NS-HDL NPs, where the NS sequence was labeled with fluorescein isothiocyanate (FITC) (Figure 4c). We visualized uptake of the fluorophore by confocal microscopy. Initially, the fluorophore was seen assembled in a punctuate pattern at the cell periphery, which then dispersed throughout the cell cytoplasm by 8 hrs. We also conducted experiments to visualize the Apo A1 component of the conjugates by using Apo A1 labeled with Alexa Fluor 488. Confocal imaging showed that Apo A1 localized to the cytosol after 8 hours and remained detectable in the cells for up to 24 hours (Figure S6). Collectively, TEM and confocal microscopy suggest that NS-HDL NPs are internalized by target cells. They are initially localized to punctate microvesicular structures. Later at least some of the gold particles, nucleic acid, and Apo A1 can be found in the cytosol. Our data are consistent with uptake of the nanoparticle conjugate via endocytosis followed by at least partial delivery into the cell cytoplasm. Our data (vide infra) demonstrate that SR-B1 expression is necessary for this internalization and regulation of gene expression by HDL NPs. Similar observations were reported by other groups [3, 4, 6] investigating nucleic acid transport and delivery by synthetic and natural HDLs.
Figure 4.
Uptake and function of ON-HDL NP conjugates is dependent upon SR-B1 expression. (a) Cellular gold content, normalized to extracted cellular protein from HUVECs, is shown with and without transient knockdown of SR-B1 expression. ICP-MS was used to quantify cellular gold content. (b) Transmission electron micrographs of HUVECs treated with NS-HDL NPs reveal gold NPs in the cytosol (scale bars clockwise from top left, 5 μm, 1 μm, 50 nm, 200 nm). (c) FITC-tagged NS ONs were conjugated to HDL NPs and their localization in HUVECs monitored at several time points post treatment by fixing the cells and imaging on a confocal microscope. FITC signal is shown in green and Hoechst stained cell nuclei are shown in blue (scale bar, 30 μm). (d) The siSR-B1 transfected HUVECs were either left untreated (NT) or treated with HDL-NPs (HDL) and their conjugates with RNAi (RNAi-HDL) and NS (NS-HDL). Also, free RNAi and NS (NS) ONs were added with (LIP) and without Lipofectamine RNAiMax. Western blots of VEGFR2, with ß-Actin as loading control, for each of the treatments (48 hrs) are shown.
SR-B1 is a high-affinity receptor for mature spherical HDLs and has been shown to mediate the uptake of natural and synthetic HDLs.[3, 4] Since ECs express SR-B1 we examined its role in the uptake of HDL NP conjugates and their capacity for target knockdown. In HUVECs, SR-B1 levels are comparable to those in hepatocytes and macrophages, both known to strongly express SR-B1 (Figure S7).[32] We reduced SR-B1 expression using conventional siRNA knockdown (KD) (Figure S7) and delivered RNAi against VEGFR2 to the wild type or SR-B1 KD HUVECs by conventional transfection or by means of RNAi-HDL NPs. Importantly, RNAi-HDL NPs were not effective upon knockdown of SR-B1 expression (Figure 4d), while RNAi delivered by Lipofectamine RNAiMax was effective in both SR-B1 positive and knockdown cells. Further, ICP-MS showed a significant reduction of gold NP uptake in the SR-B1 knockdown cells (Figure 4a). Together, data indicate that similar to natural HDLs, which act as vehicles for nucleic acids, SR-B1 expression is required for the uptake and efficacy of RNAi-HDL NPs as well. Finally, our results are consistent with published data that suggest natural HDLs bind SRB1 and deliver microRNAs that are partially buried in the surface layers of HDL.[4]
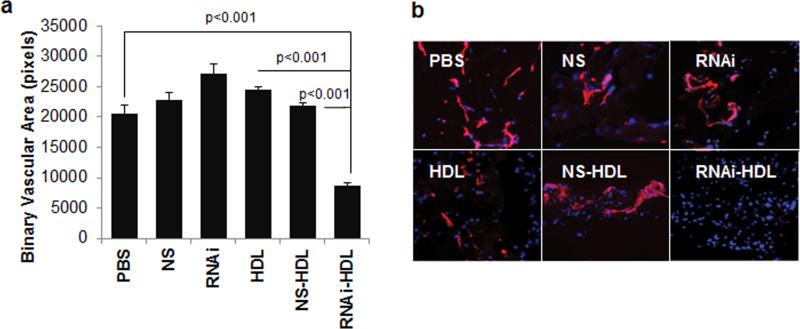
To determine whether functional changes measured in vitro would translate into in vivo inhibition of angiogenesis, we employed a Matrigel plug angiogenesis assay where VEGF-A is incorporated into a subcutaneous Matrigel plug to induce neovascularization (see Methods and Supporting Information). We utilized RNAi-HDL NPs to inhibit angiogenesis in the Matrigel plugs in three ways. First, we pre-treated cultured HUVECs with RNAi-HDL NPs prior to harvesting the cells and incorporating them into the subcutaneous Matrigel plugs (Figure S8). Second, RNAi-HDL NPs were administered locally, by mixing them with the Matrigel prior to subcutaneous implantation (Figure S9). Finally, to demonstrate the feasibility of systemic treatment, mice bearing Matrigel plugs received intravenous injections of RNAi-HDL NPs (Figure 5a, b). For each of the Matrigel experiments, we used NS-HDL NPs, free HDL NPs (vehicle) and the free RNAi ONs as controls. Regardless of the delivery method, only RNAi-HDL NPs significantly decreased angiogenic response to VEGF-A across all treatment setups, as demonstrated by measuring binary vascular area in the Matrigel sections after immunostaining for the endothelial cell marker, CD31.
Figure 5.
Matrigel plug assay and in vivo function of RNAi-HDL NPs. Nude mice implanted with Matrigel plugs containing 400 ng ml−1 VEGF-A were systemically administered HDL NPs (HDL) or conjugates with RNAi (RNAi-HDL) and NS ONs (NS-HDL). In addition, free RNAi (RNAi) and NS ONs (NS) and PBS (PBS) were used as controls. (a) Binary vascular area, measured as CD31 positive pixels in fluorescent images of immunostained sections, was plotted for each of the treatment groups. (b) Representative fluorescent images showing CD31 stained regions (red) and DAPI stained cell nuclei (blue).
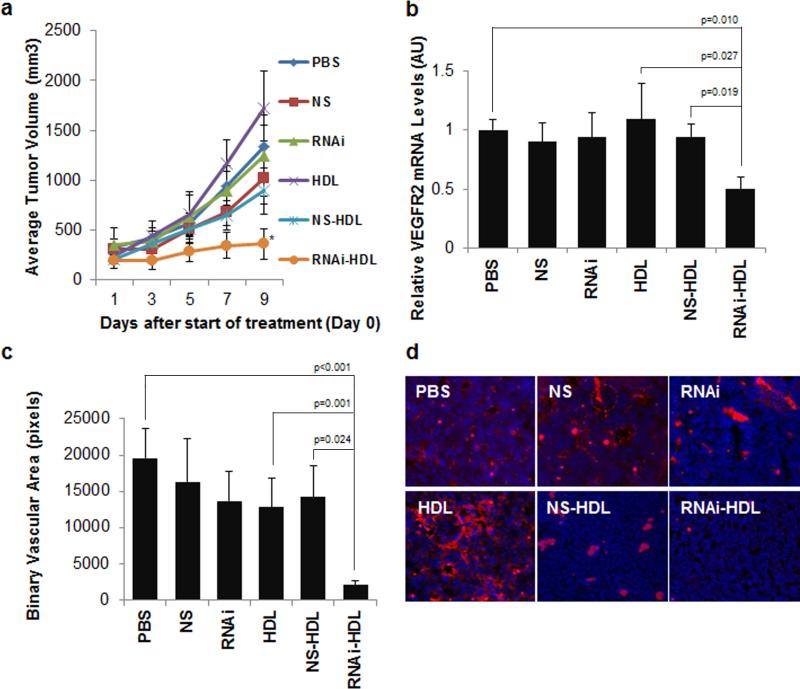
VEGF antagonists show efficacy against tumor growth in pre-clinical models and have improved cancer outcomes in the clinic.[33, 34] To test whether RNAi-HDL NPs inhibit tumor growth and angiogenesis we employed a mouse non-small cell lung carcinoma, Lewis Lung Carcinoma-1 (LLC-1), in a subcutaneous allograft model. After implantation, we allowed tumors to reach a size of 100-200 mm3 before initiating treatment. We administered RNAi-HDL NPs and control treatments via tail vein injection and measured tumor volumes throughout the experiment and tumor weight at the conclusion of the experiment (see Methods and Supporting Information). Tumor growth was significantly reduced after treatment with RNAi-HDL NPs (Figure 6a), and correspondingly the average tumor weight was significantly reduced (Figure S10). Control treated mice did not demonstrate significant reductions in tumor volumes or tumor weights. Further, RNAi delivered by the HDL NPs resulted in a significant decrease in VEGFR2 mRNA levels in the tumor tissue (Figure 6b) and reduced angiogenesis as reflected by measurements of the binary vascular area of tumor sections stained for CD31 (Figure 6c,d).
Figure 6.
Lewis lung carcinoma (LLC-1) subcutaneous tumor response to intravenous administration of RNAi-HDL NPs and control treatments. (a) Average tumor volumes are plotted for each of the treatments and there is a significant reduction in the RNAi-HDL NP (RNAi-HDL) treatment group on day 9 as compared to NS-HDL (p=0.016). (b) Tumors from the RNAi-HDL NP (RNAi-HDL) treatment group exhibit a significant reduction in VEGFR2 mRNA content relative to ß-actin mRNA. (c) Binary vascular area quantified by CD31 immunostaining and subsequent fluorescent imaging of frozen tumor sections reveals a significant reduction of neovascularization in RNAi-HDL NP treatment group as compared to controls. (d) Representative fluorescent images of tumor sections showing CD31 immunostained regions (red) and DAPI stained cell nuclei (blue).
3. Conclusions
Formulation and delivery of RNAi molecules faces several hurdles, foremost being rapid degradation by exo- and endo-nucleases and a lack of targeted delivery. Also, following cellular uptake, the interaction of the RNAi molecules with endogenous target RNA and subsequent regulation of target function are marred by sequestration in endolysosomal compartments. On the organismal level, RNAi delivery faces other major challenges, especially poor bioavailability at the target site. Here, we demonstrate that HDL NPs are able to overcome these obstacles and efficiently deliver RNAi molecules. First, HDL NPs protect ON cargo from nuclease degradation. We demonstrate in this and in earlier studies[5] that HDL NPs successfully enter cells and, at least in part, localize to the cytosol. Importantly, and in agreement with other reports,[3, 4, 6, 7] we show that fluorophore-tagged ON cargo localizes to the cytosol outside of endolysosomal compartments and is able to regulate target gene expression. Rapid clearance by the reticuloendothelial system (RES) limits the bioavailability of RNAi delivery vehicles and their RNAi cargo in target tissues. Our in vivo studies in Matrigel plug systems demonstrate that RNAi molecules conjugated to HDL NPs reach the vasculature and vascular endothelial cells in amounts sufficient to functionally inhibit angiogenesis, despite the bulk of the NPs distributing to the liver and spleen (Figure S11). Moreover, they do not cause any overt non-specific toxicity (Figure S12). Further, in a pre-clinical tumor model RNAi-HDL NPs are able to cause a significant reduction of MVD, and tumor mass which is accompanied by a reduction in VEGFR2 expression. Notably, the concomitant reduction of VEGFR2 expression and suppression of neovascularization seen in the tumor tissues suggest that the RNAi-HDL NPs are able to block VEGF signaling in tumors upon systemic administration. However, we should also point out that the reduction in angiogenesis and tumor growth may be caused in part by concomitant targeting of other tumor-associated cells like stromal fibroblasts or macrophages. For tumor associated macrophages (TAMs), prior studies have shown that VEGFR2 blockade also reduces their infiltration into the tumor tissue and associated pro-angiogenic action.[35] Most importantly, neither HDL NPs nor their conjugates with NS sequences exhibited overt non-specific cytotoxic effects.
HDL NPs as a platform for RNAi delivery appear to inherit the strengths of their natural counterparts - mature spherical HDL particles. Natural mature HDL particles have inherent capacity to adsorb, stabilize, and deliver both exogenous and endogenous ONs to cells expressing SR-B1.[3, 4] By foraying into the field of bio-mimicry, our group[5] and others [7, 10] were able to formulate synthetic biomimetics of HDL, which deliver ONs to SR-B1 positive cells. Here we provide data that clearly demonstrate the utility and therapeutic potential of this approach and unequivocal proof of the critical role that SR-B1 plays in the uptake of RNAi-HDL NPs and their therapeutic activity in endothelial cells. SR-B1 expression is normally restricted to the tissues that participate in cholesterol metabolism including the liver, adrenal glands and gonads, and to the cells relevant to the pathogenesis of atherosclerosis, such as macrophages and endothelial cells.[36] Thus, the array of healthy tissues targeted by HDL-NPs is fairly limited and molecular target specificity can be provided by the nucleic acid payload.[32, 37-41] Interestingly, many cancer types express high levels of SR-B1[6] pointing to a distinct possibility of delivering RNAi against cancer-specific targets to the primary tumors and especially occult metastases, as well as to their supporting vasculature, as shown here.
4. Experimental Section
Oligonucleotides
Oligonucleotides antisense to VEGFR2 (RNAi) and non-silencing scrambled control sequences (NS) were purchased from Integrated DNA Technology (IDT). The chimeric antisense (RNAi) and non-silencing (NS) scrambled control oligonucleotide had the following sequences: 5’-P-caaaagauggagauaauuugAA-TEGchol-3’ and 5’-P-gugaaauaguagacguaaauAA-TEGchol-3’, respectively. The nucleotides indicated in lower case are RNA bases and uppercase denotes DNA bases. Both oligos were modified with 5’ phosphorylation and a 3’ cholesteryl moiety, the latter, linked via a tetra-ethylene glycol (TEG) spacer to the 3’ end of the nucleotide sequence.
Fluorophore tagged sequences were also purchased from IDT. The sequence for the FITC tagged NS control oligo used for confocal microscopy experiments was 5’-P-gTgaaauaguagacguaaauAA-TEGchol-3’. The FITC was attached to the internal deoxy-T residue. Also, we used sequences with internal fluorescent modification using the Cy3 molecular fluorophore for quantification of HDL NP adsorbed NS and RNAi sequences. Control NS and RNAi sequences were purchased with the internal Cy3 modifier placed at the same location as the FITC-labeled deoxy T (IDT).
For silencing SCARB1 expression, Silencer select validated siRNA against SCARB1 and negative control RNA duplexes were purchased from Applied Biosystems.
HDL NP Synthesis and Characterization
Gold nanoparticles with a median diameter of 5 nm were purchased from Ted-Pella Inc. The phospholipids, (1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC); 1,2-dipalmitoyl-snglycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio)propionate (Di-S)) were purchased from Avanti Polar lipids. Purified human Apolipoprotein A1 (Apo A1, cat. A50620H) was purchased from Meridian Life Sciences. Prior to conjugation to biomolecular surface components, the gold NPs were treated with diethylpyrocarbonate (DEPC) for ~16 hrs. The colloid was then autoclaved at 121 °C and 15 p.s.i for 30 minutes. After cooling, the gold NP solution was passed through a 0.22 μm filter. The concentration of gold NPs was measured using a UV-Vis spectrophotometer (Agilent). Colloidal gold NPs have an absorbance maximum at 519 nm and an extinction coefficient (ε) of 9.696 × 106 M−1cm−1. For synthesis of HDL NPs and ON-HDL NPs, gold NPs are first incubated with a 5-fold molar excess of Apo A1 for 4 hours at room temperature (RT) with gentle shaking. Next, 100% ethanol and solutions of each of the two phospholipids dissolved in ethanol were added to the aqueous solution of ApoA1-gold NPs. Ultimately, the water:ethanol ratio was 4:1 and each of the phospholipids were added in 250-fold molar excess to the gold NP. This mixture was allowed to incubate overnight at RT with gentle shaking. To purify the HDL NP conjugates from unbound NP surface components, HDL NPs were centrifuged (16,500 g; 40 minutes; X2) and the pellet re-suspended in nuclease-free water. The final concentration of HDL NPs was determined with UV-Vis spectroscopy as previously described. To functionalize HDL NPs with cholesteryl-modified ONs, the ONs were first heated to 70°C for 3 minutes and then placed on ice for one minute. The cholesteryl-modified ONs were then added to a solution of HDL NPs in 100-fold molar excess with respect to the gold NP concentration. The mixture was allowed to gently shake for 1 hour at RT. To purify ON-HDL NPs from unbound ONs, the solution was centrifuged and the pellet re-suspended in PBS. The stability of the particles at each step of the synthesis was monitored using spectrophotometric readings and dynamic light scattering analysis on a Malvern Zetasizer.
To calculate ON loading on the HDL NPs, three HDL NP aliquots (1 μM) were incubated with Cy3 tagged NS or RNAi ONs (100 μM, final) for 1 hour with shaking at room temperature. After conjugation, the ON-HDL NPs were centrifuged (16,500 g; 40 minutes) to remove unbound components. Next, the ON-HDL NPs were washed five times with PBS using a 50 kDa MW-cutoff spin filter (Amicon Ultra, Millipore at 5,000 g × 10 minutes at RT), and then finally re-suspended in PBS and stored (1 μM) prior to use. A standard curve was generated by serially diluting Cy3 tagged NS or RNAi oligos in PBS in a 96-well plate (100 μl volume). Three dilutions (50, 10, and 5 nM) of HDL-NPs conjugated with fluorophore-labeled NS or RNAi ONs were prepared (100 μl). Then, 25 μl of 100% ethanol was added to the standards and HDL NP conjugates. A single flurometric reading of Cy3 emission immediately followed by an absorbance reading for gold NPs (at 527 nm) was carried out. Next 5μl of 1 M potassium cyanide (KCN) solution was added to each well, and mixed by repeated pipetting in order to etch away the gold core and relieve its quenching effects. Fluorimetric readings of Cy3 emission immediately followed by absorbance readings for gold NPs were repeated every 5 minutes for 30 minutes at which point the fluorescence stabilized. At each time point, the concentration of Cy3 tagged nucleotides in the HDL NP samples was calculated by referral to the standard curve. Finally, the molar loading ratio of nucleic acid on the ON-HDL NPs was calculated by dividing the calculated concentration of the ONs by the original concentration of gold nanoparticles.
For calculating ApoA1 loading of the HDL NPs and ON-HDL NPs, a similar approach was used with ApoA1 labeled with Alexa-Fluor 488.
Transmission Electron Microscopy
For visualization of the nanoparticles formed at each step of the synthesis, the nanoparticles were purified from unbound components or impurities by centrifugation, as described. Centrifugation and re-suspension in nuclease free water was also used to concentrate the particles for delivery to carbon film coated copper TEM grids (200 mesh, Electron Microscopy Sciences). In a typical experiment, 40 μl of a re-suspended particle sample was first put on the top surface of a copper TEM grid and allowed to settle for 15 minutes. Subsequently, excess liquid was removed by gentle filter paper blotting and then 40 μl of 3% (w/v) uranyl acetate solution was added and allowed to incubate for 15 minutes. Uranyl acetate solution was removed using filter paper and the grid was allowed to dry for 1 hour under a laminar flow. The grids were then imaged.
For TEM imaging of NS-HDL NPs inside HUVECs, sub-confluent HUVECs, grown on Thermonex plastic coverslips were treated overnight with NS-HDL NPs (10 nM, final) and after washing (PBS) fixed in a solution containing 2% formaldehyde/2.5% glutaraldehyde in 0.1 M sodium cacodylate (SCB). The cells were then rinsed with 0.1 M SCB and placed in secondary fixative containing 2% osmium tetroxide in 0.1M SCB. Next the HUVECs were again rinsed with distilled water and stained with 3% uranyl acetate (w/v). The HUVECs were then rinsed with distilled water and then dehydrated in ascending grades of ethanol. Propylene oxide was used as a transitional buffer, and the fixed cells were embedded in Epon 812 and Araldite resin. Subsequently the samples were placed in a 60°C oven to cure. The blocks were sectioned using an ultramicrotome and then mounted on grids for TEM imaging.
All TEM imaging experiments were carried out on a FEI Tecnai Spirit G2 instrument operated at 120 KV.
RNAse Protection Assay
Non-silencing scrambled ONs at a final concentration of 1 μM were introduced into PCR tubes containing either 100 nM HDL NPs in PBS or just PBS. The ONs were incubated for 4 hours prior to exposure to 0.4 ng μl−1 (final) of RNAseA (Bio-Rad) at 37°C. After varying times of incubation, the nuclease digestion was stopped by adding 45% (v/v, final) formamide and 0.5% (w/v, final) SDS and heating to 65 °C for 3 minutes. The reaction mixtures were then subjected to electrophoresis on a 50% urea, 15% polyacrylamide gel. Ethidium bromide staining was used to visualize the ON bands.
Cell Culture
Human umbilical vein endothelial cells (HUVEC, Lonza) were cultured in EBM-2 media supplemented with EGM-2 SingleQuot (Lonza). All HUVECs used in this study were between passages 2 to 5. Mouse LLC-1 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS).
LDH Release Assay
The LDH Release Assay kit was purchased from Roche Applied Science. HUVECs, were plated at 1.5 × 105 cells ml−1 in 12-well plates, 500 μl per well. Cells were treated overnight with PBS, free NS ON, HDL NPs and NS-HDL NP conjugates in OptiMEM (Invitrogen) containing 2% FBS (final concentrations of HDL NPs and NS-HDL NPs were 1, 10, 15 nM, for free NS ONs they were 100, 1000 and 1500 nM). Subsequently, the cell culture media was replaced with EBM-2 supplemented with EGM-2 Singlequot (Lonza). The next day the cell culture media were collected and spun down at 300g for 10 minutes to remove cell debris. Thereafter, the working reagent was formulated as per manufacturer's instructions and the LDH colorimetric assay was carried out in 96-well plates after adding 100 μl of the working reagent to a 1:20 diluted solution of the media. For dilutions and for the blank sample, RPMI-1640 containing no phenol red was used. For a positive control, three wells containing untreated cells were lysed by adding Triton X-100 (1% v/v, final). For negative control, the supernatant of three wells containing untreated cells was used.
Apoptosis Assay
An Annexin V/ Propidium Iodide (AV/PI) apoptosis kit (cat. 88-8005-74) for flow cytometric quantification of apoptotic cell populations was purchased from eBiosciences. For measuring the apoptotic response to increasing dosages of NPs, HUVECs were plated in 12-well plates at a density of 1.5 × 105 cells ml−1, 500 μl per well, and allowed to attach overnight. The next day, PBS was added to three wells (NT) and 1, 10 and 30 nM (final) of HDL NPs were added to the 48-hour treatment wells (3 wells/ dose), 24-hours later, the same was done for the 24-hour treatment wells (3 wells/ dose). The cells were incubated for an additional 24-hours where-after the cells were collected by trypsinization and re-suspension in EBM-2 containing 10% FBS. Harvested cells were washed twice with ice-cold PBS before being re-suspended in ice-cold anti-Annexin V/PI working solution. The working solution of the anti-Annexin V/PI was prepared according to manufacturer's protocols. After incubation with the working solution, cells were subject to flow cytometric analysis. A total of 10,000 gated events were recorded.
For assaying the change in VEGF-A mediated anti-apoptotic response, HUVECs were plated in 12-well plates at a density of 1.5×105 cells ml−1, 500 μl per well, and allowed to attach overnight. The cells were then treated with PBS, HDL NPs, NS-HDL NPs and RNAi-HDL NPs (10 nM, final concentrations of HDLNPs and conjugates). After an overnight incubation, the media was replaced. After additional 24 hours, HUVECs were exposed to serum starvation by removing the standard growth medium and replacing with OptiMEM (Invitrogen) supplemented with 5% FBS, with or without 20 ng ml−1 of purified VEGF-A (Millipore). Cells were cultured overnight under serum starvation, and then detached via trypsinization, washed (PBS), and then exposed to anti-Annexin V/ PI working solution as per manufacturer's protocol. The cells were then subjected to flow cytometric analysis and a total of 10,000 gated events were recorded.
Flow cytometry data was collected on a Beckman Coulter CyAn flow cytometer, and the final population partition and extraction of relevant statistical data was carried out using FlowJo (Version 10).
Transient Transfection
For transient transfections, Lipofectamine RNAiMax (Invitrogen) was used according to protocol. EBM-2 media free of supplements or antibiotics was used to dilute Lipofectamine RNAiMax reagent as well as siRNA or cholesterylated ONs. Lipofectamine RNAiMax was mixed with the appropriate nucleic acid according to the protocol. Mixtures were added to cells for 3 hr at 37°C. Subsequently, the transfection media was removed, the cells washed once with warm PBS, and then the cells were re-fed full growth media. For transient transfection with the chimeric oligos, the final concentration of the oligos was 250 nM and 10 μl of Lipofectamine RNAiMax reagent was used per 600 μl final transfection volume. For transient SCARB1 knockdown, we used validated silencer select siRNA (cat. s2649), from Applied Biosystems, at a final concentration of 50 nM and 1 μl of Lipofectamine RNAiMax reagent per 600 μl final transfection volume.
RT-qPCR
RT-qPCR was used to quantify mRNA levels of VEGFR2. For reverse transcription, we used a TaqMan Gold RT Kit (Applied Biosystems) as per manufacturer's instructions. For in vitro experiments using HUVECs, 200 ng of extracted total RNA was used per 20 μl RT reaction, for tumor RNA extracts, 1 μg of total RNA per 20 μl RT reaction was used. For qPCR, TaqMan Primer/Probe sets for human as well as mouse VEGFR2 and β-actin were purchased from Applied Biosystems. The delta delta Ct method was employed for analyzing the data using β-actin mRNA levels as the internal control.
Immunoblotting
We used antibodies against human VEGFR2 (cat. 55B11) and β-Actin (cat. 13E5) from Cell Signaling and SCARB1 (cat. ab52629) from AbCam. Secondary HRP-tagged goat anti-mouse and anti-rabbit antibodies were from Bio-Rad. Cell lysates were run on 4-20% Tris-Glycine-SDS polyacrylamide gels (Bio-Rad) and transferred onto nitrocellulose membranes (Bio-Rad). The membranes were blocked with 2% BSA in PBS-T (PBS, 0.05% Tween-20). Antibody dilutions were optimized for each target protein.
Thymidine Incorporation Assay
For measuring cell proliferation, HUVECs were plated into 12-well plates and allowed to attach, as previously described. HUVECs were treated with PBS, HDL NPs, NS-HDL NPs and RNAi-HDL NPs (10 nM, final concentrations of HDL NPs and conjugates). Following an overnight incubation with treatments, the media was removed, cells washed (PBS), and fresh EBM-2 supplemented with 5% FBS was added. The cells were thus incubated for another overnight duration. Subsequently, media were again replaced with fresh EBM-2 containing 5% FBS with or without 20ng ml−1 VEGF-A and the HUVECs were incubated for 4 hours before spiking in 1.0 μCi/well of 3H-Thymidine (Perkin Elmer; from a 100 μCi ml−1 stock solution prepared in EBM-2). The plates were incubated overnight, and the following morning the media was removed and the cells washed three times with ice-cold PBS and subjected to DNA extraction using Qiagen DNEasy Blood and Tissue Kit as per the manufacturer's protocols. Finally 130 μg of extracted DNA from each treatment sample was subjected to scintillation counting for quantification of incorporated 3H-Thymidine.
Wound Healing Assay
To measure the rate of VEGF-A dependent endothelial cell chemotaxis, HUVECs were plated at a density of 1.5 × 105 cells ml−1 in T-25 flasks, 5ml/ flask, to achieve a 50% confluent cell monolayer following overnight attachment. Then, the HUVECs were treated with PBS, HDL NPs, NS-HDL NPs and RNAi-HDL NPs (10 nM, final concentrations of HDL NPs and conjugates). After overnight incubation, the HUVECs were trypsinized and re-plated into a 12-well plate at a concentration of 5.0 × 105 cells ml−1, 500 μl/ well and allowed to attach overnight. Next, cell culture media was changed to EBM-2 supplemented with 5% FBS. The monolayer was pre-treated for 2 hr with Mitomycin-C (Roche, 10 μg ml−1) to eliminate proliferation component of the wound closure rate. The HUVEC monolayer was then scratched with a 20 μl pipette tip (Rainin) and the medium was replaced with fresh EBM-2 supplemented with 5% FBS with or without VEGF-A (20 ng ml−1) to induce migration. Initial images were taken at three separate positions per scratch using an inverted microscope (Nikon Eclipse TE2000-U). A total of three scratches (in 3 wells) per treatment were analyzed. The second set of images was taken after 4 hr incubation. Gap pixel areas were measured using ImageJ and percentage gap closure calculated.
Matrigel Tube Formation Assay
HUVECs were plated into 6-well plates at a density of 1.5 × 105 cells ml−1, 2 ml/ well, allowed to attach overnight, and then treated with PBS, HDL NPs, NS-HDL NPs and RNAi-HDL NPs (10 nM, final concentrations of HDL NPs and conjugates). Following overnight incubation, the cell culture media was replaced with fresh growth media and the cells were incubated for another 48 hours. Next, phenol red free and growth factor reduced Matrigel (15 mg ml−1, BD Biosciences) was diluted to a concentration of 10 mg ml−1 with phenol-red free, serum free RPMI-1640 supplemented with 100 ng ml−1 (final) of VEGF-A (Millipore). The supplemented Matrigel was placed on ice and then dispensed into a pre-chilled 24-well plate (300 μl/well). The Matrigel mixture was allowed to solidify by incubating the plate at 37°C for 1 hour. Subsequently, treated HUVECs were trypsinized, washed once with PBS and re-suspended in phenol-red free RPMI-1640 with 5% FBS at a final density of 5 × 105 cells ml−1. Aliquots of the cell suspensions (300 μL) were then pipetted onto the Matrigel surfaces. The plate was incubated overnight at 37°C. Next, the remaining media were aspirated and the surface rinsed twice with warm Hank's Balanced Salt Solution (HBSS). Calcein AM dye (BD Biosciences) in HBSS (8 μg ml−1) was prepared as per protocol. 300 μl of the Calcein AM dye solution was dispensed into each well and the plate was incubated at 37°C for 30 minutes after which the Calcein AM solution was removed and the gel surface was washed twice with pre-warmed HBSS. The wells were then imaged for green fluorescence signal to visualize tubes formed by the migrating live HUVEC using a Nikon Eclipse TE2000-U inverted microscope. Branch points were identified visually and the density per section calculated.
ICP-MS
For quantifying cellular gold NP uptake, HUVECs were plated at a density of 1.5 × 105 cells ml−1 in 6-well plates (2ml/ well). After overnight incubation, cells were washed and re-suspended in EBM-2 media containing 2.5% FBS and treated with 10 nM NS-HDL NPs. Similar to wild-type cells, the same experimental setup was used with HUVECs following transient knockdown of SCARB1. Following 24-hour treatment, cell lysates were obtained and the protein concentration was determined using a BCA assay (Thermo Scientific). 100 μg of total protein per sample was then lyophilized and subject to acid dissolution using 3% anhydrous hydrocholoric acid in anhydrous nitric acid (300 uL). The volume of acid dissolved samples was then brought to 5 ml with ICP-MS running matrix (3% hydrochloric acid, 3% nitric acid in deionized water) and included 5 ppb indium for use as an internal control. Gold standards were formulated by setting up serial dilutions from a 1 ppm gold standard stock solution into the ICPMS matrix which was spiked with 5 ppb indium in a final volume of 10 ml. Samples and standards were subjected to ICP-MS quantification and the amount of gold in the cell lysates was calculated using the standard calibration curve.
For ICP-MS of the organs harvested from the Matrigel plug assay experiments, tissue samples from the organs were placed in pre-weighed microcentrifuge tubes and dessicated by heating in a warming oven to 55 °C. The tubes were then re-weighed to measure the dry tissue weight. The dessicated tissues were then subjected to acid dissolution and ICP-MS as previously described. The content of gold per dry tissue weight was calculated.
Confocal Microscopy
To visualize internalization of the Apo A1 component of NS-HDL NPs, 1 mg of Alexa Fluor 488 labeled Apo A1 was prepared using an Alexa Fluor 488 protein labeling kit (Invitrogen) according to the manufacturer's protocol. The labeled Apo A1 was used for the synthesis of HDL NPs and NS-HDL NPs, as described above. To visualize the internalization of the ON cargo component of NS-HDL NPs, FITC tagged NS ONs were used for the synthesis of NSHDL NPs, as described above.
HUVECs were plated in 6-well plates on sterile glass coverslips pre-coated with vitronectin (0.5%, 37°C overnight) at 1.5 × 105 cells ml−1, 2ml/ well. The HUVECs were allowed to attach to the coverslips, overnight. The next day, HUVECs were treated with NSHDL NPs (10 nM) with either tagged Apo A1 or NS ONs. The cells were fixed at indicated time-points by replacing culture media with 3.7% formaldehyde in PBS. After 20 min, the samples were gently washed with PBS, the nuclei counterstained with Hoechst-33258 (Santa Cruz Biotechnology, Inc.), and the slides mounted with Immu-Mount (Thermo-Scientific) medium.
The images were generated using a Nikon UV-LSM 510 Meta with an optical slice thickness of 1.0 μm.
Matrigel Plug Angiogenesis Assay
IACUC approval was obtained for animal experiments. Phenol red free, growth factor depleted Matrigel (15 mg ml−1, BD-Biosciences) was used for the following experiments. Three different methods of treatment were employed. First, HUVECs were plated at a density of 1.5 × 105 cells ml−1 in T-25 flasks (5mls/flask). The cells were pre-treated with PBS, free RNAi ON, HDL NPs, NS-HDL NPs and, RNAi-HDL NPs (HDL NPs and conjugates at 10 nM and free RNAi ON at 1 μM final concentrations). Forty-eight hours later, the cells were trypsinized, washed with PBS, and re-suspended in phenol red free, serum-free RPMI-1640. Cell suspensions were then mixed with chilled Matrigel yielding a final Matrigel concentration of 10 mg ml−1 and a final cell density of 5.0 × 105 cells ml−1. VEGF-A (Millipore) and heparin (Sigma-Aldrich) were added, to 400 ng ml−1 (final) and 60 μg ml−1 (final), respectively. Matrigel plugs were introduced into athymic nude mice (Hsdnu/nu, Harlan) by subcutaneous injection (300-400 μl/ mouse, n=4/group). Second, HUVECs at a density of 5.0 × 105 cells ml−1 and PBS, RNAi-HDL NPs (5, 10 and 20 nM) or NS-HDL NPs (10 nM) were added to Matrigel (10 mg ml−1 Matrigel, 400 ng ml−1 VEGFA, and 60 μg ml−1 heparin, final concentrations) prior to subcutaneous implantation (400μl/mouse, n = 4/group). Finally, we implanted cell-free Matrigel plugs supplemented with VEGF-A and heparin, as above. Three days later, the mice were given NS ON, RNAi ON, HDL NPs, NS-HDL NPs and RNAi-HDL NPs via tail vein injections (n = 4-5/ group, 100 μl/ injection, the free ONs at 50 μM and the HDL NPs and conjugates at 500 nM). PBS was used as a negative control. The injections were repeated 3 times, 48 hours apart. In all cases, nine days after treatment initiation, the Matrigel plugs were collected, snap frozen, and their cryosections analyzed by immunofluorescent staining for the endothelial marker, CD31. Cell nuclei were counter-stained using DAPI. Imaging and quantitative analysis were performed using Nikon Diaphot 2000 and Elements Software (Nikon). In parallel, select organs were harvested and analyzed by ICP-MS.
Tumor Induction and Systemic Treatment
IACUC approval was obtained for mouse experiments. Mouse lung cancer cells (Lewis Lung Carcinoma, LLC-1) were grown to 80% confluence, harvested, re-suspended in HBSS (107 cells ml−1) and injected subcutaneously in the flanks of athymic nude mice (Hsdnu/nu, Harlan) (106 cells in 0.1 ml). After 7 days, when the tumors reached 100-200 mm3 volume, the mice were divided into six groups (n=5/ group) and treated with PBS, NS ON, RNAi ON, HDL NPs, NS-HDL NPs and RNAi-HDL NPs (100 μl/ mouse, the free ONs were used at 25 μM and the HDL NPs at 1 μM) and injected a total of 3 times via tail vein with a time interval of 72 hours between injections. The mice were sacrificed 9 days following the first treatment. Tumor volumes were measured every two days and volumes calculated as 0.52(length × width2). Tumors were resected, weighed, and then divided in half; one half was used for biochemical analysis and the other part embedded into optimum cutting temperature (OCT) compound (Tissue-Tek) for sectioning.
RNA was extracted using Qiagen RNeasy Mini Kit following manufacturer's protocol and converted into cDNA using TaqMan Gold RT kit, with 1 μg total RNA per 20 μl reaction mix. Mouse VEGFR2 and β-actin mRNAs were amplified using TaqMan primer/probe sets as per manufacturer's protocols on an ABI-7500 Fast machine.
Cryosections (4 μM) were fixed by serial incubations at −20°C in acetone, 1:1 acetone-chloroform mix and acetone (5 min each). The fixed sections were washed with PBS (3 × 5 min at RT) and blocked for 1 hr with 2% normal rat serum (eBioscience) in PBS at RT. After blocking, the sections were incubated for 1 hr in FITC-conjugated rat anti-mouse CD31 antibody (cat. 11-0311-82, eBioscience) (1:50 dilution in blocking solution). The sections were then washed 3 × 5 min with PBS and mounted with Fluormount G mounting medium with DAPI (eBioscience) for nuclear counterstaining. Imaging was performed using Nikon Eclipse TE2000-U to visualize the CD31 positive blood vessels in each section. The binary vascular area was assessed using Elements software.
Statistical Analysis
All of the error bars within this study, unless otherwise mentioned, reflect standard error of the study sample set, which is calculated as σ/√n where σ refers to standard deviation of the sample group and n refers to the number of replicates in the sample group. Statistical significance was calculated using two-tailed Student's T-test unless otherwise stated. The differences with p-values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
C.S.T. would like to thank the Howard Hughes Medical Institute (HHMI) for a Physician Scientist Early Career Award, grant funding from the Department of Defense/Air Force Office of Scientific Research (FA95501310192), and grant funding from the National Institutes of Health/National Cancer Institute (U54CA151880 and R01CA167041). O.V. acknowledges NEI R24 EY022883 and R01 CA172669. Imaging work was performed at the Northwestern University Cell Imaging Facility supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
Footnotes
Supporting Information.
Supporting Table S1, Supporting Figures S1 – S12. This material is available from the Wiley Online Library.
References
- 1.Davidson BL, McCray PB., Jr. Nature Reviews Genetics. 2011;12(5):329–340. doi: 10.1038/nrg2968. DOI 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitehead KA, Langer R, Anderson DG. Nature Reviews Drug Discovery. 2010;9(5):412–412. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Nature Biotechnology. 2007;25(10):1149–1157. doi: 10.1038/nbt1339. DOI 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 4.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Nature Cell Biology. 2011;13(4):423–U182. doi: 10.1038/ncb2210. DOI 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMahon KM, Mutharasan RK, Tripathy S, Veliceasa D, Bobeica M, Shumaker DK, Luthi AJ, Helfand BT, Ardehali H, Mirkin CA, Volpert O, Thaxton CS. Nano Letters. 2011;11(3):1208–1214. doi: 10.1021/nl1041947. DOI 10.1021/nl1041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahzad MMK, Mangala LS, Han HD, Lu CH, Bottsford-Miller J, Nishimura M, Mora EM, Lee JW, Stone RL, Pecot CV, Thanapprapasr D, Roh JW, Gaur P, Nair MP, Park YY, Sabnis N, Deavers MT, Lee JS, Ellis LM, Lopez-Berestein G, McConathy WJ, Prokai L, Lacko AG, Sood AK. Neoplasia. 2011;13(4):309–U142. doi: 10.1593/neo.101372. DOI 10.1593/neo.101372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Chen J, Ding L, Jin H, Lovell JF, Corbin IR, Cao W, Lo P-C, Yang M, Tsao M-S, Luo Q, Zheng G. Small. 2010;6(3):430–437. doi: 10.1002/smll.200901515. DOI 10.1002/smll.200901515. [DOI] [PubMed] [Google Scholar]
- 8.Lin QY, Chen J, Jin HL, Ng KK, Yang M, Cao WG, Ding LL, Zhang ZH, Zheng G. Nanomedicine. 2012;7(12):1813–1825. doi: 10.2217/nnm.12.73. DOI 10.2217/nnm.12.73. [DOI] [PubMed] [Google Scholar]
- 9.Lin Q, Chen J, Zhang Z, Zheng G. Nanomedicine. 2014;9(1):105–120. doi: 10.2217/nnm.13.192. DOI 10.2217/nnm.13.192. [DOI] [PubMed] [Google Scholar]
- 10.Lin Q, Chen J, Jin H, Ng KK, Yang M, Cao W, Ding L, Zhang Z, Zheng G. Nanomedicine. 2012;7(12):1813–1825. doi: 10.2217/nnm.12.73. DOI 10.2217/nnm.12.73. [DOI] [PubMed] [Google Scholar]
- 11.Rohrer L, Ohnsorg PM, Lehner M, Landolt F, Rinninger F, von Eckardstein A. Circulation Research. 2009;104(10):1142–U38. doi: 10.1161/CIRCRESAHA.108.190587. DOI 10.1161/circresaha.108.190587. [DOI] [PubMed] [Google Scholar]
- 12.Dev IK, Dornsife RE, Hopper TM, Onori JA, Miller CG, Harrington LE, Dold KM, Mullin RJ, Johnson JH, Crosby RM, Truesdale AT, Epperly AH, Hinkle KW, Cheung M, Stafford JA, Luttrell DK, Kumar R. British Journal of Cancer. 2004;91:1391–1398. doi: 10.1038/sj.bjc.6602109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strawn LM, McMahon G, App H, Schreck R, Kuchler WR, Longhi MP, Hui TH, Tang C, Levitzki A, Gazit A, Chen I, Keri G, Orfi L, Risau W, Flamme I, Ullrich A, Hirth KP, Shawver LK. Cancer Res. 1996;56(15):3540–3545. [PubMed] [Google Scholar]
- 14.Wicki A, Rochlitz C, Orleth A, Ritschard R, Albrecht I, Herrmann R, Christofori G, Mamot C. CLINICAL CANCER RESEARCH. 2012;18:454–464. doi: 10.1158/1078-0432.CCR-11-1102. DOI 10.1158/1078-0432.CCR-11-1102. [DOI] [PubMed] [Google Scholar]
- 15.Folkman J. SEMINARS IN ONCOLOGY. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. DOI 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 16.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. Journal of Cell Biology. 2003;161(6):1163–1177. doi: 10.1083/jcb.200302047. DOI 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrara N. The oncologist. 2004;9(Suppl 1):2–10. doi: 10.1634/theoncologist.9-suppl_1-2. [DOI] [PubMed] [Google Scholar]
- 18.Csaky K, Do DV. American journal of ophthalmology. 2009;148(5):647–56. doi: 10.1016/j.ajo.2009.06.014. DOI 10.1016/j.ajo.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Dawood S, Shaikh AJ, Buchholz TA, Cortes J, Cristofanilli M, Gupta S, Gonzalez-Angulo AM. Cancer. 2012;118(11):2780–6. doi: 10.1002/cncr.26579. DOI 10.1002/cncr.26579. [DOI] [PubMed] [Google Scholar]
- 20.Omuro AM. Curr Opin Investig Drugs. 2008;9(12):1287–95. [PubMed] [Google Scholar]
- 21.Larkin JM, Chowdhury S, Gore ME. Nature clinical practice. Oncology. 2007;4(8):470–9. doi: 10.1038/ncponc0901. DOI 10.1038/ncponc0901. [DOI] [PubMed] [Google Scholar]
- 22.Batchelor TT, Sorensen AG, di Tomaso E, Zhang W-T, Duda DG, Cohen KS, Kozak KR, Cahill DP, Chen P-J, Zhu M, Ancukiewicz M, Mrugala MM, Plotkin S, Drappatz J, Louis DN, Ivy P, Scadden DT, Benner T, Loeffler JS, Wen PY, Jain RK. Cancer cell. 2007;11(1):83–95. doi: 10.1016/j.ccr.2006.11.021. DOI 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood JM, Bold G, Buchdunger E, Cozens R, Ferrari S, Frei J, Hofmann F, Mestan J, Mett H, O'Reilly T, Persohn E, Rosel J, Schnell C, Stover D, Theuer A, Towbin H, Wenger F, Woods-Cook K, Menrad A, Siemeister G, Schirner M, Thierauch KH, Schneider MR, Drevs J, Martiny-Baron G, Totzke F, Marme D. Cancer Res. 2000;60(8):2178–2189. [PubMed] [Google Scholar]
- 24.Wedge SR, Ogilvie DJ, Dukes M, Kendrew J, Curwen JO, Hennequin LF, Thomas AP, Stokes ESE, Curry B, Richmond GHP, Wadsworth PF. Cancer Res. 2000;60(4):970–975. [PubMed] [Google Scholar]
- 25.Bair SM, Choueiri TK, Moslehi J. Trends in Cardiovascular Medicine. 2013;23(4):104–113. doi: 10.1016/j.tcm.2012.09.008. DOI http://dx.doi.org/10.1016/j.tcm.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.des Guetz G, Uzzan B, Chouahnia K, Morere JF. Targeted oncology. 2011;6(4):197–202. doi: 10.1007/s11523-011-0204-7. DOI 10.1007/s11523-011-0204-7. [DOI] [PubMed] [Google Scholar]
- 27.Izzedine H, Massard C, Spano JP, Goldwasser F, Khayat D, Soria JC. Eur J Cancer. 2010;46(2):439–48. doi: 10.1016/j.ejca.2009.11.001. DOI 10.1016/j.ejca.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Roodhart JM, Langenberg MH, Witteveen E, Voest EE. Current clinical pharmacology. 2008;3(2):132–43. doi: 10.2174/157488408784293705. [DOI] [PubMed] [Google Scholar]
- 29.Bukowski RM. Frontiers in oncology. 2012;2:13. doi: 10.3389/fonc.2012.00013. DOI 10.3389/fonc.2012.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabebe E, Wakelee H. Current treatment options in oncology. 2007;8(1):15–27. doi: 10.1007/s11864-007-0022-4. DOI 10.1007/s11864-007-0022-4. [DOI] [PubMed] [Google Scholar]
- 31.Hassan HH, Denis M, Krimbou L, Marcil M, Genest J. The Canadian journal of cardiology. 2006;22(Suppl B):35B–40B. doi: 10.1016/s0828-282x(06)70985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norata GD, Ongari M, Uboldi P, Pellegatta F, Catapano AL. International journal of molecular medicine. 2005;16(4):717–22. [PubMed] [Google Scholar]
- 33.Katoh M. International journal of molecular medicine. 2013;32(4):763–7. doi: 10.3892/ijmm.2013.1444. DOI 10.3892/ijmm.2013.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellou S, Pentheroudakis G, Murphy C, Fotsis T. Cancer letters. 2013;338(2):219–28. doi: 10.1016/j.canlet.2013.05.015. DOI 10.1016/j.canlet.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Roland CL, Dineen SP, Lynn KD, Sullivan LA, Dellinger MT, Sadegh L, Sullivan JP, Shames DS, Brekken RA. Molecular Cancer Therapeutics. 2009;8(7):1761–1771. doi: 10.1158/1535-7163.MCT-09-0280. DOI 10.1158/1535-7163.mct-09-0280. [DOI] [PubMed] [Google Scholar]
- 36.Valacchi G, Sticozzi C, Lim Y, Pecorelli A. Annals of the New York Academy of Sciences. 2011;1229:E1–7. doi: 10.1111/j.1749-6632.2011.06205.x. DOI 10.1111/j.1749-6632.2011.06205.x. [DOI] [PubMed] [Google Scholar]
- 37.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Science (New York, N.Y.) 1996;271(5248):518–20. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 38.Feng M, Cai Q, Shi X, Huang H, Zhou P, Guo X. Journal of drug targeting. 2008;16(6):502–8. doi: 10.1080/10611860802200938 . DOI 10.1080/10611860802200938. [DOI] [PubMed] [Google Scholar]
- 39.Fenske SA, Yesilaltay A, Pal R, Daniels K, Barker C, Quinones V, Rigotti A, Krieger M, Kocher O. J Biol Chem. 2009;284(9):5797–806. doi: 10.1074/jbc.M808211200. DOI 10.1074/jbc.M808211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mooberry LK, Nair M, Paranjape S, McConathy WJ, Lacko AG. Journal of drug targeting. 2010;18(1):53–8. doi: 10.3109/10611860903156419. DOI 10.3109/10611860903156419. [DOI] [PubMed] [Google Scholar]
- 41.Rigotti A, Miettinen HE, Krieger M. Endocrine reviews. 2003;24(3):357–87. doi: 10.1210/er.2001-0037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.