Abstract
The in vivo analysis of Drosophila using genetics, with almost a hundred year history, has produced an immense body of knowledge about biology. In vitro analysis, while arguably the poor cousin to its in vivo relative, has a utility—in biochemical analyses and in cell-based screening, for example, with RNAi. A major block to the development of in vitro analysis has been the lack of an efficient genetic method to derive cell lines from mutant Drosophila strains. We recently discovered that expression of activated Ras (RasV12) provides cells in vitro with both a survival and a proliferative advantage and hence promotes the generation of cell lines.1 In this addendum, we provide new data describing the genesis of seven cell lines corresponding to a rumi mutant, which demonstrate that the method can be used to derive lines and study genetic mutants in vitro.
Keywords: Drosophila, cell line, cell culture, in vitro, rumi, fibroblast, Ras
The first Drosophila melanogaster cell lines were established in the period between 1969 and 1972, and include the widely used Kc and S2 lines.2–4 These and other lines arose spontaneously from primary cultures of embryos, presumably from rare cells that acquired mutations and continued to proliferate. This process entails a long latency period and only a small fraction of primary cultures progress to cell lines. For these reasons, there is a clear need to develop directed genetic approaches for making fly cell lines. We discovered that expression of activated Ras (RasV12) jumpstarts the production of cell lines and also increases the probability that primary cultures develop into cell lines.1 This makes it possible to design and create cell lines for specific purposes, including those from mutant genotypes. Here we provide a proof of the method by highlighting a case in which we derived seven cell lines corresponding to a null mutant of rumi, which encodes a glycosyltransferase required for Notch signaling in Drosophila..5
Recombinant chromosomes were created in which the null rumi26 mutation (FBal0216685) was combined with an Act5C-Gal4 (FBti0012292) transgene or a UAS-RasV12 transgene.6 Heterozygous flies carrying these chromosomes were crossed and the embryos were used to establish seven primary cultures (Fig. 1). The primary cultures were composed of cells with four different genotypes, but homozygous mutant rumi26 embryos are the only ones in which RasV12 is expressed (box in Fig. 1). Expression of RasV12 clearly provides a strong advantage for the rumi26 homozygous mutant cells, because in the seven lines we made only one line had rare non-mutant cells (see below). With a GFP marker on the balancer chromosomes and an embryo sorter, the desired embryos could be selected, but in practice it seems this step is unnecessary.
Figure 1.
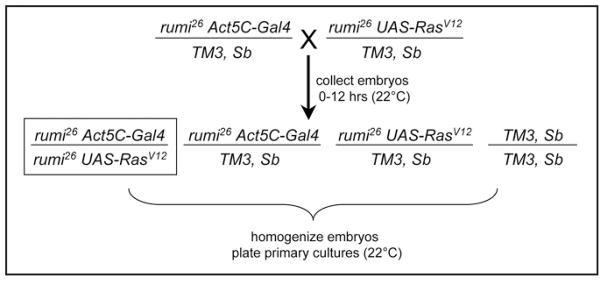
Genetic cross to produce rumi26 mutant embryos expressing activated Ras (RasV12). Primary cultures were established from a mixture of embryos of all four genotypes. The boxed embryos are the desired genotype. The UAS-RasV12 insertion site was mapped to 3R:7,394,981 (it is accompanied by a 151 bp deletion) within the 5′ region of CG14709. The Act-GAL4 insertion site was mapped to 3R:25,584,989 between CG1973 and kayak. The Act5C-GAL4 transgene is inserted in sequence corresponding to a transposable element and is currently being mapped further genetically.
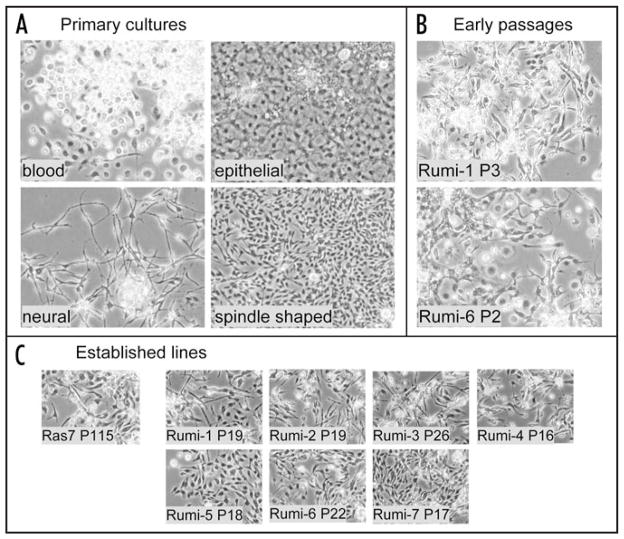
We charted the course of the cultures over time and found that like primary cultures of RasV12 alone,1 the rumi26 RasV12 primary cultures became confluent in about three weeks and all went on to produce cell lines (Fig. 2). Confluent primary cultures were split 1 in 2 and grown to confluence again. We continued to split the cells in this way, as higher dilutions sometimes resulted in poor growth. The time between passages was more variable initially but became similar by about passage 10 (Fig. 2). This could reflect either the accumulation of additional genetic changes or selection for cell types that are well adapted to growth in vitro. The variety of cell types seen in the cultures did indeed change with time. Although many cell types were present in primary cultures (Fig. 3A), even in early passages spindle-shaped cells were dominant (Fig. 3B), with only rare patches of epithelial cells. Each of the seven established cell lines was comprised of spindle-shaped cells (Fig. 3C).
Figure 2.
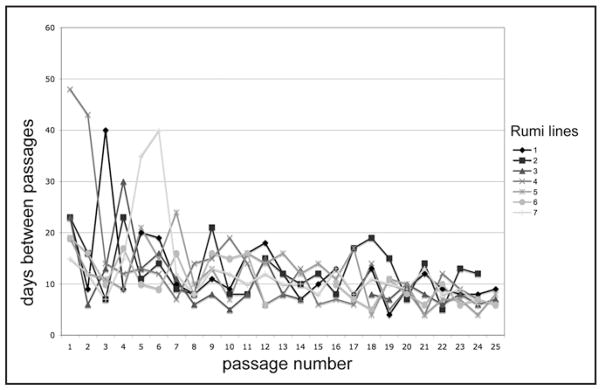
Landmarks in passaging history. The plot shows when each line (Rumi 1–7) was passaged starting at the time the primary culture was confluent (Passage 1). The primary cultures were confluent in about three weeks (line 4 took longer because the primary culture was sparse). Cells in confluent cultures were harvested and half were seeded into a new flask (1 in 2 dilution). The number of days between passages was more variable in early passages (P1–10) than in later passages. All lines have now been passaged 24 or more times and the time between passages is 5–12 days. In the lines that have now been passaged 25 or more times the cells can be split at higher dilutions (1 in 3 or 1 in 4).
Figure 3.
Evolution of primary cultures to cell lines of spindle-shaped cells. (A) Examples of cell types from primary cultures. (B) In early passages, spindle-shaped cells are the prevalent type. (C) Established Rumi cell lines and a control (Ras7) cell line are comprised of similar spindle-shaped cells. (P, passage; all panels are phase contrast images at the same magnification).
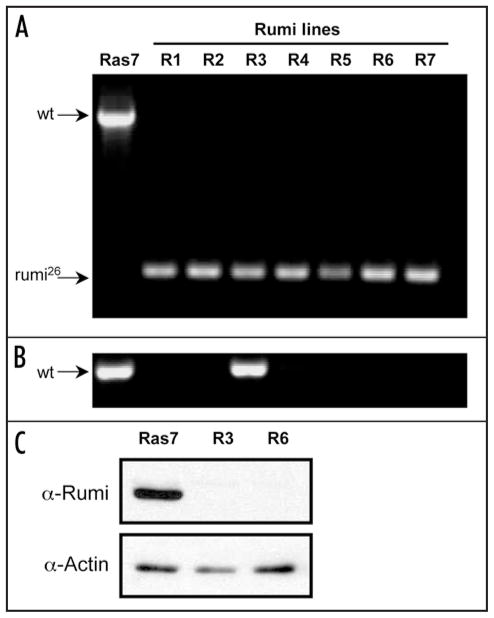
The primary cultures were initiated from cells of four genotypes (Fig. 1) but PCR-based genotype analysis showed that with one exception, Rumi-line 3, the cells in the lines were rumi26 homozygotes (Fig. 4A and B). Even in Rumi-line 3, western analysis showed that Rumi protein was undetectable, suggesting that non-mutant cells must form a small fraction of the cell population (Fig. 4C). Two other lines tested by western blotting were also negative for Rumi (Fig. 4C and not shown). Thus we conclude that even without an embryo selection step prior to setting up the primary cultures, essentially pure lines of mutant cells are produced, most probably because the desired genotype has a significant growth advantage due to the expression of RasV12.
Figure 4.
Cell lines are deficient for Rumi. Diagnostic PCR tests and Western blotting were used to genotype cells in the Rumi lines and determine their protein expression levels. Together the results show that 6/7 Rumi lines were pure populations of rumi26 homozygous cells. One line had a fraction of non-mutant cells but this fraction is assumed to be low because the cells had undetectable levels of Rumi protein. (A) Control (Ras7) cells have a diagnostic band corresponding to the wild-type rumi gene (1832 bp), whereas, each of the 7 Rumi lines (R-1 through R-7) have a 344 bp band diagnostic for the rumi26 deletion. This shows that most cells in the Rumi lines are homozygous for rumi26. (B) In order to detect rare non-homozygous mutant cells, another PCR test was performed using primers that only amplify the wild-type gene. Both control Ras7 and Rumi line 3 cells had the expected 615 bp fragment. This shows that some fraction of cells in Rumi line-3 have a wild type rumi gene. (C) Western blotting was used to analyze Rumi protein in control (Ras7) and Rumi line 3 and 6 cells (R3 and R6). Only the control cells expressed detectable levels of Rumi protein. Actin was used as a control to show that similar levels of protein were loaded.
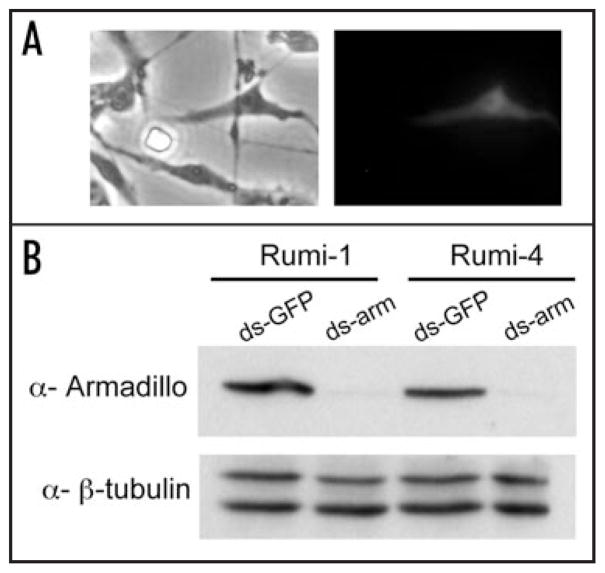
Importantly, we also found the cells could be transfected and are susceptible to RNAi. The five lines we tested for transfection all showed expression from a MAPK-YFP plasmid (Fig. 5A and not shown) and the two lines we tested for RNAi showed substantial knockdown of Armadillo using a dsRNA directed against the gene (Fig. 5B).7
Figure 5.
Rumi cells can be transfected and are susceptible to RNAi. (A) The left panel shows a phase contrast image and the right panel shows a fluorescent image. One Rumi line-5 cell in the field is expressing MAPK-YFP. Cells were transfected with an Act5C-ERK-YFP plasmid8 using Effectene (Qiagen) and assayed for GFP expression 48 hours after transfection. (B) Western blot of Rumi-1 and Rumi-4 cells treated with either a control ds-GFP RNA or ds-arm RNA.7 The ds-arm RNA substantially reduces the level of Armadillo protein in cells from both cell lines. β-tubulin levels show similar amounts of proteins were loaded. PCR products corresponding to armadillio and GFP were used in an in vitro transcription reaction (Megascript, Ambion) to generate double stranded RNA. Rumi cells were seeded in 6-well plates 24 hrs prior to transfection with 30 μg dsRNA using Effectene (Qiagen). Cells were assayed after 48 hrs by Western blotting with monoclonal antibodies against Armadillo (1/100, N2 7A1, Developmental Studies Hybridoma Bank) and polyclonal antibodies against β-tubulin (1/2000 SC33749, Santa Cruz Biotechnology).
This study shows that cell lines of mutant genotype can be efficiently established in Drosophila. The method should be useful for any cell-viable mutant (and presumably also for temperature-sensitive alleles of cell-lethal mutations) and will allow an investigator to examine mutant phenotypes in vitro beyond the normal lifespan of the mutant animal, much in the way that mouse embryonic fibroblasts (MEFs) corresponding to knock out mutants are used. These cultured fly cells will provide homogenous samples that can be manipulated with precision and produced on a large scale for biochemistry and RNAi screens. The timeframe to derive lines that can produce cells in sufficient numbers for use in such experiments is about six months.
MEFs are derived from mouse embryos that have been dissected to remove major organs, the head, and as much blood as possible. The cells that grow out are fibroblasts, which can be passaged about 5–10 times. MEFs can also be immortalized either spontaneously, as in the rare cells surviving crisis, or directly by manipulating oncogenes or tumor suppressors. In primary cultures of Drosophila wild-type embryos, patches of proliferating fibroblast-like cells populate the cultures after a few weeks, but these are transient. In rare, later stage cultures, a new population of cells appears and can give rise to continuous lines. Given the similarities, we suggest that these cells are the fly equivalent of MEFs and that expression of activated Ras promotes their survival and extends their life in vitro. This ‘diary’ of seven rumi mutant cell lines demonstrates one practical application of the method—designer cell lines. But it also poses new questions: are these cells indeed fly fibroblasts (DEFs)? And if yes, what is their origin and biology?
Acknowledgments
We thank Adam Friedman and Norbert Perrimon for the Act5C-MAPK-YFP plasmid, the Bloomington Stock center and Gerry Rubin for flies, and Yi-Dong Li for technical assistance. The anti-Armadillo monoclonal antibody developed by Eric Wieschaus was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was supported by the NIH (GM071856 to Amanda Simcox and GM084135 to Hamed Jafar-Nejad). Work in Hamed Jafar-Nejad’s lab is also supported by a Basil O’Connor Starter Scholar Research Award from the March of Dimes.
References
- 1.Simcox A, Mitra S, Truesdell S, Paul L, Chen T, Butchar JP, Justiniano S. Efficient genetic method for establishing Drosophila cell lines unlocks the potential to create lines of specific genotypes. PLoS Genet. 2008;4:1000142. doi: 10.1371/journal.pgen.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–65. [PubMed] [Google Scholar]
- 3.Echalier G, Ohanessian A. In vitro culture of Drosophila melanogaster embryonic cells. In Vitro. 1970;6:162–72. doi: 10.1007/BF02617759. [DOI] [PubMed] [Google Scholar]
- 4.Echalier G. Drosophila cells in culture. New York: Academic Press; 1997. [Google Scholar]
- 5.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–58. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Baeg GH, Selva EM, Goodman RM, Dasgupta R, Perrimon N. The Wingless morpho-gen gradient is established by the cooperative action of Frizzled and Heparan Sulfate Proteoglycan receptors. Dev Biol. 2004;276:89–100. doi: 10.1016/j.ydbio.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–4. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]