SUMMARY
MADS-box transcription factor AGL21 is responsive to several phytohormones as well as environmental cues and positively regulates auxin accumulation in lateral root primordia and lateral roots by enhancing local auxin biosynthesis, thus stimulating lateral root initiation and growth. Therefore, AGL21 may be involved in various environmental and physiological signals-mediated lateral root development.
Key words: MADS, root system architecture, lateral root, AGL21, auxin, nitrate, sulfate.
Abstract
Plant root system morphology is dramatically influenced by various environmental cues. The adaptation of root system architecture to environmental constraints, which mostly depends on the formation and growth of lateral roots, is an important agronomic trait. Lateral root development is regulated by the external signals coordinating closely with intrinsic signaling pathways. MADS-box transcription factors are known key regulators of the transition to flowering and flower development. However, their functions in root development are still poorly understood. Here we report that AGL21, an AGL17-clade MADS-box gene, plays a crucial role in lateral root development. AGL21 was highly expressed in root, particularly in the root central cylinder and lateral root primordia. AGL21 overexpression plants produced more and longer lateral roots while agl21 mutants showed impaired lateral root development, especially under nitrogen-deficient conditions. AGL21 was induced by many plant hormones and environmental stresses, suggesting a function of this gene in root system plasticity in response to various signals. Furthermore, AGL21 was found positively regulating auxin accumulation in lateral root primordia and lateral roots by enhancing local auxin biosynthesis, thus stimulating lateral root initiation and growth. We propose that AGL21 may be involved in various environmental and physiological signals-mediated lateral root development and growth.
INTRODUCTION
Root systems are crucial for plant survival, responsible for acquisition of water and mineral nutrients and anchorage, and contributing to competitive fitness in the changing environment (Lloret and Casero, 2002). In order to adapt to the changing environment, plant root system architecture (RSA) is highly plastic, responding to various environmental cues, such as soil matrix heterogeneity (Hodge, 2006), distribution of nutrients in the soil (Leyser and Fitter, 1998), and biotic interactions (Osmont et al., 2007). These extrinsic signals trigger intrinsic molecular mechanisms that have profound impacts on RSA through regulating cell division and cell differentiation processes within the root (Malamy and Ryan, 2001; Wolters and Jurgens, 2009). This allows the immobile plants to initiate root growth, such as root-hair formation, primary root (PR) growth, and lateral root (LR) formation, to greatly increase the total surface area and mechanical strength of the root system and allow the plant to efficiently adapt to environmental constraints (Deak and Malamy, 2005; Malamy, 2005; Ariel et al., 2010). Well-developed root systems have been reported beneficial for enhancing plant water and nutrient uptake and dehydration avoidance, thus increasing yield under soil-related stresses (Lynch, 2007; Serraj et al., 2009). The design of sustainable cropping systems with high yield can be achieved if sufficient knowledge about root development is available.
In response to diverse environmental signals, plants adjust their growth and development through the perception and integration of external signals into the signaling pathways of plant hormones, such as auxin, cytokinin, abscisic acid (ABA), and jasmonic acid (JA) (Lopez-Bucio et al., 2003; Malamy, 2005; De Smet et al., 2006b; Kazan, 2013). As physiological signals and hormonal factors interact with each other to modulate root development in which auxin, its polar transport, and local biosynthesis appear to have emerged as central regulators (De Smet et al., 2006a; Fukaki and Tasaka, 2009; Lavenus et al., 2013). Recent studies have shown that proper auxin transport, biosynthesis, and signaling control various steps of LR development from priming to initiation, patterning, and emergence (Fukaki et al., 2007; Lavenus et al., 2013).
Transcription factors (TFs) are known to be important for root development (Montiel et al., 2004). The MADS (MCM1/AGAMOUS/DEFICIENS/SRF) box TF family genes play important roles in controlling plant and animal development (Messenguy and Dubois, 2003). These TFs have been classified into two types (type I and type II) based on sequence relationships and structural features (Alvarez-Buylla et al., 2000b). There are 45 type II genes, which are also referred to as MIKC TFs for the four domains (M, I, K, C) they contain (Alvarez-Buylla et al., 2000b; Kaufmann et al., 2005). Plant MIKC TFs have been mostly characterized as regulators of the flowering time (Samach et al., 2000) and flower, seed, and fruit development (Saedler et al., 2001; Pinyopich et al., 2003; de Folter et al., 2006; Ripoll et al., 2011). However, some of these TFs are also expressed in various organs and vegetative tissues, such as endosperm, pollen, guard cells, trichomes, and roots, where they may have more specific functions (Alvarez-Buylla et al., 2000a; Arora et al., 2007). It has been reported that at least 50 MADS-box genes are expressed in Arabidopsis roots (Burgeff et al., 2002; Parenicova et al., 2003). However, the functions of these TFs in roots are largely unknown. Recently, XAL1/AGL12 and XAL2/AGL14 were reported to have roles in regulating PR growth (Tapia-Lopez et al., 2008; Garay-Arroyo et al., 2013). ANR1 is so far the only member of the family reported as being involved in LR development (Zhang and Forde, 1998). Gan et al. (2005) compared the responsiveness of ANR1 and 11 other root-expressed MADS-box genes to the availability of nitrogen (N), phosphorus (P), and sulfur (S), and found that seven of the them responded to N in a manner similar to ANR1 but less strongly, suggesting possible roles of these genes in nutritional regulation of LR growth.
In this study, we report that the MADS-box gene AGL21 is involved in LR development. agl21 mutant alleles had fewer and shorter LRs, while overexpression of AGL21 increased both the number and length of LRs compared with the wild-type. Further analyses showed that AGL21 was expressed in silique, flower, and seed, but mainly in roots, with higher levels during LR formation, from young lateral root primordia (LRPs) to emerged LRs. Furthermore, AGL21 responds to many hormones, including indole-3-acetic acid (IAA), methyl jasmonate (MeJA), ABA, as well as many environmental stresses such as nutrient starvations. More importantly, our data show that AGL21 positively regulates auxin accumulation and cell division activities in LRPs and LRs, suggesting that AGL21 is likely to regulate LR formation and growth by integrating multiple external and physiological signals into auxin signaling.
RESULTS
AGL21 Is a Positive Regulator of LR Development
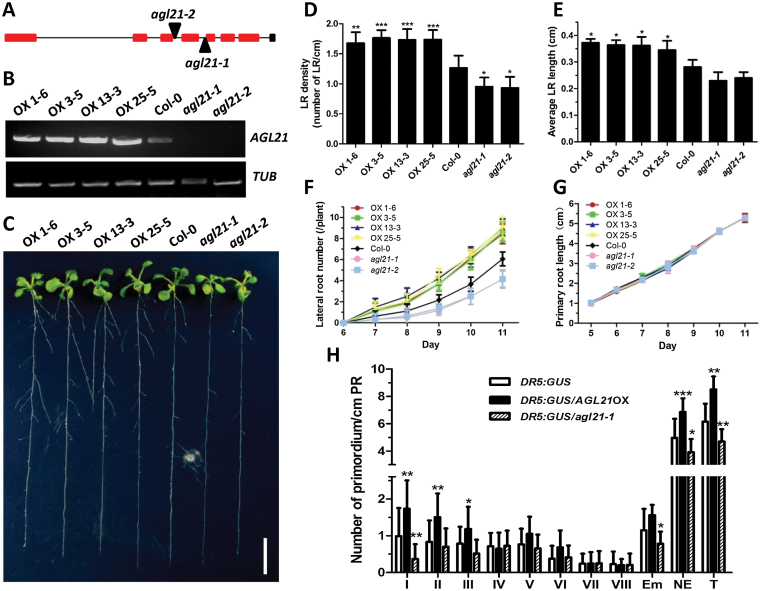
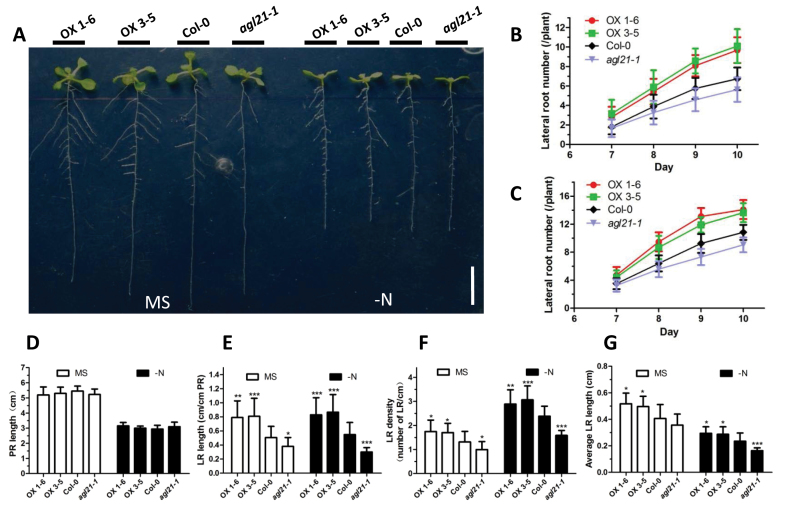
To gain insight into the function of AGL21, we generated 35S::AGL21 transgenic Arabidopsis plants, and obtained two T-DNA insertion mutants: CS118325 (agl21-1) and GK_157C08 (agl21-2) (Figure 1A and 1B). The effect of AGL21 on the development of the root system was examined using 35S::AGL21 (OX), agl21 mutants, and the wild-type plants vertically grown on Murashige and Skoog (MS) medium. After 12 d of growth, the AGL21 overexpression plants produced significantly better-developed root system than wild-type plants with higher LR density and longer average LR length, while the mutants produced much less and shorter LRs than wild-type plants (Figure 1C–1E). Time-course data also showed that AGL21 overexpression plants possessed an apparent advantage over LR development compared with the wild-type and mutant plants (Figure 1F). However, no obvious differences in PR length were observed between the AGL21 overexpression plants, wild-type, and the mutants (Figure 1G).
Figure 1.
AGL21 Is Involved in LR Development.
The seeds were germinated for 5 d on MS medium, and the seedlings were then transferred to MS medium for vertical growth.
(A) AGL21 gene structure with the sites of T-DNA insertion. Squares correspond to exons while lines represent introns.
(B) AGL21 transcript levels in the transgenic lines and mutants by RT–PCR analysis. TUBULIN (TUB) was used as the internal control.
(C) Root systems of 12-day-old 35S::AGL21, agl21 mutants and wild-type (WT) (Col-0) seedlings (bar = 1cm).
(D) Density of visible LRs of 12-day-old plants. Density of visible LRs is defined as visible LR number per cm PR. Values are mean ± standard deviation (SD) of three independent experiments each containing 15–20 plants per genotype. Asterisks denote Student’s t-test significance compared with WT plants: * P < 0.05; ** P < 0.01; *** P < 0.001.
(E) Average LR length of 12-day-old plants. Average LR length is defined as the ratio of total LR length over LR number. Values are the mean ± SD of three independent experiments each containing 15–20 plants per genotype. Asterisks denote Student’s t-test significance compared with WT plants: * P < 0.05.
(F, G) LR and PR growth curves of WT, agl21 mutants, and 35S::AGL21 plants.
(H) Numbers of LRP of 8-day-old seedlings at given stages. Stages of primordia were based on the classification by Malamy and Benfey (1997). Values are mean ± SD of three independent experiments each containing 15 plants per genotype and asterisks denote Student’s t-test significance compared with WT plants: * P < 0.05; ** P < 0.01; *** P < 0.001. NE, non-emerged LR; E, emerged LR; T, NE + E
For more detailed analysis, we introduced DR5:GUS reporter (Ulmasov et al., 1997) into 35S::AGL21 and agl21 mutant background by crossing and performed quantitative LPR and LR growth analysis by quantifying the number of GUS-stained loci of primordia and emerged LRs of 8-day-old seedlings. The results showed that AGL21-overexpressing plants had significantly increased GUS-stained LRP loci compared with the wild-type and mutant plants (Supplemental Figure 1). Further analysis demonstrated that AGL21 mainly affected the early stages of LRP development. The number of LRP at I to III stages was much higher in the AGL21-overexpressing plants, while the corresponding figures in the agl21 mutant background were much lower. Consequently, overexpression of AGL21 increased the numbers of non-emerged LR as well as emerged LR (Figure 1H). On the contrary, AGL21 knockout negatively affected LR development (Figure 1C–1H). These results suggest that AGL21 is a positive regulator of LR initiation and growth.
Expression Pattern of AGL21 and Subcellular Localization of AGL21 Protein
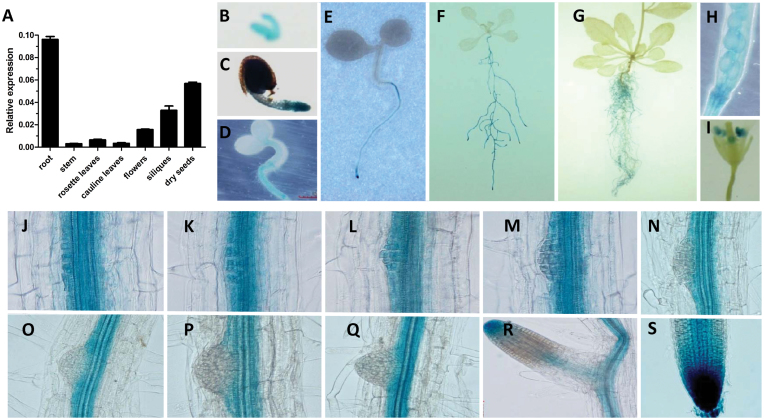
In order to investigate the expression patterns of the AGL21 in more detail in Arabidopsis, we first measured its expression levels by quantitative real-time PCR (qRT–PCR) in various organs. We detected the expression of this gene mostly in root, flowers, siliques, and dry seeds, with the strongest expression in root (Figure 2A). This result was further confirmed by using plants expressing a GUS reporter gene placed under the control of the 2.6-kb AGL21 promoter region (pAGL21::GUS). Histochemical analyses showed similar results to qRT–PCR. AGL21 was primarily expressed in the root of seedlings from germination to mature stage (Figure 2C–2G), and the expression of AGL21 in the root is mostly confined to central cylinder of the whole PR with much higher expression levels in the root tip and meristem (Figure 2E). In addition, AGL21 transcript was also detected in embryo and silique, and its expression in flower was confined to stamen-anther (Figure 2B, 2H, and 2I). More careful observation revealed that AGL21 was expressed at a higher level during LR formation, from young LRPs with one single cell layer to about three to four cell layers (Figure 2J–2M). However, hardly any expression was detected in LRPs at stages V to VIII (Figure 2N–2Q), which is consistent with the result that the effects of AGL21 on LR initiation was mainly on the early stages of LRP (Figure 1F). In emerged LR, its expression was focused on the apex and proliferative zone (Figure 2R). In the PR tip, AGL21 was expressed in all cell layers and had the strongest expression in the quiescent center (QC) area (Supplemental Figure 2). These results agree with previous reports (Burgeff et al., 2002; Parenicova et al., 2003). Taken together, our results imply that AGL21 may be involved in LR development and consistent with the phenotypes we detected in the root of AGL21 overexpression and knockout plants.
Figure 2.
AGL21 Expression Pattern.
(A) Analysis of the AGL21 expression pattern in different organs by qRT–PCR. UBQ5 was used as an internal control. Values are mean ± SD of three replica experiments.
(B–I) The expression pattern of AGL21, as revealed by promoter–GUS fusion analyses in pAGL21::GUS transgenic seedlings. GUS activity was observed in embryo (B), seedling of 2-day-old (C), seedling of 3-day-old (D), seedling of 4-day-old (E), seedling of 14-day-old (F), seedling of 35-day-old (G), silique (H), and flower (I).
(J–S) The pAGL21::GUS expressed in LRPs at stages I to VIII (J–Q), emerged LR (R), and PR tip (S). Eight-day-old pAGL21::GUS transgenic lines were used for GUS reaction for 12h.
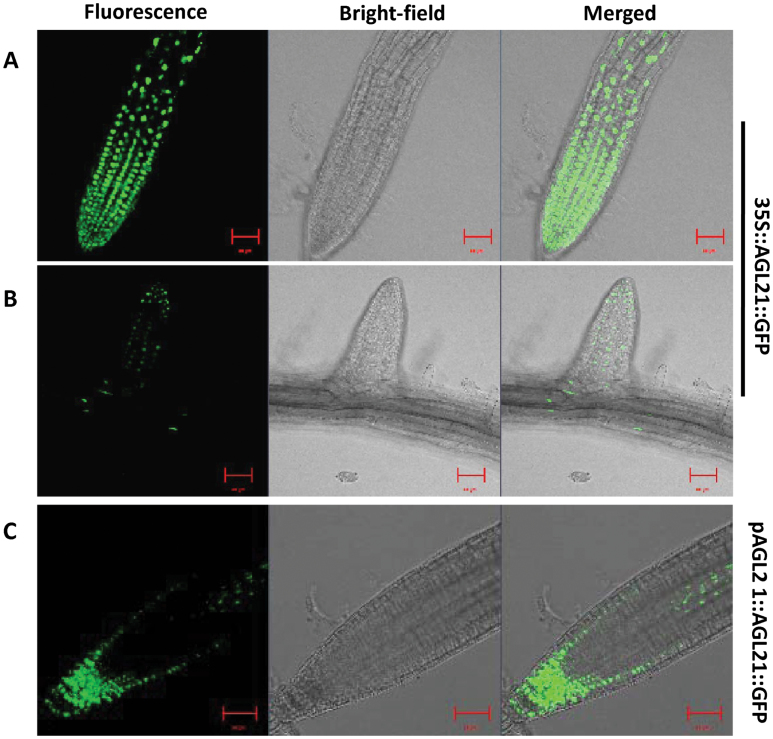
To investigate the localization of AGL21 protein in plant cell, we generate transgenic plants expressing green fluorescent protein (GFP) fusion protein under the control of 35S promoter (35S::AGL21::GFP) and its own promoter (pAGL21::AGL21::GFP). Figure 3A and 3B show that AGL21 protein targeted to the nucleus of the root cells of both PR and LR. Under the control of its own promoter, AGL21 protein is strongly expressed in the PR tip region, especially in the QC area. In the meristematic region, AGL21 protein is mainly restricted to the epidermal cell layers, while, in the elongation zone, AGL21 protein is focused in the central cylinder (Figure 3C). These data agree well with the gene expression pattern of AGL21 and the function of AGL21 in root development.
Figure 3.
Subcellular Localization of AGL21 Protein.
(A, B) Fluorescence in the root cells of transgenic plants expressing AGL21–GFP under the control of the CaMV 35S promoter (bar = 50 μm).
(C) Fluorescence in the root cells of transgenic plants expressing AGL21–GFP under the control of the 3.6-kb AGL21 promoter (bar = 50 μm).
AGL21 Is Responsive to Multiple Plant Hormones and Nutrient Deficiency
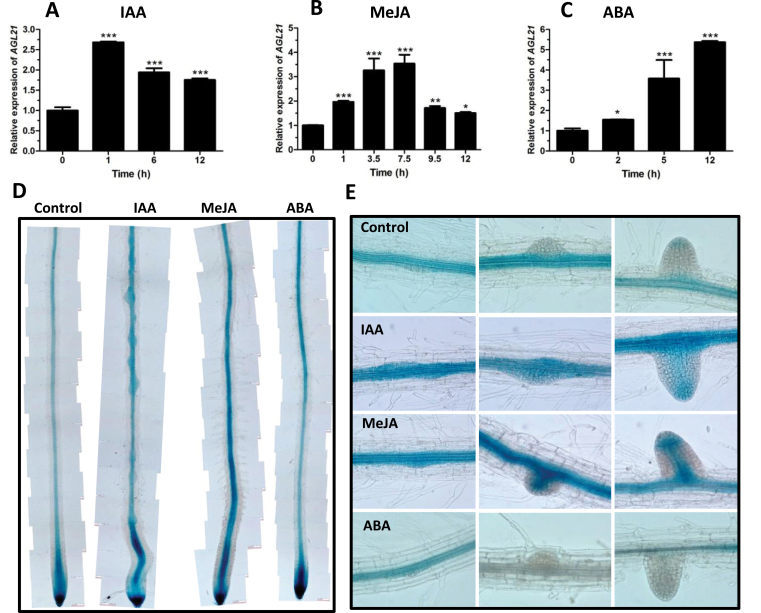
Several types of cis-acting elements, including auxin response element (AuxRE), ABA response element like (ABRE-like), G-box, GCC-box like, and JA-responsive cis-element (JARE) were found in the AGL21 promoter (Supplemental Figure 2), implicating that AGL21 could respond to various plant hormones and environmental stresses. We thus performed qRT–PCR to test this. The results showed that the expression of AGL21 was indeed up-regulated by IAA, MeJA, and ABA (Figure 4A–4C). To confirm these results, we treated the pAGL21::GUS reporter line with IAA, MeJA, and ABA. GUS staining showed that IAA and MeJA clearly induced the expression of AGL21 in the meristematic and elongation zones of PRs (Figure 4D). IAA and MeJA could enhance the expression of AGL21 in early stages of LPRs and emerged LRs and even the later stages of LRPs, where it was not expressed without hormone treatments (Figure 4E). The response of AGL21 expression to IAA was particularly intense in LRPs (Supplemental Figure 3). Interestingly, ABA could induce AGL21 expression in the PR tips, meristematic zone, and elongation zone (Figure 4D). However, in the middle and upper differentiation zone, GUS activity was diminished (Figure 4E). It should be noted that, in roots, AGL21 was preferentially enhanced by IAA and MeJA in the central cylinder, root tips, and LRPs.
Figure 4.
AGL21 Expression Is Regulated by Hormones.
(A–C) qRT–PCR analyses of AGL21 expression in wild-type seedlings during the time course after IAA (A), MeJA (B), or ABA (C) treatment. Eight-day-old Col-0 seedlings were incubated in MS liquid cultures with 10 μM IAA, 50 μM MeJA, and 20 μM ABA, respectively, and whole seedlings were harvested at indicated time points for RNA extraction and qRT–PCR analyses. The transcript levels of AGL21 were normalized to the UBQ5 expression. Values are mean ± SD and asterisks denote Student’s t-test significance compared with untreated plants: * P < 0.05; ** P < 0.01; *** P < 0.001.
(D, E) IAA, MeJA, and ABA-induced pAGL21::GUS expression in the primary root (D), LRP, and LR (E). Seven-day-old seedlings of pAGL21::GUS transgenic line grown on MS agar medium were transferred either to hormone-free MS agar medium or to MS agar medium supplemented with 10 μM IAA, 10 μM MeJA, or 10 μM ABA for 1 d, respectively. The seedlings were harvested for GUS staining for 8h.
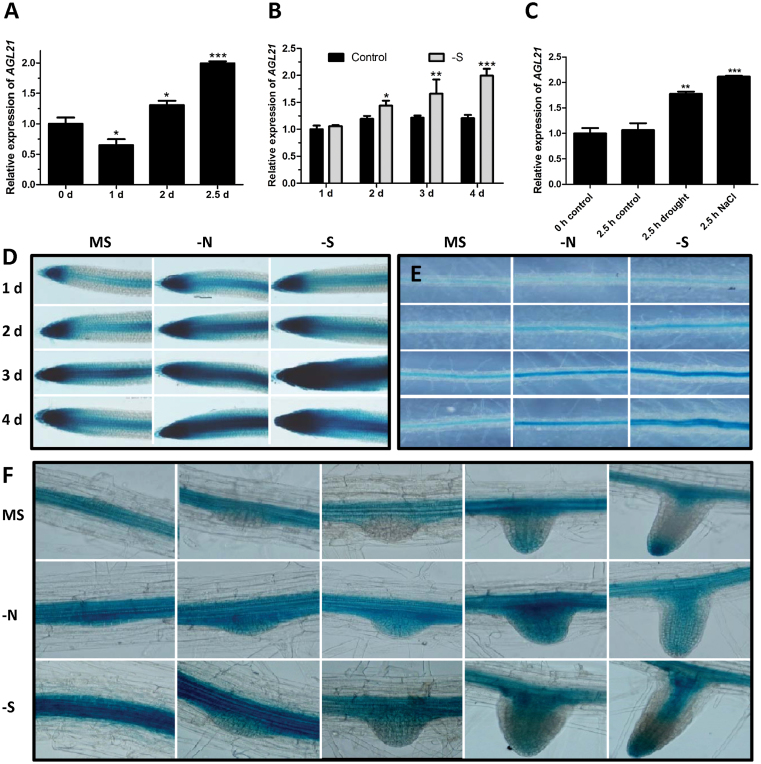
To analyze the response of AGL21 to external stresses, we performed qRT–PCR and GUS staining of pAGL21::GUS reporter line to examine the response of AGL21 to different stresses. Figure 5A and 5B indicate that AGL21 expression was induced by N or S deprivation. Interestingly, AGL21 expression was also responsive to drought and NaCl treatment (Figure 5C). Moreover, results of GUS staining demonstrated that the expression of AGL21 was clearly up-regulated in the root after 1–4 d of N or S starvation as shown in Figure 5D and 5E. Surprisingly, we found AGL21 was strongly induced in all stages of LRP and LRs after 3 d of N starvation, even the later stages of LRP where AGL21 did not express under normal conditions (Figure 5F). Induced expression of AGL21 in LRPs was also detected after 3 d of S deprivation, although less strongly compared with N deprivation treatment (Figure 5F). These results indicate that AGL21 is an important TF, at which multiple hormones and stress signals are converged to regulate LR development.
Figure 5.
Response of AGL21 to Multiple Environmental Stresses.
(A) Response of AGL21 to N starvation. Seven-day-old Col-0 seedlings were transferred to N-free nutrient solution and harvested at indicated time points for RNA extraction and qRT–PCR analyses.
(B) Response of AGL21 to S starvation. Seven-day-old Col-0 seedlings were transferred to S-free agar medium for vertical growth. Complete nutrient medium was used as control. Roots were harvested at indicated time points for RNA extraction and qRT–PCR analyses.
(C) qRT–PCR analyses of AGL21 expression in 8-day-old Col-0 seedlings after 2.5h of drought or 120mM NaCl treatment. The transcript levels of AGL21 were normalized to the UBQ5 expression. Values are mean ± SD of three replica experiments and asterisks denote Student’s t-test significance compared with the 0-h control plants: * P < 0.05; ** P < 0.01; *** P < 0.001.
(D–F) Effects of N deprivation and S deprivation on pAGL21::GUS expression in the roots. Expression of AGL21 is induced by N deprivation and S deprivation in the PR tips (D), differential zone (E) after 1–4 d of N or S starvation, and LRPs after 3 d of N or S starvation (F). Five-day-old pAGL21::GUS transgenic seedlings grown on MS medium were transferred to N- or S-free medium for 1–4 d and seedlings were harvested at the indicated time points for GUS staining for 8h.
AGL21 Is an Important Factor to Sustain LR Development under Low-N Conditions
To further study whether AGL21 is involved in environmental signals regulating LR development, we analyze the role of AGL21 in LR development in response to N availability. Six-day-old seedlings of the mutant and AGL21-overexpressing plants were transferred from MS to N-free medium and vertically grew. Visible LR number was monitored in the following days. The results clearly demonstrated that there was a significant increase in LR number per plant in the AGL21-overexpressing plants compared with the wild-type plants under both N-free and -rich conditions. In contrast, the LR number of the agl21 mutant was significantly reduced (Figure 6A–6C). However, no obvious difference was observed in the PR length (Figure 6D). Meanwhile, we also found that AGL21 affected the LR elongation under both N-rich and -free conditions. There was a slight increase in LR length per centimeter (cm) PR under the N-free conditions compared with the N-rich conditions, except in the mutant, in which the corresponding figure decreased instead (Figure 6E). However, LR length per centimeter PR is related with both LR density and LR length. Thus we checked the LR density and average LR length. Compared with the wild-type, LR density and average LR length of AGL21 overexpression plants increased by 29.6%–33.0% and 22.1%–27.2%, respectively, under N-rich conditions, 21.1%–28.4% and 22.2%–25.4%, respectively, under N-free conditions. In agl21 mutant plants, LR density decreased by 21.1%, and average LR length decreased by 12.5.0% under N-rich conditions, but, under N-free conditions, the corresponding figures of LR density and average LR length reduced by 25.1% and 30.4% compared with wild-type, respectively (Figure 6F and 6G). These results imply that AGL21 is important to both LR initiation and LR elongation, but mainly affecting LR elongation under N-restricted conditions.
Figure 6.
AGL21 Is Important for Sustaining LR Development under Low-N Conditions.
Five-day-old seedlings grown on MS medium were transferred to N-free medium and grown vertically. During the vertical growth stage, the root morphological parameters were measured.
(A) Phenotype of 11-day-old seedlings of AGL21-overexpressing, Col-0, and AGL21 knockout (bar = 1cm).
(B, C) Numbers of visible lateral roots of AGL21-overexpressing, Col-0, and AGL21 knockout plants on MS medium (B) and N-free medium (C) from the 7th day to the 10th day. Values are mean ± SD of three independent experiments each containing 15–20 plants per genotype.
(D) Average PR length of the 11-day-old plants. Values are mean ± SD of three independent experiments each containing 15–20 plants per genotype.
(E–G) Length of visible LRs per cm PR length (E), LR density (F), and average LR length (G). Average LR length defined as the ratio of total LR length over LR number. Values are mean ± SD of three independent experiments each containing 15–20 plants per genotype and asterisks denote Student’s t-test significance compared with the wild-type plants: * P < 0.05; ** P < 0.01; *** P < 0.001.
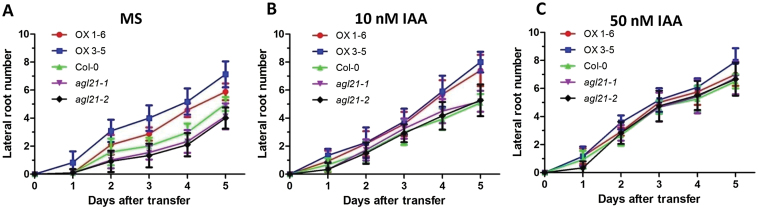
Auxin Can Rescue the Phenotype of agl21 Mutant
To test whether AGL21 regulates LR development through changing auxin concentration in the LRPs and LRs, we assayed the root phenotype by adding exogenous IAA in the medium. Without exogenous IAA, AGL21-overexpressing plants developed more LRs while the agl21 mutant plants had much fewer LRs compared with the wild-type plants (Figure 7A). However, after adding 10nM IAA, the differences in LR number between the wild-type and knockout plants diminished (Figure 7B). When exogenous IAA concentration was increased to 50nM, no differences in root number were observed (Figure 7C). These results suggest that AGL21 may regulate LR development by altering endogenous auxin concentration.
Figure 7.
LR Phenotype Is Rescued by Exogenous IAA.
Five-day-old plants grown on MS vertical agar plates were transferred to MS agar plates containing different concentrations of IAA to grow vertically for 6 d. Values are mean ± SD of three independent experiments each containing 15 plants.
(A) Time course of LR development on MS medium.
(B) Time course of LR development on MS medium supplemented with 10nM IAA.
(C) Time course of LR development on MS medium supplemented with 50nM IAA.
AGL21 Regulates Auxin Accumulation in the LRPs by Affecting Local Auxin Biosynthesis
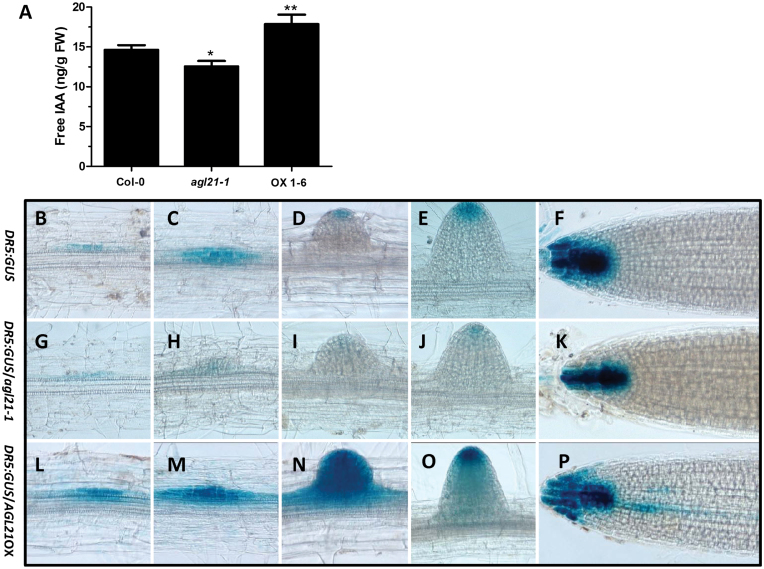
The LR phenotypes of agl21 mutant and the overexpression lines implicate that root auxin level might be affected. To verify whether endogenous auxin content was changed, we measured endogenous IAA in the root. The results in Figure 8A show that IAA content was significantly increased in the overexpression line and reduced in the knockout mutant compared with that in the wild-type.
Figure 8.
AGL21 Regulates Auxin Accumulation in the LRPs and LRs.
(A) Quantification of free IAA content in the root of 9-day-old plants. Values are mean ± SD of three replica experiments and asterisks denote Student’s t-test significance compared with the wild-type plants: * P < 0.05; ** P < 0.01.
(B–F) Expression of DR5:GUS (N = 20 plants) in three different stages of LRP (B–D), LR, and PR tips (E, F) of Col-0 plants. Nine-day-old plants grown on MS medium were used for GUS staining for 6h.
(G–K) Expression of DR5:GUS in three different stages of LRP (G–I), LR, and PR tips (J, K) of agl21 knockout plants.
(L–P) Expression of DR5:GUS in three different stages of LRP (L–N), LR, and PR tips (O, P) of AGL21-overexpressing plants.
In addition, we introduced the auxin-responsive DR5:GUS marker line into AGL21-overexpressing and agl21 knockout background by crossing to indicate endogenous auxin distribution in roots (Ulmasov et al., 1997). In the wild-type background, DR5:GUS reporter was stained in foci in LRPs and tips of PR (Figure 8B–8F). In pre-emerging and young wild-type LRs, DR5:GUS was expressed exclusively in the apex (Figure 8D and 8E). Notably, DR5:GUS in the LRPs and young LR was markedly repressed in agl21 mutant background (Figure 8G–8J). On the other hand, a dramatically increased level of GUS staining was observed in the LRPs and emerged LRs in AGL21-overexpression background (Figure 8L–8O), especially in the emerging LRs (Figure 8N). Overexpression of AGL21 did not apparently alter DR5:GUS expression in the PR tips (Figure 8P). However, the expression was slightly reduced in the PR tips of agl21 mutant (Figure 8K). Moreover, GUS activities were clearly strengthened in the leaves. In agl21 mutant background, the DR5:GUS expression was limited to the margin of young leaves but, in the AGL21 overexpression background, GUS activity extended to veins (Supplemental Figure 4). From these results, it appears that AGL21 acts as a positive regulator of auxin accumulation in the LRPs and LRs, thus resulting in more LRP initiation and faster LR growth.
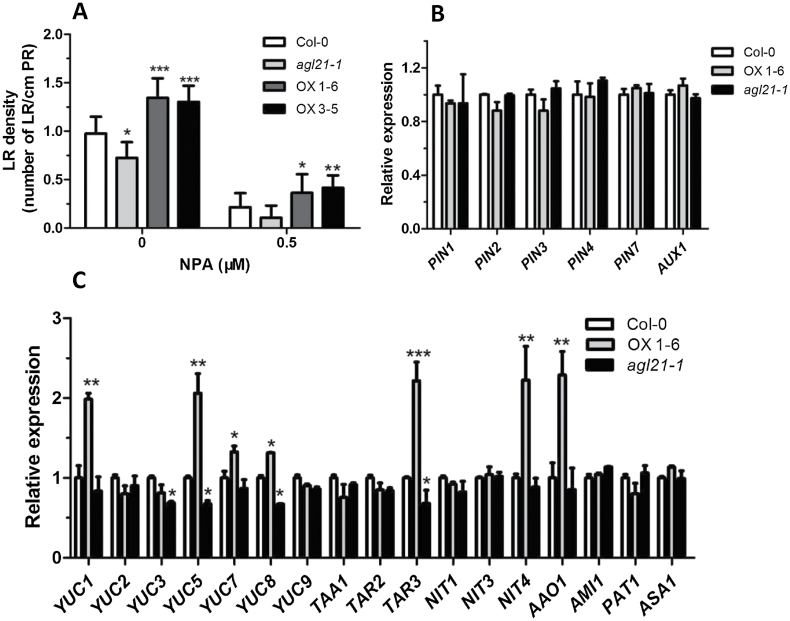
Tempo-spatial auxin accumulation is regulated by local auxin biosynthesis or/and polar transport. In order to elucidate which pathway AGL21 is involved in regulating auxin accumulation in the root, we firstly treated the seedlings with auxin transport inhibitor N-1-naphthylphthalamic acid (NPA). Results showed that NPA severely reduced LR initiation in wild-type plants and agl21 mutant plants compared with plants grown on NPA-free medium, as previously demonstrated (Reed et al., 1998). However, the AGL21 overexpression plants developed more LRs even after transfer to medium containing NPA (Figure 9A). These results indicate that the function of AGL21 in LR development is independent of polar auxin transport. The expression levels of auxin transport genes in the root also support this conclusion (Figure 9B). We then analyzed the expression levels of many auxin biosynthesis genes, including YUCCA family, TAA1 family, NIT family, and several other genes. Three of these genes, including YUC5, YUC8, and TAR3, were found to be significantly up-regulated in AGL21-overexpressing plants and down-regulated in the mutant. However, expression levels of YUC1, YUC7, NIT4, and AAO1 increased significantly in the root of AGL21 plants, but were not so significantly down-regulated in the mutant (Figure 9C). These data indicate that AGL21 may increase auxin content in LRs and LRPs via local auxin biosynthesis.
Figure 9.
AGL21 Increases Auxin Accumulation in the Root through Local Biosynthesis.
(A) Effects of auxin transport inhibitors NPA on LR initiation in wild-type, AGL21-overexpressing and mutant plants. Five-day-old seedlings were transferred to medium supplemented with DMSO alone or the auxin transport inhibitor NPA (0.5 μM) dissolved in DMSO. After 7 d of growth, the LRs produced in the new growth were counted on 30 seedlings. Values are mean ± SD of three replica experiments and asterisks denote Student’s t-test significance compared with the wild-type plants: * P < 0.05; ** P < 0.01.
(B, C) Relative expression levels of auxin transport genes (B) and auxin biosynthesis genes (C) in the roots of 9-day-old plants. The transcript levels of auxin transport or biosynthesis genes were normalized to the UBQ5 expression. The expression levels of each gene in the wild-type were set as 1.0. Values are mean ± SD of three replica experiments and asterisks denote Student’s t-test significance compared with the wild-type plants: * P < 0.05; ** P < 0.01.
AGL21 Promotes Cell Division Activities in LRs and LRPs
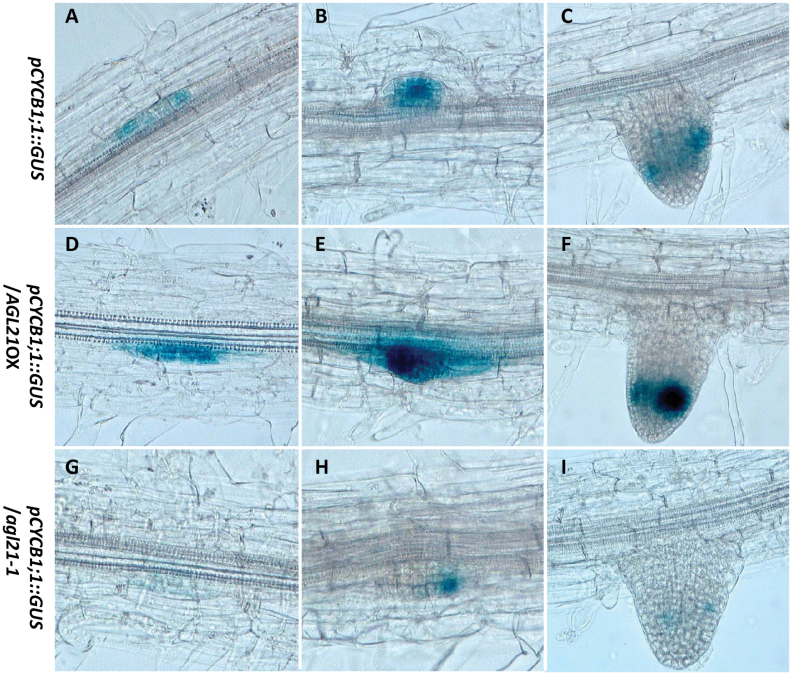
In order to determine whether AGL21 regulates cell division during the LR development, we introduced the pCYCB1;1::GUS reporter into the agl21 mutant and 35S::AGL21 background through genetic crossing. The pCYCB1;1::GUS marks the cell divisions in pericycle during LR initiation and serves as a good marker to visualize the site of LRP initiation and development (Beeckman et al., 2001; Himanen et al., 2002). Results in Figure 10 indicate that AGL21 positively regulates CYCB1;1 expression in the root. Specifically, in the AGL21-overexpressing background, GUS was strongly expressed in the LPRs (Figure 10D and 10E) and LR tips (Figure 10F) compared with that in the wild-type background (Figure 10A–10C). However, GUS expression was only found weakly in LPRs and LR tips of the agl21 mutant (Figure 10G–10I), implicating impaired initial anticlinal division of pericycle cells leading to LRP initiation and LR growth in the mutant. Therefore, we propose that AGL21 positively regulates cell division activities in the LRPs and LRs, thus promoting LR development to some extent.
Figure 10.
AGL21 Affects pCYCB1;1::GUS Expression in the LRPs and LRs.
Eight-day-old seedlings were harvested for GUS staining. The seedlings were immersed in GUS staining buffer and applied vacuum for 2min, and then incubated at 37°C overnight.
(A–C) pCYCB1;1::GUS expression in LRPs (A, B) and LR (C) of Col-0 background seedlings.
(D–F) pCYCB1;1::GUS expression in LRPs (D, E) and LR (F) of AGL21-overexpressing background seedlings.
(G–I) pCYCB1;1::GUS expression in LRPs (G, H) and LR (I) of agl21 knockout background seedlings.
DISCUSSION
Expression Pattern of AGL21 Supports Its Role in LR Development
The MADS-box gene family is generally subdivided into several well-defined monophyletic clades with typical similar expression pattern and highly related function (Becker and Theissen, 2003). In Arabidopsis, AGL17, AGL21, ANR1, together with AGL16 belong to the AGL17 clade, which are preferentially expressed in roots (Alvarez-Buylla et al., 2000a; Burgeff et al., 2002). ANR1 is the only gene in this clade known for being involved in nitrate stimulated LR development (Zhang and Forde, 1998). In Oryza sativa, four of the five AGL17-like clade genes are expressed in the central cylinder of roots, indicating potential functions in root development (Puig et al., 2013).
Like ANR1, AGL21 is primarily expressed during LR formation and embryogenesis. In situ hybridization experiments showed that AGL21 expressed in the central cylinder in the differentiated zone of the PR and young LPRs up to stage III or IV as well as emerged LRs. AGL21 was also detected in embryos from the globular stage up to the torpedo stage (Burgeff et al., 2002). In this study, we used qRT–PCR and pAGL21::GUS reporter line to analyze the expression pattern of AGL21. Our results not only agree with previous reports, but also revealed the spatiotemporal expression pattern (Figure 2), which supports that AGL21 plays an important role in LR initiation and growth.
AGL21 Regulates LR Initiation and Growth through Increasing Auxin Accumulation and Promoting Cell Division in the LRPs and LRs
The expression pattern of AGL21 implicated that it may be involved in LR development. Indeed, our subsequent study demonstrated that AGL21 is important for LR initiation and growth. Overexpression of AGL21 increases LR number and length. In contrast, AGL21 knockout results in less and shorter LRs (Figures 1 and 6). Further analysis of the GUS staining of DR5:GUS reporter plants in AGL21-overexpressing and knockout genetic background shows that AGL21 positively regulates LRP initiation, especially the I and II stage LRPs (Figure 1H). These results are in agreement with the expression pattern of AGL21 in central cylinder and young LRPs.
During LR development, auxin accumulation is one of the most important events for LR initiation as well as post-initiation events including emergence (Benkova et al., 2003; Peret et al., 2009). Our results suggest that AGL21 promotes LR development through increasing auxin accumulation during LRP initiation and in newly emerged LRs. At first, expression level of AGL21 dramatically affected LRP initiation and LR growth (Figures 1 and 6)—a process depending on the auxin-mediated establishment and activity of a new meristem (Himanen et al., 2002; Osmont et al., 2007; Nibau et al., 2008). Furthermore, the changes in auxin concentration in both LRPs and young LRs of AGL21 overexpression and knockout plants were clearly confirmed by both localization of DR5:GUS activity and IAA content measurement (Figure 8). Finally, exogenous IAA was able to rescue the phenotypes of agl21 mutant plants (Figure 7). Therefore, we propose that AGL21 affects auxin homeostasis in the LRPs and LRs in two different ways, either by increasing polar auxin transport to the initiated primordia or more likely by enhancing the local auxin biosynthesis of newly formed LRPs and LRs according to the ‘fountain’ model proposed by Benkova et al. (2003). The auxin transport inhibitor NPA can arrest LR development through blocking auxin redistribution in the root (Casimiro et al., 2001). However, AGL21-overexpressing plants still developed more LRs after NPA treatment and the transcript level of AGL21 did not affect the expression levels of auxin transport genes in the root (Figure 9A and 9B), indicating that AGL21’s promoting LR development may be not through affecting auxin transport. Instead, several auxin biosynthesis pathway genes were found up-regulated in the roots of AGL21-overexpressing plants and down-regulated to some extent in the mutant roots (Figure 9C). Therefore, taken together, our data indicate that AGL21 can enhance local auxin biosynthesis in the root to regulate LR initiation and growth.
Cell cycle activation and activity during early LR initiation are known to be regulated by auxin (Stals and Inze, 2001; Himanen et al., 2002). Thus, we crossed pCYCB1;1::GUS line with AGL21-overexpressing and knockout plants and analyzed the cell cycle activities of the offspring. Cell division activities in the LRPs and LRs of the AGL21-overexpressing plants are much higher than that of the wild-type and mutant plants (Figure 10). Taken together, these results indicate that AGL21 positively regulates the auxin accumulation in the LRPs and LRs, thus stimulating the cell proliferation activity. So we could observe the phenotypes that AGL21-overexpressing lines had more and longer LRs than the wild-type, while the mutant had opposite phenotype in the root (Figures 1 and 6).
AGL21 Responds to Multiple External and Physiological Signals and Is Likely Involved in LR Development in Response to Environmental Constraints
It is generally believed that root is the main organ to collect signals and information from the environment and incorporate them into decisions about growth and development in order to adapt to the changing environmental conditions (Comstock, 2002; Lopez-Bucio et al., 2003; Malamy, 2005; Osmont et al., 2007). So far, several genes have been reported as possible regulators of RSA to various environment signals, such as N nutrient (Zhang and Forde, 1998; Malamy and Ryan, 2001; Engineer and Kranz, 2007; Krouk et al., 2010; Vidal et al., 2010), P nutrient (Ticconi et al., 2004; Svistoonoff et al., 2007), S nutrient (Kutz et al., 2002), and osmotic stress (Deak and Malamy, 2005; Yu et al., 2013). Besides external signals, plant development also responds to all kinds of internal signals, especially the plant hormones. In fact, plants can perceive and integrate many exogenous signals into the signaling pathways of plant hormones, resulting in root architecture change (Lopez-Bucio et al., 2002; Malamy, 2005; Achard et al., 2006; Jovanovic et al., 2007). However, the underlying mechanisms controlling root system development in response to different environmental constraints are not well understood.
In Arabidopsis, several root-expressed MADS-box TFs have been reported responding to N change in a manner similar to ANR1. SOC1 was additionally found to respond to changes in the P and S supply (Gan et al., 2005). In addition, the AGL17-like clade genes in Oryza sativa had been found responding to osmotic stress, nitrate, and various hormonal treatments (Puig et al., 2013). Recently, the XAL1/AGL12 and XAL2/AGL14 genes had been proved to respond to auxin treatment (Tapia-Lopez et al., 2008; Garay-Arroyo et al., 2013). In our study, we found the expression of AGL21 was induced by hormones, such as IAA, MeJA, and ABA (Figure 4). Moreover, we found many cis-acting elements in its promoter, including AuxRE, G-box, JARE, GCC-like box, and ABRE-like (Supplemental Figure 2), which are essential for genes responding to auxin, JA, and ABA, respectively (Menkens et al., 1995; Sessa et al., 1995; Grill and Himmelbach, 1998; Ulmasov et al., 1999; Xu and Timko, 2004). Through detailed analysis of pAGL21::GUS reporter line, we revealed that IAA and MeJA treatment could dramatically up-regulate the expression of AGL21 in the root central cylinder as well as in the LRPs and LRs, even in the later-stage LRPs where AGL21 is not expressed under normal conditions (Figure 4D and 4E). The elevated AGL21 in these places may activate local auxin biosynthesis to favor LR development. However, ABA can increase AGL21 expressed in the PR tip, proliferation zone, and elongation zone, but reduces its expression in the middle and top of the differentiation zone of the PR (Figure 4D and 4E). These results are consistent with the positive roles of auxin and JA and negative roles of ABA in LR development (Woodward and Bartel, 2005; Sun et al., 2009; Raya-Gonzalez et al., 2012). Therefore, our results indicate that AGL21 may be involved in LR development regulated by hormone signals which are stimulated by environmental cues.
In addition, AGL21 expression also found responding to N and S starvations by qRT–PCR and GUS staining analyses (Figure 5). N deficiency is known to stimulate PR and particularly LR elongation but not LR initiation (Linkohr et al., 2002). We thus examine the root phenotypes of AGL21-overexpressing and mutant plants on N-rich and -free medium. The results showed that AGL21-overexpressing lines had longer LRs than wild-type plants under both these conditions, while LR growth was restrained in the mutant plants, especially under N-starved conditions. More specifically, under N-free conditions, LR length per cm PR had a slight increase in the wild-type and AGL21-overexpressing lines compared with that under N-rich conditions. However, compared with N-rich conditions, the LR length per cm PR of the mutant under N-free conditions apparently reduced (Figure 6E). Furthermore, the average LR length of the mutant decreased dramatically under N-free conditions compared with wild-type, while there was only a slight arrest under N-rich conditions, indicating that AGL21 plays some role in sustaining LR elongation in response to N availability. These results agree with the conference abstract of Suzuki et al. (2009), who reported that AGL21 is an essential factor to sustain LR growth under low-nitrate conditions. Therefore, our data demonstrate that AGL21 may play some role in N control of LR development. This is a good example of the function of AGL21 in LR development in response to environmental constraints. Furthermore, we also found AGL21 expression was up-regulated by abiotic stresses, such as drought and high salinity (Figure 5C). Collectively, it is reasonable to propose that AGL21 may play some roles in the regulation of RSA plasticity in response to various environmental and intrinsic signals.
Taken together, our results show that AGL21 expression can be induced by multiple environmental signals and internal hormones, and the up-regulated AGL21 strengthens auxin accumulation in the LRPs and LRs by regulating local auxin biosynthesis in the root, thus increasing cell division activity and stimulating LR initiation and elongation. AGL21 is also found positively regulated by auxin. Therefore, there is a positive feedback loop between auxin levels and auxin biosynthesis regulation via AGL21, which, in turn, affects auxin levels and distribution in the LPRs and LRs. As reported previously, auxin has been regarded as an integrator of diverse biotic and abiotic environmental signals and other hormonal signals to plant root development (Teale et al., 2008; Fukaki and Tasaka, 2009; Kazan, 2013; Lee and Cho, 2013). Hence, we propose that AGL21 may be a key factor to integrate the external and internal signals to auxin signals to regulate LRP initiation and LR growth, thus adapting to the environment more effectively. Our study also suggests that AGL21 may be a promising candidate gene for improving RSA in crop improvement.
METHODS
Plant Material and Growth Conditions
Seeds were surface-sterilized for 10min in 15% bleach, washed five times with sterile water, stratified at 4°C for 2 d, and plated on MS solid medium containing 1% (w/v) sucrose and 0.6% (w/v) agar at 22°C under 16-h light/8-h dark photoperiod. N-free medium was based on MS basal salt solution by replacing 20mM KNO3 and 20mM NH4NO3 with 20mM KCl. S-free medium was prepared as described previously (Wu et al., 2010).
Identification of the AGL21 Knockout Mutants
Two T-DNA insertion lines (CS118325 and GK_157C08) were obtained from the Arabidopsis Biological Resource Center (ABRC). CS118325 homozygotes were identified by genomic PCR with three primers: Spm32, CS118325 LP, and CS118325 RP. The homozygous mutant plants were confirmed by RT–PCR using gene-specific primers AGL21 LP and AGL21 RP, and β-Tubulin8 (TUB) was used as control with specific primers. All the primers used are shown in Supplemental Table 1.
Constructs and Generation of Transgenic Plants
For generation of AGL21-overexpressing plants, the 35S::AGL21 overexpression construct was made by inserting the coding region of AGL21 amplified by PCR using AGL21-attb-LP and AGL21-attb-RP into pCB2004 (Lei et al., 2007) via the GATEWAY cloning system. For promoter analysis, a pAGL21::GUS construct was produced by inserting a 2.6-kb promoter fragment amplified using forward primer AGL21-Pro-LP and reverse primer AGL21-Pro-RP into pCB308R (Lei et al., 2007). For protein localization, an AGL21 full-length coding sequence amplified by RT–PCR using specific primers AGL21-attb-LP1 and AGL21-attb-RP1 was inserted between the 35S promoter and EGFP sequences in pGWB5 (Nakagawa et al., 2007) to get pGWB5::AGL21 by the GATEWAY cloning system. To get the native promoter–gene fusion construct, a fragment containing AGL21 promoter and coding region amplified by genomic PCR with primers AGL21-attb-LP2 and AGL21-attb-RP2 was cloned into pMDC110 to fuse with GFP (Curtis and Grossniklaus, 2003). All the primers used are shown in Supplemental Table 1.
The constructs described were used to transform Arabidopsis using the Agrobacterium-mediated floral-dip method (Clough and Bent, 1998). Glufosinate-resistant T2 transgenic plants were obtained for functional analysis.
Quantitative Real-Time PCR (qRT–PCR)
qRT–PCR was performed as described previously (Yu et al., 2013). The transcript levels of AGL21 were examined using specific primers AGL21-qPCR LP and AGL21-qPCR RP. UBQ5 was used as the internal control, using specific primers UBQ5 LP and UBQ5 RP. All the primers used are shown in Supplemental Table 1. The results were based on the average of three parallel experiments.
Histochemical Detection of GUS Activity and GFP Imaging
The GUS activity staining was conducted as described previously (Xi et al., 2012). After incubating at 37°C for 2–12h in the dark, individual representative seedlings were photographed. Fluorescence of GFP in the transgenic plants was observed using a confocal microscope (Carl Zeiss LSM710, www.leica.com/).
Morphological Characterization of Roots
Root morphology was examined on MS medium solidified with 0.6% agar. Briefly, seeds were germinated on MS medium and 5-day-old seedlings were transferred to MS medium, MS medium containing hormones, or other nutrition lacking medium plates and grown vertically for a few days. Visible LR number was counted every day from the transfer day, and pictures of the plate were taken. Digital images of plants were used for root length measurement by hand using ImageJ software (NIH). LRPs and LRs of DR5:GUS and pCYCB1;1::GUS plants in different genetic backgrounds were photographed or counted using HIROX’s KH-7700 digital microscope. Classification of LRP developmental stages was performed according to Malamy and Benfey (1997).
Hormone and Abiotic Stress Treatments
For the qRT–PCR experiment, 8-day-old plants grown on MS agar medium were transferred to MS nutrient solution supplemented with hormones or N/S-free nutrient solution for different times as indicated and harvested for RNA extraction. Drought and high-salinity treatments were carried out as reported previously (Seki et al., 2002).
For GUS staining analysis, 7-day-old plants grown on MS agar medium were transferred to MS agar medium supplemented with hormones or N/S-free agar medium for the indicated time points and harvested for GUS staining.
IAA Content Measurement
The free total IAA content was measured by ELISA as described by Lin et al. (2005).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the Chinese Academy of Science (grant no. KSCX3-YW-N-007), the Ministry of Science and Technology of China (grant no. 2012CB114304), and the National Nature Science Foundation of China (grant no. 30830075).
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhen Wang for critical reading of the manuscript. We also thank the ABRC for providing T-DNA insertion lines used in this study. No conflict of interest declared.
REFERENCES
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science. 311, 91–94. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Liljegren S.J., Pelaz S., Gold S.E., Burgeff C., Ditta G.S., Vergara-Silva F., Yanofsky M.F. (2000a). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martinez-Castilla L., Yanofsky M.F. (2000b). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl Acad. Sci. U S A. 97, 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F., Diet A., Verdenaud M., Gruber V., Frugier F., Chan R., Crespi M. (2010). Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell. 22, 2171–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 8, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. [DOI] [PubMed] [Google Scholar]
- Beeckman T., Burssens S., Inze D. (2001). The peri-cell-cycle in Arabidopsis . J. Exp. Bot. 52, 403–411. [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Burgeff C., Liljegren S.J., Tapia-Lopez R., Yanofsky M.F., Alvarez-Buylla E.R. (2002). MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta. 214, 365–372. [DOI] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R.P., Beeckman T., Dhooge S., Swarup R., Graham N., Inze D., Sandberg G., Casero P.J., et al. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell. 13, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Comstock J.P. (2002). Hydraulic and chemical signalling in the control of stomatal conductance and transpiration. J. Exp. Bot. 53, 195–200. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta . Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S., Shchennikova A.V., Franken J., Busscher M., Baskar >R., Grossniklaus U., Angenent G.C., Immink R.G. (2006). A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis . Plant J. 47, 934–946. [DOI] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inze D., Beeckman T. (2006a). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60, 871–887. [DOI] [PubMed] [Google Scholar]
- De Smet I., Zhang H., Inze D., Beeckman T. (2006b). A novel role for abscisic acid emerges from underground. Trends Plant Sci. 11, 434–439. [DOI] [PubMed] [Google Scholar]
- Deak K.I., Malamy J. (2005). Osmotic regulation of root system architecture. Plant J. 43, 17–28. [DOI] [PubMed] [Google Scholar]
- Engineer C.B., Kranz R.G. (2007). Reciprocal leaf and root expression of AtAmt1.1 and root architectural changes in response to nitrogen starvation. Plant Physiol. 143, 236–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Tasaka M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69, 437–449. [DOI] [PubMed] [Google Scholar]
- Fukaki H., Okushima Y., Tasaka M. (2007). Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 256, 111–137. [DOI] [PubMed] [Google Scholar]
- Gan Y., Filleur S., Rahman A., Gotensparre S., Forde B.G. (2005). Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana . Planta. 222, 730–742. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A., Ortiz-Moreno E., de la Paz Sanchez M., Murphy A.S., Garcia-Ponce B., Marsch-Martinez N., de Folter S., Corvera-Poire A., Jaimes-Miranda F., Pacheco-Escobedo M.A., et al. (2013). The MADS transcription factor XAL2/AGL14 modulates auxin transport during Arabidopsis root development by regulating PIN expression. EMBO J. 32, 2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E., Himmelbach A. (1998). ABA signal transduction. Curr. Opin. Plant Biol. 1, 412–418. [DOI] [PubMed] [Google Scholar]
- Himanen K., Boucheron E., Vanneste S., de Almeida Engler J., Inze D., Beeckman T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 14, 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge A. (2006). Plastic plants and patchy soils. J. Exp. Bot. 57, 401–411. [DOI] [PubMed] [Google Scholar]
- Jovanovic M., Lefebvre V., Laporte P., Gonzalez‐Rizzo S., Lelandais‐Brière C., Frugier F., Hartmann C., Crespi M. (2007). How the environment regulates root architecture in dicots. Adv Botanical Res. 46, 35–74. [Google Scholar]
- Kaufmann K., Melzer R., Theissen G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 347, 183–198. [DOI] [PubMed] [Google Scholar]
- Kazan K. (2013). Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 1655–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., Lacombe B., Bielach A., Perrine-Walker F., Malinska K., Mounier E., Hoyerova K., Tillard P., Leon S., Ljung K., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell. 18, 927–937. [DOI] [PubMed] [Google Scholar]
- Kutz A., Muller A., Hennig P., Kaiser W.M., Piotrowski M., Weiler E.W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana . Plant J. 30, 95–106. [DOI] [PubMed] [Google Scholar]
- Lavenus J., Goh T., Roberts I., Guyomarc’h S., Lucas M., De Smet I., Fukaki H., Beeckman T., Bennett M., Laplaze L. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458. [DOI] [PubMed] [Google Scholar]
- Lee R.D., Cho H.T. (2013). Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front Plant Sci. 4, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z.Y., Zhao P., Cao M.J., Cui R., Chen X., Xiong L.Z., Zhang Q.F., Oliver D.J., Xiang C.B. (2007). High-throughput binary vectors for plant gene function analysis. Journal of Integrative Plant Biology. 49, 556–567. [Google Scholar]
- Leyser O., Fitter A. (1998). Roots are branching out in patches. Trends Plant Sci. 3, 203–204. [Google Scholar]
- Lin W.H., Wang Y., Mueller-Roeber B., Brearley C.A., Xu Z.H., Xue H.W. (2005). At5PTase13 modulates cotyledon vein development through regulating auxin homeostasis. Plant Physiol. 139, 1677–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr B.I., Williamson L.C., Fitter A.H., Leyser H.M. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis . Plant J. 29, 751–760. [DOI] [PubMed] [Google Scholar]
- Lloret P.G., Casero P.J. (2002). Lateral root initiation. In Plant roots: the hidden half, 3rd ed., Waisel, Y., Eshel, A., Kafkafi, U., eds. (New York: Marcel Dekker; ), pp. 127–156. [Google Scholar]
- Lopez-Bucio J., Cruz-Ramirez A., Herrera-Estrella L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287. [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J., Hernandez-Abreu E., Sanchez-Calderon L., Nieto-Jacobo M.F., Simpson J., Herrera-Estrella L. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J.P. (2007). Roots of the second green revolution. Aust. J. Bot. 55, 493–512. [Google Scholar]
- Malamy J.E. (2005). Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28, 67–77. [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Development. 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Ryan K.S. (2001). Environmental regulation of lateral root initiation in Arabidopsis . Plant Physiol. 127, 899–909. [PMC free article] [PubMed] [Google Scholar]
- Menkens A.E., Schindler U., Cashmore A.R. (1995). The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. (2003). Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 316, 1–21. [DOI] [PubMed] [Google Scholar]
- Montiel G., Gantet P., Jay-Allemand C., Breton C. (2004). Transcription factor networks: pathways to the knowledge of root development. Plant Physiol. 136, 3478–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., Tabata R., Kawai T., Tanaka K., Niwa Y., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nibau C., Gibbs D.J., Coates J.C. (2008). Branching out in new directions: the control of root architecture by lateral root formation. New Phytol. 179, 595–614. [DOI] [PubMed] [Google Scholar]
- Osmont K.S., Sibout R., Hardtke C.S. (2007). Hidden branches: developments in root system architecture. Annu. Rev. Plant Biol. 58, 93–113. [DOI] [PubMed] [Google Scholar]
- Parenicova L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 15, 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret B., De Rybel B., Casimiro I., Benkova E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. (2009). Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408. [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Puig J., Meynard D., Khong G.N., Pauluzzi G., Guiderdoni E., Gantet P. (2013). Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr. Patterns. 13, 160–170. [DOI] [PubMed] [Google Scholar]
- Raya-Gonzalez J., Pelagio-Flores R., Lopez-Bucio J. (2012). The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana . J. Plant Physiol. 169, 1348–1358. [DOI] [PubMed] [Google Scholar]
- Reed R.C., Brady S.R., Muday G.K. (1998). Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis . Plant Physiol. 118, 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll J.J., Roeder A.H., Ditta G.S., Yanofsky M.F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development. 138, 5167–5176. [DOI] [PubMed] [Google Scholar]
- Saedler H., Becker A., Winter K.U., Kirchner C., Theissen G. (2001). MADS-box genes are involved in floral development and evolution. Acta Biochim. Pol. 48, 351–358. [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis . Science. 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., Kamiya A., Nakajima M., Enju A., Sakurai T., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Serraj R., Kumar A., McNally K.L., Slamet-Loedin I., Bruskiewich R., Mauleon R., Cairns J., Hijmans R.J. (2009). Improvement of drought resistance in rice. Adv. Agron. 103, 41–99. [Google Scholar]
- Sessa G., Meller Y., Fluhr R. (1995). A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol. Biol. 28, 145–153. [DOI] [PubMed] [Google Scholar]
- Stals H., Inze D. (2001). When plant cells decide to divide. Trends Plant Sci. 6, 359–364. [DOI] [PubMed] [Google Scholar]
- Sun J., Xu Y., Ye S., Jiang H., Chen Q., Liu F., Zhou W., Chen R., Li X., Tietz O., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 21, 1495–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Miyamoto M., Forde Brian G., Takahashi H. (2009). Nitrogen-regulated transcription factor, AGL21, controls lateral root development in Arabidopsis [abstract]. 20th International Conference on Arabidopsis Research, Edinburgh, UK.
- Svistoonoff S., Creff A., Reymond M., Sigoillot-Claude C., Ricaud L., Blanchet A., Nussaume L., Desnos T. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39, 792–796. [DOI] [PubMed] [Google Scholar]
- Tapia-Lopez R., Garcia-Ponce B., Dubrovsky J.G., Garay-Arroyo A., Perez-Ruiz R.V., Kim S.H., Acevedo F., Pelaz S., Alvarez-Buylla E.R. (2008). An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis . Plant Physiol. 146, 1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W.D., Ditengou F.A., Dovzhenko A.D., Li X., Molendijk A.M., Ruperti B., Paponov I., Palme K. (2008). Auxin as a model for the integration of hormonal signal processing and transduction. Mol. Plant. 1, 229–237. [DOI] [PubMed] [Google Scholar]
- Ticconi C.A., Delatorre C.A., Lahner B., Salt D.E., Abel S. (2004). Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 37, 801–814. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E.A., Araus V., Lu C., Parry G., Green P.J., Coruzzi G.M., Gutierrez R.A. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana . Proc. Natl Acad. Sci. U S A. 107, 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H., Jurgens G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10, 305–317. [DOI] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhao Q., Gao L., Yu X.M., Fang P., Oliver D.J., Xiang C.B. (2010). Isolation and characterization of low-sulphur-tolerant mutants of Arabidopsis . J. Exp. Bot. 61, 3407–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Xu P., Xiang C.B. (2012). Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana . Plant J. 69, 782–791. [DOI] [PubMed] [Google Scholar]
- Xu B., Timko M. (2004). Methyl jasmonate induced expression of the tobacco putrescine N-methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol. Biol. 55, 743–761. [DOI] [PubMed] [Google Scholar]
- Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q., Li S., Xiang C. (2013). Arabidopsis Enhanced Drought Tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 162, 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Forde B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 279, 407–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.