Abstract
This review highlights emerging areas of interest in public health genomics. First, recent advances in newborn screening (NBS) are described, with a focus on practice and policy implications of current and future efforts to expand NBS programs (e.g., via next-generation sequencing). Next, research findings from the rapidly progressing field of epigenetics and epigenomics are detailed, highlighting ways in which our emerging understanding in these areas could guide future intervention and research efforts in public health. We close by considering various ethical, legal and social issues posed by recent developments in public health genomics; these include policies to regulate access to personal genomic information; the need to enhance genetic literacy in both health professionals and the public; and challenges in ensuring that the benefits (and burdens) from genomic discoveries and applications are equitably distributed. Needs for future genomics research that integrates across basic and social sciences are also noted.
Keywords: Newborn screening, epigenetics, epigenomics, bioethics, health education
INTRODUCTION
Ongoing advances in genomic sciences and technology have important implications for the understanding, prevention, and treatment of human disease. Most often, such developments are considered within the realm of medicine, with applications in pharmacogenomics and, more recently, ‘precision therapy’ in oncology being commonly cited examples (26, 81, 82, 102). Yet the field of public health is also witnessing important transformations brought about by the advent of new biotechnologies (e.g., next-generation sequencing) and the increasing incorporation of genomics into population health sciences. In this review, we highlight what we view as important trends and emerging issues within the changing landscape of public health genomics.
Public health genomics is a broad interdisciplinary enterprise that defies succinct description or definition. It includes within its purview many longstanding disciplines—such as genetic epidemiology, biostatistics, health policy, and health education—as well as state-funded programs focused on surveillance and prevention of birth defects and heritable disorders (18, 37). Some traditional public health activities have begun to more fully integrate genomics. For example, environmental health studies now feature more intensive consideration of genomics to better understand populations that might be particularly vulnerable to toxic environmental exposures (7, 11, 39). The field of statistical genetics has also undergone dramatic changes in recent years, with the development of innovative analytic approaches to harness the “Big Data” now being generated by whole genome exploration (28, 73, 79, 93). Given its wide-ranging goals and activities, public health genomics requires complex structures and processes to integrate knowledge from genomics-based research and ultimately translate these findings for improved population health. A useful framework for conceptualizing these functions has been developed by Burke and colleagues (14) in Figure 1. This framework highlights the importance of engaging a diverse range of stakeholders, both within and outside of the scientific and public health communities, to support core activities including 1) informing public policy; 2) development and evaluation of preventive and clinical health services; 3) communication and stakeholder engagement; and 4) education and training.
Figure 1.
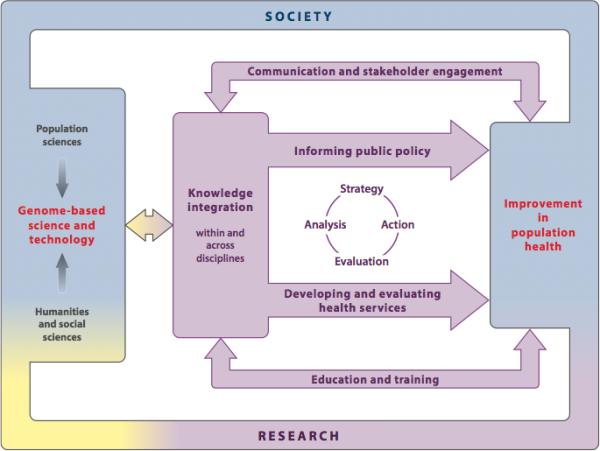
Components of the public health genomics enterprise. Adapted from Burke et al (2006)
Given the vast scope of this enterprise, it should be clear that comprehensive review of its components is beyond the reach of this article (see Mikail (2008) for a more extensive overview of the field). Our focus is instead on areas that we view as presenting particularly interesting and important emerging challenges and opportunities for applying genomics in public health. First, we will consider recent advances in newborn screening (NBS). Although NBS is a public health program that has been in effect for half a century, it is now being considered as a platform for population-based whole genome sequencing, raising new questions about the potential benefits and harms of expanding its mission for both research and practice purposes. Second, we will discuss the burgeoning science of epigenomics, considering its potential both for advancing the understanding of complex disease and providing new avenues for public health intervention and prevention. Finally, we will consider the “ELSI” issues (ethical, legal and social implications) brought about by advances in these and related areas of public health genomics. Although we recognize that public health genomics is increasingly involving international efforts, our focus will primarily be on activities within the United States (U.S.).
ADVANCES IN NEWBORN SCREENING
Background
Each year, nearly all of the 4 million infants born in the U.S. are screened by state NBS programs to identify and initiate treatment for rare diseases. During the past 50 years, NBS has saved thousands of children's lives and prevented disability in many more (86). Population-based NBS first started in Massachusetts in 1963. Now, every state in the U.S. provides mandatory NBS that is usually coordinated through each state's department of health. Programs screen for a variety of disorder types including inborn errors of metabolism, endocrine disorders, congenital heart disorders, cystic fibrosis and hearing loss. What these disorders have in common is that they are lifelong conditions without cure. Furthermore, if not tested for immediately after birth, these disorders may progress unnoticed in newborns until irreversible damage, such as cognitive impairment or death, results. Each year, NBS programs identify over 12,000 children with rare inherited disorders and connect them with life-saving interventions before irreversible health effects or death occurs (19).
NBS programs function at the intersection of public health, public policy and clinical care. Although federal guidance is provided by the Maternal Child Health Bureau (MCHB) of the Health Resources and Services Administration (HRSA), decisions about how to implement programs are made by each state's NBS program (78, 110). Each NBS program oversees both the laboratory testing and the follow-up of positive results through coordination of hospitals, state laboratories, and healthcare providers (primary care and specialty) who assure that NBS results are communicated effectively to families and that children with positive screens receive appropriate diagnostic evaluations (91). As a result, the process for delivering care in the time period between the identification of an initial positive screening result and subsequent confirmatory testing may vary by state.
There is an ongoing federal effort to minimize variation and maximize quality among state programs. In 2004, HRSA funded the organization of seven regional collaboratives (RCs). Each state is a member of one of the RCs, the goal of which is “to promote the identification of shared areas of need, as well as data collection and information sharing among member states (69).” In addition, the Secretary of Health and Human Services oversees the Discretionary Advisory Committee on NBS and Heritable Disorders in Newborns and Children (DACHDNC), which provides policy recommendations to state NBS programs. The most well recognized responsibility of this committee is the development of a panel of disorders recommended for screening in all states, called the Recommended Uniform Screening Panel (RUSP), which currently comprises over 50 disorders (50). It should be noted that the RUSP defines a floor for the content of state NBS panels. States can, and have, added disorders that have either not been reviewed by the DACHDNC or after review have been deemed not appropriate for recommendation to the RUSP, often because of insufficient existing evidence to justify population-based screening (37). Therefore, despite the influence of the RCs and the DACHDNC, the process and delivery of services in state NBS programs continue to be strongly influenced by state policies.
The expansion of NBS programs
Historically, the addition of disorders to state NBS panels has been slow and incremental. However, in the last ten years, advances in technology and the introduction of tandem mass spectrometry into NBS programs has led to a rapid and prodigious increase in the number of disorders screened. In 1995, prior to the advent of tandem mass spectrometry, states screened for an average of five disorders. By 2005 that number had increased to 19 disorders, with some states mandating screening for as many as 52 disorders (112). Given continued advances in the realm of screening technology, it is likely that the number of disorders screened will continue to increase in the coming years (83).
Next generation sequencing (NGS) is an emerging genomic technology with the potential to transform numerous aspects of NBS programs. The hope is that NGS will improve the quality of screening for current NBS conditions by helping to increase the predictive value of NBS results (77). Although some platforms, like cystic fibrosis NBS (23), incorporate genomic analysis into their screening test algorithms, it is unlikely that whole genomic sequencing will become the sole testing platform in NBS programs (113). In addition, NGS raises a number of programmatic and ethical issues for NBS programs. For example, NGS could uncover additional, unanticipated information (e.g., incidental findings including mutations associated with high risk for hereditary cancer syndromes) for infants who may have normal NBS results (109). Parents may not have consented to receive such information on their child, and some of that information, like carrier results, may not be immediately actionable for the child (109). Moreover, some of the information may be predictive, not diagnostic, and related to the onset of adult conditions. The potential harms on children and families of divulging this information need to be examined further (113) before programs incorporate NGS into their testing protocols and algorithms.
An emerging life course perspective in NBS
As the number of disorders detected by NBS increases, the types of disorders screened for and the care provided by NBS programs is also shifting. For much of its existence, NBS has focused on the identification and treatment of inherited disorders in infants and children. The subtle, but consistent, move towards a life course perspective in NBS evokes a number of public health, ethical and policy challenges. Increasing recognition of the need for lifelong treatment of many of these disorders is one area in which a life course perspective has emerged. The first disorder to be screened through NBS programs—phenylketonuria, or PKU—is a classic example of this perspective shift. Treatment for PKU requires consumption of a diet that is low in phenylalanine (an amino acid found in protein and other food products such as aspartame). Since clinicians long believed that the most important impact of the diet was on childhood development, the diet was initially prescribed only during childhood. Then data from international longitudinal studies revealed that elevated phenylalanine levels in mothers with PKU were associated with increased risk of having a child with birth defects and cognitive impairment (i.e., maternal PKU syndrome) (97). As a result, the recommendation for a phenylalanine-restricted diet expanded to include girls and women of childbearing age (24). Since the initial recommendation was made, evidence has shown improved outcomes in adolescents and adults treated with this diet, and the recommendation now calls for individuals to remain on a phenylalanine-restricted diet for life (70).
The recommendations for lifelong treatment for conditions like PKU are not without ethical and policy challenges. Metabolic conditions like PKU and galactosemia that require special diets are expensive and sometimes lack coverage by third-party payors (9, 17). As a result, many adults who were diagnosed with an inherited, lifelong by a mandatory public health program have to forego treatment because they cannot afford to pay for it out-of-pocket. In some states, the NBS program pays for the formulas and medical foods that make up this diet. However, this service is becoming increasingly difficult to provide as state public health budgets are decreasing. The extent to which these challenges will be addressed by the recently established Affordable Care Act remain uncertain.
This shift towards a life course perspective in NBS raises the additional ethical and policy issue of how long the NBS program is responsible for the oversight of the care individuals diagnosed with NBS disorders. After newborns receive a positive NBS result, they undergo additional testing and evaluation to determine whether or not they have the disorder in question. If they are subsequently diagnosed with a NBS condition, they go on to receive care in the clinical healthcare system (although some clinics may be funded, in part, by NBS program funds). Recommendations have been made that these children be followed for sufficient time after their initial diagnosis to ensure that they receive appropriate and high quality clinical care that maximizes health outcomes (i.e., long-term follow-up surveillance) (51). However, given the increasing recognition that many of these disorders require treatment into adulthood, it is unclear whether NBS programs have the resources and funds to conduct extensive long-term follow-up surveillance of these children (53).
Screening beyond childhood onset disorders
Although some disorders might require that treatment extend into adulthood, others may have limited health implications until adulthood. While NBS has screened largely for disorders with symptoms that develop in childhood, subtle shifts in this paradigm have also been occurring. Perhaps the first shift began with the addition of disorders in which unaffected carriers are identified (e.g., sickle cell disease, cystic fibrosis). The greatest individual benefits to unaffected carriers of knowing their status is for future reproductive planning, which is not an immediate concern for childhood health. In fact, parents receive the most immediate benefits from identification of newborns as carriers. When a child is diagnosed as a carrier for a disorder, parents may be alerted to the fact that one or both of them is also a carrier, alerting parents to a risk of having future children who are affected with the disorder. Screening of this type is a departure from the primary mission of NBS, which is to screen for disorders that pose emergent health risks in childhood (44).
An additional change in mission has occurred recently with the introduction of screening for adult-onset disorders. In September of 2013, DACHDNC recommended that Pompe disorder be added to the RUSP (50). Pompe, a lysosomal storage disease, is the first disorder recommended for addition to the RUSP that has a distinct adult onset form (59). The addition of Pompe represents a shift from this focus of NBS on the screening of disorders that develop in childhood, to screening for disorders that may manifest initial symptoms later in the life course. This situation creates a group of individuals, called “patients-in-waiting”, who are diagnosed with a disorder but are “waiting” for symptoms of their disorder to develop (65, 116). Data on the psychological and emotional effects of living with a looming diagnosis of a disorder discovered by NBS is lacking.
Screening for disorders with an adult-onset of symptoms also raises the issue of long-term follow-up (LTFU) responsibilities. After being diagnosed with a condition that may onset in adulthood, the infant must be consistently followed to identify signs and symptoms of disease as soon as they occur so that treatment can be initiated. It will be a challenge to track these infants as they move into adulthood to ensure that they have appropriate clinical follow-up. While some NBS programs engage in LTFU activities (52), such a systematic, long-term surveillance of an asymptomatic group of infants diagnosed through NBS has little precedent.
Against these emerging developments, one should not lose sight of the fact that NBS is a mandatory program. This means that state programs require that infants be screened and do not require parental consent or notification (71). The legal framework for mandatory screening is parens patriae, whereby the state steps in to make decisions in the best interest of the child (41). It should also be noted that screening is not mandatory in all countries. Few non-U.S. programs mandate screening of newborns for genetic disease. For example, NBS in the Netherlands has always been voluntary, but prior to the recent expansion of screening, parents were provided with limited information about the tests and didn't know about the option of declining screening. In France, written consent is required before any specimen is collected for DNA analysis (45). Given the movement towards screening for adult onset conditions and disorders that do not meet traditional screening criteria (1), the President's Council on Bioethics has suggested a two-tiered approach to screening that would involve mandatory screening for conditions that fulfill the traditional public health criteria and a voluntary pilot screening program for selected conditions that do not yet meet these criteria (10). However, this poses the theoretical risk that tiered screening (i.e., voluntary and mandatory) would confuse parents and cause them to seek exemption from NBS altogether.
It should be clear that state NBS programs, a longstanding public health enterprise, are undergoing rapid transformation with numerous implications for practice and policy. We now shift to an active area of research within public health genomics: epigenetics and epigenomics. Although public health programs based on this emerging science are not yet undertaken, the field holds significant promise for better understanding, and intervening on, population health risks.
EPIGENETICS AND PUBLIC HEALTH
Epigenetic modifications comprise the epigenome
The term ‘epigenetics’ was popularized in the 1940s by developmental biologist Conrad Waddington to explain “the interactions of genes with their environment, which bring the phenotype into being (120).” In the 1970s, Holliday and Pugh first proposed covalent DNA modifications, such as methylation of cytosine-guanine (CpG) dinucleotides, as the molecular mechanism to explain Conrad's hypothesis (54). In the 1990's and beyond, the Human Genome Project inspired the investigation of genome-wide rather than local gene analyses, and the term “epigenomics” was coined to describe all of the chemical modifications that are added to the genome to regulate gene expression and activity (84). “Epi-” is Greek for “above,” and thus the epigenome is defined as the entirety of the modifications to the genome, including those modifications directly to DNA as well as modifications that attach to nucleosomes, the proteins around which DNA is wrapped. While all of the different cell types in the body share the same DNA sequence (the genome), the epigenome controls whether cells become liver, lung, skin, or heart cells.
External influences on the epigenome
Environmental factors, including social, chemical and nutritional exposures, are viewed as influential predictors of subsequent phenotypes and disease risk in later life. In particular, the “developmental origins of health and disease” (DOHaD) hypothesis posits that gene-environment interactions during early life result in long-lasting effects and points to epigenetic inheritance as a prime underlying mechanism (6). Epigenetics is the study of changes in gene expression that are heritable from cell to cell, hence through cell lineage development, or in rarer cases, transgenerationally from parent to child to grandchild (132). Because epigenetic changes are dynamic and, unlike genetic changes, potentially reversible, they hold promise for public health as targets for preventive and therapeutic interventions. For example, in a mouse model, epigenetic alterations associated with perinatal chemical exposure can be counteracted by maternal nutritional supplementation with methyl donors (e.g. folic acid) or components of soy (30). Similarly, in rats in utero choline supplementation negated alcohol-induced alterations in epigenetic modification (8). Although it is unclear whether and how these animal model findings generalize to humans, they nevertheless suggest great potential for identifying at-risk populations and strategies for epigenetic intervention.
Increasingly, we are recognizing that environmental influences on the epigenome are diverse and include dietary (total caloric intake, specific micro- and macronutrient levels, phytochemicals), physical (temperature, species density), social (stress, behavior, socioeconomic), chemical (toxins, endocrine disruptors, drugs), or unknown (stochastic, random) effects. Extensive reviews on environmental epigenetics have recently been published (5, 22, 35); here we wish to highlight a few seminal studies relating to life course epigenetic effects. Because diet-derived methyl donors and co-factors are necessary for the synthesis of S-adenosylmethionine (SAM), the methyl group donor responsible for DNA methylation, environmental factors that alter early nutrition and/or SAM synthesis can potentially influence life course phenotype. For example, persistent epigenetic adaptations occur early in development in response to maternal nutritional factors, like folic acid (122) or genistein (31), and are associated with increased disease susceptibility later in life among genetically identical mice. Consequently, aberrant epigenetic gene regulation has been proposed as a mechanism of action for the discordance of disease susceptibility in monozygotic twins (95). Esteller and colleagues compared epigenetic profiles in sets of monozygotic twins at different ages, as well as from those raised in different environments (36). Epigenetic profiles exhibited greater divergence in older twins as well as in twins who had spent more than 50% of their lives apart, implicating lifestyle choices and/or environmental exposures as contributing factors to divergent epigenomes. Studies also associate pharmaceutical exposures with an altered epigenome and increased disease risk. For example, increased incidence of uncommon disorders was also observed in the granddaughters and grandsons of pregnant women prescribed diethylstilbestrol (DES), suggesting epigenetic multi-generational inheritance (reviewed in (90)). Taken together, these findings across animal models and human populations implicate epigenetic modifications as mechanisms by which stressful environments and toxic exposures can have single and multi-generational adverse health effects.
Critical periods and vulnerable populations
In order to understand the role of environmental factors on plasticity through epigenetic mechanisms, both exposure timing and epigenetic drift across time must be considered. DNA methylation and other epigenetic patterns are prone to change throughout the life course, but are especially vulnerable during reprogramming events associated with embryogenesis and early development. This is because the epigenome undergoes extensive reprogramming during two key time-points in early gestation with the purpose of establishing cell- and tissue-specific gene expression (49). In male primordial germ cells, which eventually become sperm, methylation is initiated during gestations whereas, in female primordial germ cells, which will become eggs, methylation occurs after birth in mature oocytes. A second period of epigenetic reprogramming occurs at fertilization, which helps in the establishment of individual cell-specific methylation patterns (58). As individuals age, DNA methylation is gradually lost genome-wide, concurrent with gene-specific increases in DNA methylation, leading to genome instability or gene-specific suppression, respectively. Whether early or life-course environmental exposures influence the rate of epigenetic drift with time is an active area of interest (35, 115).
As the epigenome is most vulnerable to external factors during development, women and men of childbearing age, pregnant women, and fetuses serve as vulnerable populations for epigenetic reprogramming that is associated with life course health outcomes (128). Exposure levels, in addition to the absorption, distribution, metabolism, and elimination of exposed factors, may vary across individuals due to several factors such as genetics, occupation, dietary practices, and health status. Additionally, the fetus is more susceptible to chemical, physical, and biological exposures due to the ability of the exposure to cross the placental barrier into the uterine environment (57, 60, 64, 92). Fetuses and pregnant women also exhibit altered metabolizing machinery and biotransformation capacity for chemical exposures as compared to non-pregnant adults (75, 85, 118), thereby, potentially increasing the risk of abnormal epigenetic reprogramming resulting from environmental exposures. These findings suggest a need for enhancement of maternal and child public health programs to protect against harmful exposures to the developing and potentially fragile fetus.
Emerging technologies and analytic approaches
Akin to the increased use of genome-wide association studies (GWAS) in genetics research, the field of epigenomics has recently experienced a rapid advancement in technology allowing for the characterization of multiple epigenetic marks across the genome. Until recently, most attempts to elucidate the effects on the epigenome following environmental exposures were either candidate gene-driven or based on epigenetic techniques with limited genome coverage/sensitivity. We are now experiencing a rapid advancement in technology, methodology, and data acquisition that allow for the identification of the constellation of genomic loci with altered epigenetic status (25, 66, 106). To fully succeed in identifying the role of epigenetic mechanisms across the life course, scientists must integrate animal model, human clinical, and human population approaches, paying close attention to windows of vulnerability, environmental and nutritional assessment, and cell-specific epigenetic patterns (29). Animal models, in which exposures are well controlled and characterized, will continue to help inform the evaluation of dose-response effects on the epigenome as well as vulnerable time periods, including gestation and multi- or trans-generational effects (72, 94, 108). Further, animal models and clinical samples are often useful for proteomic and chromatin structure evaluation (32), which may be limited in human population approaches due to sample storage and processing limitations. Finally, as the epigenome (in contrast to the genome) is particularly subjected to animal model and human clinical studies, in which cell-type specificity is more readily evaluated, this could provide important proof-of-principle approaches to evaluate the use of peripheral tissue (e.g. blood and saliva) in human epigenetic epidemiology studies.
Implications for public health intervention and areas for future research
The evaluation of the epigenome in humans has important implications for reducing health risks by identifying potentially modifiable risk factors. First, environmental exposures are good candidates for modifiable risk factors as they can be effectively regulated at the personal behavioral as well as regulatory policy level. Second, through epigenomic profiling, we can facilitate the identification of biomarkers of exposure, enabling clinicians to identify at-risk individuals prior to disease onset. Unlike genetic mutations, which are static and non-modifiable, epigenetic marks are dynamic and potentially modifiable. Therefore, if environmental exposures are affecting health outcomes via epigenetics, we may be able to use nutritional and/or pharmacological approaches to counteract adverse effects, not to mention broader policy approaches to reduce the prevalence of toxic exposures in a given state or municipality. Once epigenetic biomarkers of exposure associated with health outcomes have been established, the future of epigenomics holds tremendous potential not only for individualized reproductive health care, but also for population-wide disease diagnostic, screening, and prevention strategies.
Although the investigation of epigenetics as a mechanism of DOHaD has made substantial progress over the past decade, there remains a distinct need for more studies that: (1) begin exposure prior to conception to ensure coverage of the initial epigenetic reprogramming events, (2) characterize exposures and health outcomes over time, with corresponding measures of potential epigenetic drift, (3) investigate tissue and cell-specific differences in epigenetic responses to environmental exposures, and (4) examine exposure mixtures that more closely approximate real-world exposures that people face. In regards to 1-4 above, epigenetic studies may be particularly successful in identifying mechanisms associated with exposures and disease risk by incorporating neonatal blood spots (sometimes accessible at the population level from state NBS described earlier), which are important resources for both exposure assessment in utero and for DNA and gene expression studies (13, 123). In addition to the general needs noted above, the realization that many chemical exposures act as endocrine disrupting compounds (EDC) necessitates the evaluation of multiple doses in toxicological studies, as well as advance exposure modeling and statistical methods in human studies. Investigation of sex/gender differences in epigenetic alterations and their downstream health effects are also critical.
ETHICAL, LEGAL AND SOCIAL IMPLICATIONS (ELSI)
The expansion of NBS programs, and the potential future application of findings from epigenetics and epigenomics studies—among other emerging trends in public health genomics—raise numerous ELSI issues that will need to be addressed. As already noted, expanded NBS efforts raise delicate issues around informed consent, while insights from epigenetics studies may suggest a more urgent ethical duty to enhance public health interventions early in the life course. In this section, we consider a variety of ELSI issues relevant for current and future efforts in public health genomics, including 1) health policies that determine access to genomic information, 2) challenges in providing health education and understanding behavioral responses to genetic test results, and 3) procedural and distributive issues in public health genomics.
Health policies regarding access to personal genomic information
Many prominent health policy challenges in public health genomics involve the regulation of access to genomic information generated by emerging technologies. For example, as discussed previously, integration of next-generation sequencing into NBS programming could generate incidental findings of uncertain value to parents, children and clinicians. Typically, policies regarding access to genetic information with health implications have conformed to a medical model, with clinical experts as the gatekeepers of new information. According to this approach, most genomic information would currently be deemed unsuitable for disclosure given limitations in predictive value and clinical utility (i.e., available treatment options), difficulties in conveying information in accurate and easily understood terms, and possible psychological and social harms in individuals receiving such information. However, many individuals, particularly those with family histories of certain diseases, are curious about their genetic profiles. Both national surveys and clinical research suggest that individuals are generally interested in genetic risk information (even for severe, incurable conditions like Alzheimer's disease), with the benefits of testing seen as outweighing its limitations and risks due to its perceived value in informing advance planning, monitoring treatment developments, and coping with uncertain risk status (89, 99). Survey respondents have claimed that they would be willing to pay significant out-of-pocket costs for genetic risk information, even if it is of modest predictive value (62, 88). Reflecting a broader trend toward patients asserting their “right to know” personal health information, many believe they should have access to their own genomic information if desired.
Some commentators (98, 105) have argued that the research ethics principle of respect for participants means that greater access to such information should generally be provided to individuals engaged in genomic research. This could prove a daunting challenge, particularly for investigators using data from NBS research repositories and other large population-based datasets. Wolf et al (2012) have developed guidelines for large biobank research systems to discharge potential responsibilities to disclose individual genomic research results to participants (130). Recommendations include proactive identification of results warranting disclosure, incorporation of participant preferences regarding return of results into initial consent processes, and development of processes and procedures whereby primary and secondary researchers communicate about the potential need to return individual research results. Still, given the increasing use of techniques to interrogate the whole genome, researchers could easily be overwhelmed by the need to sift through incidental findings and guard against the possibility of disclosing false positive results (61).
The issue of incidental findings in clinical use of whole-genome and whole-exome sequencing has received much recent attention, in large part due to controversial guidelines issued by the American College of Medical Genetics (43). These recommendations assert a more expansive duty to disclose genetic findings of potential clinical utility, even if they are not the intended focus of testing. Some commentators (15) have argued that the guidelines are contrary to standard practice in that they a) mandate disclosure regardless of patient preferences and b) endorse the provision of risk information for adult-onset conditions regardless of patient age. The latter recommendation would have particular relevance if, as discussed earlier, NBS programs begin to incorporate whole-genome sequencing approaches into their efforts (40). Leading professional organizations in clinical genetics have issued policy statements that genetic testing should be deferred until adulthood unless determination of carrier status results in medical benefit in childhood (4, 33). A main ethical justification for withholding this information is that there are no immediate clinical benefits to outweigh potential harms of receiving distressing and potentially stigmatic information. Some also note the value of preserving autonomy, suggesting that genetic risk information could infringe on the child's right to an “open future” and the corresponding opportunity to make testing decisions as a consenting adult (27). Yet other commentators have argued for a more flexible stance against testing minors for adult-onset conditions, noting that in the absence of proven (versus speculative) harms from genetic risk disclosure, parental authority in such decision-making should be respected (129).
Another area of controversy concerns direct access to personal genomic information via commercially available services. Multiplex genetic testing can now be conducted at a fraction of its prior cost, using saliva samples that can be shipped from one's home. Test results for scores of conditions can be delivered simultaneously via sophisticated web-based user interfaces that provide relevant and tailored education. Direct-to-consumer genetic testing (DTC-GT) has raised ELSI concerns ever since its inception. Commentators have noted that many company sites claim exaggerated benefits of testing and may not fully disclose risks of testing (particularly privacy and familial implications). For example, a content analysis of 23 DTC websites found six times as many statements about test benefts as risk and limitation statements (107). DTC-GT services typically do not screen out psychologically vulnerable consumers or collect detailed family history information, and most do not provide genetic counseling, which could increase the potential for inadequate understanding of the meaning and/or implications of test results. Leading professional organizations representing health professionals and genetics researchers have expressed concerns regarding DTC-GT and its potential harms (3, 55), and others have suggested that risk information gained from DTC-GT services will lead to more, and potentially unnecessary, health care utilization and screening (80). Yet many proponents of consumer genomics view direct access to one's genome as an individual right, noting potential benefits of learning more about one's predisposition to disease and likelihood of response to specific medications. Findings to date suggest that neither the health benefits promoted by DTC-GT proponents (e.g., improvements in positive health behaviors) nor the worst-case scenarios envisioned by its critics (e.g., catastrophic psychological distress and misunderstanding of test results, undue burden on the healthcare system) have materialized to date (100). However, research on the benefits and harms of DTC-GT is in its early stages and possesses numerous key limitations (e.g., lack of comparison groups, use of non-representative convenience samples). In the meantime, some countries (e.g., Germany) have banned the practice of DTC-GT entirely, while in the US regulators have recently issued cease-and-desist style warnings to leading personal genome service companies like 23andMe (47).
Another concern about the provision of genetic susceptibility test results involves potential discrimination from insurers and employers. Although proven cases of genetic discrimination are relatively rare, a U.S. survey of individuals at-risk for Huntington's disease suggested numerous examples of perceived discrimination in insurance, employment, health care, and social settings (12). In the U.S., the federal Genetic Information Nondiscrimination Act (GINA) was passed in 2008, prohibiting health insurers and employers from using genetic information (including family history) to inform decisions about coverage, premiums, or hiring (63). However, GINA does not cover life, disability, or long-term care (LTC) insurance, which is important to note given the availability of genetic susceptibility testing for disorders like Alzheimer's disease (AD) in DTC-GT and other formats. In a series of randomized clinical trials, disclosure of genetic susceptibility test results (in this case, APOE genotype) to individuals at-risk for AD appeared to prompt changes to participants’ LTC insurance plans (133). Given that AD accounts for a significant proportion of LTC costs, insurers may be within their legal rights to address potential adverse selection by consumers who know they are at increased risk for AD (114). Should insurers increase premiums or deny coverage based on APOE or other genetic test results, an expansion of GINA protections could be in order to address potential genetic discrimination in LTC and other insurance domains. It is unclear, however, whether current GINA protections would cover results from epigenomic analyses like those considered in the prior section of the paper (101). The legislation was drafted before the emergence of this area of public health genomics and thus does not specifically include epigenetics in its definition of genetic information.
Health education in public health genomics
Educating the public about genomic information poses numerous challenges, not least of which is that scientists are still determining its meaning across various contexts relevant for human health. Take, for example, genetic risk assessment for health conditions. The complexity of most medical disorders makes it difficult to integrate genetic information with the multiple other factors that influence disease expression, such as health behaviors, environmental exposures, comorbid conditions, and social determinants of health. Within the realm of genetics, factors including penetrance, variable expressivity and genotype-phenotype correlations can impact the expression of gene mutations, and we are still relatively ignorant about the role of gene-gene and gene-environment interactions in human diseases. Even when data are available on these factors, sampling biases may limit the generalizability of results. For example, allele frequencies in genes associated with various diseases are known to differ by racial and ethnic groups, but the populations enrolled in studies of disease risk often lack diversity on this dimension (16). Furthermore, extrapolating from aggregate data to make inferences at the individual level can be problematic.
Health education challenges are faced not only in the interpretation of genomic information, but also its disclosure. Conveying genetic information effectively requires an ability to translate complicated findings to individuals who often lack the advanced skills in health literacy, numeracy, and genetic literacy that would be required to fully appreciate the meaning and implications of results (56). Further complicating matters is that tendencies toward genetic exceptionalism and essentialism may mean that recipients treat information differently merely by virtue of its label as ‘genetic’. These biases could result in information being misconstrued in either fatalistic or falsely reassuring terms. As discussed earlier, NBS is a context in which parents often receive information about disorders they have never even heard of, resulting from a test that they may never have even consented to or been educated about. This raises the important issue of expectations regarding health information; bad health news is much more likely to pose psychological harms when it is unanticipated. Even in the best of circumstances, the post-partum period can be an overwhelming time for parents. When parents receive the news of a positive NBS result, they are often in a vulnerable state both in terms of their prior knowledge of NBS, which is suspected to be suboptimal, and their emotional status. Studies have shown that the prevalence of post-partum anxiety disorders may be higher than that of post-partum depression (124–126). Thus, during the post-partum period, parents may not be emotionally prepared for the stressful news of the detection of a potential serious illness in their newborn (117).
When parents are unprepared to hear potentially stressful news, they may fail to understand the significance of the positive initial screening result, the process of evaluation and the ultimate designation of the positive screening result as true or false. Failure to understand the implications of a positive screening result, whether it is later confirmed as true or deemed false, has been associated with increased utilization of healthcare that is unrelated to the NBS disorder tested, as well as dysfunctional parent-child relationships (121). The extent of parental misunderstanding is significant. In one study, only 1/3 of parents were able to correctly report the reason for the repeat screening that occurred (46). Against the backdrop of expanded NBS for unfamiliar disorders, it is essential to deliver information to parents about NBS results in a way that optimizes both parental understanding of the child's health status and parental healthcare seeking behavior for the child.
Given these nuances and sensitivities, genomic information would ideally be delivered by a trained genetic counselor with expertise in human genetics, health education, and interpersonal practice. Yet the traditional genetic counseling model—as developed for heritable, high penetrance adult-onset disorders—poses feasibility challenges given the need for extensive case preparation, comprehensive review of family and medical history information, and pre- and post-test education and counseling. This model may not be practical from a public health perspective, especially as greater numbers of people require genetic services. There are over 3,100 genetic counselors (GCs) board certified in the U.S., (2), with most of these professionals geographically concentrated in urban areas and working in prenatal, pediatrics, and cancer genetics (87). To meet anticipated increased future demands for genetic counseling, leaders in the field have called for the development of alternative models of genetic service delivery and recognized the need for increased involvement of non-genetics health care professionals, use of educational media, and briefer protocols (48). These efforts should draw upon what has been learned in recent years about effective strategies for health risk communication. These include the following: (a) use of natural frequencies (i.e., not only percentages) to communicate risk estimates (38), (b) supplementing verbal disclosure of risk information with graphical representations (e.g., pictograph) (68), (c) use of printed take-home education materials to reinforce information presented in person (131), and (d) provision of strategies for coping with risk, such as possible options for risk reduction and resources for additional information and support.
Health behavior responses to genomic information
From a public health perspective, the value of genomic information lies primarily in its potential for spurring disease prevention efforts. Proponents of genetic susceptibility testing, for example, view it as a means to promote healthy behaviors among high-risk subpopulations to reduce chances of disease onset. However, genetic risk assessment has not generally added value when the behaviors in question are complex and difficult to change, such as smoking cessation and improved diet and exercise habits (76). A few studies (74, 96) suggest that genetic susceptibility testing may enhance preferences for biological interventions (e.g., medications) over health behavior changes (e.g., lifestyle change) when both are viable options (104). Such a phenomenon was observed in studies of genetic susceptibility testing for Alzheimer's disease, where the most common health behavior change reported by participants receiving high-risk results was the addition of vitamins or nutritional supplements (often vitamin E), even though our education materials noted this was not a proven means of AD risk reduction (20, 119).
Genomics and social justice
Public health ethics has long placed an emphasis on issues of justice, with a concern that health benefits and burdens be distributed fairly across the population (42). Yet many current genomic technologies used in health care are only available via specialty medical centers and not always covered by health insurers, limiting their access to significant proportions of the population, even in wealthy countries. Although the financial costs of whole genome sequencing have dropped dramatically over time (127), their applications require intensive resources both in terms of sequence analysis and clinical interpretation. This is not to mention the aforementioned LTFU costs implicated in expanded approaches to NBS programs. ELSI scholars have further reflected justice concerns by noting how applications of genomics technologies could exacerbate, rather than ameliorate, health disparities between racial and ethnic groups (103) by focusing on biological causes of disease instead of more compelling social and environmental risk factors. Emerging findings from epigenetics could help bridge the gap here, by suggesting potential underlying mechanisms by which differential exposures to extreme environmental stressors translate into adverse health outcomes.
Public health ethics can also be viewed through the lens of procedural justice, where notions of responsible stewardship would require transparency to the general public regarding policy changes and research initiatives. From this standpoint, recent efforts in certain U.S. states to repurpose NBS samples for research without notification or consent of families would be problematic (67, 111). The resulting legal actions taken in Texas and Minnesota to halt ongoing research and even destroy research repositories, while extreme to some, have hopefully resulted in a greater appreciation for the openness and public engagement necessary to maintain trust in the public health genomics enterprise. Public trust may also be at stake when forecasts are made about the potential future benefits of applying genomic discoveries in medicine and public health. Many predictions about the ways in which personalized medicine would transform health care, for example, look wildly optimistic in hindsight, threatening to undermine the credibility of its proponents. Some leaders in the field (34) have called for more realistic promises and responsible claims about the ‘revolutionary’ potential of genomics for medicine and public health.
CONCLUSION
These are exciting times in public health genomics, as witnessed by the potential to integrate powerful new technologies like next-generation sequencing into public health efforts, and the promise of rapidly advancing scientific disciplines like epigenomics. Clearly, however, much research is needed to harness the potential of genomic discoveries to improve human health and to secure public trust in ongoing public health genomic initiatives. To have impact beyond the specialty clinic, and at more of a population health level, we may also need to think in broader and more creative terms about transdisciplinary approaches to research. We are intrigued with Geronimus’ (2013) call for ‘deep integration’ of epigenetic science with social sciences focused on the ways in which socially structured, repeated activation of stress processes enhance disease vulnerability in marginalized groups. Such integration would seemingly allow public health genomics a larger stake in addressing the chronic disease burdens that account for an increasing proportion of public health costs. Another research development we would advocate is the increased integration of ELSI research within clinical and basic sciences investigations. This approach has been championed by the National Human Genome Research Institute in several of its recent grant programs, including the Clinical Sequencing Exploratory Research (CSER) consortium (21).
We have highlighted in this review some areas that we believe to be of emerging importance in the diverse, dynamic field of public health genomics. We anticipate that this enterprise will continue to rapidly evolve in the years ahead, with the implications and challenges for clinical, research and public policy activities continuing to expand in turn.
ACKNOWLEDGEMENTS
J.S.R. is supported by National Human Genome Research Institute grants R01HG02213, R01HG005092, and UM1HG006508. D.C.D. is supported by National Institutes of Health grant ES017524 and the University of Michigan National Institute of Environmental Health Science (NIEHS) Core Center P30ES017885. B.A.T is supported by a K23 Mentored Patient-Oriented Research Career Development Award from the National Institute for Child Health and Human Development (K23HD057994).
The authors wish to thank Shaila Chhibba for her assistance with manuscript preparation.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
J. Scott Roberts, Department of Health Behavior & Health Education University of Michigan School of Public Health.
Dana Dolinoy, Department of Environmental Health Sciences University of Michigan School of Public Health.
Beth Tarini, Child Health Evauation & Research Unit Division of Pediatrics University of Michigan Health System.
LITERATURE CITED
- 1.Alexander D, van Dyck PC. A vision of the future of newborn screening. Pediatrics. 2006;117(5 Pt 2):S350–4. doi: 10.1542/peds.2005-2633O. [DOI] [PubMed] [Google Scholar]
- 2.American Board of Genetic Counseling, Inc. 2013 http://www.abgc.net/ABGC/AmericanBoardofGeneticCounselors.asp.
- 3.American College of Medical Genetics ACMG Statement on Direct-to-Consumer Genetic Testing. 2008 doi: 10.1097/01.GIM.0000106164.59722.CE. [DOI] [PubMed] [Google Scholar]
- 4.American Society of Human Genetics Board of Directors and American College of Medical Genetics Board of Directors Points to consider: ethical, legal, and psychosocial implications of genetic testing in children and adolescents. American journal of human genetics. 1995;57(5):1233–41. [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and dna methylation. The Journal of nutritional biochemistry. 2012;23(8):853–59. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker DJP, Eriksson JG, Forsén T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. International journal of epidemiology. 2002;31(6):1235–39. doi: 10.1093/ije/31.6.1235. [DOI] [PubMed] [Google Scholar]
- 7.Barouki R, Gluckman PD, Grandjean P, Hanson M, Heindel JJ. Developmental origins of non-communicable disease: implications for research and public health. Environmental Health. 2012;11:42–51. doi: 10.1186/1476-069X-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, dna methylation, and proopiomelanocortin (pomc) gene expression in β-endorphin-producing pomc neurons of the hypothalamus. Alcoholism, clinical and experimental research. 2013;37(7):1133–42. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry SA, Brown C, Grant M, Greene CL, Jurecki E, et al. Newborn screening 50 years later: access issues faced by adults with pku. Genetics in medicine : official journal of the American College of Medical Genetics. 2013;15(8):591–99. doi: 10.1038/gim.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bioethics PC on . The changing moral focus of newborn screening: an ethical analysis by the president's council on bioethics. Washington DC: 2008. [Google Scholar]
- 11.Bollati V, Baccarelli A. Environmental epigenetics. Heredity. 2010;105(1):105–12. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bombard Y, Veenstra G, Friedman JM, Creighton S, Currie L, et al. Perceptions of genetic discrimination among people at risk for huntington's disease: a cross sectional survey. BMJ (Clinical research ed.) 2009;338:b2175. doi: 10.1136/bmj.b2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botkin JR, Goldenberg AJ, Rothwell E, Anderson RA, Lewis MH. Retention and research use of residual newborn screening bloodspots. Pediatrics. 2013;131(1):120–27. doi: 10.1542/peds.2012-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke W, Khoury M, Stewart A, Zimmern R. Bellagio group.(2006). the path from genome-based research to population health: development of an international public health genomics network. Genetics in Medicine. 2006;8(7):451–58. doi: 10.1097/01.gim.0000228213.72256.8c. [DOI] [PubMed] [Google Scholar]
- 15.Burke W, Matheny Antommaria AH, Bennett R, Botkin J, Clayton EW, et al. Recommendations for returning genomic incidental findings? we need to talk! Genetics in medicine : official journal of the American College of Medical Genetics. 2013 doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011;475(7355):163–65. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camp KM, Lloyd-Puryear MA, Huntington KL. Nutritional treatment for inborn errors of metabolism: indications, regulations, and availability of medical foods and dietary supplements using phenylketonuria as an example. Molecular genetics and metabolism. 2012;107(1-2):3–9. doi: 10.1016/j.ymgme.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CDC Pediatric Genetics: Newborn Screening. 2013 http://www.cdc.gov/ncbddd/pediatricgenetics/newborn_screening.html.
- 19.Cdc grand rounds: newborn screening and improved outcomes MMWR. Morbidity and mortality weekly report. 2012;61(21):390–93. [PubMed] [Google Scholar]
- 20.Chao S, Roberts JS, Marteau TM, Silliman R, Cupples LA, Green RC. Health behavior changes after genetic risk assessment for alzheimer disease: the reveal study. Alzheimer disease and associated disorders. 2008;22(1):94–97. doi: 10.1097/WAD.0b013e31815a9dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical Sequencing Exploratory Research. http://www.genome.gov/sequencingcosts.
- 22.Collotta M, Bertazzi PA, Bollati V. Epigenetics and pesticides. Toxicology. 2013;307:35–41. doi: 10.1016/j.tox.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Comeau AM, Accurso FJ, White TB, Campbell PW, Hoffman G, et al. Guidelines for implementation of cystic fibrosis newborn screening programs: cystic fibrosis foundation workshop report. Pediatrics. 2007;119(2):e495–518. doi: 10.1542/peds.2006-1993. [DOI] [PubMed] [Google Scholar]
- 24.Committee on Genetics Maternal phenylketonuria. PEDIATRICS. 2008;122(2):445–49. [Google Scholar]
- 25.Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Human genetics. 2012;131(10):1565–89. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crews KR, Hicks JK, Pui C-H, Relling MV, Evans WE. Pharmacogenomics and individualized medicine: translating science into practice. Clinical pharmacology and therapeutics. 2012;92(4):467–75. doi: 10.1038/clpt.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis DS. Genetic dilemmas and the child's right to an open future. The Hastings Center report. 1997;27(2):7–15. [PubMed] [Google Scholar]
- 28.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. A framework for variation discovery and genotyping using next-generation dna sequencing data. Nature genetics. 2011;43(5):491–98. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolinoy DC, Faulk C. Introduction: the use of animals models to advance epigenetic science. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2012;53(3-4):227–31. doi: 10.1093/ilar.53.3-4.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol a-induced dna hypomethylation in early development. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):13056–61. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects avy mouse offspring from obesity by modifying the fetal epigenome. Environmental health perspectives. 2006;114(4):567–72. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolinoy DC, Weinhouse C, Jones TR, Rozek LS, Jirtle RL. Variable histone modifications at the a(vy) metastable epiallele. Epigenetics : official journal of the DNA Methylation Society. 2010;5(7):637–44. doi: 10.4161/epi.5.7.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.European Society of Human Genetics Genetic testing in asymptomatic minors: recommendations of the european society of human genetics. European Journal of Human Genetics. 2009;17(6):238–51. doi: 10.1038/ejhg.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans JP, Meslin EM, Marteau TM, Caulfield T. Genomics. deflating the genomic bubble. Science (New York, N.Y.) 2011;331(6019):861–62. doi: 10.1126/science.1198039. [DOI] [PubMed] [Google Scholar]
- 35.Faulk C, Dolinoy DC. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics : official journal of the DNA Methylation Society. 2011;6(7):791–97. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(30):10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genetic Alliance Baby's First Test: Conditions Screened by State. 2013 http://www.babysfirsttest.org/newborn-screening/states.
- 38.Gigerenzer G, Edwards A. Simple tools for understanding risks: from innumeracy to insight. BMJ (Clinical research ed.) 2003;327(7417):741–44. doi: 10.1136/bmj.327.7417.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease — nejm. The New England journal of medicine. 2008;359(1):61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldenberg AJ, Sharp RR. The ethical hazards and programmatic challenges of genomic newborn screening. JAMA : the journal of the American Medical Association. 2012;307(5):461–62. doi: 10.1001/jama.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gostin L. Public Health Law: Power, Duty, Restraint. University of California Press; 2000. [Google Scholar]
- 42.Gostin LO, Powers M. What does social justice require for the public's health? public health ethics and policy imperatives. Health affairs (Project Hope) 2006;25(4):1053–60. doi: 10.1377/hlthaff.25.4.1053. [DOI] [PubMed] [Google Scholar]
- 43.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, et al. Acmg recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in medicine : official journal of the American College of Medical Genetics. 2013;15(7):565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grosse SD, Boyle CA, Kenneson A, Khoury MJ, Wilfond BS. From public health emergency to public health service: the implications of evolving criteria for newborn screening panels. Pediatrics. 2006;117(3):923–29. doi: 10.1542/peds.2005-0553. [DOI] [PubMed] [Google Scholar]
- 45.Grosse SD, Rogowski WH, Ross LF, Cornel MC, Dondorp WJ, Khoury MJ. Population screening for genetic disorders in the 21st century: evidence, economics, and ethics. Public health genomics. 2010;13(2):106–15. doi: 10.1159/000226594. [DOI] [PubMed] [Google Scholar]
- 46.Gurian EA, Kinnamon DD, Henry JJ, Waisbren SE. Expanded newborn screening for biochemical disorders: the effect of a false-positive result. Pediatrics. 2006;117(6):1915–21. doi: 10.1542/peds.2005-2294. [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez A. 23andMe, Inc. 11/22/13. 2013 http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/2013/ucm376296.htm.
- 48.Guttmacher AE, Jenkins J, Uhlmann WR. Genomic medicine: who will practice it? a call to open arms. American journal of medical genetics. 2001;106(3):216–22. doi: 10.1002/ajmg.10008. [DOI] [PubMed] [Google Scholar]
- 49.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, et al. Epigenetic reprogramming in mouse primordial germ cells. Mechanisms of development. 2002;117(1-2):15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 50.Health Resources and Services Administration Discretionary Advisory Committee on Heritable Disorders in Newborns and Children. 2013 http://www.hrsa.gov/advisorycommittees/mchbadvisory/heritabledisorders/
- 51.Hinton CF, Feuchtbaum L, Kus CA, Kemper AR, Berry SA, et al. What questions should newborn screening long-term follow-up be able to answer? a statement of the us secretary for health and human services’ advisory committee on heritable disorders in newborns and children. Genetics in medicine : official journal of the American College of Medical Genetics. 2011;13(10):861–65. doi: 10.1097/GIM.0b013e3182209f09. [DOI] [PubMed] [Google Scholar]
- 52.Hoff T, Ayoob M, Therrell BL. Long-term follow-up data collection and use in state newborn screening programs. Archives of pediatrics & adolescent medicine. 2007;161(10):994–1000. doi: 10.1001/archpedi.161.10.994. [DOI] [PubMed] [Google Scholar]
- 53.Hoff T, Hoyt A, Therrell B, Ayoob M. Exploring barriers to long-term follow-up in newborn screening programs. Genetics in medicine : official journal of the American College of Medical Genetics. 2006;8(9):563–70. doi: 10.1097/01.gim.0000237790.54074.3d. [DOI] [PubMed] [Google Scholar]
- 54.Holliday R, Pugh JE. Dna modification mechanisms and gene activity during development. Science. 1975;187(4173):226–32. [PubMed] [Google Scholar]
- 55.Hudson K, Javitt G, Burke W, Byers P. Ashg statement* on direct-to-consumer genetic testing in the united states. Obstet Gynecol. 2007;110:1392–95. doi: 10.1097/01.AOG.0000292086.98514.8b. [DOI] [PubMed] [Google Scholar]
- 56.Institute of Medicine . Health literacy: a prescription to end confusion. national academy of sciences; washington dc: 2004. Washington DC. [Google Scholar]
- 57.Jiménez-Díaz I, Zafra-Gómez A, Ballesteros O, Navea N, Navalón A, et al. Determination of bisphenol a and its chlorinated derivatives in placental tissue samples by liquid chromatography-tandem mass spectrometry. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2010;878(32):3363–69. doi: 10.1016/j.jchromb.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 58.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nature reviews. Genetics. 2007;8(4):253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kemper AR, Hwu W-L, Lloyd-Puryear M, Kishnani PS. Newborn screening for pompe disease: synthesis of the evidence and development of screening recommendations. Pediatrics. 2007;120(5):e1327–34. doi: 10.1542/peds.2007-0388. [DOI] [PubMed] [Google Scholar]
- 60.King E, Shih G, Ratnapradipa D, Quilliam DN, Morton J, Magee SR. Mercury, lead, and cadmium in umbilical cord blood. Journal of environmental health. 2013;75(6):38–43. [PubMed] [Google Scholar]
- 61.Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA : the journal of the American Medical Assocation. 2006;296(2):212–15. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- 62.Kopits IM, Chen C, Roberts JS, Uhlmann W, Green RC. Willingness to pay for genetic testing for alzheimer's disease: a measure of personal utility. Genetic testing and molecular biomarkers. 2011;15(12):871–75. doi: 10.1089/gtmb.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korobkin R, Rajkumar R. The genetic information nondiscrimination act--a half-step toward risk sharing. The New England journal of medicine. 2008;359(4):335–37. doi: 10.1056/NEJMp0804352. [DOI] [PubMed] [Google Scholar]
- 64.Krueger C, Horesh E, Crossland BA. Safe sound exposure in the fetus and preterm infant. Journal of obstetric, gynecologic, and neonatal nursing : JOGNN / NAACOG. 2012;41(2):166–70. doi: 10.1111/j.1552-6909.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwon JM, Steiner RD. “I'm fine; i'm just waiting for my disease”: the new and growing class of presymptomatic patients. Neurology. 2011;77(6):522–23. doi: 10.1212/WNL.0b013e318228c15f. [DOI] [PubMed] [Google Scholar]
- 66.Laird PW. Principles and challenges of genomewide dna methylation analysis. Nature reviews. Genetics. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 67.Lewis MH, Goldenberg A, Anderson R, Rothwell E, Botkin J. State laws regarding the retention and use of residual newborn screening blood samples. Pediatrics. 2011;127(4):703–12. doi: 10.1542/peds.2010-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lipkus IM. Numeric, verbal, and visual formats of conveying health risks: suggested best practices and future recommendations. Medical decision making : an international journal of the Society for Medical Decision Making. 2007;27(5):696–713. doi: 10.1177/0272989X07307271. [DOI] [PubMed] [Google Scholar]
- 69.Lloyd-Puryear MA, Tonniges T, van Dyck PC, Mann MY, Brin A, et al. American academy of pediatrics newborn screening task force recommendations: how far have we come? Pediatrics. 2006;117(5 Pt 2):S194–211. doi: 10.1542/peds.2005-2633B. [DOI] [PubMed] [Google Scholar]
- 70.Macleod EL, Ney DM. Nutritional management of phenylketonuria. Annales Nestle [English ed.] 2010;68(2):58–69. doi: 10.1159/000312813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mandl KD, Feit S, Larson C, Kohane IS. Newborn screening program practices in the united states: notification, research, and consent. Pediatrics. 2002;109(2):269–73. doi: 10.1542/peds.109.2.269. [DOI] [PubMed] [Google Scholar]
- 72.Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PloS one. 2012;7(2):e31901. doi: 10.1371/journal.pone.0031901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nature reviews. Genetics. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 74.Marteau T, Senior V, Humphries SE, Bobrow M, Cranston T, et al. Psychological impact of genetic testing for familial hypercholesterolemia within a previously aware population: a randomized controlled trial. American journal of medical genetics. Part A. 2004;128A(3):285–93. doi: 10.1002/ajmg.a.30102. [DOI] [PubMed] [Google Scholar]
- 75.Martinez-Arguelles DB, Campioli E, Culty M, Zirkin BR, Papadopoulos V. Fetal origin of endocrine dysfunction in the adult: the phthalate model. The Journal of steroid biochemistry and molecular biology. 2013;137:5–17. doi: 10.1016/j.jsbmb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 76.McBride CM, Koehly LM, Sanderson SC, Kaphingst KA. The behavioral response to personalized genetic information: will genetic risk profiles motivate individuals and families to choose more healthful behaviors? Annual review of public health. 2010;31:89–103. doi: 10.1146/annurev.publhealth.012809.103532. [DOI] [PubMed] [Google Scholar]
- 77.McCandless SE, Chandrasekar R, Linard S, Kikano S, Rice L. Sequencing from dried blood spots in infants with “false positive” newborn screen for mcad deficiency. Molecular genetics and metabolism. 2013;108(1):51–55. doi: 10.1016/j.ymgme.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCann PJC. Agency discretion and public health service delivery. Health services research. 2009;44(5 Pt 2):1897–1908. doi: 10.1111/j.1475-6773.2009.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature reviews. Genetics. 2008;9(5):356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 80.McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA : the journal of the American Medical Association. 2008;300(22):2669–71. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. England Journal of Medicine. 2012:2012–14. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 82.Motsinger-Reif AA, Jorgenson E, Relling MV, Kroetz DL, Weinshilboum R, et al. Genome-wide association studies in pharmacogenomics: successes and lessons. Pharmacogenetics and genomics. 2013;23(8):383–94. doi: 10.1097/FPC.0b013e32833d7b45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moyer VA, Calonge N, Teutsch SM, Botkin JR. Expanding newborn screening: process, policy, and priorities. The Hastings Center report. 2008;38(3):32–39. doi: 10.1353/hcr.0.0011. [DOI] [PubMed] [Google Scholar]
- 84.Murrell A, Rakyan VK, Beck S. From genome to epigenome. Human molecular genetics. 2005;14:R3–R10. doi: 10.1093/hmg/ddi110. Spec No. [DOI] [PubMed] [Google Scholar]
- 85.Nahar MS, Liao C, Kannan K, Dolinoy DC. Fetal liver bisphenol a concentrations and biotransformation gene expression reveal variable exposure and altered capacity for metabolism in humans. Journal of biochemical and molecular toxicology. 2013;27(2):116–23. doi: 10.1002/jbt.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.National Newborn Screening and Genetics Resource Center . National newborn screening report - 2000. Austin, TX: 2003. [Google Scholar]
- 87.National Society of Genetic Counselors Learn about Genetic Counselors. 2013 http://nsgc.org/p/cm/ld/fid=43.
- 88.Neumann PJ, Cohen JT, Hammitt JK, Concannon TW, Auerbach HR, et al. Willingness-to-pay for predictive tests with no immediate treatment implications: a survey of us residents. Health economics. 2012;21(3):238–51. doi: 10.1002/hec.1704. [DOI] [PubMed] [Google Scholar]
- 89.Neumann PJ, Hammitt JK, Mueller C, Fillit HM, Hill J, et al. Public attitudes about genetic testing for alzheimer's disease. Health affairs (Project Hope) 2001;20(5):252–64. doi: 10.1377/hlthaff.20.5.252. [DOI] [PubMed] [Google Scholar]
- 90.Newbold RR. Lessons learned from perinatal exposure to diethylstilbestrol. Toxicology and applied pharmacology. 2004;199(2):142–50. doi: 10.1016/j.taap.2003.11.033. [DOI] [PubMed] [Google Scholar]
- 91.Newborn screening expands: recommendations for pediatricians and medical homes--implications for the system Pediatrics. 2008;121(1):192–217. doi: 10.1542/peds.2007-3021. [DOI] [PubMed] [Google Scholar]
- 92.Nguyen CP, Goodman LH. Fetal risk in diagnostic radiology. Seminars in ultrasound, CT, and MR. 2012;33(1):4–10. doi: 10.1053/j.sult.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 93.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and snp calling from next-generation sequencing data. Nature reviews. Genetics. 2011;12(6):443–51. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova MI, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PloS one. 2012;7(5):e36129. doi: 10.1371/journal.pone.0036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petronis A. Epigenetics and twins: three variations on the theme. Trends in genetics : TIG. 2006;22(7):347–50. doi: 10.1016/j.tig.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 96.Phelan JC, Yang LH, Cruz-Rojas R. Effects of attributing serious mental illnesses to genetic causes on orientations to treatment. Psychiatric services (Washington, D.C.) 2006;57(3):382–87. doi: 10.1176/appi.ps.57.3.382. [DOI] [PubMed] [Google Scholar]
- 97.Platt LD, Koch R, Hanley WB, Levy HL, Matalon R, et al. The international study of pregnancy outcome in women with maternal phenylketonuria: report of a 12-year study. American journal of obstetrics and gynecology. 2000;182(2):326–33. doi: 10.1016/s0002-9378(00)70219-5. [DOI] [PubMed] [Google Scholar]
- 98.Ravitsky V, Wilfond BS. Disclosing individual genetic results to research participants. The American journal of bioethics : AJOB. 6(6):8–17. doi: 10.1080/15265160600934772. [DOI] [PubMed] [Google Scholar]
- 99.Roberts JS, LaRusse SA, Katzen H, Whitehouse PJ, Barber M, et al. Reasons for seeking genetic susceptibility testing among first-degree relatives of people with alzheimer disease. Alzheimer disease and associated disorders. 2003;17(2):86–93. doi: 10.1097/00002093-200304000-00006. [DOI] [PubMed] [Google Scholar]
- 100.Roberts JS, Ostergren J. Direct-to-consumer genetic testing and personal genomics services: a review of recent empirical studies. Current genetic medicine reports. 2013;1(3):182–200. doi: 10.1007/s40142-013-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rothstein M a. Epigenetic exceptionalism. The Journal of law, medicine & ethics : a journal of the American Society of Law, Medicine & Ethics. 2013;41(3):733–36. doi: 10.1111/jlme.12083. [DOI] [PubMed] [Google Scholar]
- 102.Roychowdhury S, Chinnaiyan AM. Advancing precision medicine for prostate cancer through genomics. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(15):1866–73. doi: 10.1200/JCO.2012.45.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sankar P, Cho MK, Condit CM, Hunt LM, Koenig B, et al. Genetic research and health disparities. JAMA : the journal of the American Medical Association. 2004;291(24):2985–89. doi: 10.1001/jama.291.24.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Senior V, Marteau TM. Causal attributions for raised cholesterol and perceptions of effective risk-reduction: self-regulation strategies for an increased risk of coronary heart disease. Psychology & Health. 2007;22(6):699–717. [Google Scholar]
- 105.Shalowitz DI, Miller FG. Communicating the results of clinical research to participants: attitudes, practices, and future directions. PLoS medicine. 2008;5(5):e91. doi: 10.1371/journal.pmed.0050091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, et al. Genome-wide profiling of dna methylation reveals a class of normally methylated cpg island promoters. PLoS genetics. 2007;3(10):2023–36. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singleton A, Erby LH, Foisie KV, Kaphingst KA. Informed choice in direct-to-consumer genetic testing (dtcgt) websites: a content analysis of benefits, risks, and limitations. J Genet Couns. 2012;21(3):433–39. doi: 10.1007/s10897-011-9474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of endocrine disruptors. Reproductive toxicology (Elmsford, N.Y.) 2011;31(3):337–43. doi: 10.1016/j.reprotox.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Solomon BD, Pineda-Alvarez DE, Bear KA, Mullikin JC, Evans JP. Applying genomic analysis to newborn screening. Molecular syndromology. 2012;3(2):59–67. doi: 10.1159/000341253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stoddard JJ, Farrell PM. State-to-state variations in newborn screening policies. Archives of pediatrics & adolescent medicine. 1997;151(6):561–64. doi: 10.1001/archpedi.1997.02170430027005. [DOI] [PubMed] [Google Scholar]
- 111.Tarini BA. Storage and use of residual newborn screening blood spots: a public policy emergency. Genetics in medicine : official journal of the American College of Medical Genetics. 2011;13(7):619–20. doi: 10.1097/GIM.0b013e31822176df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tarini BA, Christakis DA, Welch HG. State newborn screening in the tandem mass spectrometry era: more tests, more false-positive results. Pediatrics. 2006;118(2):448–56. doi: 10.1542/peds.2005-2026. [DOI] [PubMed] [Google Scholar]
- 113.Tarini BA, Goldenberg AJ. Ethical issues with newborn screening in the genomics era. 2012 doi: 10.1146/annurev-genom-090711-163741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taylor DH, Cook-Deegan RM, Hiraki S, Roberts JS, Blazer DG, Green RC. Genetic testing for alzheimer's and long-term care insurance. Health affairs (Project Hope) 2010;29(1):102–8. doi: 10.1377/hlthaff.2009.0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Human molecular genetics. 2013;22(R1):R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Timmermans S, Buchbinder M. Patients-in-waiting: living between sickness and health in the genomics era. Journal of health and social behavior. 2010;51(4):408–23. doi: 10.1177/0022146510386794. [DOI] [PubMed] [Google Scholar]
- 117.Tluczek A, Koscik RL, Farrell PM, Rock MJ. Psychosocial risk associated with newborn screening for cystic fibrosis: parents’ experience while awaiting the sweat-test appointment. Pediatrics. 2005;115(6):1692–1703. doi: 10.1542/peds.2004-0275. [DOI] [PubMed] [Google Scholar]
- 118.Vandenberg LN, Colborn T, Hayes TB, Heindel JJ, Jacobs DR, et al. Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocrine reviews. 2012;33(3):378–455. doi: 10.1210/er.2011-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vernarelli JA, Roberts JS, Hiraki S, Chen CA, Cupples LA, Green RC. Effect of alzheimer disease genetic risk disclosure on dietary supplement use. The American journal of clinical nutrition. 2010;91(5):1402–7. doi: 10.3945/ajcn.2009.28981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Waddington CH. Organisers & genes. Cambridge University Press; Cambridge, UK: 1940. [Google Scholar]
- 121.Waisbren SE, Albers S, Amato S, Ampola M, Brewster TG, et al. Effect of expanded newborn screening for biochemical genetic disorders on child outcomes and parental stress. JAMA : the journal of the American Medical Association. 2003;290(19):2564–72. doi: 10.1001/jama.290.19.2564. [DOI] [PubMed] [Google Scholar]
- 122.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Molecular and cellular biology. 2003;23(15):5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wei C, Lu Q, Khoo SK, Lenski M, Fichorova RN, et al. Comparison of frozen and unfrozen blood spots for gene expression studies. The Journal of pediatrics. 2013 doi: 10.1016/j.jpeds.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wenzel A, Gorman LL, O'Hara MW, Stuart S. The occurrence of panic and obsessive compulsive symptoms in women with postpartum dysphoria: a prospective study. Archives of Women's Mental Health. 2001;4(1):5–12. [Google Scholar]
- 125.Wenzel A, Haugen EN, Jackson LC, Brendle JR. Anxiety symptoms and disorders at eight weeks postpartum. Journal of anxiety disorders. 2005;19(3):295–311. doi: 10.1016/j.janxdis.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 126.Wenzel A, Haugen EN, Jackson LC, Robinson K. Prevalence of generalized anxiety at eight weeks postpartum. Archives of women's mental health. 2003;6(1):43–49. doi: 10.1007/s00737-002-0154-2. [DOI] [PubMed] [Google Scholar]
- 127.Wetterstrand K. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) http://www.genome.gov/sequencingcosts.
- 128.Wigle DT, Arbuckle TE, Walker M, Wade MG, Liu S, Krewski D. Environmental hazards: evidence for effects on child health. Journal of toxicology and environmental health. Part B, Critical reviews. 2007;10(1-2):3–39. doi: 10.1080/10937400601034563. [DOI] [PubMed] [Google Scholar]
- 129.Wilfond B, Ross LF. From genetics to genomics: ethics, policy, and parental decision-making. Journal of pediatric psychology. 2009;34(6):639–47. doi: 10.1093/jpepsy/jsn075. [DOI] [PubMed] [Google Scholar]
- 130.Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, et al. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genetics in medicine : official journal of the American College of Medical Genetics. 2012;14(4):361–84. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woloshin S, Schwartz LM, Welch HG. Know Your Chances: Understanding Health Statistics. University of California Press; 2008. [PubMed] [Google Scholar]
- 132.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annual review of genomics and human genetics. 2008;9:233–57. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 133.Zick CD, Mathews CJ, Roberts JS, Cook-Deegan R, Pokorski RJ, Green RC. Genetic testing for alzheimer's disease and its impact on insurance purchasing behavior. Health affairs (Project Hope) 2005;24(2):483–90. doi: 10.1377/hlthaff.24.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]


