Abstract
We engineered patterned co-cultures of primary neurons and astrocytes on polyelectrolyte multilayer (PEM) films without the aid of adhesive proteins/ligands to study the oxidative stress mediated by astrocytes on neuronal cells. A number of studies have explored engineering co-culture of neurons and astrocytes predominantly using cell lines rather than primary cells owing to the difficulties involved in attaching primary cells onto synthetic surfaces. To our knowledge this is the first demonstration of patterned co-culture of primary neurons and astrocytes for studying neuronal metabolism. In our study, we used synthetic polymers, namely poly(diallyldimethylammoniumchloride) (PDAC) and sulfonated poly(styrene) (SPS) as the polycation and polyanion, respectively, to build the multilayers. Primary neurons attached and spread preferentially on SPS surfaces, while primary astrocytes attached to both SPS and PDAC surfaces. SPS patterns were formed on PEM surfaces, either by microcontact printing SPS onto PDAC surfaces or vice-versa, to obtain patterns of primary neurons and patterned co-cultures of primary neurons and astrocytes. We further used the patterned co-culture system to study the neuronal response to elevated levels of free fatty acids as compared to the response in separated monoculture by measuring the level of reactive oxygen species (ROS; a widely accepted marker of oxidative stress). The elevation in the ROS levels was observed to occur earlier in the patterned co-culture system than in the separated monoculture system. The results suggest that this technique may provide a useful tool for engineering neuronal co-culture systems, that may more accurately capture neuronal function and metabolism, and thus could be used to obtain valuable insights into neuronal cell function and perhaps even the pathogenesis of neurodegenerative diseases.
1. Introduction
This work describes the engineering of patterned co-cultures of primary neurons and astrocytes on polyelectrolyte multilayer (PEM) films without the aid of adhesive proteins/ligands. Cell-cell communication between primary neurons and astrocytes is crucial for the development, repair and metabolism of neuronal systems.[1] A co-culture system allows for the neuronal responses that may be mediated by the astrocytes. Although several studies have explored co-cultures of neurons and astrocytes to study neuronal metabolism or the pathogenesis of neurodegenerative diseases, using predominantly trans-well, conditioned media or random co-cultures,[2–5] none thus far has explored patterned co-culture of primary neurons and astrocytes. We developed a patterned co-culture system using PEM films by first controlling the attachment of primary neurons to produce patterns of neurons. This was subsequently used to develop patterned co-cultures of primary neurons and astrocytes. In the current study we evaluated the effect of saturated free fatty acids (FFAs) on co-culture systems as compared with monocultures of neurons and astrocytes. The results obtained with a patterned co-culture system could provide insights into neuronal cell function and in understanding the pathogenesis of neurodegenerative diseases.
In vitro assays employed to monitor neuronal function and response to particular molecules often involve culturing neuron-like cell lines in the presence of immobilized protein substrata or gradients of purified proteins.[6–8] Significant insight has been gained from these studies with respect to intrinsic properties of guidance molecules,[9] growth rate, and signaling cascades within the growth cone.[10] Nevertheless, neurons and astrocytes function as interdependent networks with bidirectional communication during development. Heterotypic interactions between neurons and astrocytes exist in brain tissues, so it is important to mimic this cell-cell communication when studying the neuronal system. During the perinatal period, astrocytes mediate intercellular communications which are important for neuronal differentiation and plasticity.[11] Astrocytes have also been shown to mediate numerous functions of neurons in brain.[11–13] In this study, we used PEMs to pattern primary neurons and astrocytes to allow neuronal responses to be studied in the context of a complex cellular environment, namely, when in direct contact with astrocytes.[14–17]
Cell-cell interactions are central to the function of many tissues, e.g., blood vessels form when endothelial cells are allowed to interact with smooth muscle cells[18] and nervous system function depends upon proper interactions between neuronal and glia cells.[19] The ability to mimic such interactions in vitro is important in cell biology studies as well as tissue engineering applications. Neuronal function is mediated in part by the complex interactions among different cell types including astrocytes. A strocytes are glial cells that are in close proximity to the neurons and play multiple roles in the functioning of the brain. They can sense neuronal activity through neurotransmitter receptors[20] and provide direct neurotrophic factors to support the neurons.[21] Previous studies have shown that astrocytes mediate both positive and negative responses in neuronal cells. Primary neurons co-cultured randomly with astrocytes showed reduced toxicity to ammonia[22] but an increased sensitivity to the toxicity of glutamate[23] as compared to pure neuronal cultures. Thus the ability to create and maintain neuronal and astrocyte co-cultures are crucial in developing neuronal systems to study neuron function and interactions between neurons and glial cells.[1,4,24]
Different approaches have been used to create spatially defined co-cultures of two different cell types.[25,26] However, these techniques have certain limitations. They 1) typically require adhesive proteins for cell attachment,[25] 2) are limited to specific parallel geometries defined by laminar flow patterns,[26] or 3) have been used predominantly with cell lines rather than primary cells due to the difficulties involved in attaching primary cells onto synthetic surfaces.[27–31] PEMs have been shown to be excellent candidates for biomaterial applications[31–39] and provide flexibility in building complex three-dimensional architectures.[40] The PEM surfaces also provide an ability to control the arrangement of multiple cell types with subcellular resolution.[27,35,36,41–43] We previously reported that primary hepatocytes attached and spread on PEM films.[42,44] Here we report that primary neurons can be cultured on PEM films and further co-cultured with astrocytes in patterned co-cultures and used for studying the effect of saturated FFAs on neuronal cell function.
2. Results and Discussion
To investigate the effects of PEM films on primary neuronal cells and astrocytes, we assessed the response of primary neurons and astrocytes over continuous culture. We used synthetic polymers, namely poly(diallyldimethylammonium chloride) (PDAC) and poly(4-styrenesulfonic acid) (SPS), as the polycation and polyanion, respectively, to build the PEM films. The PEM surfaces used for the cell studies were not coated with adhesive proteins or ligands.
2.1. Primary Neurons and Astrocyte Adhesion on Various PEMs
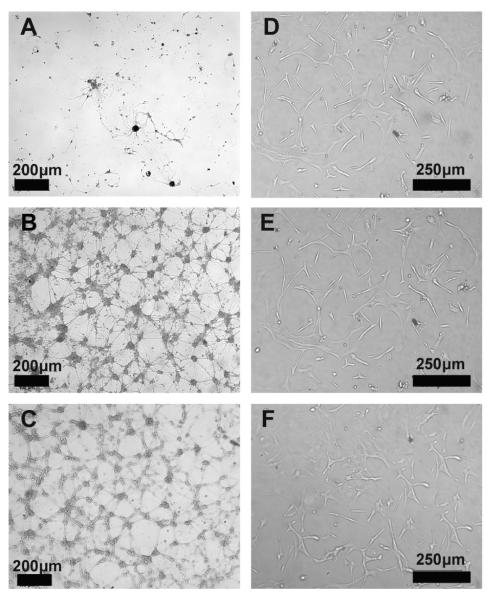
Figure 1 and Table 1 compare the adhesion of primary neurons and astrocytes on PEM surfaces to PLL coated TCPS control. The difference in the projected cell area for primary neu rons and astrocytes on the different surfaces is shown in Table 1. The number of primary neurons that attached on the SPS, LPEI and BPEI surfaces on Day 7 (234, 241, 210 cells/mm2, respectively) were comparable to the number of neurons that attached on the PLL coated TCPS control surfaces on Day 7 (250 cells/mm2), see Table 1. In contrast, fewer cells attached and spread on PEM films with PDAC as the topmost surface (Fig. 1 and Table 1). By day 7 most of the primary neurons (10 cells/mm2) lifted off the PDAC surfaces. Primary neurons attached and spread on SPS surfaces and the morphology of the cells were comparable to the control, see Figure 1B and C. Astrocytes attached to both PDAC and SPS surfaces as shown in Figure 1D–F. The number of astrocytes attached to both PDAC (178 cells/mm2) and SPS surfaces (183 cells/mm2) were comparable to the control surfaces (192 cells/mm2), see Figure 1D–F, Table 1. Astrocytes, unlike primary neuronal cells, proliferate, therefore their growth on the PEMs vs. PLL coated TCPS control surfaces are reported in Table 2.
Figure 1.
Phase contrast images of primary neurons and astrocytes after 7 days and 3 days in culture, respectively, on PEM surfaces. Primary neurons on A) (PDAC/SPS)10.5 – PDAC topmost surface, B) (PDAC/SPS)10 – SPS topmost surface, C) poly(lysine) (PLL)-control surfaces. Astrocytes on D) (PDAC/SPS)10.5 – PDAC topmost surface, E) (PDAC/SPS)10 – SPS topmost surface, F) poly(lysine)-control surfaces (Scale bars: A–C: 100 μm, D–F: 250 μm).
Table 1.
Primary neurons and astrocytes cell numbers on the projected area on the different surfaces used in the study after 5 days in culture. Student's t-test was used for analyzing the differences between the cell number on the various surfaces.
| Surfaces | Primary neurons [cells mm−2] (2 × 106/substrate initial cone) | Astrocytes [cells mm−2] (4 × 105/substrate initial cone) |
|---|---|---|
| PLL coated TCPS | 250 ± 19 | 792 ± 19 |
| PDAC | 10 ± 2[a] | 778 ± 20 |
| SPS | 234 ± 18 | 783 ± 17 |
| LPEI | 241 ± 23 | 761 ± 21 |
| BPEI | 210 ± 15 | 791 ± 14 |
p<0.05 compared with cell adhesion on poly(lysine) (PLL) coated TCPS control.
Table 2.
Astrocyte numbers on the projected area on the different surfaces used in the study after 1, 3, and 5 days.
| Surfaces | Astrocytes [cells mm−2] (4×105, initial concentration) |
Rate of proliferation [cells on projected area mm−2 h−1] | ||
|---|---|---|---|---|
| 1 day | 3 days | 5 days | ||
| PLL coated TCPS | 176 ± 8 | 401 ± 11 | 792 ± 19 | 6.42 ± 0.7 |
| PDAC | 164 ± 9 | 398 ± 17 | 778 ± 20 | 6.39 ± 0.4 |
| SPS | 169 ± 7 | 381 ± 13 | 783 ± 17 | 6.40 ± 0.5 |
| LPEI | 166 ± 10 | 407 ± 11 | 761 ± 21 | 6.20 ± 0.9 |
| BPEI | 155 ± 11 | 400 ± 15 | 791 ± 14 | 6.62 ± 0.5 |
The choice of SPS and PDAC was based on previous studies, wherein the latter material was shown to be resistant to attachment by primary hepatocytes,[44] smooth muscle cells,[45] and primary neuronal cells,[46] while SPS was cytophilic for all the cell types evaluated. The combination of these two polymers facilitated the patterning of co-cultures of primary neurons and astrocytes, thus allowing direct contact between the cells, more akin to the cell-cell contact which exists in the brain than can be achieved with random co-cultures.[14–17]
In addition, primary neurons were grown on various positive surfaces, such as (LPEI/SPS)10.5 and (BPEI/SPS)10.5, to evaluate whether the difference in cell adhesion and spreading was due to the electrostatic charge. Unlike the PDAC surfaces, primary neurons attached and spread on the positively charged LPEI and BPEI surfaces, suggesting that the electrostatic charge was not likely the mechanism for the difference in cell adhesion observed on the PDAC substrates. The functional group involved in enhancing cell adhesion on the LPEI and BPEI surfaces may be the primary and secondary amine groups. It is notable that primary and secondary amine groups are present in cell adhesion ligands and proteins such as fibronectin, collagen and poly-l-lysine, which contain primary and secondary amines, whereas PDAC contains a quaternary amine group. This may account in part for the difference in cell adhesion observed among these polyelectrolytes. Nevertheless, this does not preclude the possibility of other effects that may contribute to the observed differences in adhesion and spreading.
2.2. Patterned versus Random Co-Culture
We capitalized upon the cell adhesive and resistive property of SPS and PDAC, respectively, to create patterns of primary neurons and co-cultures of neurons and astrocytes as shown in Figure 2. Figure 3 illustrates the random and patterned primary neuron monocultures and co-cultures of primary neurons and astrocytes. Figure 3A and B shows the fluores cent images of random mono-culture of primary neurons and random co-culture of primary neurons and astrocytes, respectively, on top of non-patterned SPS surfaces. Figure 3C and D illustrates the fluorescent images of patterned primary neurons and patterned co-culture of neurons and astrocytes, respectively. PDAC patterns were formed on SPS surfaces (or vice-versa) and subsequently seeded with neuronal cells. Primary neurons preferentially attached on the SPS patterns when they were seeded on the PEM surfaces (Fig. 3C). Astrocytes were subsequently seeded onto the patterns of neurons and the cells attached onto the open PDAC regions and resulted in patterned co-cultures of neurons and astrocytes (Fig. 3D). Since primary neurons do not proliferate but form neurites which leaves areas that are exposed and unoccupied (gaps) and astrocytes, being small (in red ~ 10–20 micrometer as compared to 40–60 micrometer for neurons), less selective and proliferating, can easily fill these gaps. As observed in Figure 3D, the neurons are on the SPS surfaces while the astrocytes are on the PDAC surfaces, although some are present on the SPS surfaces, resulting in patterned co-cultures. This effect is commonly observed and is very similar to the cell patterns seen in other patterned co-culture studies.[27,47] Studies show that random, dissociated neurons in culture develop physiological responses to neurotransmitters[48,49] and self-organize into neuronal networks,[49–52] however, they lack the structure normally present within the nervous system, where the neurons reside in specific regions with numerous network connectivity. The advantage of patterned co-cultures of neurons and astrocytes is that it can allow for precise control of both the direction of neurite extension and degree of contact between the neurons and astrocytes, unlike the random co-culture, which is important for repair and regeneration of the nervous systems.[53] It has also been demonstrated by Wheeler and co-workers that patterned neurons synapse with each other, release neurotransmitters and develop electrical activity.[54–56] Furthermore, restricting neurons to patterns has been shown to enhance the cellular activity such as glutamine secretion and electrical activity as compared to random monocultures of neurons.[57,58]
Figure 2.
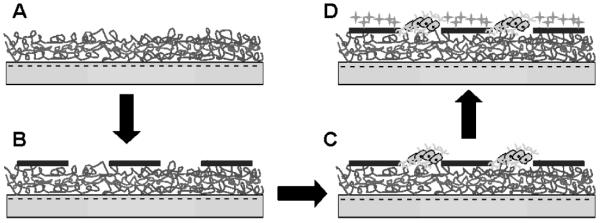
Scheme illustrating the approach for engineering patterned co-culture of primary neurons and astrocytes on PEM surfaces. A) First, (PDAC/SPS)10 PEMs were built on top of the TCPS surfaces with SPS as the topmost surface. B) Second, patterns of PDAC were formed on PEM surfaces by microcontact printing (μCP) PDAC onto the PEM surfaces. C) Third, patterns of primary neurons were formed by capitalizing on the preferential attachment of neurons to SPS surfaces. D) Fourth, since astrocytes, unlike the primary neurons, attached to both surfaces, astrocytes were subsequently seeded onto the patterns of neurons and attached onto the open PDAC regions and resulted in patterned co-cultures of neurons and astrocytes. (The scheme is not drawn to represent the true nature of the co-culture system.)
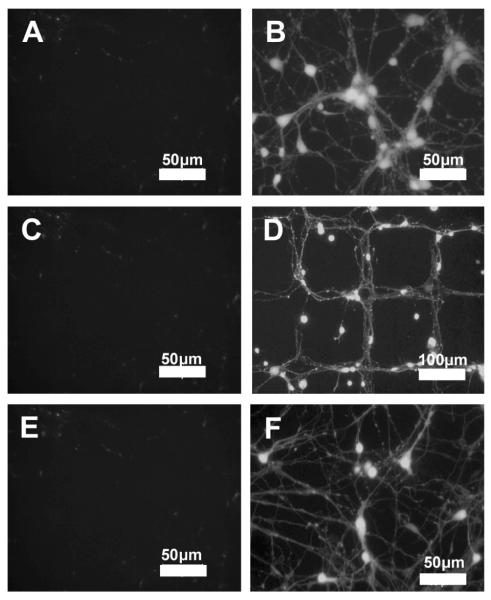
Figure 3.
Fluorescent images of primary neurons and astrocytes co-culture on SPS surfaces A) Random neuron monocultures (green) after 7 days in culture, B) Random co-culture of neurons (green) and astrocytes (red) after seeding astrocytes. C) Patterned primary neurons on SPS patterns after 7 days in culture (D) Patterned co-culture of neurons (green) and astrocytes (red) after seeding astrocytes (Scale bars: 200 m).
2.3. Advantages of Our Co-Culture System over Other Systems
In the present study, we developed patterned neuron-astrocyte co-culture system using PEM films and μCP which has several advantages over the methods generally used for patterning surfaces for co-cultures. The advantages include its high fidelity, ease of duplication, ability to print a variety of molecules with nanometer resolution and without the need for dust-free environments and harsh chemical treatments.[59–61] In addition, we were able to achieve primary neuron-astrocyte interactions without adhesive proteins, which was not possible with the other methods.[27,62]
Numerous studies use monocultures of neurons and astrocytes to study neuronal systems, however, monocultures of neurons and astrocytes do not accurately capture the diverse biological responses of living brain tissue.[63] Transwells or neurons cultured onto cover slips that are subsequently added to the center of a confluent culture of astrocytes, are the most common forms of co-cultures of neurons and astrocytes.[2,3] However, these culture systems imposed an artificial boundary that precluded cell-cell interactions. Finally, previous studies with patterned surfaces predominantly used cell lines rather than primary cells to engineer patterned co-cultures of neurons and astrocytes.[27,31,64]
2.4. Reactive Oxygen Species (ROS) Studies on Co-Culture Systems
We assessed the behavior of the neurons by evaluating their response to elevated levels of saturated FFAs, namely the accumulation of ROS. Various studies have implicated the involvement of saturated fatty acids in the pathogenesis of Alzheimer's disease (AD).[5,65] Saturated FFAs, palmitic and stearic acids, have been shown to cause increased ROS and in turn increased amyloidogenesis and tau hyperphosphorylation in primary rat cortical neurons, the two hallmarks of AD.[5,66] Previously, we showed that intracellular levels of ROS were elevated when the neurons were cultured with conditioned media from astrocytes treated with palmitic acid (PA) as compared to controls (i.e., cultured with conditioned media from astrocytes treated with 5 % bovine serum albumin (BSA) media).[5,66] This previous work was performed using conditioned media, whereby neurons and astrocytes were cultured separately, which is not representative of in vivo conditions. The goal of this study was to assess whether the neuronal behavior mediated by the astrogial FFA metabolism altered when the two types of cells were brought in direct contact. Thus, we compared the astrocyte-mediated response of neurons in the patterned co-culture system with the previous, more common, method of culture.
The earlier study used random neuronal monocultures to evaluate the effect of FFAs on primary neurons.[5,67] As shown in Figure 4A and B, the intracellular levels of ROS were elevated in the neurons cultured in conditioned media from astrocytes treated with PA (Fig. 4B) as compared to the controls (Fig. 4A). The astrocytes were treated with 0.2 mM PA for 12 h prior to transfer of the conditioned media to neuronal cultures for 24 h (Fig. 4B). By contrast, random monocultures of neurons treated directly with PA did not show oxidative stress-induced effects in the neurons (Fig. 4A), which we have also shown previously.[5,67] This suggests, as we have found previously, that FFA-induced oxidative stress observed in the neurons are mediated by astrocytes. Therefore, we exposed co-cultures, both patterned and random, of neurons and astrocytes to elevated levels of FFAs. When the co-cultures were treated with BSA, an elevation in ROS was not observed (Fig. 4C and E). In Figure 4D, we treated the co-culture system with 0.2 mM PA for 12 h and then performed the ROS measurements. The patterned neuron-astrocyte co-culture systems treated with fatty acids for 12 h had higher levels of ROS (Fig. 4D) compared to the neurons cultured for 24 h with conditioned media from astrocytes treated with fatty acids (separated monoculture system, Fig. 4B). We also measured the ROS level in a random co-culture system (Fig. 4F) and found the ROS level was higher in the patterned co-culture than in the random co-culture system. The level of ROS was further quantified by measuring the green fluorescence intensity of DCF from microscopic images using National Institutes of Health ImageJ software by pixel quantification (available at rsb.nih.gov/ij) as shown in Figure 5. This method of quantification has been used to quantify the fluorescence intensity in numerous studies.[68,69] The level of ROS on the patterned neuron-astrocyte co-culture system treated with fatty acids for 12h was higher compared to the random co-culture of primary neurons and astrocytes (Fig. 5). In both co-culture systems, since both the astrocytes and the neuronal cells were on the same surface, the elevation in the ROS levels was observed earlier than in the separated monoculture system. The earlier response (i.e., faster elevation of ROS levels in the co-cultures) may be because both types of cells were in direct contact and in the same culture media, which allowed the neuronal cells to response to soluble factors, such as oxidative-stress inducing cytokines or intermediate me tabolites, as they secrete from the astrocytes. Patterned co-culture of primary neurons and astrocytes, however is physiologically more relevant than the random co-culture of these cell types since neurons and astrocytes are arranged in a highly ordered cytoarchitecture in vivo in the brain.[14–17] In further sup port of the pattern co-culture system, previous studies have found that random co-cultures lack diversity of homotypic interactions and are unable to engineer variable local contact with the other cell type.[70,71] Although random, dissociated neurons in culture develop physiological responses to neuro-transmitters,[48,72] they lack the structure normally present with in the nervous system, where the neurons reside in specific regions and project their processes in a specific manner. Furthermore, previous studies have shown that restricting neurons to patterns appeared to enhance their recordable network and cellular activity as compared to random neuronal monoculture.[56,57,73] Taken together, this suggests that there is a need for spatial arrangement of cells in a controlled environment that incorporates cell-cell interactions.
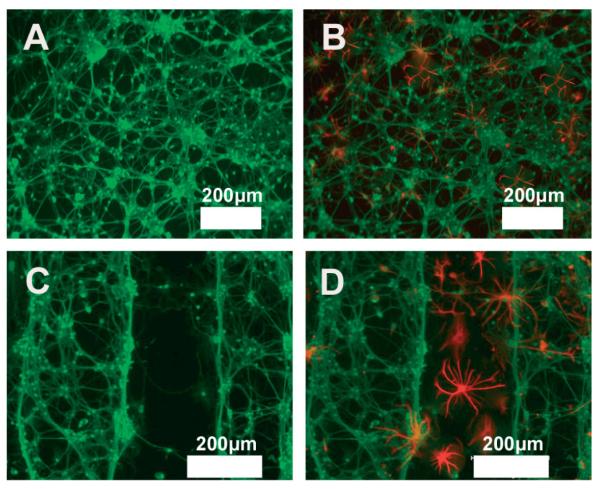
Figure 4.
Intracellular accumulation of ROS in neurons. Fluorescent images of ROS accumulation in primary neurons monoculture system treated with astrocytes-conditioned media for 24 h after treating astrocytes for 12 h with A) 5 % BSA (control) and B) 0.2 mM of PA. Fluorescent images of ROS accumulation in patterned primary neuron-astrocytes co-culture system treated for 12 h with C) 5 % BSA (control) and D) 0.2 mM of palmi-tate (PA). E) Fluorescent image of ROS accumulation in primary neurons directly treated with 0.2 mM of PA. F) Fluorescent image of ROS accumulation in random co-culture of neurons-astrocytes treated with 0.2 mM of PA for 12 h (Scale bars: A–C, E, F: 50 μm, D: 100 μm).
Figure 5.
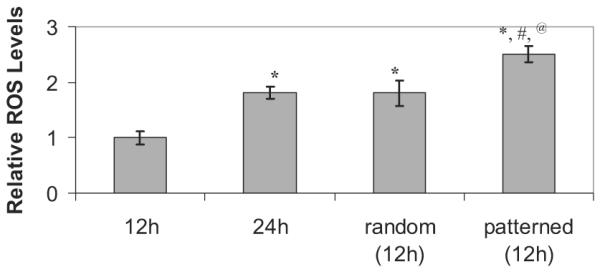
Quantification of intracellular accumulation of ROS in neurons using ImageJ software. The ROS levels were normalized to the total number of neuronal cells seeded. Data represents mean ± S.E. of three independent experiments (* p < 0.05 compared with 12h treatment of mono-cultures of neurons, # p < 0.05 compared with 12h treatment of random co-cultures of neurons and astrocytes, @ p < 0.05 compared with 24h treatment of monocultures of neurons).
3. Conclusions
In conclusion, the present work illustrates a method for controlling primary neuronal cell adhesion on synthetic surfaces. PEMs were used to produce defined cell-resistant and cell-adhesive surfaces depending on the topmost surface and the type of cell used. We observed that primary neurons attached and spread onto PEM films with SPS as the top most surface. We also demonstrated that the layer-by-layer deposition and μCP of ionic polymers formed templates for patterned co-cultures of primary neurons with astrocytes. We used the patterned co-culture system where the neurons are in direct contact with astrocytes to study the neuronal response to elevated levels of FFAs. The neuronal cells in the co-culture responded quicker than in the separated monoculture system. Taken together, this technique provides a useful tool for engineering neuronal co-culture systems, which may more accurately capture neuronal function and metabolism in normal versus diseased states.
4. Experimental
Materials
Poly(diallyldimethylammonium chloride) (PDAC) (Mw ~ 100000–200000) as a 20 wt % solution, sulfonated poly(styrene), sodium salt (SPS) (Mw ~ 70000), linear polyethyleneimine (LPEI), branched polyethyleneimine (BPEI), fluorosilanes, poly-L-lysine( (PLL) and sodium chloride were purchased from Aldrich (Milwaukee, WI). All polymers were used without further purification. Poly(dimethylsiloxane) (PDMS) from the Sylgard 184 silicone elastomer kit (Dow Corning, Midland, MI) was used to prepare stamps. The PDMS stamps were used for microcontact printing [74]. Dulbecco's Modified Eagle's Medium (DMEM) and all other media were purchased from Invitrogen, CA, USA, 10 % horse serum, 25 mM glucose, 10 % fetal calf serum (FCS), 10 mM HEPES, 100 IU ml−1 penicillin, and 0.1 mg ml−1 streptomycine were purchased from Sigma, MO, USA, 2 mM glutamine (BioSource International, CA, USA), cytosine-β-arabinofuranoside (Ara-C) was purchased from Calbiochem, CA, USA.
For neurons and astrocytes immunostaining, neurofilament (Sigma, MO, USA) for neurons and GFAP (Dako, CA, USA) for astrocytes was used. Primary antibodies were detected with Fluorescein conjugated and rhodamine conjugated (Chemicon, CA, USA) secondary antibodies for neurons and astrocytes, respectively. Intracellular reactive oxygen species (ROS) were detected by staining with the oxidant-sensitive dye 5-(6)-chloromethyl-2',7'-dichlorodihydrofluoresceindiacetate (CM-H2DCFDA, from Molecular Probes, CA, USA).
Preparation of Polyelectrolyte Multilayers
PDAC and SPS polymer solutions were prepared with deionized (DI) water at concentrations of 0.02 M and 0.01 M respectively, (based on the repeating unit molecular weight) with the addition of 0.1 M NaCl salt. Polyelectrolyte dipping solutions were prepared with DI water supplied by a Barnstead Nano-pure-UV 4 stage purifier (Barnstead International Dubuque, Iowa), equipped with a UV source and final 0.2 μm filter. Solutions were filtered with a 0.45 μm Acrodisc syringe filter (Pall Corporation) to remove particulates. The tissue culture polystyrene (TCPS) surfaces were subjected to a Harrick plasma cleaner (Harrick Scientific Corporation, Broading Ossining, NY) for 10 min at 0.15 Torr and 50 sccm flow of O2 in a plasma chamber. The layer-by-layer process was carried out in an automatic dipping machine (HMS programmable slide stainer from Zeiss Inc.). To form the first bilayer, the TCPS were immersed for 20 min in a polycation solution. Following two sets of 5 min rinses with agitation, the TCPS were subsequently placed in a poylanion solution and allowed to deposit for 20 min. Afterwards, the 6 well plates were rinsed twice for 5 min each. The samples were cleaned for 3 min in an ultrasonic cleaning bath after depositing a layer of polycation/polyanion pair. The sonication step removed weakly bounded polyelectrolytes on the substrate, forming uniform bilayers. This process was repeated to build multiple layers. All experiments were performed using ten (i.e., 20 layers) or ten and half bilayers (i.e., 21 layers).
Preparation of PDMS Stamps
An elastomeric stamp was made by curing PDMS on a microfabricated silicon master, which acts as a mold, to allow the surface topology of the stamp to form a negative replica of the master. The PDMS stamps were made by pouring a 10:1 solution of elastomer and initiator over a prepared silicon master. The silicon master was pretreated with fluorosilanes to facilitate the removal of the PDMS stamps from the silicon master. The mixture was allowed to cure overnight at 60 °C. The masters were prepared in the BioMEMS facilities at MGH East and consisted of various features (squares and lines).
Microcontact Printing of Polyelectrolytes
The polyelectrolytes were stamped onto the multilayer system using the polymer-on-polymer stamping process developed by Hammond and co-workers [75]. Aqueous solutions of 20 mM PDAC (or 10mM SPS) and 0.1 M NaCl in water were used to stamp the polymer from aqueous solution. Briefly, the PDMS stamps were placed in air plasma for 20 s before inking. The polymer solution, or ink (PDAC or SPS), was then applied using a cotton swab wet with the ink to the stamp surface. This inked PDMS stamp was then dried with N2 flow, and the stamp was placed on the multilayer platform and allowed to sit 20min. Following the stamping process, the patterned surface was rinsed thoroughly with deionized (DI) water applied directly to the film surface from a solvent squeeze bottle to remove any excess unbound polyelectrolyte. The 6-carboxyfluorescein (6-CF) green dye was used to visualize the PDAC patterns on PEM following the stamping and rinsing processes. The dye, which is negatively charged, preferentially stained the positively charged PDAC surface. We observed patterns of green regions providing evidence for the presence of PDAC patterns (data not shown).
Animals and Neuron Isolation
Primary cerebellar neurons were prepared from 8-day-old Sprague–Dawley rat pups (Charles River, Sulzfeld, Germany) as described previously [76]. Cells were dissociated from freshly dissected cerebella by mechanical disruption in the presence of trypsin and DNase and then plated in poly-L-lysine-precoated or PEM-coated six-well plates. Cells were seeded at a density of 2 × 106 cells/well in DMEM supplemented with 10 % fetal calf serum, 2 mM glutamine, and 20 μg mL−1 gentamycin. Three days after incubation (37 °C, 5 % CO2), the medium was subsequently replaced with 2 ml of cerebellum medium supplemented with 5 μm Arac to arrest the growth of non-neuronal cells. After 2 days, the neuronal culture was switched back to cerebellum medium without Ara-C. The experiments were performed on 6- to 7-day-old culture. Cultures generated by this method have been shown to contain > 95 % cerebellar granule neurons [77]. Astrocytes were prepared from 7-day-old Sprague–Dawley rat pups as described previously [78]. The cells were seeded in poly-L-ly-sine-precoated or PEM-coated six-well plates (4 × 105cells/well) in culture medium (90 % DMEM, 10 % FCS, 20 U mL−1 penicillin and 20 μg mL−1 streptomycin sulfate), and cultivated in an incubator (humidified, 10 % CO2). The cerebellum neurons were used in co-culture studies.
Primary cortical neurons were isolated from one-day-old Sprague–Dawley rat pups and cultured according to the published methods as described in Chandler et al. [79]. The cells were plated on poly-D-ly-sine-coated (control) or PEM coated, six-well plates at a concentration of 2 × 106 cells per well in fresh cortical medium (DMEM supplemented with 10 % horse serum, 25 mM glucose, 10 mM HEPES, 2 mM gluta-mine, 100 IU ml−1 penicillin, and 0.1 mg ml−1 streptomycine). Three days after incubation (37 °C, 5 % CO2), the medium was subsequently replaced with 2 ml of cortical medium supplemented with 5 μm Arac. After 2 days, the neuronal culture was switched back to cortical medium without Ara-C. The experiments were performed on 6- to 7-day-old culture. To obtain primary cultures of astroglial cells, the cortical cells from one-day-old Sprague–Dawley rat pups were cultured in DMEM/ Ham's F12 medium (1:1), 10 % fetal bovine serum (Biomeda, CA, USA), 100 IU ml−1 penicillin, and 0.1 mg ml−1 streptomycine. The cells were plated on poly-D-lysine or PEM coated, 6-well plates at a concentration of 2 × 106 cells per well. Cells were grown for 8–10 days (37 °C, 5 % CO2) and culture medium was changed every 2 days. Twenty-four hours prior to treatment with fatty acids, the medium was changed to neuronal cell culture medium. The cortical neurons were used in co-culture and ROS studies. Primary cortical neurons were used for the ROS studies as the oxidative stress-induced effects are higher in the affected regions (cortex and hippocampus) as compared to the unaffected areas (cerebellum) [80].
Neuron and Astrocytes Culture System
Primary neurons and astrocytes were cultured on PEM coated 6-well tissue culture polystyrene surfaces (TCPS). All the multilayer coated TCPS were sterilized by spraying with 70 % ethanol and exposing them to UV light before culturing the cells onto these surfaces. Poly-D-lysine-coated TCPS were used as controls in these studies. Samples were kept in the incubator where the temperature and humidity were properly controlled. A Leica inverted phase contrast microscope with Soft RT 3.5 software was used to capture images of cell density, morphology, and spreading on the multilayer surfaces.
Immunostaining of Primary Neurons and Astrocytes
To perform confocal immunofluorescence microscopic study, neurons and astrocytes cultures were fixed for 20 min in 4 % paraformaldehyde and permeabilized with 0.1 % Triton X-100 and 5 % goat serum (Invitrogen) in PBS. Cells were then labeled overnight at 4 °C with appropriate primary antibodies [1:50 neurofilament for neurons and 1:1000 glial fibrillary acidic protein (GFAP) for astrocytes) in 5 % goat serum in PBS. After three PBS washes, primary antibodies were detected with Fluorescein conjugated and rhodamine conjugated secondary antibodies for neurons and astrocytes, respectively. The cells were visualized with Leica fluorescent microscope.
Determination of Cell Size and the Number of Cells on the Projected Area
The Soft RT 3.5 software was used on the phase contrast images of the cells to determine the average area occupied by astrocytes. The surface area occupied by a typical cell on TCPS surfaces was measured from five different areas and repeated on three different substrates and then averaged for each surface. The number of cells on the projected cell area on the different surfaces was measured using the Image J software (Table 2). The projected cell area refers to the area occupied by the cells as seen under the microscope. The number of cells per unit projected area was plotted over time for the various surfaces. The surfaces that supported proliferation showed a linear increase in the number of cells attached per unit area over time. The slope of this plot provided the rate of cell proliferation. Statistics was performed using the Student's t-test. A p value of 0.05 or lower was considered to be significant.
Reactive Oxygen Species (ROS) Studies
Intracellular reactive oxygen species (ROS) were detected by staining with the oxidant-sensitive dye CM-H2DCFDA. H2DCFDA is cleaved of the ester groups by intracellular esterases and converted into membrane impermeable, non-fluorescent derivative H2DCF. Oxidation of H2DCF by ROS results in highly fluorescent 2,7-dichlorofluorescein (DCF) [81]. The cells were incubated for 30 min at 37 °C with 2 μM CM-H2DCFDA in Hanks' Balanced Salt Solution without phenol red (Invitrogen, CA, USA). The cells were then washed three times with PBS and analyzed with Leica fluorescent microscopy. Fluorescent images were taken in at least 10 different regions and fluorescence quantification was performed by using National Institutes of Health Image J software (available on the World Wide Web at rsb.nih.gov/ij/). Statistics was performed using the Student's t-test. A p value of 0.05 or lower was considered to be significant.
Footnotes
The authors thank the MSU Foundation, Michigan Economic Development Corporation, NIH 1R01GM079688-01, NSF BES 0331297, NSF BES 0425821 and NSF CTS 0609164, EPA RD83184701, and the Whitaker Foundation. The authors of this paper would like to thank Lufang Sheng and Deebika Balu for isolating the primary neurons and astrocytes.
References
- [1].Feng Y, Walsh CA. Nat. Rev. Neurosci. 2001;2:408. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- [2].Park LC, Zhang H, Gibson GE. Mech. Ageing Devel. 2001;123:21. doi: 10.1016/s0047-6374(01)00336-0. [DOI] [PubMed] [Google Scholar]
- [3].Chen Y, Vartiainen NE, Ying W, Chan PH, Koistinaho J, Swanson RA. J. Neurochem. 2001;77:1601. doi: 10.1046/j.1471-4159.2001.00374.x. [DOI] [PubMed] [Google Scholar]
- [4].Desagher S, Glowinski J, Premont J. J. Neurosci. 1996;16:2553. doi: 10.1523/JNEUROSCI.16-08-02553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Patil S, Chan C. Neurosci. Lett. 2005;384:288. doi: 10.1016/j.neulet.2005.05.003. [DOI] [PubMed] [Google Scholar]
- [6].Baier H, Bonhoeffer F. Science. 1992;255:472. doi: 10.1126/science.1734526. [DOI] [PubMed] [Google Scholar]
- [7].Snow DM, Letourneau PC. J. Neurobiol. 1992;23:322. doi: 10.1002/neu.480230311. [DOI] [PubMed] [Google Scholar]
- [8].Steinmetz MP, Horn KP, Tom VJ, Miller JH, Busch SA, Nair D, Silver DJ, Silver J. J. Neurosci. 2005;25:8066. doi: 10.1523/JNEUROSCI.2111-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Goodman CS. Annu. Rev. Neurosci. 1996;19:341. doi: 10.1146/annurev.ne.19.030196.002013. [DOI] [PubMed] [Google Scholar]
- [10].Wen Z, Zheng JQ. Curr. Opin. Neurobiol. 2006;16:52. doi: 10.1016/j.conb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- [11].Fields RD, Stevens-Graham B. Science. 2002;298:556. doi: 10.1126/science.298.5593.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Horner PJ, Palmer TD. Trends Neurosci. 2003;26:597. doi: 10.1016/j.tins.2003.09.010. [DOI] [PubMed] [Google Scholar]
- [13].Ye ZC, Sontheimer H. Glia. 1998;22:237. doi: 10.1002/(sici)1098-1136(199803)22:3<237::aid-glia3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- [14].Giaume C, Kirchhoff F, Matute C, Reichenbach A, Verkhratsky A. Cell Death Differ. 2007;14:1324. doi: 10.1038/sj.cdd.4402144. [DOI] [PubMed] [Google Scholar]
- [15].Kirchhoff F, Dringen R, Giaume C. Eur. Arch. Psychiatry Clin. Neurosci. 2001;251:159. doi: 10.1007/s004060170036. [DOI] [PubMed] [Google Scholar]
- [16].DeFelipe J, Alonso-Nanclares L, Arellano JI. J. Neurocytol. 2002;31:299. doi: 10.1023/a:1024130211265. [DOI] [PubMed] [Google Scholar]
- [17].Sherwood CC, Stimpson CD, Raghanti MA, Wildman DE, Uddin M, Grossman LI, Goodman M, Redmond JC, Bonar CJ, Erwin JM, Hof PR. Proc. Natl. Acad. Sci. USA. 2006;103:13606. doi: 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].van Breemen C, Skarsgard P, Laher I, McManus B, Wang X. Clin. Exp. Pharmacol. Physiol. 1997;24:989. doi: 10.1111/j.1440-1681.1997.tb02737.x. [DOI] [PubMed] [Google Scholar]
- [19].Schmidt C, Leach J. Annu. Rev. Biomed. Eng. 2003;5:293. doi: 10.1146/annurev.bioeng.5.011303.120731. [DOI] [PubMed] [Google Scholar]
- [20].Haydon PG. Nat. Rev. Neurosci. 2001;2:185. doi: 10.1038/35058528. [DOI] [PubMed] [Google Scholar]
- [21].Martin ED, Araque A, Buno W. J. Neurophysiol. 2001;86:2878. doi: 10.1152/jn.2001.86.6.2878. [DOI] [PubMed] [Google Scholar]
- [22].Rao KVR, Panickar K, Jayakumar AR, Norenberg MD. Neurochem. Res. 2005;30:1311. doi: 10.1007/s11064-005-8803-2. [DOI] [PubMed] [Google Scholar]
- [23].Brown DR. Mol. Cell. Neurosci. 1999;13:379. doi: 10.1006/mcne.1999.0751. [DOI] [PubMed] [Google Scholar]
- [24].Taylor DL, Diemel LT, Pocock JM. J. Neurosci. 2003;23:2150. doi: 10.1523/JNEUROSCI.23-06-02150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bhatia SN, Yarmush ML, Toner M. J. Biomed. Mater. Res. 1997;34:189. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- [26].Takayama S, McDonald JC, Ostuni E, Liang MN, Kenis PJA, Ismagilov RF, Whitesides GM. Proc. Natl. Acad. Sci. USA. 1999;96:5545. doi: 10.1073/pnas.96.10.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang IH, Co CC, Ho CC. J. Biomed. Mater. Res. Part A. 2005;75:976. doi: 10.1002/jbm.a.30509. [DOI] [PubMed] [Google Scholar]
- [28].Reyes DR, Perruccio EM, Becerra SP, Locascio LE, Gaitan M. Langmuir. 2004;20:8805. doi: 10.1021/la049249a. [DOI] [PubMed] [Google Scholar]
- [29].Dunn JC, Tompkins RG, Yarmush ML. Biotechnol. Prog. 1991;7:237. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
- [30].Langer R, Vacanti J. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- [31].Yang IH, Co CC, Ho CC. Biomaterials. 2005;26:6599. doi: 10.1016/j.biomaterials.2005.04.024. [DOI] [PubMed] [Google Scholar]
- [32].Mendelsohn JD, Yang SY, Hiller J, Hochbaum AI, Rubner MF. Biomacromolecules. 2003;4:96. doi: 10.1021/bm0256101. [DOI] [PubMed] [Google Scholar]
- [33].Lvov Y, Ariga K, Ichinose I, Kunitake T. J. Am. Chem. Soc. 1995;117:6117. [Google Scholar]
- [34].Decher G. Science. 1997;277:1232. [Google Scholar]
- [35].Vodouhe C, Schmittbuhl M, Boulmedais F, Bagnard D, Vautier D, Schaaf P, Egles C, Voegel JC, Ogier J. Biomaterials. 2005;26:545. doi: 10.1016/j.biomaterials.2004.02.057. [DOI] [PubMed] [Google Scholar]
- [36].Boura C, Muller S, Vautier D, Dumas D, Schaaf P, Voegel JC, Stoltz JF, Menu P. Biomaterials. 2005;26:4568. doi: 10.1016/j.biomaterials.2004.11.036. [DOI] [PubMed] [Google Scholar]
- [37].Peretz H, Talpalar AE, Vago R, Baranes D. Tissue Eng. 2007;13:461. doi: 10.1089/ten.2005.0522. [DOI] [PubMed] [Google Scholar]
- [38].Co CC, Wang Y-C, Ho C-C. J. Am. Chem. Soc. 2005;127:1598. doi: 10.1021/ja044382a. [DOI] [PubMed] [Google Scholar]
- [39].Kidambi S, Udpa N, Schroeder SA, Findlan R, Lee I, Chan C. Tissue Eng. 2007;13:2105. doi: 10.1089/ten.2006.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kidambi S, Chan C, Lee I. J. Am. Chem. Soc. 2004;126:4697. doi: 10.1021/ja039359o. [DOI] [PubMed] [Google Scholar]
- [41].Khademhosseini A, Suh Kahp Y, Yang Jen M, Eng G, Yeh J, Levenberg S, Langer R. Biomaterials. 2004;25:3583. doi: 10.1016/j.biomaterials.2003.10.033. [DOI] [PubMed] [Google Scholar]
- [42].Kidambi S, Sheng L, Yarmush ML, Toner M, Lee I, Chan C. Macromol. Biosci. 2007;7:344. doi: 10.1002/mabi.200600205. [DOI] [PubMed] [Google Scholar]
- [43].Fukuda J, Khademhosseini A, Yeh J, Eng G, Cheng J, Farokhzad Omid C, Langer R. Biomaterials. 2006;27:1479. doi: 10.1016/j.biomaterials.2005.09.015. [DOI] [PubMed] [Google Scholar]
- [44].Kidambi S, Lee I, Chan C. J. Am. Chem. Soc. 2004;126:16286. doi: 10.1021/ja046188u. [DOI] [PubMed] [Google Scholar]
- [45].Li MY, Kondabatni KK, Cui TH, McShane MJ. IEEE Trans. Nanotechnol. 2004;3:115. [Google Scholar]
- [46].Mohammed JS, DeCoster MA, McShane MJ. Biomacromolecules. 2004;5:1745. doi: 10.1021/bm0498631. [DOI] [PubMed] [Google Scholar]
- [47].Khademhosseini A, Suh KY, Yang JM, Eng G, Yeh J, Levenberg S, Langer R. Biomaterials. 2004;25:3583. doi: 10.1016/j.biomaterials.2003.10.033. [DOI] [PubMed] [Google Scholar]
- [48].Gramowski A, Schiffmann D, Gross GW. Neurotoxicology. 2000;21:331. [PubMed] [Google Scholar]
- [49].Gross RA, Uhler MD, Macdonald RL. Brain Res. 1990;535:214. doi: 10.1016/0006-8993(90)91603-e. [DOI] [PubMed] [Google Scholar]
- [50].Kamioka H, Maeda E, Jimbo Y, Robinson HPC, Kawana A. Neurosci. Lett. 1996;206:109. doi: 10.1016/s0304-3940(96)12448-4. [DOI] [PubMed] [Google Scholar]
- [51].Jimbo Y, Robinson HPC, Kawana A. IEEE Trans. Biomed. Eng. 1998;45:1297. doi: 10.1109/10.725326. [DOI] [PubMed] [Google Scholar]
- [52].Shahaf G, Marom S. J. Neurosci. 2001;21:8782. doi: 10.1523/JNEUROSCI.21-22-08782.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Corey JM, Feldman EL. Exp. Neurol. 2003;184:S89. doi: 10.1016/s0014-4886(03)00392-3. [DOI] [PubMed] [Google Scholar]
- [54].Ravenscroft MS, Bateman KE, Shaffer KM, Schessler HM, Jung DR, Schneider TW, Montgomery CB, Custer TL, Schaffner AE, Liu QY, Li YX, Barker JL, Hickman JJ. J. Am. Chem. Soc. 1998;120:12169. [Google Scholar]
- [55].Liu QY, Coulombe M, Dumm J, Shaffer KM, Schaffner AE, Barker JL, Pancrazio JJ, Stenger DA, Ma W. Development Brain Res. 2000;120:223. doi: 10.1016/s0165-3806(00)00014-6. [DOI] [PubMed] [Google Scholar]
- [56].Ma W, Liu QY, Jung D, Manos P, Pancrazio JJ, Schaffner AE, Barker JL, Stenger DA. Development Brain Res. 1998;111:231. doi: 10.1016/s0165-3806(98)00142-4. [DOI] [PubMed] [Google Scholar]
- [57].Chang JC, Brewer GJ, Wheeler BC. Biosens. Bioelectron. 2001;16:527. doi: 10.1016/s0956-5663(01)00166-x. [DOI] [PubMed] [Google Scholar]
- [58].Chang JC, Brewer GJ, Wheeler BC. Biomaterials. 2003;24:2863. doi: 10.1016/s0142-9612(03)00116-9. [DOI] [PubMed] [Google Scholar]
- [59].Xia Y, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:550. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- [60].Quake SR, Scherer A. Science. 2000;290:1536. doi: 10.1126/science.290.5496.1536. [DOI] [PubMed] [Google Scholar]
- [61].Kumar A, Whitesides GM. Appl. Phys. Lett. 1993;63:2002. [Google Scholar]
- [62].Fukuda J, Khademhosseini A, Yeh J, Eng G, Cheng J, Farokhzad OC, Langer R. Biomaterials. 2006;27:1479. doi: 10.1016/j.biomaterials.2005.09.015. [DOI] [PubMed] [Google Scholar]
- [63].Araque A, Carmignoto G, Haydon PG. Annu. Rev. Physiol. 2001;63:795. doi: 10.1146/annurev.physiol.63.1.795. [DOI] [PubMed] [Google Scholar]
- [64].Sorribas H, Padeste C, Tiefenauer L. Biomaterials. 2001;23:893. doi: 10.1016/s0142-9612(01)00199-5. [DOI] [PubMed] [Google Scholar]
- [65].Mattson MP. Nature. 2004;430:631. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Patil S, Sheng LF, Masserang A, Chan C. Neurosci. Lett. 2006;406:55. doi: 10.1016/j.neulet.2006.07.015. [DOI] [PubMed] [Google Scholar]
- [67].Blazquez C, Galve-Roperh I, Guzman M. FASEB J. 2000;14:2315. doi: 10.1096/fj.00-0122com. [DOI] [PubMed] [Google Scholar]
- [68].Samper E, Nicholls DG, Melov S. Aging Cell. 2003;2:277. doi: 10.1046/j.1474-9728.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- [69].Jacobson J, Duchen MR, Hothersall J, Clark JB, Heales SJ. J. Neurochem. 2005;95:388. doi: 10.1111/j.1471-4159.2005.03374.x. [DOI] [PubMed] [Google Scholar]
- [70].Bhatia SN, Balis UJ, Yarmush ML, Toner M. FASEB J. 1999;13:1883. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- [71].Westergaard N, Fosmark H, Schousboe A. J. Neurochem. 1991;56:59. doi: 10.1111/j.1471-4159.1991.tb02562.x. [DOI] [PubMed] [Google Scholar]
- [72].Gross GW, Gramowski A, Schiffmann D. Eur. J. Cell Biol. 1997;74:36. [Google Scholar]
- [73].Cornish T, Branch DW, Wheeler BC, Campanelli JT. Mol. Cell. Neurosci. 2002;20:140. doi: 10.1006/mcne.2002.1101. [DOI] [PubMed] [Google Scholar]
- [74].Kumar A, Biebuyck HA, Whitesides GM. Langmuir. 1994;10:1498. [Google Scholar]
- [75].Jiang X, Zheng H, Gourdin S, Hammond PT. Langmuir. 2002;18:2607. [Google Scholar]
- [76].Marini AM, Paul SM. Proc. Natl. Acad. Sci. USA. 1992;89:6555. doi: 10.1073/pnas.89.14.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nicoletti F, Wroblewski JT, Novelli A, Alho H, Guidotti A, Costa E. J. Neurosci. 1986;6:1905. doi: 10.1523/JNEUROSCI.06-07-01905.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hamprecht B, Loeffler F. Methods Enzymol. 1985;109:341. doi: 10.1016/0076-6879(85)09097-8. [DOI] [PubMed] [Google Scholar]
- [79].Chandler LJ, Newsom H, Sumners C, Crews F. J. Neurochem. 1993;60:1578. doi: 10.1111/j.1471-4159.1993.tb03326.x. [DOI] [PubMed] [Google Scholar]
- [80].Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Proc. Natl. Acad. Sci. USA. 2004;101:2070. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sauer H, Klimm B, Hescheler J, Wartenberg M. FASEB J. 2001;15:2539. doi: 10.1096/fj.01-0360fje. [DOI] [PubMed] [Google Scholar]