Abstract
To gain insight into liver and pancreas development, we investigated the target of 2F11, a monoclonal antibody of unknown antigen, widely used in zebrafish studies for labeling hepatopancreatic ducts. Utilizing mass spectrometry and in vivo assays, we determined the molecular target of 2F11 to be Annexin A4 (Anxa4), a calcium binding protein. We further found that in both zebrafish and mouse endoderm, Anxa4 is broadly expressed in the developing liver and pancreas, and later becomes more restricted to the hepatopancreatic ducts and pancreatic islets, including the insulin producing β-cells. Although Anxa4 is a known target of several monogenic diabetes genes and its elevated expression is associated with chemoresistance in malignancy, its in vivo role is largely unexplored. Knockdown of Anxa4 in zebrafish leads to elevated expression of caspase 8 and Δ113p53, and liver bud specific activation of Caspase 3 and apoptosis. Mosaic knockdown reveal that Anxa4 is required cell-autonomously in the liver bud for cell survival. This finding is further corroborated with mosaic anxa4 knockout studies using the CRISPR/Cas9 system. Collectively, we identify Anxa4 as a new, evolutionarily conserved hepatopancreatic factor that is required in zebrafish for liver progenitor viability, through inhibition of the extrinsic apoptotic pathway. A role for Anxa4 in cell survival may have implications for the mechanism of diabetic β-cell apoptosis and cancer cell chemoresistance.
Keywords: AnnexinA4, 2F11, hepatopancreatic duct, pancreas, liver, cell survival
Introduction
Extensive genetic and morphogenetic similarities between the mammalian and fish hepatopancreatic system validate the zebrafish as a practical vertebrate model system for studying liver and pancreas development. Discovering new factors expressed in the developing zebrafish hepatopancreatic system may therefore yield insight into human liver and pancreas development. The monoclonal antibody (mAb) 2F11 is widely used to mark the developing and adult zebrafish hepatopancreatic ducts (HPDs), as well as enteroendocrine cells (Crosnier et al., 2005; Curado et al., 2010; Delous et al., 2012; Dong et al., 2007; EauClaire et al., 2012; Lancman et al., 2013; Manfroid et al., 2012; Matthews et al., 2011; Ninov et al., 2012). Particularly, 2F11 marks the intestinal secretory cells, extra-HPDs (common bile duct, cystic duct, hepatopancreas ampulla, extra-hepatic duct and extra-pancreatic duct), gallbladder, intra-pancreatic (Dong et al., 2007), and intra-hepatic ducts (Lorent et al., 2010; Matthews et al., 2008). However, the target antigen bound by this mAb is not known – as 2F11 was among other randomly raised mAbs against the lysate of adult zebrafish gut (Crosnier et al., 2005). Identifying the target bound by 2F11 will shed new light on published works in which this mAb was employed, contribute to a short list of genes that are specifically expressed in the hepatopancreatic ductal system and intestinal secretory cells, and yield new perspectives into hepatopancreas development.
In this study, we demonstrated that the antigen of 2F11 is zebrafish Anxa4. Anxa4 belongs to the Annexin family of calcium- and phospholipid-binding proteins, which from cell-based studies, have been implicated in multiple cellular functions including endocytosis, exocytosis, and ion fluxes (Gerke and Moss, 2002). Zebrafish Anxa4 is distinct from other zebrafish Annexins in that it has a short, highly conserved N-terminal domain that is more similar to mammalian Anxa4 orthologues (Farber et al., 2003). Human ANXA4 is predominantly detected in the epithelial cells of several organs, including the liver and pancreas (Dreier et al., 1998). In vitro studies suggest that it inhibits Ca2+ activated Cl− conductance (Chan et al., 1994) and reduces the water and proton permeability of the membrane possibly through modulating membrane rigidity (Hill et al., 2003). ANXA4 expression is increased in many cancer types, including cancers of renal, gastric, colonic, ovarian, and cervical origins (Duncan et al., 2008; Lin et al., 2008; Miao et al., 2009; Shen et al., 2004; Toyama et al., 2012; Zimmermann et al., 2004). Its expression has been associated with loss of cell-to-cell adhesion, increased metastasis, and chemo-resistance, and therefore is now regarded as a potential cancer diagnostic and therapeutic target (Kim et al., 2010; Masuishi et al., 2011). In vitro studies suggest that in response to cytotoxic stress, ANXA4 exhibits an anti-apoptotic effect (Han et al., 2000; Kim et al., 2009) by activating NF-κB transcriptional activity (Jeon et al., 2010; Sohma et al., 2003). Also, pancreas expression of ANXA4 is downstream of several monogenic diabetes genes, including PDX1, HNF1A, and HNF4A (Bolotin et al., 2010; Servitja et al., 2009; Svensson et al., 2007), suggesting potential roles for ANXA4 in diabetes.
Only limited in vivo functional analysis of Anxa4 has been reported. Specific loss of Anxa4 from the mouse urothelium caused no detectable phenotypes. However other Annexins were suggested to have redundant roles (Hill et al., 2008). In Xenopus laevis, there are two paralogs of this gene, anxa4a and anxa4b. Knockdown of anxa4a, which is expressed in the pronephros but not in the liver, affects pronephros development (Masse et al., 2007; Seville et al., 2002). The in vivo role of anxa4b, which is expressed in the liver, was not described. Therefore, Anxa4 function during foregut endoderm development has yet to be investigated in animal models.
In our present study, we determined that the 2F11 mAb targets Anxa4 in zebrafish, demonstrating an efficient approach for identifying the antigen of an orphan antibody. We show that hepatopancreas expression of Anxa4 in zebrafish and mouse is conserved. Our in vivo functional studies using zebrafish embryos, suggest that Anxa4 plays a role in maintaining liver cell survival by inhibiting the extrinsic apoptotic pathway. Mosaic studies suggest that Anxa4 functions cell-autonomously to block apoptosis specifically in the liver bud. This is the first in vivo investigation to link Anxa4 to cell viability, consistent with previous in vitro studies.
Results
The antigen of mAb 2F11 is expressed in hepatopancreatic progenitors
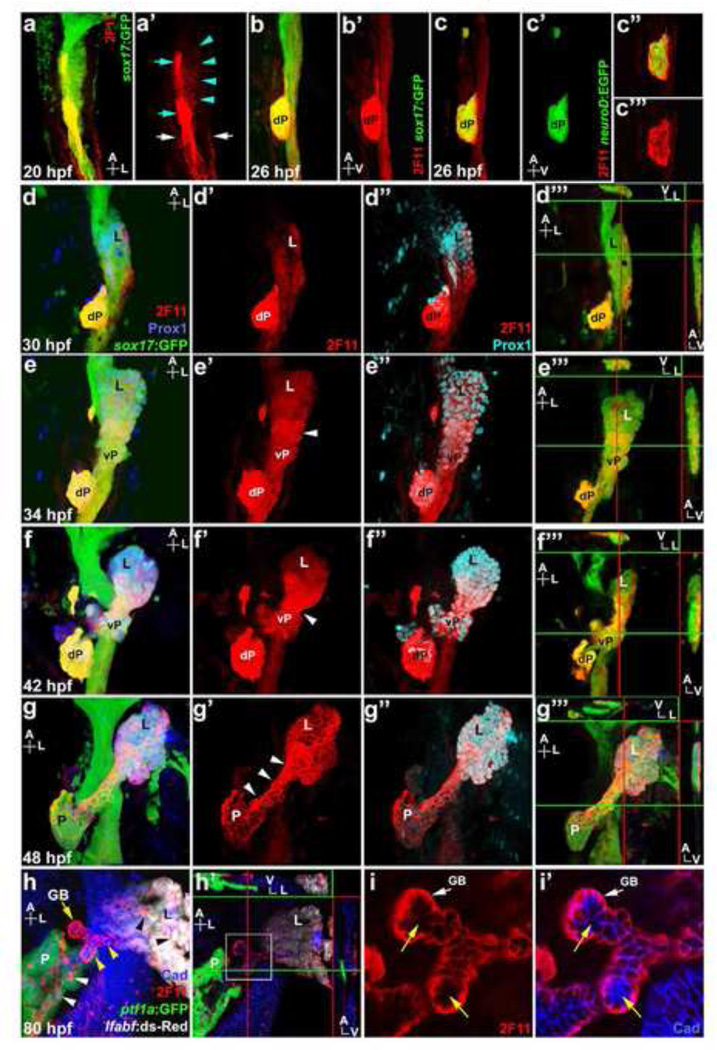
It has been well documented that the 2F11 mAb is a robust marker for hepatopancreatic ducts and intestinal secretory cells in zebrafish. To gain further insight into the 2F11 target antigen, we examined its expression pattern at different stages of hepatopancreas development in Tg(sox17:GFP) embryos, which express GFP in the early endoderm (Mizoguchi et al., 2008). Initial budding of the zebrafish liver occurs at 28 hpf (hours post fertilization) (Field et al., 2003b). As with mammals, the mature zebrafish pancreas arises from the fusion of the dorsal and ventral pancreas. The dorsal pancreas emerges from the dorsal foregut endoderm at 24 hpf. At 32 hpf, the ventral pancreas appears posterior to and contiguous with the liver bud. By 40 hpf, the ventral pancreas begins to fuse with the dorsal pancreas, which has separated from the dorsal endoderm (Field et al., 2003a). At 20 hpf, 2F11 marks the GFP positive foregut endoderm, in a region posterior to the pharyngeal endoderm (Fig. 1a). At this stage, low level 2F11 staining with membrane localization is observed broadly in the foregut endoderm. Within this domain, the dorsal pancreatic cells are strongly marked, with labeling throughout the cell. The pronephric ducts are also labeled by 2F11. By 26 hpf, while low level 2F11 remains broad along the foregut, high level 2F11 becomes restricted to the dorsal pancreas bud (Fig. 1b). The zebrafish dorsal pancreas is composed primarily of endocrine cells, which can be visualized using Tg(neuroD:EGFP) embryos (Obholzer et al., 2008). We found that the high level 2F11 staining is coincident with the dorsal pancreas endocrine cells in 26 hpf Tg(neuroD:EGFP) embryos (Fig. 1c). All neuroD:EGFP cells in the dorsal pancreas appears to have varying levels of 2F11 (Fig. 1c”-c”’), indicating that 2F11 marks all endocrine cell types at this stage.
Figure 1. 2F11 mAb labels hepatopancreas progenitors during organogenesis.
(a, a’) Ventral view of a 20 hpf Tg(sox17:GFP) foregut showing broad low 2F11 labeling (turquoise arrowheads), and higher labeling in dorsal pancreas cells (turquoise arrows) and the pronephric ducts (white arrows). (b–c’) Lateral view of 26 hpf Tg(sox17:GFP) (b) and Tg(neuroD:EGFP) (c) foregut showing 2F11 labels the dorsal pancreas (dP) with a single focal plane shown in c” and c”’. (d–g”’) Ventral view of the Tg(sox17:GFP) foregut with Prox1 and 2F11 labeling at 30, 34, 42 and 48 hpf; (d”’, e”’, f”’, g”’) digital sections along X, Y, and Z axes showing higher 2F11 staining on the ventral side of the endoderm (green lines indicate the positions of the X-axis sections, right insets; red lines indicate the positions of the Y-axis sections, top insets, a larger image of f”’ is shown in Figure S1). (d–d”) 2F11 staining increases in the Prox1+ budding liver (L) at 30 hpf. (e–e”) At 34 hpf, 2F11 labeling increases in the Prox1+ ventral pancreas (vP) with broad cellular localization. Note the elevated staining in cells between the liver and ventral pancreas (white arrowhead). (f–f”) At 42 hpf, 2F11 labeling increases further in the cells between liver and ventral pancreas with broad cellular localization. (g–g”) At 48 hpf, 2F11 labels the entire hepatopancreas system with elevated staining and non-nuclear localization (white arrowheads) in cells between the liver and pancreas (P). (h) Ventral view of the Tg(ptf1a:GFP);Tg(lfabf:ds-Red) foregut at 80 hpf showing 2F11 labeling becomes restricted to the gallbladder (GB; yellow arrow) and ducts, including intrapancreatic ducts (white arrowheads), intrahepatic ducts (black arrowheads), and extra hepatopancreatic ducts (yellow arrowheads). (h’) Digital sections along X, Y, and Z axes showing higher 2F11 staining in the ventral region of the ducts. (i–i’) Higher magnification of h’ (white square inset) showing 2F11 basal-lateral cellular localization in ductal columnar epithelium where it does not overlap with apically restricted Cadherin (yellow arrows).
At 30 hpf, as the liver bud becomes prominent, 2F11 staining is markedly increased in the liver bud, exhibiting broad intracellular localization (Fig. 1d). The ventral pancreas bud also shows elevated 2F11 labeling when it initially appears at 34 hpf (Fig. 1e). At these stages, low-level 2F11 labeling persists broadly throughout the foregut. In the liver and ventral pancreas buds, 2F11-marked cells also express Prox1, one of the earliest liver and pancreas progenitor markers in zebrafish (Ober et al., 2006), confirming that the expression of the antigen of 2F11 is elevated in the early developing liver and ventral pancreas. Also, 2F11 labeling is stronger on the ventral side of the foregut endoderm (Fig. 1d”’ and 1e”’; larger image of 1f”’ is shown in Fig. S1). As the liver and pancreas grow out laterally, 2F11 continues to broadly label the entire hepatopancreas region, and becomes heightened in cells between the Prox1 positive liver and ventral pancreas domains (Fig. 1e and 1f), suggesting that the antigen of 2F11 is specifically up-regulated in the presumptive extra-HPDs. From 42 to 48 hpf, as 2F11 staining continues to be elevated in the ventral endoderm, the broad labeling in the presumptive extra-HPDs becomes more localized to the plasma membrane (Fig.1f and 1g). After the dorsal pancreas has fused with the ventral pancreas, 2F11 staining of the endocrine cells in the principal islets is decreased (Fig. 1g).
By 80 hpf, the epithelial cells between the liver and pancreas become columnar and organize into ducts. 2F11 staining remains high in these ducts, showing lateral-basal localization (Fig. 1h). Within the liver and pancreas, 2F11 marks the intrapancreatic ducts and intrahepatic biliary ducts, as previously reported (Dong et al., 2007; Lorent et al., 2010; Matthews et al., 2008). Neogenic endocrine cells found outside the principal islet become more apparent after 60 hpf, presumably arising from the 2F11 labeled ducts. In Tg(neuroD:EGFP) embryos, a subset of these GFP labeled new endocrine cells are also positive for 2F11 (Fig. S2, a–b’), consistent with 2F11 labeling new endocrine cells in juvenile zebrafish (Matsuda et al., 2013). We also observed that in the liver and pancreas, there is moderate to low level 2F11 labeling of non-ductal cell types, particularly in peripheral cells along the proximal regions of these organs (not shown).
Our analysis of 2F11 labeling in the developing zebrafish endoderm reveals that its antigen is dynamically expressed during hepatopancreas organogenesis. It appears broadly in the foregut and becomes gradually restricted to the hepatopancreas progenitors and then to hepatopancreatic ducts. Its intracellular localization is also dynamic, initially in whole cells and then becoming more restricted to the membrane. This progression of intracellular localization may correlate with the differentiation state of the cell and suggests the target factor may be involved in multiple cellular processes. This intriguing expression pattern led us to seek the identity of the 2F11 antigen.
Annexin A4 is the antigen of 2F11 mAb
To identify a candidate target for 2F11, we used a mass spectrometry approach. The 2F11 antibody was used to immuno-precipitate possible antigens from the lysate of zebrafish embryos at 72 hpf, when 2F11 foregut labeling is robust. The proteins captured were then separated by PAGE gel electrophoresis under reducing conditions, where a clear band was resolved that was not the immunoglobulin light or heavy chains (Fig. 2a). That band, appearing between 28 and 39 kDa, was cut out and in-gel digested into small peptides by Trypsin. These peptides were then separated by HPLC and analyzed by tandem Mass spectrometry (MS/MS). All peptide sequences identified precisely matched various regions of the zebrafish Annexin A4 protein (Anxa4) (Fig. 2b). Anxa4 is a 35.6 kDa protein, consistent with the size of the band analyzed. Anxa4 belongs to the Annexin family of Ca2+ and phospholipid binding proteins and is often associated with the inner leaflet of the plasma membrane (Gerke et al., 2005). This intracellular localization is consistent with our immunohistochemical data showing that 2F11 can mark the plasma membrane. Annexins are thought to form multimers (Gerke et al., 2005), which may explain the multiple higher bands previously observed on a 2F11 western blot run under non-reducing conditions (Crosnier et al., 2005). These findings collectively support Anxa4 as a strong candidate target of 2F11.
Figure 2. Anxa4 is bound by 2F11 and is expressed in tissues labeled by 2F11.
(a–b) A 2F11 immuno-precipitated band (arrow in a) was analyzed by peptide mass fingerprinting, yielding peptide sequences (colored text in b) that match zebrafish Anxa4. (c–g’)Anxa4 is expressed in the developing zebrafish foregut (orange arrow), pronephric ducts (magenta arrowheads), dorsal pancreas (yellow arrows), liver bud (red arrowheads), lateral lines (turquoise arrowheads), and hatching gland (green arrowheads). (h–k’) The ventral view of Tg(sox17:GFP) with anxa4 transcript (h, j) or 2F11 labeling (i, i’, k, k’) of the liver (L), ventral pancreas (vP), and dorsal pancreas (dP) at 34 hpf (h–I’) and 47 hpf (j–k’). (l–m’) At 80 hpf, both anxa4 transcripts (l) and 2F11 (m, m’) are found in the gallbladder (GB), hepatopancreatic ducts, including extrahepatic duct (EHD), common bile duct (CBD), extrapancreatic duct (EPD), intrapancreatic duct (IPD) and intrahepatic duct (IHD), intestinal secretory cells (yellow arrowheads), and mesenchymal cells around the swim bladder (turquoise arrowheads).
We next assessed whether 2F11 labeling corresponds with anxa4 transcript expression. In situ hybridization (ISH) studies revealed that anxa4 mRNA was expressed in the floor plate, hypochord, and pronephric ducts at 24 hpf, as well as in the liver and gallbladder at 3 dpf (days post fertilization) (Farber et al., 2003). These domains of anxa4 mRNA expression strongly coincide with tissues labeled by 2F11, including intrahepatic biliary ducts (Lorent et al., 2010; Matthews et al., 2008), extra-hepatopancreatic ducts, and gallbladder at 3 dpf (Dong et al., 2007). We found 2F11 also labels the floor plate, the hypochord and pronephric ducts (Fig. 1 and data not shown). Therefore the reported expression of anxa4 mRNA corresponds well with 2F11 labeling. We further examined the global expression of anxa4 mRNA during development via whole mount ISH. At 18 hpf, anxa4 transcripts can be detected in the region of the foregut endoderm and pronephric duct precursors (Fig. 2c). By 26 hpf, high levels of anxa4 transcripts appear in the dorsal pancreas (Fig. 2d, 2e). Similar to 2F11 labeling, anxa4 is expressed in the liver bud and its expression domain extends posteriorly towards the dorsal pancreas from 30 to 36hpf (Fig. 2f–g). In reference to GFP expression in Tg(sox17:GFP) embryos, anxa4 transcripts appear within the liver, ventral pancreas and dorsal pancreas regions of the endoderm (Fig 2h–2i’). By 47 hpf, anxa4 transcripts are found throughout most of the hepatopancreas domain with the proximal liver exhibiting stronger signals (Fig. 2j). 2F11 staining has a similar pattern except that it is higher in the extra-HPD region (between liver and pancreas) than in liver or pancreas (Fig. 2k, 2k’). Differences between Anxa4 protein and transcript production and turnover may account for this discrepancy between protein and transcript expression levels. We conclude that during early organogenesis, anxa4 transcripts are expressed in the same foregut endoderm tissue that is labeled by 2F11, including the dorsal pancreas, liver, ventral pancreas, and presumptive ducts connecting these organs.
At 80 hpf, anxa4 transcripts appear elevated in the extra-hepatopancreatic ducts and gallbladder. Within the pancreas and liver, anxa4 expression becomes more restricted to the intrapancreatic ducts and intrahepatic ducts, respectively (Fig. 2l). The anxa4 transcripts are expressed in a ring of cells around the principal islet in the pancreatic head and in a strip of cells in the middle of the pancreatic tail, correlating to the developing intrapancreatic ducts. In the liver, it is expressed in a distinct branched pattern, consistent with the structure of intrahepatic biliary ducts. Therefore anxa4 probe appears to label the hepatopancreatic ducts, consistent with 2F11 (Fig. 2m, 2m’). We found anxa4 is also expressed in individual cells scattered in the gut, consistent with intestine secretory cells, which are also labeled by 2F11 (Crosnier et al., 2005). Outside of the endoderm, anxa4 transcripts appear in the mesenchymal cells around the swim bladder, similar to 2F11 labeling. In summary, the temporal-spatial expression pattern of anxa4 transcripts, in both endoderm and non-endoderm tissues, coincides with 2F11 labeling, further supporting that Anxa4 is the target of the 2F11 mAb.
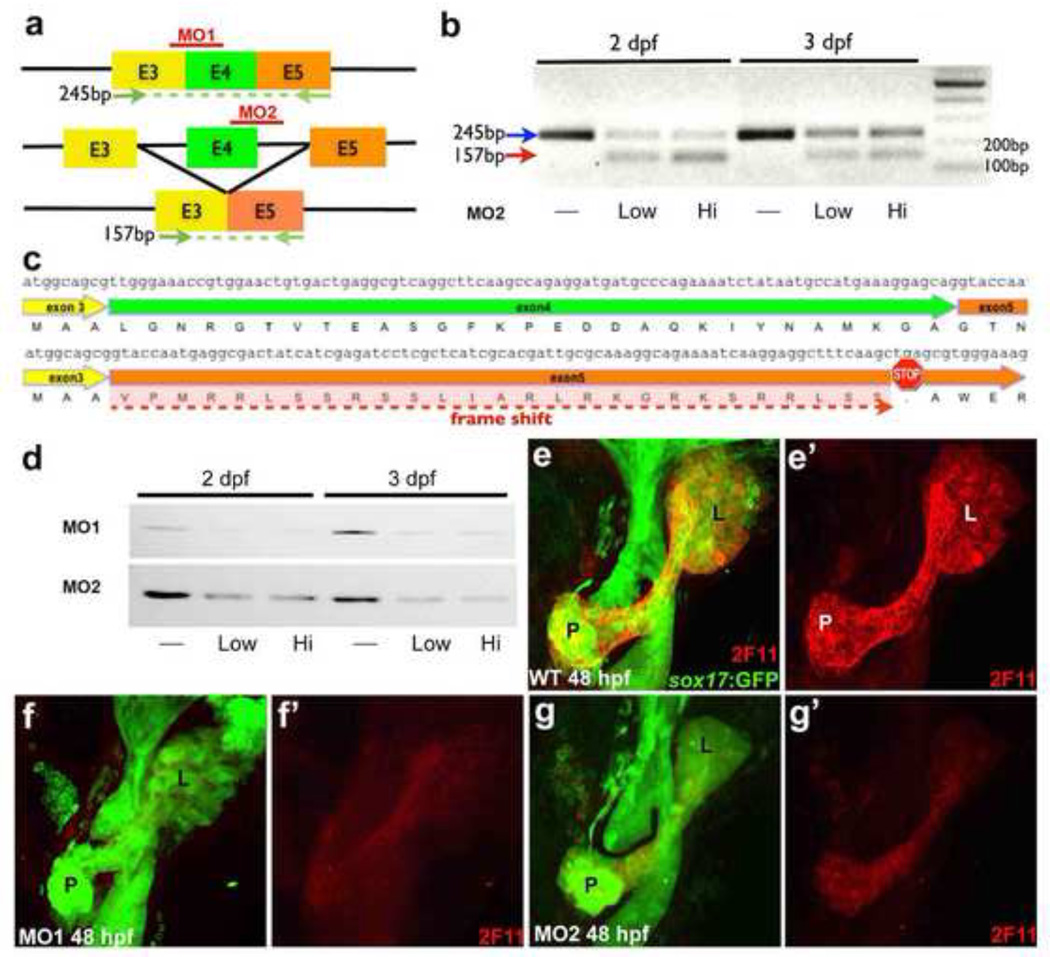
To functionally test whether Anxa4 is recognized by 2F11, we examined whether knockdown of Anxa4 translation by antisense morpholinos (MO) leads to a reduction of 2F11 labeling. We used two different MOs to disrupt Anxa4 protein expression. MO1 blocks Anxa4 translation initiation, whereas MO2 disrupts normal splicing of anxa4 by blocking the splicing donor site of Intron 4. Disruption of this splicing donor site is predicted to cause skipping of Exon 4, leading to translation from Exon 3 directly into Exon 5 (Fig. 3a). This would result in a translational frame-shift, leading to a premature stop codon in Exon 5. The effectiveness of MO2 was assessed by RT-PCR, which demonstrated the presence of a mis-spliced transcript and a reduction of the normally spliced transcripts in the MO2 morphants (MO injected animals) (Fig. 3b). The sequence of the mis-spliced transcript lacks Exon 4 and reads directly from Exon 3 into Exon 5, consistent with our prediction (Fig. 3c). Translation of anxa4 transcript starts in Exon 3, which codes for the first 3 amino acids of Anxa4. With only the first 3 amino acids from Exon 3 in frame, it is unlikely that the resulting protein will contain the epitope recognized by 2F11. Following injection of these anxa4 MOs, we used western blot analysis to examine 2F11 bands from the whole embryo lysate of the 48 hpf and 72 hpf morphants. We found 2F11 bands were markedly reduced for morphants as compared to uninjected controls. Further, immunohistochemical analysis of morphant Tg(sox17:GFP) embryos injected with either MO show significantly reduced 2F11 labeling in their hepatopancreas system indicating that the 2F11 target antigen is reduced when anxa4 translation is comprised (Fig. 3e–g’). The severe loss of 2F11 labeling caused by anxa4 specific antisense MOs in both whole mount immuno-staining and western blot provides functional evidence that Anxa4 is the target of 2F11 antibody. Together with similar expression patterns between anxa4 mRNA and 2F11 antibody staining, and the identification of the protein pulled down by 2F11 via peptide mass fingerprinting, we conclude that the antigen recognized by the mAb 2F11 is zebrafish Anxa4.
Figure 3. Knockdown of Anxa4 reduces 2F11 staining.
(a) Diagram of anxa4 transcripts to depict targets of MOs and RT-PCR primers (green arrows). MO1 is targeted to block translation initiation in Exon 3 and MO2 is targeted to block the splice donor site at the Exon 4 / Intron 4 boundary, predicted to cause skipping of Exon 4, leading translation from Exon 3 directly to Exon 5. (b) RT-PCR (with primers indicated in a) of control and MO2 morphant embryos showing the predicted 157bp PCR product from the mis-spliced transcript (red arrow) and a reduction of the 245bp PCR product from the normally-spliced transcripts (blue arrow) in morphants. (c) The sequence of the mis-spliced transcript confirms Exon 4 skipping and indicates a premature stop codon in Exon 5. (d) Both MOs lead to reduced 2F11 staining in a western blot of whole embryo lysate. (e–g’) Ventral views of 48 hpf Tg(sox17:GFP) showing that Anxa4 morphants have reduced 2F11 labeling in the entire hepatopancreas area compared to the control. L, liver; P, pancreas; dP, dorsal pancreas.
Conserved Anxa4 expression in the vertebrate liver and pancreas
To assess the expression of Anxa4 during mammalian liver and pancreas development, we examined its protein expression in paraffin sections of mouse embryos. From embryonic day 12 (E12) to E14, we found Anxa4 is broadly co-expressed with Prox1, a hepatoblast marker (Sosa-Pineda et al., 2000). In the E12 pancreas, Anxa4 appears to be expressed in the entire pancreatic epithelium, including Sox9 and Glucagon expressing cells (Fig 4b–b”). At E14, Anxa4 remains expressed in both the high Sox9 positive cells in the primitive ducts and the low Sox9 positive cells near the tips of the branches. It is also expressed in the islet, including the Glucagon expressing cells (Fig 4d–d”). These findings suggest that similar to zebrafish, mouse Anxa4 is broadly expressed in the developing liver and pancreas.
Figure 4. Conserved zebrafish and mouse Anxa4 expression in the liver and pancreas.
(a–g’) Immunofluorescent labeling of Anxa4 in the liver and pancreas of E12, E14, and P25 mice. From E12 to E14, Anxa4 is detected in Prox1+ liver cells (a, c), and in all pancreatic cells (b–b”; d–d”), including Sox9+ or Glucagon+ cells and cells negative for both Sox9 and Glucagon (white arrowheads in b–b”). At E14, both the Sox9 high (yellow arrowheads) and low (blue arrowheads) expression cells are positive for Anxa4. In adult mice, Anxa4 is expressed the intrahepatic biliary ducts (e, e’; white arrowheads) and intrapancreatic ducts (f, f’; white arrowheads), co-localizing with ductal expression of E-Cadherin. Anxa4 is also expressed in pancreatic endocrine islets (g–g’), with localization to the membrane (white arrowhead) and nuclear envelope (yellow arrowhead). Low level Anxa4 is expressed in acinar cells (blue arrowheads) (h–j’) Immunofluorescent labeling of Anxa4 in the zebrafish liver (h) and pancreas (I) of one-year old Tg(ptf1a:GFP);Tg(lfabf:ds-Red), and pancreas of 2-month old Tg(ins:dsRed);Tg(gcga:GFP), showing Anxa4 is expressed in the networks of intrahepatic (h) and intrapancreatic (i–j’) ducts as well as pancreatic islets (j, j’ single optical section, white arrowheads).
In the mouse liver and pancreas of the postnatal day 25 (P25), Anxa4 is expressed in cells with high levels of E-Cadherin, suggesting that elevated Anxa4 expression is restricted to the intrahepatic ducts and intrapancreatic ducts (Fig. 4e, 4f). Anxa4 expression is also observed in pancreatic islets, showing co-expression with Glucagon in α-cells and with Insulin in (β-cells. In islets, Anxa4 appears to be localized to the cell membrane, and in certain cells, the nuclear envelope (Fig. 4g–g’). Nuclear envelope localization of Anxa4 was also previously observed in mouse urothelium (Hill et al., 2008). Further, lower expression of Anxa4 is detectable at the cell membrane of the acinar cells (Fig. 4g–g’). In one-year old zebrafish liver and pancreas, Anxa4 is markedly expressed in the ductal networks (Fig. 4h, 4i). In 2-month old fish, Anxa4 is also expressed in zebrafish pancreatic islets, including a subset of the cells expressing Glucagon or Insulin (Fig. 4j–j’). Interestingly, in the human liver and pancreas, ANXA4 is also expressed in ducts and islets (Dreier et al., 1998), (www.proteinatlas.org), (Ahmed et al., 2005). Similar hepatopancreas expression of Anxa4 in zebrafish, mouse, and human suggests that foregut expression of ANXA4 is conserved, supporting zebrafish as a suitable model for investigating ANXA4 function.
Annexin A4 is required cell-autonomously for cell survival in the developing liver
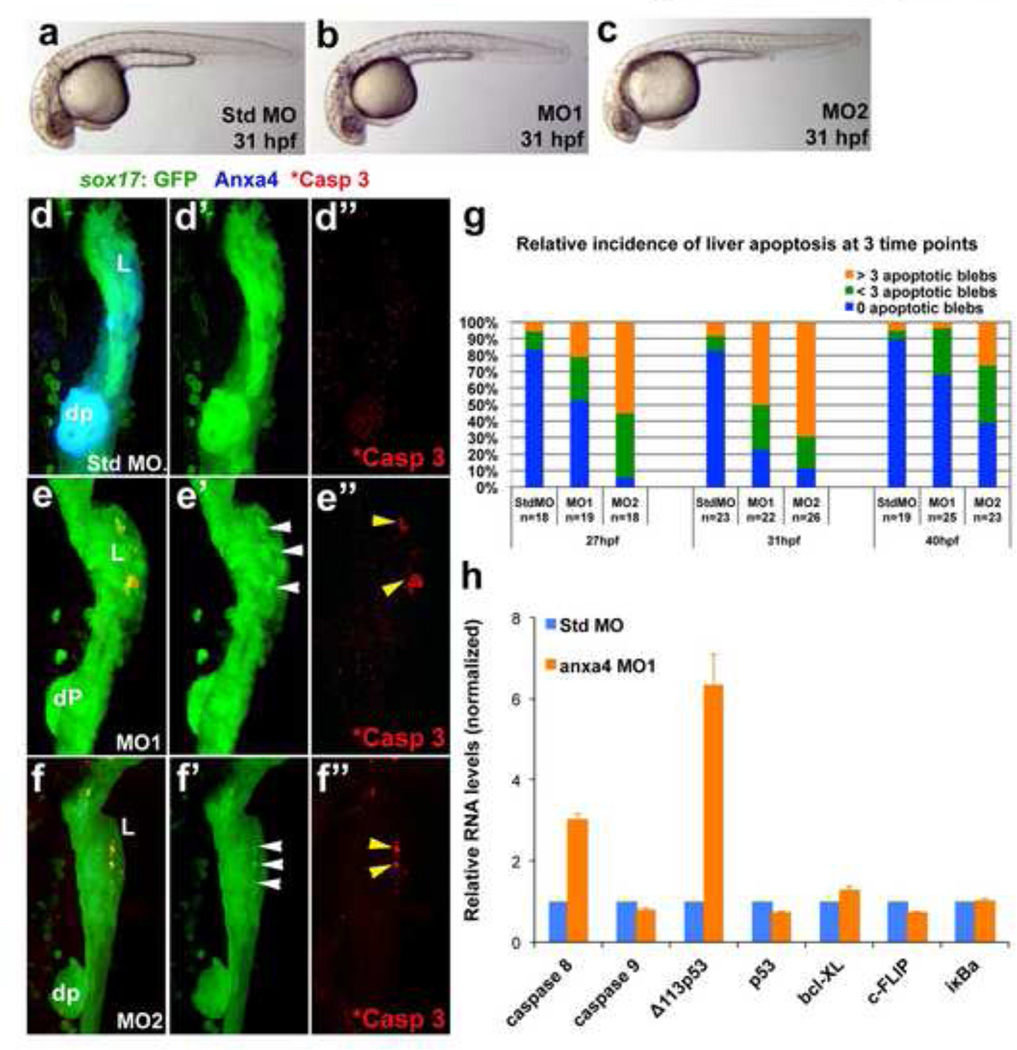
To investigate the role of Anxa4 in foregut endoderm development, we further examined anxa4 morphants. Based on bright-field microscopy, 31 hpf Anxa4 morphants (3ng of MO1 or 1.5ng of MO2 per embryo) are slightly shorter, with less pigmentation compared to the standard control morphants, suggesting a mild developmental delay (Fig. 5a–c). No other obvious body phenotypes were observed. The pancreas and liver in Anxa4 morphants are hypoplastic at 48 hpf. This defect may be due to the developmental delay, a decrease in proliferation, and/or an apoptotic defect (Fig. 3e–g’). By 66 hpf, endocrine cells still appear outside the principal islet in the Anxa4 morphants (Fig. S2 c–c’), suggesting that endocrine neogenesis is not lost in Anxa4 morphants. Since Anxa4 shows elevated expression in the intra-pancreatic ducts and intra-hepatic ducts, we looked for ductal defects in the Tg(Tp1:eGFP) background, in which the ductal cells of the liver and pancreas are marked by GFP expression (Parsons et al., 2009). We found the GFP+ cells in the liver and pancreas of Anxa4 morphants Tg(Tp1:eGFP) embryos forms ductal networks with high expression of Alcam, similar to those in the standard control morphants. Although the ductal compartment is reduced in size compared to controls, this reduction appears proportional to the overall reduction of the entire liver and pancreas, suggesting that the smaller ductal compartment in the Anxa4 morphants is not a specific phenotype (Fig. S3 a–b’). However, 2F11 labeling shows less effective knockdown of Anxa4 by this stage. Although higher doses of MO2 (>1.5ng) can more effectively knock down Anxa4 expression, it can also lead to more severe developmental delay, suggesting the MO2 may have greater off target effects than MO1. Proliferation assays based on phosphorylated Histone 3 expression in the developing liver and pancreas did not show a significant difference between the Anxa4 morphants (both MO1 and MO2) and the standard control morphants at 31 hpf (not shown). In the Tg(sox17:GFP) background, patches of blebs with elevated GFP are present in the 31 hpf Anxa4 morphant liver bud but not in surrounding endodermal tissue or in control morphants (Fig. 5d–f”). These GFP blebs appear within the Prox1 expressing hepatopancreatic domain (Fig. S4). The Prox1 domain also overlaps with the Pdx1 expression domain, suggesting that pancreatic progenitors may reside in this region. Cellular blebs are a hallmark of apoptosis (Coleman et al., 2001). Due to cytoplasmic shrinkage, GFP in these apoptotic blebs becomes more concentrated, and thus appears brighter. Because of restricted intracellular localization of activated Caspase 3 (Luo et al., 2010), a direct regulator of apoptosis and blebbling (Sebbagh et al., 2001), only a subset of the GFP blebs are positive for activated Caspase 3 (Fig. 5e”, 5f”). Increased activated Caspase 3 was not observed in tissues that do not express Anxa4. Further, in Anxa4 morphants, there is no significant detectable cell death in other Anxa4-expressing tissue such as the dorsal pancreas, which show almost no GFP blebs (>98% in MO1 morphants, n=53). These findings indicate that there is liver bud specific apoptotic cell death in Anxa4 morphants and suggest that liver progenitor cell survival is particularly sensitive to decreased Anxa4 expression. Among either MO1 or MO2 morphants, more than 60% showed patches of blebs in the liver buds. Stage specificity of the liver bud apoptosis is also similar for both morphants. Although liver bud apoptosis can be found between 27 and 40 hpf, the peak penetrance occurs at 31 hpf (Fig. 5g). This stage correlates with the increase of Anxa4 expression in the developing liver buds (Fig. 1d,e). Because apoptosis is restricted to the liver bud only 1–2 hours before the ventral pancreas progenitors become detectable, it is not clear why the pancreas in Anxa4 morphants is also hypoplastic. One possibility is that pancreas progenitors are co-opted to adopt liver fate to help compensate for liver cell loss. Alternatively, it is possible that some apoptotic cells in the liver bud are pancreas progenitors. Although ventral pancreatic progenitors are believed to arise from an area immediately adjacent to the liver bud, whether some pancreas progenitors also reside within the liver bud is unknown. Consistently, earliest markers of the liver bud, prox1 and hhex, are not specific to the liver but also are expressed in the ventral pancreas.
Figure 5. Anxa4 knockdown causes stage and tissue specific apoptosis in the developing foregut.
(a–c) Bright field microscopy of 31 hpf standard control MO (a), Anxa4 MO1 (b) and Anxa4 MO2 (c) morphant zebrafish embryos showing a mild developmental delay in the Anxa4 morphants. (d–f”) The ventral views of 31 hpf Tg(sox17:GFP) foregut stained for Anxa4 and cleaved Caspase 3 (*Casp 3). In Anxa4 morphants (MO1 in e–e”; MO2 in f–f”), but not in control morphants (d–d”), Anxa4 labeling is mostly lost (e, f) and patches of GFP+ apoptotic blebs can be found restricted to the Anxa4 morphant liver buds (white arrowheads). A subset of these GFP blebs are also positive for cleaved Caspase 3 (yellow arrowheads). (g) Graph summarizing relative incidence of liver apoptosis in anxa4 morphants, which peaks at 31 hpf. (h) Graph of a representative set of real-time RT-PCR results showing that in 27 hpf Anxa4 MO1 morphants, there is elevated expression of caspase 8 and Δ113p53.
To elucidate the molecular mechanism of apoptosis in Anxa4 morphants, we examined the effect of Anxa4 knockdown on the expression of key apoptosis related genes by real-time RT-PCR. We found that caspase 8 but not caspase 9 transcripts in MO1 morphants were increased by nearly 2 fold over standard control morphants (Fig. 5h). This suggests that in Anxa4 morphants, cell death occurs through the extrinsic, and not the intrinsic, apoptotic pathway (Elmore, 2007). Interestingly, we found that the transcripts of the Δ113p53 isoform increase more than 5 fold. Δ113p53, an anti-apoptotic isoform of p53 in zebrafish, has been reported to antagonize p53-mediated apoptosis via Bcl-xL under stress conditions, specifically in the developing digestive system (Chen et al., 2009; Chen et al., 2005; Dong et al., 2007; Lorent et al., 2010; Matthews et al., 2008). The increased expression of the anti-apoptotic Δ113p53 transcripts may indicate a foregut specific compensatory mechanism in response to the liver specific apoptosis caused by Anxa4 knockdown. Although our results showed a consistent decrease of p53 transcripts in Anxa4 morphants, the level of reduction was mild and insignificant. Despite the fact that in vitro studies suggest that Anxa4 interacts directly with the NFκB p50 subunit to affect its transcriptional targets (Jeon et al., 2010), we observed no significant difference for NFkB target genes, such as bcl-XL, c-Flip and iκBα (bcl2l, cflara and nfkbiaa in zebrafish respectively), between Anxa4 morphants and controls.
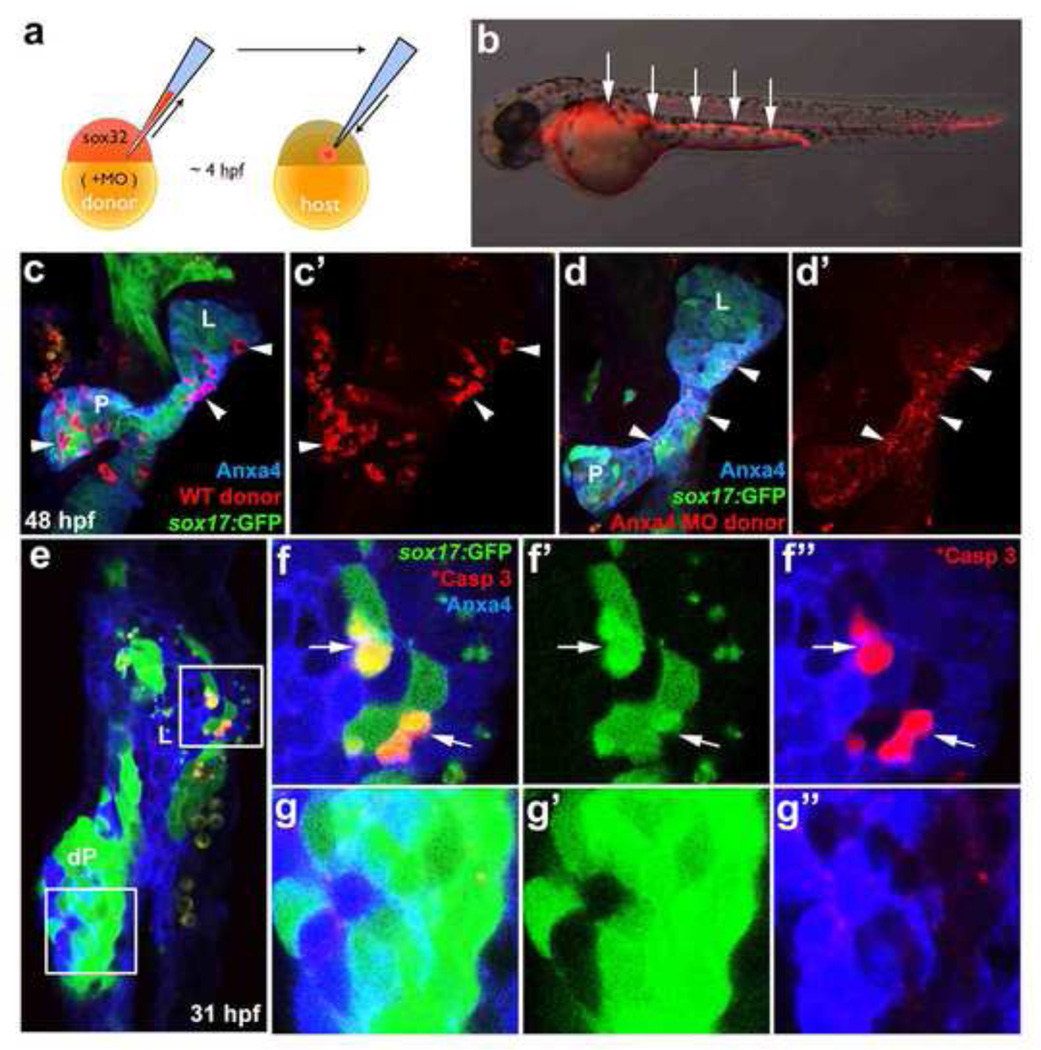
It is possible that the whole-embryo knockdown of Anxa4 function via MO disrupts liver development indirectly by affecting other tissues. Anxa4 transcripts are expressed broadly before and during gastrulation in zebrafish. Further, Anxa4 is also expressed specifically at 24hpf in the floor plate and hypochord (data not shown), which are important midline signal sources for patterning the surrounding tissue (Crosier et al., 2002; Placzek and Briscoe, 2005). To determine whether Anxa4 function is required in the endoderm to cell-autonomously support liver cell survival, we utilized an endoderm transplantation technique to knock down Anxa4 expression only in the endoderm. This cell transplantation technique utilizes sox32 mRNA to induce endoderm fate in the entire donor embryo (Stafford et al., 2006). Transplanted donor cells, labeled with rhodamine dextran, will then contribute predominantly to the host endoderm (Fig. 6a, 6b). By co-injecting sox32 mRNA and anxa4 MOs into donor embryos, the resulting mosaic host embryos will contain donor derived endoderm cells with reduced Anxa4 expression.
Figure 6. Anxa4 is cell-autonomously required for liver cell viability.
(a) Diagram depicts endoderm transplantation strategy. Donor embryo is injected with sox32 mRNA to drive endoderm fate, rhodamine dextran for labeling, and MOs to knock down gene expression. At 4 hpf, donor cells are transplanted into a host embryo, where they will contribute primarily to the host endoderm. (b) The mosaic 35 hpf host embryo obtained after the endoderm transplantation contains rhodamine dextran labeled donor endoderm cells (white arrows). (c–d’) The ventral view of 48 hpf Tg(sox17:GFP) foregut with rhodamine dextran labeled WT (white arrowheads in c, c’) or anxa4 morphant donor cells (d, d’). L, liver; P, pancreas. Note that rhodamine labeling (white arrowheads in d, d’) appears scattered and not restricted within individual cells in the host embryo with Anxa4 morphant donor cells. (e–g”) A single optical section (ventral view) of a 31 hpf mosaic host embryo with Anxa4 morphant cells from a Tg(sox17:GFP) donor embryo showing GFP blebs in the liver (upper inset and f–f”) but not dorsal pancreas (lower inset and g–g”). Subsets of GFP positive morphant donor cells are also positive for the cleaved Caspase 3 (white arrows) in the liver but not the dorsal pancreas. L, liver; dP, dorsal pancreas.
Wild type control donor cells without anxa4 MOs appeared morphologically normal in 48 hpf Tg(sox17:GFP) host endoderm (Fig. 6c, 6c’), with the rhodamine signal showing clear cell shapes. However at this stage, in the host embryos with donor cells containing anxa4 MO1 (3ng), the rhodamine signal was no longer focused into distinct cell shapes, but appeared diffuse throughout the host foregut endoderm (Fig. 6d, 6d’). We posit that the scattered rhodamine signal may be indicative of remnants of the donor cells that have undergone apoptosis at an earlier stage. Consistent with this postulation, we noted above that liver apoptosis in Anxa4 morphants is most penetrant at 31 hpf, leading us to examine mosaic morphants at this earlier stage. To be more certain that donor cells are of endoderm origin, anxa4 MO1 was injected into Tg(sox17:GFP) embryos and transplanted into non-transgenic wild-type host embryos. In the resulting mosaic embryo, the GFP positive endoderm cells are from anxa4 MO1 injected donors. At 31 hpf, as expected, GFP positive morphant donor cells lack Anxa4 expression in contrast to surrounding wild-type host endoderm cells (Fig. 6f”, g”), with most of those morphant cells still intact. In these host embryos, GFP positive blebs were detected in the liver (100% of the mosaic livers, n=9) but not dorsal pancreas (0% of the mosaic dorsal pancreas, n=7). Consistently, cleaved Caspase 3 can be detected in a subset of the GFP positive cells in the liver but not in either the dorsal pancreas or in GFP negative surrounding cells (Fig. 6e–g”), further supporting that liver bud cell survival is more sensitive to decreased Anxa4 function than other tissues. Although only a subset of the morphant clones in the liver appears to be apoptotic at this early stage, our observation that in most cases, intact morphant donor cells are not found in the liver at later stages, suggests that most morphant clones within the liver are gradually lost as foregut development proceeds.
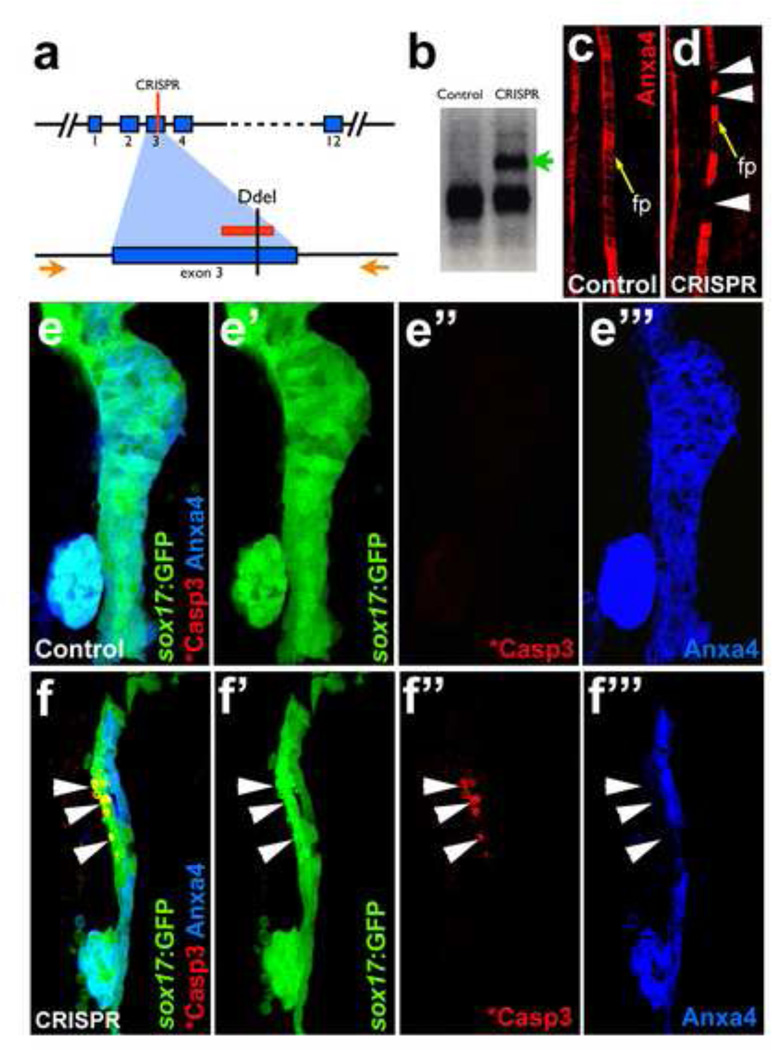
As a complementary approach for the Anxa4 mosaic knockdown studies, mosaic knockout of anxa4 was also examined after a transient application of the CRISPR/Cas9 nuclease targeting system. Injection of CRISPR/Cas9 can lead to frequent bi-allelic disruption of the targeted genetic sequence in random cells of an embryo, thereby achieving a genetically mosaic animal (Jao et al., 2013; Jinek et al., 2012). In taking this approach, one guide RNA was designed to target a region in Exon 3 of anxa4, which contains a restriction site for the enzyme Ddel (Fig. 7a). Co-injection of this guide RNA with cas9 mRNA into embryos leads to disruption of this restriction site (Fig. 7b). To confirm if the induced mutations lead to a significant disruption of the Anxa4 protein expression in a mosaic manner, 2F11 staining was assessed in the floor plate, where Anxa4 expression is consistent. In control embryos, Anxa4 is expressed broadly throughout the floor plate in a continuous stripe pattern. However, injected embryos exhibit gaps of Anxa4 expression in the floor plate (Fig. 7c–d), suggesting that certain clones of cells have lost any detectable Anxa4 expression, further validating the effectiveness of the guide RNA designed. Upon examination of the foregut of Tg(sox17:GFP) embryos injected with anxa4 guide RNA and cas9 mRNA, apoptotic GFP blebs and cleaved Caspase 3 expression were observed (25% of anxa4 CRISPR injected embryos, n=12). The apoptotic cell death occurs in the Anxa4 negative region of the liver anlagen but not in other areas of the foregut endoderm, consistent with the mosaic knockdown studies (Fig. 7e–f”). Similar results were found with two other guide RNAs targeting either Exon 2 or 4 of anxa4 (data not shown). Together, these mosaic knockdown and knockout studies suggest that Anxa4 is required cell-autonomously for cell survival in the early developing liver.
Figure 7. Mosaic disruption of the anxa4 gene by CRISPR/Cas9 leads to apoptosis in the developing liver.
a. The CRISPR guide RNA is designed to target a region in Exon 3 of anxa4, which contains a DdeI restriction enzyme site. The red lines indicate the guide RNA target site. Orange arrows indicate the primers for the genotyping PCR. (b–d) Effective anxa4 disruption by CRISPR/Cas9 was confirmed by DdeI digestion of the PCR products (b) and by Anxa4 antibody labeling of the floor plate (c–d). An uncut band (green arrow in b) and the floor plate cells without Anxa4 expression (white arrowheads in d) are found only in injected embryos. Yellow arrows point to floor plate (fp) in c–d. (e–f”) Confocal projections showing that apoptosis, indicated by GFP blebs and cleaved Caspase3 labeling, occurs in a subset of liver cells where Anxa4 expression is disrupted in the CRISPR injected embryos (white arrows) but not in control embryos.
Discussion
We have definitively identified the molecular target bound by the 2F11 mAb, demonstrating an effective strategy for determining the target of an antibody with an unknown antigen. Via peptide mass fingerprinting, we initially identified a strong candidate target for 2F11 mAb. By knocking down the candidate protein expression, we functionally confirmed the identity of the factor recognized by 2F11 mAbs. This approach may be generally useful for determining the targets of other orphan antibodies. With the efficiency of this strategy, it may now be more practical to screen for new factors by generating random monoclonal antibodies against extracts from cells of interest and determining their targets using the approach described here. Coupled with the use of traditional MO knockdown approaches and modern genome editing tools such as TALENs (Zu et al., 2013) and CRISPR/Cas (Hwang et al., 2013) for mutant functional studies of the newly identified factors, as demonstrated in this work, an antibody screen should now be considered a more practical approach for gene discovery.
The 2F11 mAb has become an essential reagent for labeling zebrafish hepatopancreatic ducts and intestinal secretory cells. By uncovering Anxa4 as the target of 2F11, we shed new light on published studies and future studies utilizing this antibody. With our findings that mAb 2F11 labels the hepatopancreas anlagen and pancreas islets, and other tissues including the pronephric ducts, floor plate, hypochord, swimbladder mesenchyme, and lateral lines, we expect this reagent to be increasingly useful for the zebrafish research community. Further with our results showing conserved expression of Anxa4 in mouse, we expect that assaying for Anxa4 expression to become a new approach for robustly labeling tissues such as ducts of pancreas, liver, and kidney in mammalian models.
Conserved expression of Anxa4 among vertebrates suggests that its function may also be similar. Our investigation of Anxa4 function shows that knockdown of Anxa4 in zebrafish embryos, using either of two functionally validated MOs, leads to the same cell-autonomous, stage and tissue specific apoptotic phenotype in the developing liver bud, consistent with when and where Anxa4 is expressed. This tissue specific phenotype is not likely due to MO toxicity as observed with certain MOs. Apoptosis resulting from MO toxicity generally lacks spatiotemporal specificity and shows increased p53 transcription (Gerety and Wilkinson, 2011), which we did not observe (Fig. 5h). Further, it is unlikely that both validated MOs would cause the same tissue specific, off-target effect. Experiments to rescue morphants via over expression of wild type Anxa4 were inconclusive, likely due to the difficulty in replicating Anxa4’s high expression level and dynamic, tissue specific expression pattern. However, our transient studies using the CRISPR/Cas9 system to knockout anxa4 in random cell populations during embryogenesis provide strong corroborative data. Assuming complete knockout of anxa4 is not gastrulation lethal, depending on its function during its early broad expression, future work generating and analyzing stable anxa4 mutants will be important for comparing and contrasting the effect of mosaic anxa4 knockout during development versus complete anxa4 knockout throughout development. Although anxa4 is also expressed in other tissues such as the floor plate, lateral line, pronephric duct, and more prominently in the dorsal pancreas, we did not detect significant cell death in these tissues in Anxa4 morphants. These tissues may be less sensitive to decreased Anxa4 function, perhaps due to redundancy with other Annexins. Alternatively, complete loss of Anxa4 function may be necessary to yield detectable defects in these tissues. It is also possible that Anxa4 is not required for cell survival in these tissues, but may play other roles yet to be discovered. Its dynamic intracellular localization in different tissues at different stages may be indicative of multiple functions for Anxa4. Consistent with previous studies, liver cell survival appears to be particularly sensitive to loss of several other factors, including RelA, IKKβ, or NEMO/IKKγ of the NFκBsignal pathway (Beg et al., 1995; Rudolph et al., 2000; Tanaka et al., 1999), and SNX7 (Xu et al., 2012), suggesting that liver cell survival is a highly regulated process. It may be that these factors act together to regulate liver cell survival. However unlike many of these other factors, which exhibit broader expression within the foregut, Anxa4 shows more restricted liver bud expression, suggesting that cell survival is specifically regulated in this organ.
Although Anxa4 and other members of the Annexin family have been implicated in cell survival, our studies provide the first in vivo data to implicate Anxa4 in this process. A role for Anxa4 in cell viability may have implications for β-cell and cancer cell survival. Loss of the insulin producing, pancreatic β-cells is characteristic of nearly all forms of diabetes. Intriguingly, the expression of ANXA4 is positively regulated by several MODY (Maturity Onset Diabetes of the Young) genes, including PDX1, HNF1A, and HNF4A (Bolotin et al., 2010; Servitja et al., 2009; Svensson et al., 2007). Because our studies implicate Anxa4 in cell survival, it will be interesting to investigate whether β-cell death and decreased β-cell ANXA4 expression are characteristic of these monogenic forms of diabetes. Although we found that loss of Anxa4 did not lead to apoptosis of embryonic β-cells, it will be important to examine Anxa4 function in stressed adult β-cells, where Anxa4 may be needed for cell survival.
Elevated levels of Anxa4 have been associated with many cancers, including pancreatic ductal adenocarcinoma (Shen et al., 2004), renal clear cell carcinoma (Zimmermann et al., 2004), gastric cancer (Lin et al., 2008), ovarian clear cell carcinoma (Miao et al., 2009; Toyama et al., 2012), and colorectal carcinoma (Duncan et al., 2008), and have been suggested as a diagnostic biomarker for these cancers. Cell culture studies suggest that Anxa4 promotes cell migration (Zimmermann et al., 2004) and proliferation (Lin et al., 2008) but represses apoptosis induced by chemotherapy drugs. Our findings are consistent with an anti-apoptotic role for Anxa4, leading us to suggest future in vivo studies to determine whether elevated Anxa4 is sufficient to confer chemoresistance.
Materials and Methods
Zebrafish strains
Zebrafish were raised and maintained under standard conditions. The following transgenic lines were used: Tg(sox17:GFP)s870 (Mizoguchi et al., 2008) Tg(neuroD:EGFP)nl1 (Obholzer et al., 2008); Tg(ptf1a:GFP)jh1 (Godinho et al., 2005); Tg(lfabf:ds-Red, elaA:EGFP)gz12 (Korzh et al., 2008; Pisharath et al., 2007); Tg(ins:dsRed)m1081 (Anderson et al., 2009); Tg(gcga:GFP)ia1 (Pauls et al., 2007); Tg(Tp1bglob:eGFP)um14 (Parsons et al., 2009), abbreviated as Tg(Tp1:eGFP).
Immunohistochemistry (IHC)
IHC was performed as previously described for zebrafish (Dong et al., 2007) and for mouse paraffin sections (Lee et al., 2007). Antibodies used include mouse monoclonal 2F11 (Crosnier et al., 2005) (1:1000; gift from J. Lewis, Cancer Research UK), rabbit polyclonal against Prox-1 (1:200; Millipore), rabbit polyclonal antibody against pan-Cadherin (1:5000; Sigma); rabbit polyclonal against GFP (1:300; Torrey Pines Biolabs), rabbit polyclonal against cleaved Caspase 3 (1:50, Cell Signaling), goat polyclonal against rat Anxa4 (1:50 Santa Cruz Biotechnology), rabbit polyclonal against Sox9 (1:50 Chemicon International), rabbit monoclonal against E-Cadherin (1:200 Cell Signaling). Control and morphant embryos were stained in the same tube. Imaging was done with a Zeiss 710 confocal microscope with Zen 9 software and images were processed with Adobe Photoshop CS3.
Immunoprecipitation and proteomics
100 embryos of 72 hpf were dissolved in 1 ml lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1% deoxycholate, 1% IGEPAL, pH 7.4) supplemented with protease inhibitor mixture set III (Sigma), 1 mM phenylmethylsulfonyl fluoride and 10 mM EDTA on ice for 30 min. The lysate was then clarified by centrifugation at 10,000g for 30 min at 4°C. Then 1 ug of 2F11 antibody was added and incubated 2 hours at 4°C (with constant rotation). The precipitated protein was captured on protein G agarose beads (20 ul) at 4°C overnight, with rotation. The beads were washed 5 times with lysis buffer and the protein was eluted with PAGE sample buffer. After 4–20% PAGE, the proteins were stained with SimplyBlue SafeStain (Invitrogen). The protein band was excised from the gel and subjected to in-gel trypsin digestion with Trypsin Gold, Mass Spectrometry Grade trypsin (Promega). The digest samples were analyzed by LC/MS/MS using the LTQ XL Linear Ion Trap Mass Spectrometer (Thermo Scientific). To identify the protein MS/MS spectra were searched against the Swiss-Prot database using SEQUEST Sorcerer software.
Whole mount in situ hybridization (ISH)
Zebrafish embryo whole mount ISH was performed as previously described (Lancman et al., 2013). Following ISH, IHC for GFP was carried out as reported with minor modification (Huang et al., 2008). Imaging was conducted using a Zeiss 710 confocal microscope with AxioPlan 4.8 software. The bright field and fluorescent images of ISH and IHC, respectively, were captured with the same vision field and magnification of individual samples, and merged with Abode Photoshop CS3. The anxa4 anti-sense probe was generated corresponding to the segment from PstI to BglII in anxa4 cDNA clone (MGC: 64121).
Morpholino injections
The embryos were injected at one to two cell stages with 3ng Anxa4 MO1, 1.5ng Anxa4 MO2, or 3ng standard control MO (GENE TOOLS, LLC.). The morphonlino injection into 48 hpf embryos did not effectively reduce 2F11 antibody labeling. Anxa4 MO1 has the sequence of TTC CAC GGT TTC CCA ACG CTG CCA T and MO2 has the sequence of TGG TAT GGT CTC TCT CAC CTG CTC C. Standard control MO has the sequence of cctcttacctcagttacaatttata. The primers used in RT-PCR for testing the efficacy of Splice MO are 5’-cttcatctctacaatggcagcg-3’ and 5’-agtttccagtcagctcaga-3’.
Real-time PCR
Total RNA was extracted from 20–30 zebrafish embryos collected at 27 hpf using Tri-reagent (Molecular Research Center, Inc). 5 ug total RNA was used as the template and reverse transcribed with oligo-dT primer using Improm-II Reverse transcriptase in 20 ul reaction as indicated by the product manual. The real-time PCR was performed as described with minor modifications (Correa et al., 2005). The following primers were determined through Roche online universal probe library Assay Design Center: cflara (NM_194399, 5’-CTG TGC CTG AAA CAG GGA TT-3’ and 5’-CGT GTG CTT CAA CAT TTC TCC-3’), bcl2l (NM_131807.1, 5’-GGC TTG TTT GCT TGG TTG AC-3’ and 5’-TGG TGC AAT GGC TCA TAC C-3’), p53 (NM_131327.1, 5’-GAG GTC GGC AAA ATC AAT TC-3’, 5’-CAC CTG GGG GCT GAA TAA T-3’), caspase 8 (NM_131510.2, 5’-TCA GAG TCA CAG AGA CCA GGA A-3’, 5’-CAG ATA CAG GGT TGT TGA GAA AAA-3’), caspase 9 (NM_001007404.2, 5’-AAA GGG GGT CTT CAC TCA GG-3’, 5’-GTT GCC TGG CTT GGT CAC-3’). The primers for Δ113p53 isoform (5’-ATA TCC TGG CGA ACA TTT GGA G-3’, 5’-CCT CCT GGT CTT GTA ATG TCA-3’), nfkbiaa (5’-CAG CAC CTG CGT TCC ATT CT-3’ and 5’-GCA CGT GTG TCC GCT GTA GT-3’) and cyclophilin (5’-GAC GAT GCT GTT CGC TAC CA-3’ and 5’-CAA GCG CAC GTA GCC TTT CT-3’) are as described (Xu et al., 2012), (Correa et al., 2005). Results were normalized to cyclophilin.
Endoderm transplantation
The procedure was performed as previously described (Stafford et al., 2006). Briefly, embryos injected with sox32 mRNA (and MOs) at the one cell stage were used as donors. At 4 hpf, both donor and host embryos were dechorioned in an agarose-coated petri dish. Donor cells were drawn into a glass needle (pulled from Borosilicate Glass Capillaries, World Precision Instruments, Inc) using CellTram Air (Eppendorf). About 15 donor cells were then injected into one host embryo using CellTram Air.
Targeting the anxa4 gene with the CRISPR/Cas9 system
The targeting sites (5’-GGA ACT GTG ACT GAG GCG TC-3’ for Exon 2; 5’-GGA GGC TTT CAA GCT GAG CG-3’ for Exon 3; 5’-GGG CCT GAT GAT GCC TGC AG-3’ for Exon 4) were selected using the online application available at zifit.partners.org. The guide RNAs were produced and injected with cas9 mRNA as previously described (Jao et al., 2013). The following primer pairs and restriction enzymes were used for assessing genetic disruption: 5’-GAT CCT TGA CAC AGT CTG ATC T-3’, 5’- CTG AGT AGT TGG CGG TTC A-3’ and DdeI for Exon 2; 5’-CTT AGG CTT TGA CTG ATG CCA-3’, 5’-GGT TGC TGA AGA ACT ACT GAG A-3’ and DdeI for Exon 3; 5’-CCT CAC CCA CAC TTT GCC T-3’, 5’- GTA CAC TGA TGG ACT ACA GAC A-3’ and PstI for Exon 4.
Supplementary Material
Highlights.
Zebrafish AnnexinA4 is the molecular target of mAb 2F11.
AnnexinA4 expression in the hepatopancreas system is conserved among vertebrates.
AnnexinA4 is required cell-autonomously for zebrafish liver bud cell survival.
Acknowledgements
We thank Hongna Pei for technical support on mouse section immunostaining, Joseph Lancman, Keith Gates and Marco Maruggi for discussions and/or critical reading of the manuscript. We thank Julian Lewis (Cancer Research UK) for 2F11 antibody and Teresa Nicolson (Oregon Health & Science University) for Tg(neuroD:EGFP)nl1.
Funding: This work was supported by funds from National Institutes of Health [1DP2DK098092] and from the Sanford Children’s Health Research Center to P.D.S.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests statement: The authors declare no competing financial interests.
Author Contributions
D.Z. designed and executed experiments and assembled figures with the guidance of P.D.S.D.; P.D.S.D. oversaw studies; V.S.G. initiated mass spec analysis, performed immunoprecipitation and western blots; W.H. performed IHC for adult mouse tissue sections; Y.Z. and R.G.C. contributed to real time PCR studies; D.Z. and P.D.S.D. wrote manuscript with feed back from coauthors.
Reference
- Ahmed M, Forsberg J, Bergsten P. Protein profiling of human pancreatic islets by two-dimensional gel electrophoresis and mass spectrometry. Journal of proteome research. 2005;4:931–940. doi: 10.1021/pr050024a. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PD, Shin D, Chi NC, Shin CH, Schlegel A, Halpern M, Stainier DY. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334:213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HC, Kaetzel MA, Gotter AL, Dedman JR, Nelson DJ. Annexin IV inhibits calmodulin-dependent protein kinase II-activated chloride conductance. A novel mechanism for ion channel regulation. J Biol Chem. 1994;269:32464–32468. [PubMed] [Google Scholar]
- Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, Lane DP, Peng J. p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev. 2009;23:278–290. doi: 10.1101/gad.1761609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900–2911. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- Correa RG, Matsui T, Tergaonkar V, Rodriguez-Esteban C, Izpisua-Belmonte JC, Verma IM. Zebrafish IkappaB kinase 1 negatively regulates NF-kappaB activity. Curr Biol. 2005;15:1291–1295. doi: 10.1016/j.cub.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Crosier PS, Kalev-Zylinska ML, Hall CJ, Flores MV, Horsfield JA, Crosier KE. Pathways in blood and vessel development revealed through zebrafish genetics. The International journal of developmental biology. 2002;46:493–502. [PubMed] [Google Scholar]
- Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development. 2005;132:1093–1104. doi: 10.1242/dev.01644. [DOI] [PubMed] [Google Scholar]
- Curado S, Ober EA, Walsh S, Cortes-Hernandez P, Verkade H, Koehler CM, Stainier DY. The mitochondrial import gene tomm22 is specifically required for hepatocyte survival and provides a liver regeneration model. Disease models & mechanisms. 2010;3:486–495. doi: 10.1242/dmm.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, Farber SA, Moens CB, Stainier DY. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8:e1002754. doi: 10.1371/journal.pgen.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- Dreier R, Schmid KW, Gerke V, Riehemann K. Differential expression of annexins I, II and IV in human tissues: an immunohistochemical study. Histochemistry and cell biology. 1998;110:137–148. doi: 10.1007/s004180050275. [DOI] [PubMed] [Google Scholar]
- Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer. 2008;98:426–433. doi: 10.1038/sj.bjc.6604128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EauClaire SF, Cui S, Ma L, Matous J, Marlow FL, Gupta T, Burgess HA, Abrams EW, Kapp LD, Granato M, Mullins MC, Matthews RP. Mutations in vacuolar H+ -ATPase subunits lead to biliary developmental defects in zebrafish. Dev Biol. 2012;365:434–444. doi: 10.1016/j.ydbio.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicologic pathology. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber SA, De Rose RA, Olson ES, Halpern ME. The zebrafish annexin gene family. Genome research. 2003;13:1082–1096. doi: 10.1101/gr.479603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003a;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003b;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev Biol. 2011;350:279–289. doi: 10.1016/j.ydbio.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Gerke V, Moss SE. Annexins: from structure to function. Physiological reviews. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, Park SW, Leach SD, Wong RO. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132:5069–5079. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- Han EK, Tahir SK, Cherian SP, Collins N, Ng SC. Modulation of paclitaxel resistance by annexin IV in human cancer cell lines. Br J Cancer. 2000;83:83–88. doi: 10.1054/bjoc.2000.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Kaetzel MA, Kishore BK, Dedman JR, Zeidel ML. Annexin A4 reduces water and proton permeability of model membranes but does not alter aquaporin 2-mediated water transport in isolated endosomes. The Journal of general physiology. 2003;121:413–425. doi: 10.1085/jgp.200308803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Meyers S, von Bodungen M, Apodaca G, Dedman JR, Kaetzel MA, Zeidel ML. Studies on localization and function of annexin A4a within urinary bladder epithelium using a mouse knockout model. Am J Physiol Renal Physiol. 2008;294:F919–F927. doi: 10.1152/ajprenal.00265.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Ruan H, Aw MY, Hussain A, Guo L, Gao C, Qian F, Leung T, Song H, Kimelman D, Wen Z, Peng J. Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development. 2008;135:3209–3218. doi: 10.1242/dev.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon YJ, Kim DH, Jung H, Chung SJ, Chi SW, Cho S, Lee SC, Park BC, Park SG, Bae KH. Annexin A4 interacts with the NF-kappaB p50 subunit and modulates NF-kappaB transcriptional activity in a Ca2+-dependent manner. Cell Mol Life Sci. 2010;67:2271–2281. doi: 10.1007/s00018-010-0331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Enomoto T, Serada S, Ueda Y, Takahashi T, Ripley B, Miyatake T, Fujita M, Lee CM, Morimoto K, Fujimoto M, Kimura T, Naka T. Enhanced expression of Annexin A4 in clear cell carcinoma of the ovary and its association with chemoresistance to carboplatin. International journal of cancer. Journal international du cancer. 2009;125:2316–2322. doi: 10.1002/ijc.24587. [DOI] [PubMed] [Google Scholar]
- Kim A, Serada S, Enomoto T, Naka T. Targeting annexin A4 to counteract chemoresistance in clear cell carcinoma of the ovary. Expert Opin Ther Targets. 2010;14:963–971. doi: 10.1517/14728222.2010.511180. [DOI] [PubMed] [Google Scholar]
- Korzh S, Pan X, Garcia-Lecea M, Winata CL, Pan X, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancman JJ, Zvenigorodsky N, Gates KP, Zhang D, Solomon K, Humphrey RK, Kuo T, Setiawan L, Verkade H, Chi YI, Jhala US, Wright CV, Stainier DY, Dong PD. Specification of hepatopancreas progenitors in zebrafish by hnf1ba and wnt2bb. Development. 2013;140:2669–2679. doi: 10.1242/dev.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, Ferrara N, Nagy A, Roos KP, Iruela-Arispe ML. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130:691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LL, Chen CN, Lin WC, Lee PH, Chang KJ, Lai YP, Wang JT, Juan HF. Annexin A4: A novel molecular marker for gastric cancer with Helicobacter pylori infection using proteomics approach. Proteomics. Clinical applications. 2008;2:619–634. doi: 10.1002/prca.200780088. [DOI] [PubMed] [Google Scholar]
- Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Lu Z, Sun H, Yuan K, Zhang Q, Meng S, Wang F, Guo H, Ju X, Liu Y, Ye T, Lu Z, Zhai Z. Nuclear entry of active caspase-3 is facilitated by its p3-recognition-based specific cleavage activity. Cell research. 2010;20:211–222. doi: 10.1038/cr.2010.9. [DOI] [PubMed] [Google Scholar]
- Manfroid I, Ghaye A, Naye F, Detry N, Palm S, Pan L, Ma TP, Huang W, Rovira M, Martial JA, Parsons MJ, Moens CB, Voz ML, Peers B. Zebrafish sox9b is crucial for hepatopancreatic duct development and pancreatic endocrine cell regeneration. Dev Biol. 2012;366:268–278. doi: 10.1016/j.ydbio.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse KL, Collins RJ, Bhamra S, Seville RA, Jones EA. Anxa4 Genes are Expressed in Distinct Organ Systems in Xenopus laevis and tropicalis But are Functionally Conserved. Organogenesis. 2007;3:83–92. doi: 10.4161/org.3.2.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuishi Y, Arakawa N, Kawasaki H, Miyagi E, Hirahara F, Hirano H. Wild-type p53 enhances annexin IV gene expression in ovarian clear cell adenocarcinoma. FEBS J. 2011;278:1470–1483. doi: 10.1111/j.1742-4658.2011.08059.x. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Parsons MJ, Leach SD. Aldh1-expressing endocrine progenitor cells regulate secondary islet formation in larval zebrafish pancreas. PLoS One. 2013;8:e74350. doi: 10.1371/journal.pone.0074350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews RP, Eauclaire SF, Mugnier M, Lorent K, Cui S, Ross MM, Zhang Z, Russo P, Pack M. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53:905–914. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–131. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- Miao Y, Cai B, Liu L, Yang Y, Wan X. Annexin IV is differentially expressed in clear cell carcinoma of the ovary. Int J Gynecol Cancer. 2009;19:1545–1549. doi: 10.1111/IGC.0b013e3181a844c8. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Verkade H, Heath JK, Kuroiwa A, Kikuchi Y. Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development. 2008;135:2521–2529. doi: 10.1242/dev.020107. [DOI] [PubMed] [Google Scholar]
- Ninov N, Borius M, Stainier DY. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, Seiler C, Sidi S, Sollner C, Duncan RN, Boehland A, Nicolson T. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci. 2008;28:2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S, Zecchin E, Tiso N, Bortolussi M, Argenton F. Function and regulation of zebrafish nkx2.2a during development of pancreatic islet and ducts. Dev Biol. 2007;304:875–890. doi: 10.1016/j.ydbio.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek M, Briscoe J. The floor plate: multiple cells, multiple signals. Nature reviews. Neuroscience. 2005;6:230–240. doi: 10.1038/nrn1628. [DOI] [PubMed] [Google Scholar]
- Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia AJ, Mak TW. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- Servitja JM, Pignatelli M, Maestro MA, Cardalda C, Boj SF, Lozano J, Blanco E, Lafuente A, McCarthy MI, Sumoy L, Guigo R, Ferrer J. Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol Cell Biol. 2009;29:2945–2959. doi: 10.1128/MCB.01389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seville RA, Nijjar S, Barnett MW, Masse K, Jones EA. Annexin IV (Xanx-4) has a functional role in the formation of pronephric tubules. Development. 2002;129:1693–1704. doi: 10.1242/dev.129.7.1693. [DOI] [PubMed] [Google Scholar]
- Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res. 2004;64:9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- Sohma H, Ohkawa H, Sakai R, Hashimoto E, Ukai W, Saito T. Augmentation of ethanol-induced cell damage and activation of nuclear factor-kappa B by annexin IV in cultured cells. Alcoholism, clinical and experimental research. 2003;27:64S–67S. doi: 10.1097/01.ALC.0000078615.47704.73. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- Stafford D, White RJ, Kinkel MD, Linville A, Schilling TF, Prince VE. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development. 2006;133:949–956. doi: 10.1242/dev.02263. [DOI] [PubMed] [Google Scholar]
- Svensson P, Williams C, Lundeberg J, Ryden P, Bergqvist I, Edlund H. Gene array identification of Ipf1/Pdx1−/− regulated genes in pancreatic progenitor cells. BMC Dev Biol. 2007;7:129. doi: 10.1186/1471-213X-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- Toyama A, Suzuki A, Shimada T, Aoki C, Aoki Y, Umino Y, Nakamura Y, Aoki D, Sato TA. Proteomic characterization of ovarian cancers identifying annexin-A4, phosphoserine aminotransferase, cellular retinoic acid-binding protein 2, and serpin B5 as histology-specific biomarkers. Cancer science. 2012;103:747–755. doi: 10.1111/j.1349-7006.2012.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yin W, Xia J, Peng M, Li S, Lin S, Pei D, Shu X. An antiapoptotic role of sorting nexin 7 is required for liver development in zebrafish. Hepatology. 2012;55:1985–1993. doi: 10.1002/hep.25560. [DOI] [PubMed] [Google Scholar]
- Zimmermann U, Balabanov S, Giebel J, Teller S, Junker H, Schmoll D, Protzel C, Scharf C, Kleist B, Walther R. Increased expression and altered location of annexin IV in renal clear cell carcinoma: a possible role in tumour dissemination. Cancer letters. 2004;209:111–118. doi: 10.1016/j.canlet.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.