Abstract
Autophagy is the principal catabolic response to nutrient starvation and is necessary to clear dysfunctional or damaged organelles, but excessive autophagy can be cytotoxic or cytostatic and contributes to cell death. Depending on the abundance of enzymes involved in molecule biosynthesis, cells can be dependent on uptake of exogenous nutrients to provide these molecules. Argininosuccinate synthetase 1 (ASS1) is a key enzyme in arginine biosynthesis, and its abundance is reduced in many solid tumors, making them sensitive to external arginine depletion. We demonstrated that prolonged arginine starvation by exposure to ADI-PEG20 (pegylated arginine deiminase) induced autophagy-dependent death of ASS1-deficient breast cancer cells, because these cells are arginine auxotrophs (dependent on uptake of extracellular arginine). Indeed, these breast cancer cells died in culture when exposed to ADI-PEG20 or cultured in the absence of arginine. Arginine starvation induced mitochondrial oxidative stress, which impaired mitochondrial bioenergetics and integrity. Furthermore, arginine starvation killed breast cancer cells in vivo and in vitro only if they were autophagy-competent. Thus, a key mechanism underlying the lethality induced by prolonged arginine starvation was the cytotoxic autophagy that occurred in response to mitochondrial damage. Last, ASS1 was either low in abundance or absent in more than 60% of 149 random breast cancer bio-samples, suggesting that patients with such tumors could be candidates for arginine starvation therapy.
INTRODUCTION
Breast cancer is one of the most common cancers that kill women (1). Gene expression analyses of breast cancer have identified five intrinsic molecular “subtypes” (normal-like, luminal A, luminal B, basal, and HER2-positive), each of which has unique clinical and histological phenotypes (2, 3). Currently, breast cancers are subtyped so that different treatments can be tailored to maximize therapeutic benefit. However, it is still estimated that 39,620 women and 410 men will die of breast cancer in the United States in 2013 to 2014 (4). Therefore, it is necessary to identify new therapeutic targets, especially for treatment-refractory tumors.
Altered cellular metabolism has emerged as a common phenotype of cancers and other complex diseases (5). Cancer cells adapt their metabolic pathways to meet the high-energy demands required for their accelerated growth and proliferation and the associated metabolic stresses. Metabolomic studies have revealed that the steady-state abundance of many amino acids in stomach, colon, lung, and prostate cancers is higher than in the corresponding normal tissue, suggesting that the tumors have increased biosynthetic needs for amino acids (6, 7). For example, some tumor cells are addicted to glutamine because it supports anabolic processes and fuels proliferation (8, 9). The serine and glycine biosynthetic pathways have also been suggested to play critical roles in oncogenesis (10, 11). Here, we aimed to exploit the distinct metabolic requirements of breast cancers to identify impaired metabolic pathways that can be targeted for breast cancer treatment.
Among the metabolic adaptations that occur in cancer cells is the increased use of the amino acid arginine to fuel anabolic processes. Arginine is a nonessential amino acid in humans, but it plays a vital role in multiple metabolic pathways, including protein synthesis and the production of nitric oxide, polyamines, urea, creatine, nucleotides, proline, glutamate, and agmatine (12, 13). Arginine concentrations in cells are partly maintained by de novo synthesis from citrulline, which is converted to arginine by argininosuccinate synthetase 1 (ASS1) and argininosuccinate lyase (ASL). Accumulating evidence suggests that the endogenous production of arginine is not sufficient to meet the needs of rapidly proliferating tumor cells (14-20). Thus, arginine is now considered a semiessential amino acid under stress conditions, and arginine auxotrophs are cells that have lost the ability to synthesize arginine and are dependent on external arginine sources. Paradoxically, although there is an increased demand for arginine by tumor cells, many human tumor cells, including melanoma, lymphoma, glioma, and prostate cancer, are ASS1-deficient and become arginine auxotrophs (http://www.proteinatlas.org/ENSG00000130707). The biological mechanisms underlying this paradox are not completely understood, and it may be that arginine auxotrophs have a previously overlooked metabolic liability that could be exploited to treat many cancers, including breast cancers. Thus, our analyses could not only improve our understanding of the biology of ASS1 deficiencies in cancer development and recurrence but also lead to the development of therapies that target arginine auxotrophic breast cancers.
Arginine deiminase (ADI) is a microbial enzyme originally isolated from mycoplasma that metabolizes arginine to citrulline and is 300-fold more effective than arginase at depleting arginine from the environment (21-23). Recombinant ADI has been used to deprive arginine auxotrophic tumors of external arginine, thereby inhibiting their growth. Sensitive tumors are usually ASS1-deficient and therefore incapable of synthesizing endogenous arginine (17, 24-28). Conversely, overexpressing ASS1 in ASS1-deficient melanoma cells confers resistance to ADI (17, 29). Recombinant ADI-based therapies have been used in several clinical trials to treat advanced arginine auxotrophic melanoma and hepatocellular carcinoma, and have had acceptable response rates and minimal side effects (30-33). However, besides arginine depletion, the mechanism by which ADI mediates its effect is unknown. Recombinant pegylated ADI (ADI-PEG20) is a new form of ADI with increased safety and efficacy profiles. Nutrient starvation approach, including arginine depletion, will invariably induce autophagy in many cancer cells (34, 35); however, the biological relevance and consequence of autophagy induction in this context are not yet understood.
Here, we sought to identify the molecular determinants of sensitivity and resistance to arginine starvation therapy that could guide patient selection or the choice of agents to be given in combination. We found that arginine starvation killed ASS1-deficient breast cancer cells by impairing mitochondrial energy production and increasing the generation of reactive oxygen species (ROS), which compromised mitochondrial integrity. The accumulation of impaired mitochondria would eventually result in autophagy-dependent cell death. We determined that ASS1-deficient breast cancer cell lines and mouse xenografts were susceptible to ADI-PEG20 only if they were autophagy-competent. In addition, more than 60% of a random sample of 149 breast cancers had low abundance of ASS1 or no detectable ASS1, irrespective of the breast cancer molecular subtype. Thus, we defined the mechanisms that underlie the sensitivity of arginine auxotrophic breast cancer cells to arginine starvation and then analyzed the role of autophagy in this process.
RESULTS
ADI-PEG20 selectively inhibits the proliferation of ASS1-deficient breast cancer cells
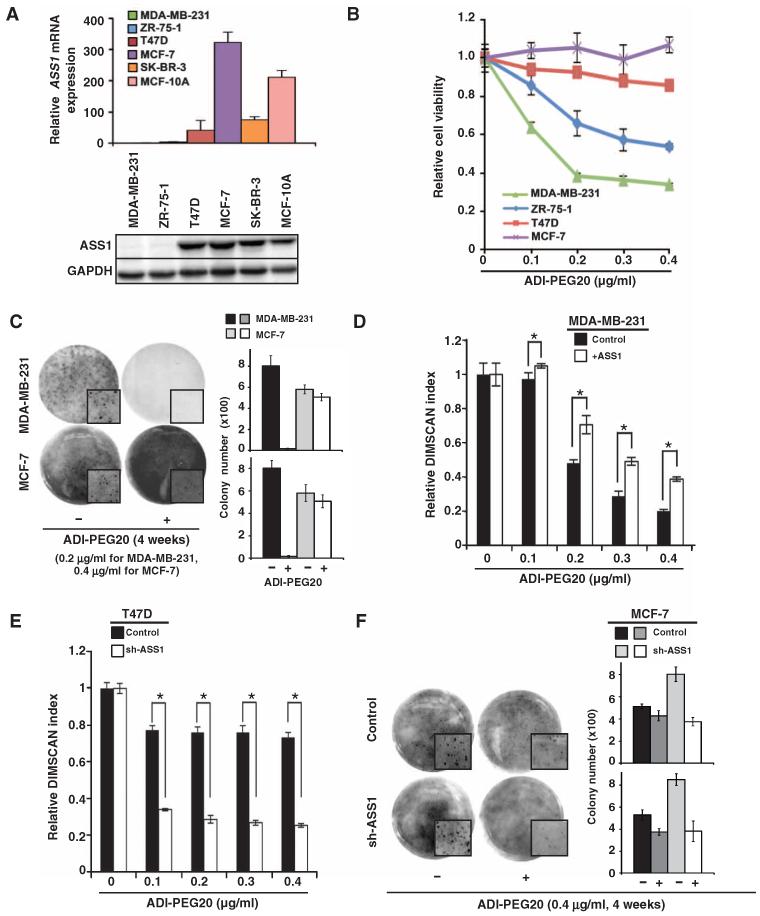
We hypothesized that reduced ASS1 abundance would increase the sensitivity of breast cancer cells to arginine starvation by treatment with recombinant ADI-PEG20. We found that ASS1 mRNA and ASS1 protein abundance was relatively low in MDA-MB-231 and ZR-75-1 cells compared to MCF-7 and T47D cells (Fig. 1A). As predicted, the relative sensitivity of these four breast cancer cells lines to ADI-PEG20–induced inhibition of proliferation was inversely correlated with the abundance of ASS1 (MDA-MB-231 > ZR-75-1 > T47D ≈ MCF-7) (Fig. 1B). The inhibition of proliferation in MDA-MB-231 cells by ADI-PEG20 was independently validated with a time course growth assay (fig. S1A) and by anchorage-independent colony formation assay (Fig. 1C). To confirm that the inhibition of proliferation by ADI-PEG20 was inversely related to ASS1 abundance, we established ASS1-overexpressing MDA-MB-231 (fig. S1B) and ASS1-knockdown T47D cells (fig. S1C). As predicted, MDA-MB-231 cells overexpressing ASS1 were more resistant to ADI-PEG20 (Fig. 1D), and the knockdown of ASS1 sensitized T47D cells to ADI-PEG20 (Fig. 1E). Together, our results indicate that recombinant ADI-PEG20 effectively inhibits the proliferation of breast cancer cells with ASS1 knockdown, which cannot synthesize sufficient endogenous arginine for their metabolic needs. Unexpectedly, we noticed that although knockdown of ASS1 promoted ADI-PEG20 sensitivity (Fig. 1E), decreased ASS1 abundance increased the proliferation (fig. S1D) and the ability to form colonies (fig. S1E) in T47D cells. We speculated that ASS1 might play a tumor suppressor role in breast cancer cells. To test this possibility, we knocked down ASS1 in MCF-7 cells, which had the greatest ASS1 abundance of the five breast cancer cell lines (Fig. 1A) and exhibited resistance to ADI-PEG20 (Fig. 1, B and C). A 50% knockdown of ASS1 (fig. S1F) increased the anchorage-independent, colony-forming ability of MCF-7 cells, as well as their sensitivity to ADI-PEG20 when grown in soft agar (Fig. 1F). These data suggest that ASS1 may have a tumor suppressor function that prevents anchorage-independent breast cancer cell growth.
Fig. 1. Arginine starvation inhibits cell proliferation of ASS1-deficient breast cancer cells.
(A) ASS1 abundance varies in different breast cancer cell lines and immortalized MCF-10A cells. ASS1 mRNA abundance was analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR; upper panel), and ASS1 protein abundance was assessed by Western blotting (lower panel). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) message or GAPDH protein abundance was used to normalize qRT-PCR results or to assure equal loading; n = 3 sets of cells. (B) Inverse correlation between ASS1 protein abundance and the cytotoxic effects of ADI-PEG20 on four breast cancer cell lines; n = 4 sets of cells. (C) Anchorage-independent growth of MDA-MB-231 cells is affected by ADI-PEG20 treatment; n = 2 sets of cells in triplicates (total 6). (D) Overexpression of ASS1 increases the IC50 (median inhibitory concentration) of ADI-PEG20 in MDA-MB-231 cells from 0.2 to 0.3 μg/ml; n = 3 sets of cells. (E) Knockdown of ASS1 abundance decreases IC50 of ADI-PEG20 in T47D cells (from greater than 0.4 to 0.075 μg/ml); n = 3 sets of cells. (F) Knockdown of ASS1 abundance sensitizes MCF-7 cells to ADI-PEG20 in colony formation assays; n = 2 sets of cells in triplicates (total 6). (C and F) Representative images show total colony area or a selected area of colony growth (enlarged; insets) on day 28. Bar graph representing means ± SD from individual set of three technical repeats is shown on the right. Viability and qRT-PCR data are shown as means ± SD; *P < 0.05.
ADI-PEG20 induces autophagy-dependent cell death
Using MDA-MB-231 cells, we investigated the mechanism underlying arginine deprivation–induced cell death of breast cancer cells with low ASS1 abundance. We first determined if ADI-PEG20 induces apoptosis in MDA-MB-231 cells, as previously suggested by studies using other cancer cells (28, 36). Flow cytometry indicated that about 13% of MDA-MB-231 cells were annexin V–positive after being treated with ADI-PEG20 or starved of arginine (Fig. 2A and fig. S2A). However, on the basis of the extent by which ADI-PEG20 inhibited cell proliferation (Fig. 1B), the apoptotic cell population was smaller than expected. It seemed likely that cell proliferation was inhibited by another pathway that was induced by ADI-PEG20.
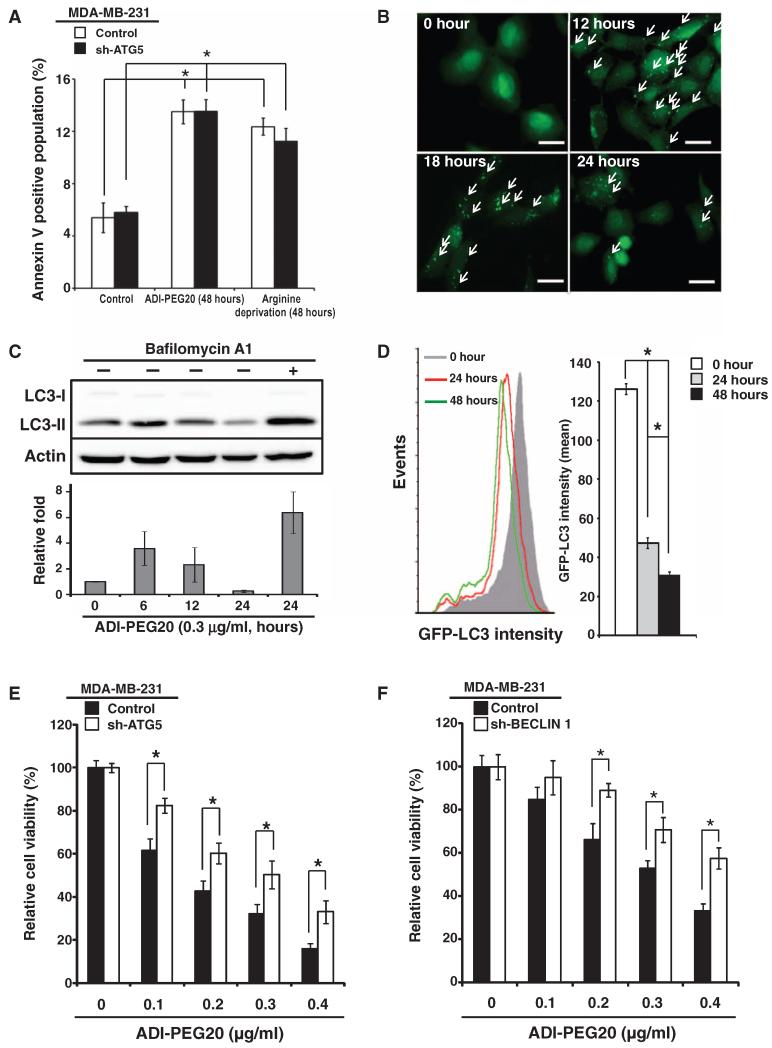
Fig. 2. ADI-PEG20 induces autophagy-dependent cell death.
(A) ADI-PEG20 treatment or arginine starvation moderately increases apoptotic cell percentage in MDA-MB-231 cells. Knockdown of ATG5 in MDA-MB-231 cells does not significantly affect response to ADI-PEG20 treatment or arginine starvation. Histograms are presented as means ± SD; n = 3 sets of cells. (B) Representative images of GFP-LC3 puncta formation (indicated by arrows) using fluorescence microscopy; n = 5 sets of cells. Scale bars, 50 mm. (C) ADI-PEG20 induces LC3 lipidation, which is reversed by bafilomycin A1. One representative Western and bar graph, presented as means ± SD, are shown; n = 3 sets of cells. (D) ADI-PEG20 induces autophagic flux. MDA-MB-231 cells overexpressing GFP-LC3 were treated with ADI-PEG20 for the indicated time periods and analyzed by flow cytometry (left panel). Data are shown as means ± SD (right panel); n = 3 sets of cells. *P < 0.05. (E and F) Knockdown of ATG5 (E) or BECLIN 1 (F) reduces ADI-PEG20-induced cytotoxicity. n = 3 sets of cells; *P < 0.05.
Arginine starvation has been reported to induce autophagy (28, 37). We first determined whether ADI-PEG20 induced autophagy in MDA-MB-231 cells. During autophagy, a cytosolic form of microtubule-associated protein 1A/1B light chain 3 (LC3-I) is conjugated to phosphatidylethanolamine to form LC3-II, which inturn is recruited to autophagosomal membranes (38-41). Formation of GFP (green fluorescent protein)–LC3–positive puncta, a marker for autophagy (39), was monitored in MDA-MB-231 cells stably transfected with GFP-LC3 and treated with ADI-PEG20. GFP-LC3 puncta were observed 12 hours after ADI-PEG20 treatment, but were decreased after 24 hours, indicating the appearance and disappearance of autophagosomes (Fig. 2B). Next, we used Western blotting to show that LC3 was lipidated (LC3-II) and that LC3-II abundance increased to about twofold at 6 hours after ADI-PEG20 treatment, and then decreased over time (Fig. 2C). Furthermore, bafilomycin A1, a vacuolar-type H+-ATPase (H+-dependent adenosine triphosphatase) inhibitor that prevents the maturation of autophagic vacuoles (42), reversed the decrease in LC3-II that occurred 24 hours after ADI-PEG20 treatment, suggesting that there was active autophagic flux (Fig. 2C). To confirm that ADI-PEG20 induced autophagic flux, we used a flow cytometry–based assay to show that GFP-LC3 intensity was significantly decreased at 24 and 48 hours after treatment with ADI-PEG20 (Fig. 2D).
To determine the role of autophagy in inhibiting cell proliferation mediated by ADI-PEG20, we used short hairpin RNA (shRNA) to knock down ATG5 and BECLIN 1 (fig. S2B), two essential components of the autophagy machinery (34, 35). Knockdown of ATG5 or BECLIN 1 decreased the sensitivity of MDA-MB-231 cells to ADI-PEG20 (Fig. 2, E and F). The size of the annexin V–positive population of MDA-MB-231 cells expressing shRNA against ATG5 was not significantly altered by ADI-PEG20 treatment or arginine starvation (Fig. 2A and fig. S2A). In addition, phase-contrast microscopy revealed that ADI-PEG20 or arginine starvation altered the overall morphology of MDA-MB-231 cells, but not cells stably transfected with ATG5 shRNA at 24 or 48 hours after treatment (fig. S2, C and D). Last, although the combination of ADI-PEG20 with rapamycin [a mammalian target of rapamycin (mTOR) inhibitor], an autophagy inducer, did not increase ADI-PEG20–mediated cell killing, addition of chloroquine, an autophagic flux inhibitor, decreased ADI-PEG20–induced cell death (fig. S2E). Therefore, we propose that autophagy is required for recombinant ADI-PEG20 to suppress MDA-MB-231 cell proliferation.
ADI-PEG20 impairs mitochondrial bioenergetics
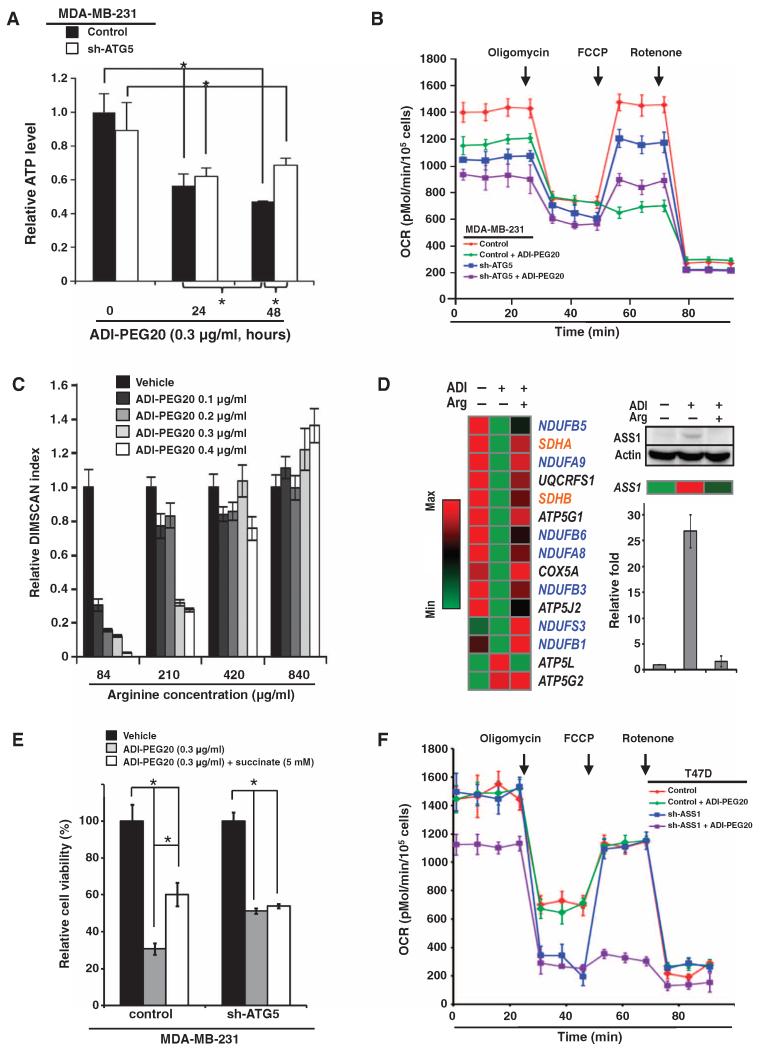
Autophagy is a catabolic process that delivers intracellular proteins or organelles sequestered in autophagosomes to lysosomes for recycling, thereby providing energy during metabolic stress (34, 35). We have shown that glutamine depletion or autophagy impairment decreases ATP (adenosine 5′-triphosphate) production (43). We then analyzed the difference of ATP concentrations during the course of ADI-PEG20 treatment in MDA-MB-231 cells stably transfected with control or ATG5 shRNA. ATP concentrations began to decrease 24 hours after ADI-PEG20 treatment and continued to decrease to less than 50% of vehicle-treated control cells at 48 hours after treatment (Fig. 3A). Knocking down ATG5 appeared to slow the reduction of ATP 48 hours after ADI-PEG20 treatment in MDA-MB-231 cells (Fig. 3A). Because mitochondria produce ATP through oxidative phosphorylation, we next analyzed whether arginine starvation by ADI-PEG20 affected the oxygen consumption rate of living cells. Oxygen consumption was assayed in the presence of the mitochondrial inhibitor oligomycin or in the presence of the mitochondrial uncoupler FCCP [carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone] to assess maximal oxidative capacity. First, we showed that MDA-MB-231 cells were already at maximal respiratory capacity for ATP synthesis under basal conditions, because oxygen consumption rate did not increase in response to FCCP treatment (Fig. 3B). Knockdown of ATG5 markedly decreased mitochondria-dependent respiration in MDA-MB-231 cells (Fig. 3B), consistent with a previous observation made in mouse embryonic fibroblasts (MEFs) (43). In cells treated with ADI-PEG20, the oxygen consumption rate for ATP synthesis was decreased and further inhibited by the ATP synthase blocker oligomycin. Moreover, the subsequent addition of FCCP failed to stimulate maximal respiration and, instead, caused it to fall below basal respiration, indicating that there was little or no uncoupled maximal respiration and reserve capacity (Fig. 3B). In contrast, FCCP partially stimulated rotenone-sensitive respiratory capacity in ADI-PEG20–treated cells with ATG5 knockdown. Likewise, the oxygen consumption rate of MDA-MB-231 cells cultured under arginine-free condition was comparable to that of cells treated with ADI-PEG20 (fig. S3A), suggesting that the lack of arginine is mainly responsible for ADI-PEG20-caused mitochondrial dysfunction. Together, these results suggest that autophagy exacerbates the impaired mitochondrial functions triggered by ADI-PEG20 treatment or arginine starvation in MDA-MB-231 cells.
Fig. 3. ADI-PEG20 impairs mitochondrial function.
(A) Autophagy-dependent ATP concentration decreases over time in ADI-PEG20–treated MDA-MB-231 cells; n = 3 sets of cells; *P < 0.05. (B) Combination of ADI-PEG20 treatment and ATG5 gene context regulates the basal and maximal mitochondrial respiration. ATG5 knockdown partially rescued the maximal respiration stimulated by FCCP in MDA-MB-231 cells treated with ADI-PEG20; n = 3 sets of cells. OCR, oxygen consumption rate. (C) Extracellular arginine reverses the reduced cell proliferation induced by ADI-PEG20 in a dose-dependent manner; n = 5 sets of cells; *P < 0.05. (D) ADI-PEG20 treatment reversibly decreases the abundance of mRNAs encoding mitochondrial respiratory chain proteins (left panel). A list of top candidates includes messages encoding proteins of complex I (blue) and complex II (orange). Other messages shown in black are UQCRFS1 (complex III), COX5A (complex IV), and ATP5G1 and ATP5J2 (complex V). Arginine supplementation rescued ASS1 message and protein abundance in ADI-PEG20–treated MDA-MB- 231 cells (right panel), thus verifying the effectiveness of arginine supplementation. Bar graph, presented as means ± SD, from three independent Western blots is shown. ATP5L (complex V) message abundance increased in response to ADI-PEG20, and then decreased after the addition of arginine. (E) Succinate rescues the cytotoxic effect of ADI-PEG20. Data are shown as means ± SD; n = 3 sets of cells; *P < 0.05. (F) Knockdown of ASS1 suppresses basal and uncoupled mitochondrial OCR in T47D cells; n = 3 sets of cells.
Increasing the extracellular concentration of arginine, but not that of glutamine, attenuated the reduction in cell proliferation induced by ADI-PEG20 (Fig. 3C and fig. S3B). Moreover, excess arginine also largely rescued the oxygen consumption rate in ADI-PEG20–treated cells in a dose-dependent manner, thus suggesting that arginine deprivation was the critical mechanism underlying the cytotoxic effects of ADI-PEG20 and ruling out potential off-target effect by ADI-PEG20 (fig. S3C). To investigate the metabolic defects caused by ADI-PEG20, we analyzed the mRNA abundance profiles of untreated, ADI-PEG20–treated, or ADI-PEG20–treated and arginine-supplemented cells on oligonucleotide microarray chips and validated the array data by qRT-PCR. The top candidates affected by ADI-PEG20 included NDUFB5, NDUFA9, NDUFB6, NDUFA8, NDUFB3, NDUFS3, and NDUFB1 from complex I; SDHA and SDHB from complex II; UQCRFS1 from complex III; COX5A from complex IV; and ATP5G1, ATP5G2, ATP5J2, and ATP5L from complex V (Fig. 3D). The heatmap analysis also confirmed that the decreased transcript abundance in ADI-PEG20–treated cells was reversed by supplementation with extracellular arginine. Notably, the ASS1 message and ASS1 protein abundance were increased by ADI-PEG20 and reduced by arginine supplementation (Fig. 3D).
During mitochondrial electron transfer to the ATP synthase, both complexes I and II are involved in shuttling electrons to complex III (44-48). We found that adding more succinate for the complex II reaction rescued the reduced proliferation of ADI-PEG20–treated MDA-MB-231 cells in a concentration-dependent manner (Fig. 3E and fig. S3D). Succinate did not noticeably rescue proliferation in ATG5-knockdown MDA-MB-231 cells (Fig. 3E and fig. S3D). Furthermore, knockdown of ASS1 decreased both basal and uncoupled oxygen consumption rate in ADI-PEG20–treated T47D cells (Fig. 3F), indicating that ASS1 status affects vulnerability of mitochondrial bioenergetics to ADI-PEG20. Together, these results suggest that the combination of loss of ASS1 and ADI-PEG20 treatment severely dampens mitochondrial respiratory function.
ADI-PEG20 increases ROS and decreases mitochondrial membrane potential in ASS1-deficient MDA-MB-231 cells
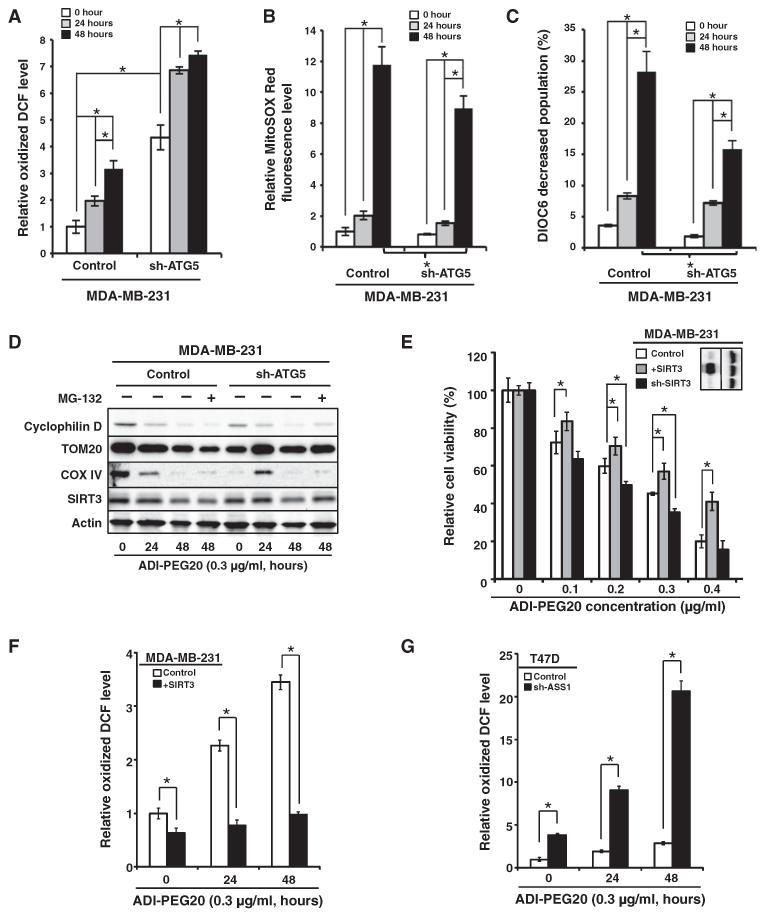
There is a complex relationship between oxidative phosphorylation, electron transport, and the production of superoxide by mitochondria. Physiological uncoupling of ATP production from electron transport decreases superoxide production, whereas inhibiting the electron transport chain increases superoxide production (49). Furthermore, simultaneously uncoupling and inhibiting electron transport increases superoxide production (50). We used the oxidation of DCFDH to DCF (51) as a first approach (because of the limitations of fluorescent dyes in measuring specific reactive species) to determine whether ADI-PEG20 increases the intracellular concentration of ROS that impairs mitochondrial respiratory function. Although the relative DCF signal in control MDA-MB-231 cells was lower than that in cells with ATG5 knockdown, ADI-PEG20 increased the relative signals of oxidized DCF in both sets of cells (Fig. 4A). The relative increase was greater in MDA-MB-231 cells (about threefold) than in MDA-MB-231 cells with ATG5 knockdown (less than twofold) (Fig. 4A). We used MitoSOX Red to further analyze the effect of ADI-PEG20 on nonmetabolic oxidation. ADI-PEG20 increased MitoSOX Red fluorescence ~12-fold in MDA-MB-231 cells, an increase that was significantly higher than the ~9-fold increase in the MDA-MB-231 cells with ATG5 knockdown (Fig. 4B). Consistently, mitochondrial membrane potential (MMP) was reduced by ADI-PEG20 to a greater extent in MDA-MB-231 cells than in ATG5-knockdown MDA-MB-231 cells (Fig. 4C). Together, these observations suggest that autophagy competence exacerbated the mitochondrial damage induced by ADI-PEG20 in MDA-MB-231 cells (Figs. 3B and 4, A to C), supporting the hypothesis that ADI-PEG20–induced cell death is autophagy-dependent (Fig. 2, E and F).
Fig. 4. ADI-PEG20 induces mitochondrial ROS.
(A to C) ADI-PEG20 increases ROS. DCFDH oxidation (A), MitoSOX Red oxidation (B), and MMP as assessed by DiOC6 staining (C) were determined by flow cytometry. Data are shown as means ± SD; n = 3 sets of cells; *P < 0.05. (D) Representative Western blot shows that autophagy promotes ADI-PEG20–induced decrease of the mitochondrial protein cyclophilin D, TOM20, COX IV, and SIRT3 abundance in MDA-MB-231 cells; n = 3 sets of cells. Equal amounts of whole-cell lysates were analyzed by Western blot for the indicated mitochondrial proteins. One representative Western is shown, and quantitative analysis of densitometric tracing, presented as means ± SD, for cyclophilin D, TOM20, COX IV, and SIRT3 is shown in fig. S4 (C to F); n = 3 sets of cells. (E) SIRT3 abundance governs ADI-PEG20 sensitivity. SIRT3 abundance was modified by lentiviral overexpression (+SIRT3) or knockdown (sh-SIRT3). Data are shown as means ± SD; n = 3 sets of cells; *P < 0.05. (F and G) Both SIRT3 and ASS1 are involved in regulating ROS production in ADI-PEG20–treated breast cancer cells. DCFDH oxidation was determined by flow cytometry in MDA-MB-231, SIRT3-overexpressing MDA-MB-231 (F), T47D, and ASS1-knockdown T47D cells (G). The positively stained population in vehicle-treated parental cells was designated as 1 to calculate the relative extent of DCF oxidization. Data are shown as means ± SD; n = 5 sets of cells; *P < 0.05.
The ADI-PEG20–induced increase in the generation of ROS (Fig. 4, A and B) could result from a decrease in the abundance of SIRT or superoxide dismutase (SOD) members. However, the mRNA abundance of SIRT and SOD family members showed no change in ADI-PEG20–treated or arginine-supplemented, ADI-PEG20–treated MDA-MB-231 cells (fig. S4, A and B). Thus, we used Western blotting to assess whether ADI-PEG20 treatment altered the abundance of mitochondrial proteins, including TOM20 (located in the mitochondrial outer membrane), COX IV (cytochrome c oxidase subunit IV; located in the mitochondrial inner membrane), cyclophilin D (located in the mitochondrial matrix), and SIRT3 [nicotinamide adenine dinucleotide (NAD)–dependent deacetylase sirtuin-3; located in the mitochondrial matrix], in control MDA-MB-231 cells and MDA-MB-231 cells with ATG5 knockdown (Fig. 4D and fig. S4, C to F). The abundance of mitochondrial-specific SIRT3 was modestly decreased at an earlier time point of ADI-PEG20 treatment in MDA-MB-231 cells (24 hours) than in the ATG5-knockdown MDA-MB-231 cells (48 hours) (Fig. 4D and fig. S4F). Other mitochondria-specific proteins, including cyclophilin D, TOM20, and COX IV, showed decreased abundance over time in response to ADI-PEG20 in MDA-MB-231 cells, supporting the involvement of mitochondrial dysfunction in response to ADI-PEG20 (Fig. 4D and fig. S4, C to E). In contrast, an increase in the TOM20 and COX IV abundance at 24 hours after ADI-PEG20 treatment in cells with reduced ATG5 was noticed. Although it was reproducible, the underlying mechanism is unclear. Furthermore, adding the proteasome inhibitor MG132 mainly rescued the abundance of SIRT3 and cyclophilin D in ATG5-knockdown MDA-MB-231 cells. SIRT3 is located within mitochondria and is implicated in regulating metabolic processes through deacetylation of transcription factors or metabolic enzymes [reviewed in (52)]. Together with bioenergetics data (Fig. 3, A and B), these results showed that ADI-PEG20 treatment disrupted mitochondrial components in MDA-MB-231 cells. Finally, we demonstrated that modifying the abundance of SIRT3 (Fig. 4E, inset) affected the sensitivity of cells to ADI-PEG20. SIRT3-overexpressing cells were more resistant to ADI-PEG20, whereas the knockdown of SIRT3 generally increased the cytotoxicity (Fig. 4E). Consistently, overexpression of SIRT3 decreased the basal concentration of ROS and attenuated its increase upon ADI-PEG20 treatment in MDA-MB-231 cells (Fig. 4F). Last, knockdown of ASS1 increased the basal concentration of ROS and sensitized T47D cells to an ADI-PEG20–triggered increase in ROS production (Fig. 4G).
ADI-PEG20 induces mitochondrial fragmentation in ASS1-deficient MDA-MB-231 cells
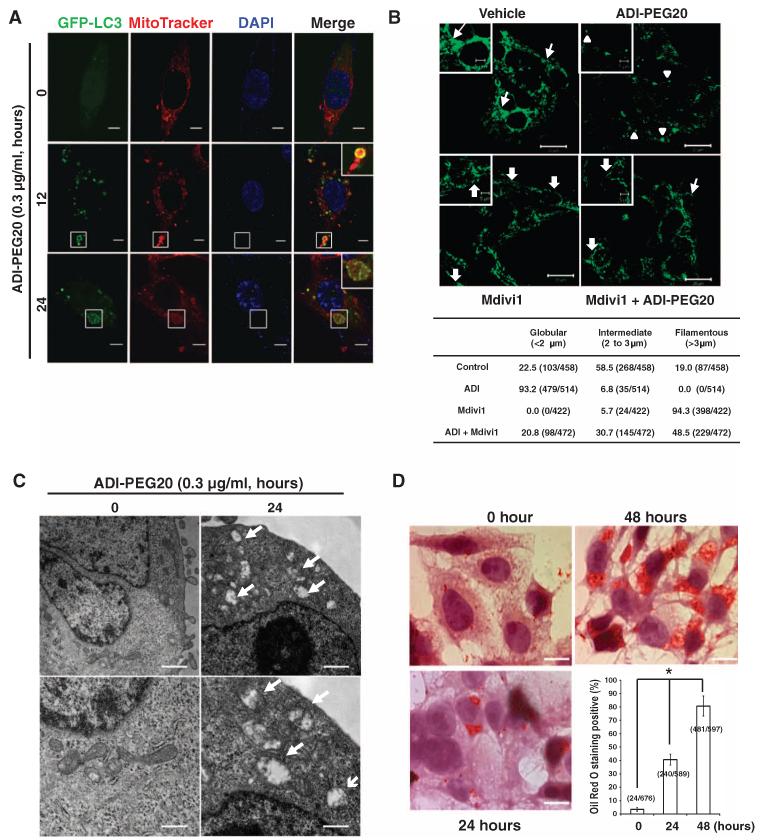
Our observation that autophagy-competent MDA-MB-231 cells were more likely to be subjected to ADI-PEG20–induced loss of mitochondrial function, such as decreased ATP production and oxygen consumption (Fig. 3, A and B), suggested an active role for autophagy in impairing mitochondrial function. Mitochondrial morphology is determined by the balance of fission and fusion (53), and using MitoTracker staining, we showed that mitochondrial syncytia were disrupted by ADI-PEG20 treatment in a time-dependent manner, suggestive of mitochondrial structural changes (fig. S5A). In MDA-MB-231 cells stably transfected with GFP-LC3, confocal microscopy revealed that GFP-LC3 and MitoTracker colocalized at 12 and 24 hours after ADI-PEG20 treatment, suggesting that mitochondria were being engulfed by autophagosomes (Fig. 5A). We then assessed how treatment of MDA-MB-231 cells with ADI-PEG20 affected the mitochondrial network. More than 50% of the cells before ADI-PEG20 treatment harbored vermiform mitochondria (fig. S5B). Exposure to ADI-PEG20 led to nearly all of the cells acquiring a fragmented mitochondrial structure (Fig. 5B). Next, cells cotreated with ADI-PEG20 and Mdivi1, an inhibitor of mitochondrial fission (54), showed a largely preserved filamentous mitochondrial morphology compared with cells treated with ADI-PEG20 alone (Fig.5B). Transmission electron microscopy (TEM) further revealed mitochondria that were engulfed in what were presumably autophagosomes in ADI-PEG20–treated MDA-MB-231 cells (fig. S5B). Moreover, TEM images also revealed swollen, fragmented mitochondria, which had lost their electron-dense staining (Fig. 5C). We propose that the swollen and decreased matrix density was caused by limited degradation and/or loss of the mitochondrial matrix and cristae, the site of the mitochondrial respiratory chain, as well as degradation or loss of numerous carrier proteins (55). These data also insinuate a role for mitochondrial structural damage in ADI-PEG20–induced mitochondrial dysfunction. Lipid droplet number was increased (fig. S5B) and Oil Red O staining confirmed that more than 70% MDA-MB-231 cells accumulated lipids 48 hours after ADI-PEG20 treatment (Fig. 5D). This increase provided additional, supporting evidence for the presence of damaged mitochondria, because mitochondria metabolize fatty acids through β-oxidation, and damage to mitochondria decreases the use of fatty acids, resulting in the accumulation of lipid droplets (56).
Fig. 5. ADI-PEG20 causes mitochondrial fragmentation.
(A) GFP-LC3 puncta colocalize with MitoTracker. The number of GFP-LC3 puncta was highest in cells treated with ADI-PEG20 for 12 hours, and then, they decreased after 24 hours of treatment. Part of GFP-LC3 and MitoTracker signals are colocalized after ADI-PEG20 treatment. Magnified images are shown in the upper right corner (Merge). n = 3 sets of cells. Scale bars, 5 μm. (B) Representative confocal images of MDA-MB-231 cells treated with a combination of ADI-PEG20 and Mdivi1 for 24 hours and stained with an anti-TOM20 antibody (upper panel); n = 3 sets of cells. Mitochondria lengths were measured and scored as follows: fragmented (<2 μm; arrowheads), intermediate (2 to 3 μm; arrows), and filamentous (>3 μm; block arrows). The percentage of cells with indicated dominant mitochondrial morphologies was determined by dividing by the total number of counted cells as indicated (lower panel); scale bars, 20 μm. Magnified images are shown in the insets (upper left); scale bars, 10 μm. (C) Representative TEM micrographs showing damaged and swollen mitochondria after ADI-PEG20 treatment (indicated by arrows). Scale bars, 1 μm (upper panels) and 0.5 μm (lower panels). n = 3 sets of paired cells. (D) Representative images of Oil Red O staining after treatment with ADI-PEG20 for indicated time periods; n = 4 sets of cells. The percentage of cells with the oil droplets was counted and determined as a percentage of the total number of examined cells. Scale bars, 10 μm; *P < 0.05.
ASS1 abundance is a prognostic factor for overall breast cancer survival
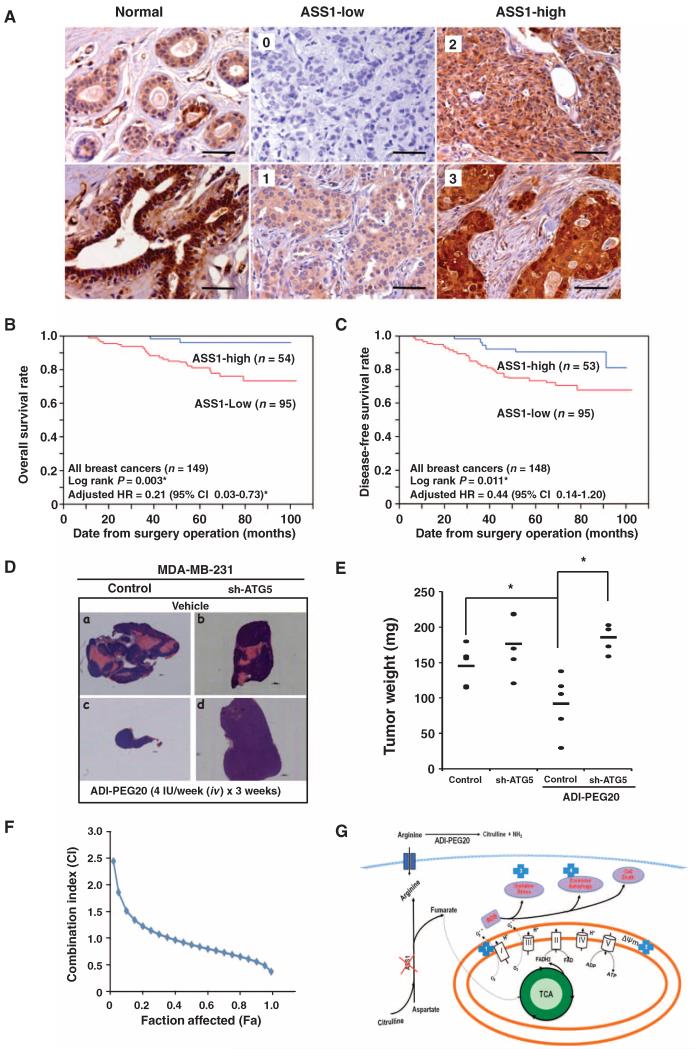
We observed that ASS1 knockdown increased the ability of breast cancer cells to grow in an anchorage-independent manner (Fig. 1F and fig. S1E). To determine whether altered ASS1 abundance affected overall survival (OS) of patients with breast cancer, we examined ASS1 abundance in 149 breast cancer samples by staining three independent multiple tissue arrays with an anti-ASS1 antibody. We found that normal mammary tissues were ASS1-positive (Fig. 6A). However, 95 (63.8%) of the 149 breast cancer samples had low ASS1 abundance, which was defined as having little or no diffuse cytoplasmic ASS1 staining (Fig. 6A). In contrast, the remaining 54 breast cancer samples (36.2%) had high ASS1 abundance because they had strong diffuse staining in the cytoplasm (Fig. 6A). Our analyses of the clinical characteristics and pathology of the tumors revealed that reduced ASS1 abundance was significantly associated with advanced tumor stage and with progesterone receptor (PR)– and Ki-67–positive breast cancers (Table 1). However, there was no significant association with lymph node metastasis, or estrogen receptor (ER), HER2, or p53 status (Table 1). Last, the frequency of low ASS1 abundance was significantly different among all of the four different molecular breast cancer subtypes (Table 1).
Fig. 6. Low ASS1 abundance is prognostic of poor breast cancer survival and predisposes MDA-MB-231 cells to ADI-PEG20–mediated tumor shrinkage in an autophagy-dependent manner in vivo.
(A) Representative ASS1 immunohistochemical staining of 149 breast cancer samples. Normal breast epithelium (left panels), breast cancer tumors with low ASS1 abundance (middle panel; score: 0 and 1), and breast cancer tumors with high ASS1 abundance (right panels; score: 2 and 3). Scale bars, 100 μm. (B) Kaplan-Meier analysis indicates that reduced ASS1 abundance in breast tumors is significantly associated with poor OS. (C) Kaplan-Meier analysis shows low ASS1 abundance in breast tumors impacts DFS. The sample number was reduced from 149 to 148 because a patient died before metastasis occurred. (D) Representative images of tumors harvested from either vehicle- or ADI-PEG20–treated mice xenografted with MDA-MB-231 or ATG5-knockdown cells. Magnification, ×50; scale bars, 1000 μm. (E) Scatter plot of tumor weight measured at the endpoint of experiment. One dot represents one mouse with total of five mice per group, and “–” represents the average (145.4 ± 28.4, 176.6 ± 42.2, 92.4 ± 28, and 185.8 ± 18.9 mg; n = 5; *P < 0.05). Statistical analysis showed a significant difference between vehicle- and ADI-PEG20–treated MDA-MB-231 groups, and between ADI-PEG20-treated and ATG5-knockdown MDA-MB-231 groups. (F) Synergistic effect of ADI-PEG20 combined with doxorubicin; n = 3 sets of cells. (G) Proposed model depicting ADI-PEG20–induced cytotoxic killing. The location of ADI-PEG20–induced alterations within mitochondrial and cell death pathways is indicated by the numbers inside blue crosses: respiration chain (1), membrane permeability (2), oxidative stress (3), and activation of excessive autophagy (4).
Table 1. Clinicopathological characteristics and ASS1 distribution of eligible patients with breast cancer.
| No. of cases | No. of patients with low ASS1 abundance (%)* |
P | |
|---|---|---|---|
| Age (years) | |||
| <40 | 12 | 9 (75) | |
| 40–49 | 53 | 34 (64.1) | |
| 50–59 | 43 | 26 (60.5) | |
| 60–69 | 23 | 15 (65.2) | |
| 70–79 | 14 | 10 (81.4) | |
| >80 | 4 | 3 (75.0) | 0.573 |
| T stage† | |||
| T0–T1 | 36 | 15 (41.7) | |
| T2–T4 | 108 | 76 (70.4) | 0.002‡ |
| N stage† | |||
| N0 | 72 | 44 (61.1) | |
| N1–N3 | 77 | 51 (66.2) | 0.516 |
| ER | |||
| Negative | 52 | 30 (57.7) | |
| Positive | 73 | 47 (64.4) | 0.448 |
| PR | |||
| Negative | 62 | 30 (48.4) | |
| Positive | 60 | 43 (71.7) | 0.009‡ |
| Ki-67 | |||
| Negative | 48 | 36 (75.0) | |
| Positive | 78 | 51 (55.3) | 0.012‡ |
| HER2 | |||
| Negative | 97 | 63 (64.9) | |
| Positive | 30 | 14 (46.7) | 0.07 |
| Molecular subtype§ | |||
| Luminal A | 36 | 27 (75) | |
| Luminal B | 43 | 26 (60.5) | |
| TNBC | 22 | 12 (54.5) | |
| HER2 | 12 | 2 (16.7) | 0.005‡ |
| p53 | |||
| Negative | 62 | 42 (67.7) | |
| Positive | 61 | 33 (54.1) | 0.12 |
Either no staining or weak cytoplasmic ASS1 staining.
Tumor node metastasis stage is according to AJCC Cancer Staging Manual, 6th Edition.
Statistical significance, P < 0.05.
Molecular subtype is categorized according to ER, PR, HER2, and Ki-67 status. Luminal A subtype is defined as ER-positive and/or PR-positive, HER2-negative, and Ki-67–negative; luminal B subtype is defined as ER-positive and/or PR-positive, HER2-negative, and Ki-67–positive; TNBC subtype is defined as ER-negative, PR-negative, and HER2-negative; HER2 subtype is defined as ER-negative, PR-negative, and HER2-positive.
Kaplan-Meier analyses indicated that reduced ASS1 abundance was a critical prognostic indicator for both OS (Fig. 6B) and DFS (disease-free survival) (Fig. 6C) for all 149 patients with breast cancer. Multivariate Cox analyses for OS rate indicated that patients with tumors with high ASS1 abundance had a lower relative risk of breast cancer–specific mortality [hazard ratio (HR), 0.21; 95% confidence interval, 0.03 to 0.73] (fig. S6A). Multivariate analysis also showed that high ASS1 abundance was associated with a lower relative risk of breast cancer relapse (HR, 0.44; 95% confidence interval, 0.14 to 1.20) (fig. S6B). Stratification analyses revealed that reduced ASS1 abundance was significantly associated with poor OS in the subgroups of patients with breast cancer who had ER-negative, PR-negative, and Ki-67–positive cancers (fig. S6, C to E). The HR for high ASS1 abundance could not be calculated in ER-negative and HER2-positive breast cancers because no deaths were reported within these two subgroups during the follow-up period (fig. S6, C and G). In comparison with reduced ASS1 abundance, the adjusted HRs of high ASS1 abundance were 0.16 (95% confidence interval, 0.01 to 0.98) and 0.23 (95% confidence interval, 0.04 to 0.88) in the subgroups of PR-negative and Ki-67–positive breast cancers, respectively (fig. S6, D and E). Uni- and multivariate analyses indicated that reduced ASS1 abundance poorly impacted survival in both HER2-negative and HER2-positive subgroups (fig. S6, F and G), indicating that the loss of ASS1 abundance was associated with poor overall patient survival independently of breast cancer molecular subgroup. Moreover, microarray analysis and associated clinical data extracted from a published study of 295 breast cancers revealed that ASS1 mRNA abundance from breast cancer samples of patients with >5-year OS or DFS was significantly higher than that in patients with <5-year OS or DFS (fig. S6H) (57, 58). Therefore, the abundance of ASS1 may be a useful biomarker for breast cancer prognosis and is consistent with the results of soft agarose growth assays, which indicated increased ability of cells with low ASS1 abundance to grow under anchorage-independent conditions (Fig. 1F and fig. S1E).
Autophagy is required for ADI-PEG20 to cause shrink tumors in vivo
For proof of principle, we determined the effects of autophagy competence and treatment with ADI-PEG20 on tumors in vivo. Female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice with xenografts formed from control MDA-MB-231 cells or MDA-MB-231 cells with ATG5 knockdown were injected intravenously with vehicle control or ADI-PEG20. The tumors from ADI-PEG20–treated mice were smaller than tumors from control mice (Fig. 6D) at 19 days after beginning treatment. Although ATG5 deficiency did not impair MDA-MB-231 tumor growth, it markedly impaired shrinking of the tumors in response to ADI-PEG20 (Fig. 6D). The average weight of ADI-PEG20–treated MDA-MB-231 tumors was significantly (50%) less than that of ADI-PEG20–treated MDA-MB-231/sh-ATG5 tumors (Fig. 6E). The data showing that MDA-MB-231 cells with ATG5 knockdown were more resistant to ADI-PEG20 in vivo than control MDA-MB-231 cells support our conclusion that tumor shrinkage induced by ADI-PEG20 is autophagy-dependent.
Last, we examined if there was a synergistic effect between ADI-PEG20 and doxorubicin, cisplatin, or taxol, which are conventionally used as therapeutic agents against breast cancer, in MDA-MB-231 cells. The combination index (CI) of ADI-PEG20 with doxorubicin was assessed as previously described (59-61). CI < 1, CI = 1, and CI > 1 indicate synergism, additive effect, and antagonism, respectively (60, 61). The calculated CI suggested a synergistic effect between ADI-PEG20 and doxorubicin (Fig. 6F). However, synergy was not observed when ADI-PEG20 was combined with Abraxane or carboplatin (fig. S7). In summary, ADI-PEG20, alone or combined with doxorubicin, may represent a promising therapeutic strategy for treating breast cancers that are arginine auxotrophs.
DISCUSSION
Understanding the metabolic differences between normal cells and tumor cells could provide the opportunity to design selective personalized therapy against breast cancer and other cancers. Considerable attention has been paid to glucose metabolism and glutamine addiction in tumor cells (5). However, the metabolism of cancer cells is complex and involves various rewiring of metabolic pathways that occurs during malignant transformation. In this regard, we report here a mechanism connecting the metabolic stress caused by arginine starvation to induce cell death of ASS1-deficient breast cancer cells (Fig. 6G). This is particularly interesting because arginine deprivation affected mitochondrial bioenergetics and integrity by decreasing the abundance of the mitochondrial inner membrane and matrix proteins and impairing mitochondrial oxidative phosphorylation and ATP production, which was accompanied by increased ROS production.
The molecular mechanisms underlying cell death induced by ADI-PEG20–mediated arginine starvation of ASS1-deficient cells are not yet fully understood. Nutrient starvation, including arginine deprivation, often leads to autophagosome induction (19, 23, 37). Paradoxically, although other reports have suggested that autophagy protects cells from dying upon the initiation of starvation, our results showed that prolonged autophagy induction by sustained ADI-PEG20 treatment or arginine starvation eventually leads to cell death. To decipher this paradox, we examined the function and fate of mitochondria in ADI-PEG20–treated MDA-MB-231 cells because mitochondria play a central role in satisfying higher demands for energy and anabolic needs during stress. Stressed cells have developed mechanisms, known as mitochondrial quality control, to deal with dysfunctional mitochondria to maintain homeostasis. Fusion between damaged mitochondria and healthy mitochondria may serve as one mechanism, either to dilute the damaged and detrimental components or to regain functional counterparts (62-64). Alternatively, sustained starvation induces the degradation of mitochondria by an autophagy-mediated system, known as mitophagy (65). Specifically, when damaged mitochondria lose their membrane potential (Fig. 4C) and/or undergo fragmentation (Fig. 5B), they are likely engulfed by autophagosomes for lysosomal degradation (65, 66). Our results suggest that the dysfunctional mitochondria, probably due to increased mitochondrial ROS production and the loss of MMP caused by disruptions in the respiratory chain (Figs. 3 and 4), resulted in cytotoxic autophagy in ADI-PEG20–treated MDA-MB-231 cells. This possibility is supported by studies in other systems in which mitophagy is triggered by the use of mitochondrial respiratory chain uncouplers (67-69).
We found that ADI-PEG20 decreased the steady-state abundance of complex I/II transcripts (Fig. 3D). It is possible that increased ROS production, presumably resulting from impaired mitochondrial function, may play a key role in favoring ADI-PEG20–induced cell death. Along the same lines, supplementing with succinate, a complex II substrate, partially rescued the viability of arginine-starved cells (Fig. 3E) by restoring portions of electron transfer capacity. Most likely, arginine starvation of cells lacking ASS1 affects mitochondrial integrity. We envision that ADI-PEG20 treatment in the context of ASS1 deficiency creates a pathological “vicious cycle,” wherein the impairment in the mitochondrial electron transport that leads to a loss of MMP is also responsible for an incomplete O2 reduction, resulting in an increase in ROS production that further amplifies the generation of ROS. Consequently, sustained and excessive autophagy exacerbates the arginine starvation–induced cytotoxic effect (Fig. 6G). The consequence of sustained autophagy is reinforced by reduced biogenesis of new mitochondria caused by decreased amounts of complex I/II, which further inhibits oxidative phosphorylation and induces cell death. In support of this, Vincow et al. have shown that in the fly, several respiratory complex I proteins are selectively routed for autophagosomal degradation (70). Moreover, our data suggest that impairing or losing the mitochondrial electron transport chain could account for the lack of mitochondrial FCCP response after ADI-PEG20 treatment in cells with reduced ASS1 (Fig. 3F).
The reduction in SIRT3 protein abundance by prolonged ADI-PEG20 treatment (Fig. 4D) is of particular interest because SIRT3, which is located in the mitochondrial matrix, is a member of an evolutionarily conserved family of NAD+-dependent deacetylases that regulates lysine acetylation of mitochondrial proteins (52). Rardin et al. have shown that an absence of SIRT3 in the livers of fasted mice leads to selective hyperacetylation at 283 sites that represent 136 of the 483 acetylated mitochondrial proteins (71). One of the largest fold changes in lysine acetylation was observed for ATP synthase, a key enzyme involved in oxidative phosphorylation. In addition, these authors reported several SIRT3-regulated sites in complexes I, II, and IV. Lysine acetylation of mitochondrial proteins has been proposed to reduce complex I activity and decrease the basal ATP concentration (72). Besides oxidative phosphorylation, the SIRT3-regulated metabolic pathways in mitochondria also include fatty acid oxidation, tricarboxylic acid cycle, and branched chain amino acid catabolism (71). Conceivably, the decrease in SIRT3 abundance, albeit modest (Fig. 4D), could exacerbate the overall metabolic stress upon prolonged ADI-PEG20 treatment. This hypothesis is supported by the fact that overexpressing SIRT3 can rescue the viability of cells treated with ADI-PEG20 (Fig. 4E) by suppressing ROS concentrations (Fig. 4F).
ASS1 catalyzes a rate-limiting step in de novo arginine biosynthesis to maintain arginine serum concentrations. Our analyses indicate that ASS1 abundance is an independent prognostic indicator of the survival of patients with breast cancer (Fig. 6, B and C). We confirmed the potential role of ASS1 in inhibiting cells from growing in an anchorage-independent manner (Fig. 1F and fig. S1E). A putative tumor suppressor role for ASS1 has been demonstrated by two independent studies (73, 74), although neither of these studies specifically examined breast cancer cells. Malignant transformation can be caused by or lead to metabolic changes because tumor cells tend to increase their glycolytic activity without a matching increase of oxidative phosphorylation and dependence on oxygen (a phenomenon known as the Warburg effect) (75, 76). The key to this metabolic rewiring is the decrease of mitochondrial respiration (77), which allows the cancer cells to grow in hypoxic conditions usually found in the interior of the tumor mass (78). However, the molecular mechanisms that suppress oxidative phosphorylation during tumorigenesis are still unclear. On the basis of our findings from the present work, we speculate that the down-modulating ASS1 abundance leads to decreased oxidative phosphorylation (namely, decreased maximal respiratory capacity), potentially promoting neoplastic transformation. Indeed, we find that ASS1 behaves like a tumor suppressor because (i) ASS1 knockdown increased the anchorage-independent proliferation of breast cancer cells in vitro, and (ii) ASS1 deficiency or low ASS1 abundance is associated with a poor prognosis for patients with breast cancers. We further speculate that decreasing ASS1 abundance could create a prooxidant state (Fig. 4G) that favors tumorigenesis because even a moderate increase in ROS production can promote cell proliferation and tumorigenesis (79). However, reduced ASS1 abundance also makes the breast cancer cells vulnerable to ADI-PEG20, which effectively kills them by starving them of arginine. This is reminiscent of the synthetic lethal interaction between BRCA1/2 deficiency and PARP1 inhibitors (80, 81).
In summary, we focused on how to take advantage of the mitochondrial dysfunction that is induced by sustained treatment of ADI-PEG20 in ASS1-deficient breast cancers. Our observations have important implications for the development of tumor therapies because conventional thoughts suggest that activating autophagy may contribute to the survival, rendering, and recurrence of refractory tumors that are resistant to conventional chemotherapy. The synergistic effect observed between doxorubicin and ADI-PEG20 supports the possibility of exploiting arginine starvation as a therapeutic strategy by targeting a specific metabolic liability due to ASS1 deficiency in breast cancer cells. The number of breast cancers and other human malignancies that have substantial subtypes that lack ASS1 abundance provides opportunities to validate our findings of a “mitochondria-targeting mechanism” in human patients with ASS1-deficient breast cancers and other types of ASS1-deficient cancers.
MATERIALS AND METHODS
Patient enrollment, follow-up, and tissue array
Patients included in this study were diagnosed with breast cancer and treated by surgical resection between January 2002 and January 2006 in the Second Affiliated Hospital of Zhejiang University (ZJU). This study was approved by the Institutional Review Board (IRB) of the Second Affiliated Hospital of ZJU (IRB 210-025). A breast cancer pathologist (J.X.) used hematoxylin and eosin (H&E)–stained slides to retrospectively review the history of all cases. The clinicopathological parameters that were evaluated included patient age at the time of diagnosis, tumor node metastasis stage, date of last follow-up, and overall patient survival. Exclusion criteria were lack of a pathologic diagnosis, presence of multiple cancers, or loss of contact after surgery. A total of 149 patients with breast cancer met the inclusion criteria. All participants were periodically followed up, and surgery relapse and death data were collected until 2010. OS rate was calculated from the date of surgery to the date of death by breast cancer–associated illness. DFS rate was calculated from date of surgery to date of local recurrence or metastasis. If no death or relapse occurred, the OS and DFS rates were calculated from date of surgery to September 2010. The formalin-fixed, paraffin-embedded breast cancer tissue samples that were collected were reassembled into multiple tissue arrays. ASS1 immunohistological signals did not correlate with storage time (likelihood, P = 0.246), indicating that it was unlikely that storage time would affect the immunohistochemical outcome.
Immunohistochemistry
ASS1 protein abundance in the 149 breast cancer samples was assessed by immunohistochemistry (IHC) using an anti-ASS1 antibody (1:75 dilution) as previously described (82). The IHC conditions for ASS1 abundance determination were preoptimized on checkerboards with multiple tissue samples. Briefly, after deparaffinization, endogenous peroxidase activity was blocked by pretreatment with 3% H2O2. The slides were incubated with normal goat serum for 20 min at room temperature (RT) to block non-specific signal, then incubated with the primary antibody for 20 min at RT. The array slides were then incubated with polymer horseradish peroxidase (HRP)–labeled secondary antibodies for 30 min at RT, and then 3,3′-diaminobenzidine (DAB)–treated [0.05 g of DAB and 100 ml of 30% H2O2 in 100 ml of phosphate-buffered saline (PBS)] for 5 and 10 min, respectively. Each slide was counterstained with Dako’s hematoxylin. For each IHC staining, the negative and positive checkerboards served as quality control. The specificity of anti-ASS1 antibody was validated by Western blotting. ASS1 staining was predominantly cytoplasmic, and ASS1 abundance was assessed using a visual grading system on the basis of the intensity of staining signals observed using a light microscope. Each sample was independently scored by two investigators (F.Q. and J.X.), including one breast pathologist (J.X.), using a double-blind design to avoid scoring bias. Discrepancies were reevaluated by joint review between the two readers. ASS1-high was defined as strong diffusive staining, and ASS1-low was defined as weak diffusive staining or no staining (Fig. 6A).
Cell lines, media, and chemicals
Human embryonic kidney (HEK) 293T/FT, MDA-MB-231, ATG5-, BECLIN 1-, or SIRT3-knockdown MDA-MB-231, ASS1, SIRT3 or GFP-LC3 stably transfected MDA-MB-231, MCF-7, ASS1-knockdown MCF-7, ZR-75-1, and MCF-10A cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Cellgro, 10-013-CV) containing fetal bovine serum (10%; Gibco, 26140), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Gibco, 15240) at 37°C and 5% CO2 in a humidified incubator. T47D and ASS1-knockdown T47D cells were cultured in RPMI 1640 medium under similar conditions. Recombinant ADI-PEG20 was a gift of Polaris Pharmaceuticals Inc. Bafilomycin A1 (B1793), chloroquine diphosphate salt (C6628), rapamycin (R8781), succinic acid (S9512), l-arginine (A8094), and MG132 (M7449) were from Sigma. l-Glutamine solution was from Gibco (25030).
Antibodies
Anti–MAP1LC3-I/II antibody (M115-3) was from Medical & Biological Laboratories, anti-SIRT3 antibody (C73E3) was from Cell Signaling Technology, anti-Tom20 antibody (sc-17764) and anti-GAPDH antibody (sc-25778) were from Santa Cruz Biotechnology, anti–cyclophilin D antibody (ab110324) and anti–COX IV antibody (ab110261) were from Abcam, and anti-actin antibody (MAB1501R) was from Millipore. Anti-human ASS1 antibody and the lentiviral sh-ASS1 construct were from L.-J. Shen (83).
Virus production and transduction
Target gene DNA in a lentiviral backbone (pLKO.puro or pSin) (21 μg), pΔ8.7 (14 μg), and pVSV-G (7 μg) were transfected using Lipofectamine 2000 (Life Technologies, 11668-019) into HEK293T/FT cells that had been seeded in a T-175 flask and had reached 60% confluence on the day of transfection. On the second day after transfection, cells were treated with sodium butyrate (10 mM) to stimulate virus production. Media containing viruses were harvested on the fifth day and filtered through a 0.45-μm filter. For viral transduction, cells were treated with media containing viruses overnight in the presence of polybrene (8.3 μg/ml), which was followed by selection with puromycin (1 μg/ml) or G418 (200 μg/ml), or analysis by fluorescence-activated cell sorting (FACS).
RNA extraction, qRT-PCR, and complementary DNA microarray analyses
Total RNA was isolated from cells using RNeasy Mini Kit (Qiagen, 74104) following the manufacturer’s instructions. The RNA concentration and purity were determined by spectrophotometry (NanoDrop Technologies Inc., LLC). One microgram of total RNA was used as the template for synthesizing complementary DNA (cDNA) using the iScript cDNA Synthesis Kit (Bio-Rad, 170-8893). Real-time quantitative PCR was performed with the Single-Color Real-Time PCR Detection System (Bio-Rad). Equal amounts of mRNA from MDA-MB-231 cells subjected to a combination of ADI-PEG20 (0.3 μg/ml) and arginine (420 μg/ml) were analyzed. The relative abundance of ASS1 mRNA in each sample was compared to that of untreated MDA-MB-231 cells, which was designated as 1. For cDNA microarray analyses, the Illumina TotalPrep RNA Amplification Kit (Ambion) was used to transcribe 500 ng of RNA by following the manufacturer’s protocol. HumanHT-12 Expression BeadChips (Illumina) used the Direct Hybridization Assay and were scanned on the BeadArray Reader to quantify signal and background intensity for each feature. The selected array data were validated by real-time PCR. All primer pair sequences are listed in table S1.
Cytotoxicity and apoptosis assays
Four methods were used to measure cytotoxicity. (i) In the DIMSCAN assay, 2500 to 5000 cells were seeded onto 96-well plates. At the end of the indicated time periods of ADI-PEG20 treatment, cells were incubated with fluorescein diacetate (40 μg/ml), which selectively accumulates in viable cells. A semiautomatic, fluorescence-based, digital image microscopy system was used to measure the viability. (ii) Real-time cell growth was monitored by using a 16× E-Plates RTCA DP Analyzer (ACEA Biosciences). (iii) Acid phosphatase (ACP) assays were used to avoid the effect of mitochondria dysfunction. Between 2500 and 5000 cells were seeded onto 96-well plates. At the end of the indicated time periods of ADI-PEG20 treatment, cells were washed twice with PBS and then incubated at 37°C for 30 min with 100 μl of pNPP (p-nitrophenol phosphate) solution (5 mM) in a buffer containing sodium acetate (0.1 M) and Triton X-100 [0.1% (v/v), pH 5.5]. The reaction was terminated by adding NaOH (1 N, 10 μl), and the absorbance was measured at 410 nm using a microplate reader. (iv) Trypan blue exclusion was used to count viable cells under a light microscope. Relative cell viability of ADI-PEG20–treated cells was calculated relative to the respective viability of the corresponding matched, vehicle-treated cells, which was designated as 1. Data were collected from at least three independent experiments and are shown as means ± SD. The interaction between drug combinations was analyzed using the CalcuSyn software program (Biosoft) to determine whether the combination was antagonistic, additive, or synergistic. This program is based on the Chou-Talalay method to calculate a CI, and CI values less than 0.9 indicate synergistic effect. The CIs were determined from cell viability DIMSCAN or ACP assays as the fraction of cells killed by individual drugs, or combination of drugs, by comparing to vehicle-treated cells. For the apoptosis assay, cells were treated with ADI-PEG20 (0.3 μg/ml) or subjected to arginine starvation for the indicated time periods. After trypsinization, cells were washed twice with cold PBS and collected by centrifugation at 1000 rpm. Cells were then resuspended in 1× binding buffer at a concentration of 1 × 106 cells/ml, and 100 μl of the suspension (1 × 105 cells) was transferred to a polystyrene round-bottom tube. Cells were then stained with FITC (fluorescein isothiocyanate)–conjugated annexin V (4 μl) and propidium iodide (50 μg/ml, 5 μl). The mixture was gently vortexed and incubated for 15 min at RT, and 1× binding buffer (400 μl) was added to each tube before analyzing by flow cytometry.
Soft agar colony assays
Cells (0.5 × 104 to 3 × 104) were mixed in 0.35% agarose/complete medium and plated on a 0.7% agarose/complete medium bottom layer in each well of a six-well plate. The culture medium contained vehicle or different concentrations of ADI-PEG20 and was changed every 2 to 3 days during the 4- to 6-week cell growth period. Colonies were stained with methylthiazolyldiphenyl-tetrazolium bromide (MTT; 0.2 mg/ml; Sigma, M5655) and were counted using a colony-counting software (VersaDoc Imaging System, Bio-Rad) or counted manually. Each experiment was performed at least twice in triplicate wells.
Whole-cell extracts and immunoblotting
Cells treated with ADI-PEG20 were harvested at the end of the incubation period and lysed on ice for 30 min in radioimmunoprecipitation assay buffer (Cell Signaling Technology, #9806) containing complete protease inhibitor cocktail (Roche, 11836145001) and PhosSTOP (Roche, 04906837001). The protein concentrations of whole-cell extracts were determined using a Bio-Rad Protein Assay Kit (500-0001). About 20 to 40 μg of protein were mixed with an equal volume of 2× SDS loading buffer, boiled for 5 min, and then separated by tris-glycine SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% nonfat milk in PBST (PBS containing 0.05% Tween 20) and incubated with primary antibodies at 4°C overnight. The membranes were then washed three times with PBST for 10 min and then incubated with HRP-labeled secondary antibodies for 2 hours at RT. Immunoblots were visualized using VersaDoc 5000 Imaging System (Bio-Rad). The intensity was determined using Quantity One (Bio-Rad) and compared to 0-hour treatment after normalizing with actin. Three independent experiments were performed, and representative blots were presented.
ATP assay
The ENLITEN ATPAssay System (Promega, FF2000) was used according to the manufacturer’s instructions. At the indicated time points, cells were harvested by scraping and resuspending in PBS (500 μl). The cell suspension was divided into unequal aliquots, and 400 μl of cell suspension was mixed with 5% trichloroacetic acid (TCA; 100 μl). The remaining cells (100 μl) were used for cell number calculation. Tris-acetate buffer (900 μl, pH 7.75) was then added to neutralize the TCA and to dilute the TCA to a final concentration of 0.1%. The extracts were further diluted 100-fold with dilution buffer (0.1% TCA, 0.08× PBS, and 0.9× tris-acetate). Diluted sample (40 μl) was added to an equal volume of rL/L Reagent (Promega, FF2000) that contained d-luciferin and recombinant luciferase, and then luminescence was measured using a TD-30e luminometer (Turner). The ATP standard (Promega, FF2000) was serially diluted to generate a regression curve for calculating ATP concentrations in individual samples. The relative ATP concentration was determined and normalized to that of vehicle-treated cells, which was designated as 1. Three independent experiments were performed, and results are presented as means ± SD.
Oxygen consumption rate
Cellular mitochondrial function was measured using the Seahorse Bioscience XF24 Extracellular Flux Analyzer. The mitochondrial function was assayed by sequential injections of oligomycin, FCCP, and rotenone to define basal oxygen consumption rate, ATP-linked oxygen consumption rate, proton leak, maximal respiratory capacity, reserve respiratory capacity, and nonmitochondrial oxygen consumption, all according to the manufacturer’s instructions. Oligomycin A inhibits ATP synthase by blocking proton channel and reduces electron flow through the electron transport chain, which is necessary for oxidative phosphorylation of ADP (adenosine 5′-diphosphate) to ATP. FCCP is a protonophore (H+ ionophore) and uncoupler of oxidative phosphorylation in mitochondria, which is capable of depolarizing plasma and mitochondrial membranes. Rotenone, an inhibitor of mitochondrial electron transport at NADH (reduced form of NAD+)/ubiquinone oxidoreductase, inhibits the transfer of electrons from iron-sulfur centers in complex I to ubiquinone and interferes with NADH. Basal oxygen consumption rate was a proxy for mitochondria function. Briefly, 2 × 104 cells were seeded onto 24-well plates and incubated overnight before sequentially adding preoptimized concentrations of oligomycin, FCCP, and rotenone, in that order. After washing the cells with 1 ml of Seahorse buffer [DMEM without phenol red containing glucose (4.5 g/liter; Sigma, G7021), sodium pyruvate (1 mM; Gibco, 11360070), and glutamine (4 mM; Gibco, 25030081)], 600 μl of Seahorse buffer plus 60 μl each of oligomycin (50 μg/ml; Sigma, 75351), FCCP (10 μM; Sigma, C2920), and rotenone (10 μM; Sigma, R8875) was automatically injected. At the end of the recording period, cells were collected and cell numbers were determined using a trypan blue exclusion assay. Oxygen consumption rate values were calculated after normalizing with the cell number and plotted as the means ± SD.
Fluorescence-based methods to measure DCFDH oxidation, MitoSOX Red oxidation, MMP, and autophagic flux
To measure DCFDH oxidation, cells were stained with DCFDA (10 μM; Sigma, D6883) for 30 min before flow cytometry analysis (Gallios, Beckman Coulter). In a second fluorescent dye oxidation measurement, cells were incubated with MitoSOX Red (5 mM; Life Technologies, M36008) for 30 min in serum-free DMEM. We used increased oxidized DCF and MitoSOX Red fluorescence as indicators of increased nonmetabolic oxidation, which is consistent with oxidative stress, although we acknowledge that dye oxidation by itself does not actually represent the increased production of ROS (51). For MMP analyses, cells were incubated in DMEM with DiOC6 (10 nM; Sigma, 318426) for 30 min before analyses. For autophagic flux analyses, MDA-MB-231/GFP-LC3 cells were harvested after ADI-PEG20 treatment for different time periods, and immediately analyzed by flow cytometry. Data were collected from three independent experiments and are presented in a histogram. The flow cytometry analyses were performed with FlowJo software (Tree Star).
Confocal microscopy and TEM
MDA-MB-231 cells stably transfected with GFP-LC3 cells were cultured in six-well plates with cover slips at a density of 1 × 105 cells per well, and then treated with ADI-PEG20 for 0, 12, and 24 hours, respectively. Cells were stained with MitoTracker Deep Red FM (300 nM; Life Technologies, M22426) for 15 min in serum-free DMEM. After washing with PBS, the cover slips were mounted over a microscope slide in ProLong Gold Antifade Reagent that contained DAPI (4′,6-diamidino-2-phenylindole) (Life Technologies, P-36931), and examined using a Stallion Digital Imaging Station (Carl Zeiss Microscopy). For mitochondria morphology characterization, MDA-MB-231 cells were cultured in six-well plates with cover slips at a density of 5 × 105 cells per well, and then treated with Mdivi1 (10 μM; Sigma, M0199), ADI-PEG20, or combination of both for 24 hours. Cells were stained with TOM20 for 1 hour at RT after fixation with 4% paraformaldehyde in PBS and permeabilized with 1% Triton X-100. After washing, secondary antibody conjugate with Alexa Fluor 488 was added and incubated for 30 min at RT. Finally, slides were mounted using ProLong Gold Antifade Reagent and imaged using confocal microscopy as described above. For TEM studies, cell pellets were fixed in glutaraldehyde {2% in 0.1 M cacodylate buffer [Na(CH3)2AsO2·3H2O], pH 7.2} at 4°C overnight. The cell pellets were then washed three times with 0.1 M cacodylate buffer (pH 7.2), postfixed with OsO4 (1% in 0.1 M cacodylate buffer) for 30 min, and then washed three times again with cacodylate buffer (0.1 M). The samples were dehydrated using a series of 60, 70, 80, and 95% ethanol, and then 100% absolute ethanol (twice) and propylene oxide (twice), and were then placed in propylene oxide/Eponate (1:1) overnight at RT. The vials were sealed for overnight incubation, and then opened the next day for 2 to 3 hours to allow the propylene oxide to evaporate. Finally, the samples were infused with 100% Eponate and polymerized at −64°C for 48 hours. Ultrathin sections (~70 nm thick) were prepared using a Leica Ultracut UCT Ultramicrotome with a diamond knife, picked up on 200-mesh copper electron microscopy grids. Grids were stained with uranyl acetate (2%) for 10 min, followed by Reynold’s lead citrate staining for 1 min. The samples were viewed using a FEI Tecnai 12 TEM equipped with a Gatan UltraScan 2k CCD (charge-coupled device) camera. Three independent experiments were performed, and representative images are shown.
Oil Red O staining
MDA-MB-231 cells were cultured in six-well plates with cover slips at a density of 5 × 105 cells per well, and then treated with ADI-PEG20 for 0, 24, and 48 hours, respectively. Cells were fixed using 4% paraformaldehyde in PBS for 30 min. Each well was gently rinsed twice with 1 ml of deionized water. Sixty percent isopropanol (1.5 ml) was added to each well for 5 min. Isopropanol was then removed and replaced with 1.5 ml of Oil Red O working solution (2 mg/ml in 60% isopropanol; Sigma, O0625) for 10 min at RT. Each well was rinsed with tap water until clear and counterstained with hematoxylin for 1 min. Last, each well was rinsed with tap water and mounted with 70% glycerol.
Xenograft studies
MDA-MB-231 (5 × 106) and ATG5-knockdown MDA-MB-231 cells (resuspended in 100 μl of serum-free DMEM) were injected subcutaneously into the right flank of 6-week-old female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) (NOD-SCID) mice. Mice were randomized at day 10 after tumor inoculation and received either vehicle or 4 IU of ADI-PEG20 (0.2 ml) weekly through tail vein injection (intravenous) for 3 weeks (n = 5 mice per group). Tumor volume was measured weekly. Mice were euthanized on day 35, 4 days after the last injection. Tumors were harvested for weight measurement and H&E staining. Animal experiments were approved by the Institutional Animal Care and Use Committee at City of Hope.
Statistical analysis
The database was created using Microsoft Excel and analyzed using JMP 8.0 (SAS Institute) and GraphPad Prism 5.0 software programs. Group comparisons for normal distribution data were done by t test (two samples) and one-way analysis of variance (ANOVA) (multiple comparisons). The nonparametric Wilcoxon test was used for continuous nonnormal distribution data. For multiple comparisons, Tukey-Kramer HSD (honestly significant difference) and Steel-Dwass methods were applied for following pairs’ comparison in ANOVA and nonparametric Wilcoxon test, respectively. Categorical variables were compared using χ2 analysis, Fisher’s exact test, or binomial test of proportions. Kaplan-Meier analysis and COX hazard proportional model were used to analyze OS and DFS. Multivariate analysis and stratification were used to reduce the confounder’s impact on the estimation of the HR. Statistical significance was set as P < 0.05, two-tailed.
Supplementary Material
Acknowledgments
We are sincerely grateful to W.-X. Ding, W.-C. Shen, E. Wang, and M. Kong for their helpful suggestions and critical reading of the manuscript. We thank B.-W. Wu, J. Bomalaski, and W.-J. Kung (Polaris) for providing quality ADI-PEG20 for the experiments; M. M. Miller, Z. Li, and R. Zerda of Electron Microscopy Core at City of Hope for TEM analyses; J. Wu, Y. Wang, and M.-Y. Hsieh of Animal Tumor Core at City of Hope for xenograft efficacy studies; B. Armstrong, M. Lee, and T. Patel of Light Microscopy Digital Imaging Core at City of Hope for confocal and fluorescence microscopy analyses; S. Loera of Pathology Core at City of Hope for immunohistochemical analyses; L. Brown of Analytical Cytometry Core at City of Hope for flow cytometry analyses; J. Wu and S. Hu for their help on the DIMSCAN analyses; B. Stiles and Y. Li for their help in oxygen consumption rate measurements; H.-M. Shih and the RNAi Consortium at Academia Sinica for providing the lentiviral constructs against human ATG5 and BECLIN 1; members of Ann’s laboratory for helpful discussions; and M. Morgan for editing.
Funding: This work was supported in part by NIH Research Grants R01DE10742 and R01DE14183 (to D.K.A.), City of Hope’s Women’s Cancers Program Award (to Y.-R.C.), and the University of California at Merced (to H.J.F.).
Footnotes
www.sciencesignaling.org/cgi/content/full/7/319/ra31/DC1
Fig. S1. Knockdown of ASS1 increases sensitivity to ADI-PEG20 treatment.
Fig. S2. Autophagy is required for ADI-PEG20–induced cell death.
Fig. S3. Arginine depletion suppresses the basal oxygen consumption rate and reserve capacity.
Fig. S4. ADI-PEG20 does not affect SOD and SIRT mRNA abundance, but decreases mitochondrial protein abundance.
Fig. S5. ADI-PEG20 causes mitochondrial fragmentation in MDA-MB-231 cells.
Fig. S6. ASS1 abundance predicts OS.
Fig. S7. Lack of synergistic effect of ADI-PEG20 combined with Abraxane or carboplatin.
Table S1. Primer sequences.
Author contributions: F.Q., Y.-R.C., H.J.F., H.-J.K., and D.K.A. designed the experiments and analyzed the data. F.Q. and Y.R.C. executed the experiments. F.Q., Y.-R.C., H.-J.K., and D.K.A. wrote the manuscript. F.Q., X.L., J.X., H.Z., S.Z., J.H., and Y.Y. analyzed breast cancer biospecimens and interpreted the data. C.-Y.C. performed microarray and RT-PCR validation analyses. S.G. analyzed the CI data. L.-J.S. provided anti-ASS1 antibody. X.L. performed statistical analyses.
Competing interests: F.Q., Y.-R.C., and D.K.A. are named on a pending patent titled “Methods for targeting breast and other cancers by targeting argininosuccinate synthetase 1-deficiency,” filed by Beckman Research Institute, City of Hope Medical Center. The other authors declare that they have no competing interests.
Data and materials availability: The use of ADI-PEG20 requires a materials transfer agreement with Polaris. The microarray data of ADI-PEG20 treatment with or without arginine supplementation in MDA-MB-231 cells have been deposited in Gene Expression Omnibus with accession number GSE53838.
REFERENCES AND NOTES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Cancer Society . Cancer Facts & Figures 2013. American Cancer Society; Atlanta, GA: 2013. [Google Scholar]
- 5.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: What do metabolic outliers teach us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 7.Kami K, Fujimori T, Sato H, Sato M, Yamamoto H, Ohashi Y, Sugiyama N, Ishihama Y, Onozuka H, Ochiai A, Esumi H, Soga T, Tomita M. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics. 2013;9:444–453. doi: 10.1007/s11306-012-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wise DR, Thompson CB. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu G, Morris SM., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998;336(Pt. 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris SM., Jr. Arginine: Beyond protein. Am. J. Clin. Nutr. 2006;83:508S–512S. doi: 10.1093/ajcn/83.2.508S. [DOI] [PubMed] [Google Scholar]
- 14.Müller HJ, Boos J. Use of l-asparaginase in childhood ALL. Crit. Rev. Oncol. Hematol. 1998;28:97–113. doi: 10.1016/s1040-8428(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 15.Husson A, Brasse-Lagnel C, Fairand A, Renouf S, Lavoinne A. Argininosuccinate synthetase from the urea cycle to the citrulline-NO cycle. Eur. J. Biochem. 2003;270:1887–1899. doi: 10.1046/j.1432-1033.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 16.Wheatley DN, Campbell E. Arginine deprivation, growth inhibition and tumour cell death: 3. Deficient utilisation of citrulline by malignant cells. Br. J. Cancer. 2003;89:573–576. doi: 10.1038/sj.bjc.6601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ensor CM, Holtsberg FW, Bomalaski JS, Clark MA. Pegylated arginine deiminase (ADI-SS PEG20,000 mw) inhibits human melanomas and hepatocellular carcinomas in vitro and in vivo. Cancer Res. 2002;62:5443–5450. [PubMed] [Google Scholar]
- 18.Huang CC, Tsai ST, Kuo CC, Chang JS, Jin YT, Chang JY, Hsiao JR. Arginine deprivation as a new treatment strategy for head and neck cancer. Oral Oncol. 2012;48:1227–1235. doi: 10.1016/j.oraloncology.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Feun L, You M, Wu CJ, Kuo MT, Wangpaichitr M, Spector S, Savaraj N. Arginine deprivation as a targeted therapy for cancer. Curr. Pharm. Des. 2008;14:1049–1057. doi: 10.2174/138161208784246199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lind DS. Arginine and cancer. J. Nutr. 2004;134:2837S–2841S. doi: 10.1093/jn/134.10.2837S. [DOI] [PubMed] [Google Scholar]
- 21.Takaku H, Takase M, Abe S, Hayashi H, Miyazaki K. In vivo anti-tumor activity of arginine deiminase purified from Mycoplasma arginini. Int. J. Cancer. 1992;51:244–249. doi: 10.1002/ijc.2910510213. [DOI] [PubMed] [Google Scholar]
- 22.Beloussow K, Wang L, Wu J, Ann D, Shen WC. Recombinant arginine deiminase as a potential anti-angiogenic agent. Cancer Lett. 2002;183:155–162. doi: 10.1016/s0304-3835(01)00793-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim RH, Bold RJ, Kung HJ. ADI, autophagy and apoptosis: Metabolic stress as a therapeutic option for prostate cancer. Autophagy. 2009;5:567–568. doi: 10.4161/auto.5.4.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szlosarek PW, Klabatsa A, Pallaska A, Sheaff M, Smith P, Crook T, Grimshaw MJ, Steele JP, Rudd RM, Balkwill FR, Fennell DA. In vivo loss of expression of argininosuccinate synthetase in malignant pleural mesothelioma is a biomarker for susceptibility to arginine depletion. Clin. Cancer Res. 2006;12:7126–7131. doi: 10.1158/1078-0432.CCR-06-1101. [DOI] [PubMed] [Google Scholar]
- 25.Yoon CY, Shim YJ, Kim EH, Lee JH, Won NH, Kim JH, Park IS, Yoon DK, Min BH. Renal cell carcinoma does not express argininosuccinate synthetase and is highly sensitive to arginine deprivation via arginine deiminase. Int. J. Cancer. 2007;120:897–905. doi: 10.1002/ijc.22322. [DOI] [PubMed] [Google Scholar]
- 26.Cheng PN, Lam TL, Lam WM, Tsui SM, Cheng AW, Lo WH, Leung YC. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007;67:309–317. doi: 10.1158/0008-5472.CAN-06-1945. [DOI] [PubMed] [Google Scholar]
- 27.Bowles TL, Kim R, Galante J, Parsons CM, Virudachalam S, Kung HJ, Bold RJ. Pancreatic cancer cell lines deficient in argininosuccinate synthetase are sensitive to arginine deprivation by arginine deiminase. Int. J. Cancer. 2008;123:1950–1955. doi: 10.1002/ijc.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim RH, Coates JM, Bowles TL, McNerney GP, Sutcliffe J, Jung JU, Gandour-Edwards R, Chuang FY, Bold RJ, Kung HJ. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res. 2009;69:700–708. doi: 10.1158/0008-5472.CAN-08-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai WB, Aiba I, Lee SY, Feun L, Savaraj N, Kuo MT. Resistance to arginine deiminase treatment in melanoma cells is associated with induced argininosuccinate synthetase expression involving c-Myc/HIF-1α/Sp4. Mol. Cancer Ther. 2009;8:3223–3233. doi: 10.1158/1535-7163.MCT-09-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ascierto PA, Scala S, Castello G, Daponte A, Simeone E, Ottaiano A, Beneduce G, De Rosa V, Izzo F, Melucci MT, Ensor CM, Prestayko AW, Holtsberg FW, Bomalaski JS, Clark MA, Savaraj N, Feun LG, Logan TF. Pegylated arginine deiminase treatment of patients with metastatic melanoma: Results from phase I and II studies. J. Clin. Oncol. 2005;23:7660–7668. doi: 10.1200/JCO.2005.02.0933. [DOI] [PubMed] [Google Scholar]
- 31.Izzo F, Marra P, Beneduce G, Castello G, Vallone P, De Rosa V, Cremona F, Ensor CM, Holtsberg FW, Bomalaski JS, Clark MA, Ng C, Curley SA. Pegylated arginine deiminase treatment of patients with unresectable hepatocellular carcinoma: Results from phase I/II studies. J. Clin. Oncol. 2004;22:1815–1822. doi: 10.1200/JCO.2004.11.120. [DOI] [PubMed] [Google Scholar]
- 32.Glazer ES, Piccirillo M, Albino V, Di Giacomo R, Palaia R, Mastro AA, Beneduce G, Castello G, De Rosa V, Petrillo A, Ascierto PA, Curley SA, Izzo F. Phase II study of pegylated arginine deiminase for nonresectable and metastatic hepatocellular carcinoma. J. Clin. Oncol. 2010;28:2220–2226. doi: 10.1200/JCO.2009.26.7765. [DOI] [PubMed] [Google Scholar]
- 33.Yang TS, Lu SN, Chao Y, Sheen IS, Lin CC, Wang TE, Chen SC, Wang JH, Liao LY, Thomson JA, Wang-Peng J, Chen PJ, Chen LT. A randomised phase II study of pegylated arginine deiminase (ADI-PEG 20) in Asian advanced hepatocellular carcinoma patients. Br. J. Cancer. 2010;103:954–960. doi: 10.1038/sj.bjc.6605856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 36.Delage B, Luong P, Maharaj L, O’Riain C, Syed N, Crook T, Hatzimichael E, Papoudou-Bai A, Mitchell TJ, Whittaker SJ, Cerio R, Gribben J, Lemoine N, Bomalaski J, Li CF, Joel S, Fitzgibbon J, Chen LT, Szlosarek PW. Promoter methylation of argininosuccinate synthetase-1 sensitises lymphomas to arginine deiminase treatment, autophagy and caspase-dependent apoptosis. Cell Death Dis. 2012;3:e342. doi: 10.1038/cddis.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4:621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 38.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol. Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Bánhegyi G, Bartholomew CR, Bassham DC, Bast RC, Jr., Batoko H, Bay BH, Beau I, Béchet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahová M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LA, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae HJ, Chai CY, Chan DC, Chan EY, Chang RC, Che CM, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SS, Chen W, Chen X, Chen X, Chen X, Chen YG, Chen Y, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CH, Cheng Y, Cheong H, Cheong JH, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Chin LS, Chiou SH, Chisari FV, Cho CH, Cho DH, Choi AM, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang TH, Chueh SH, Chun T, Chwae YJ, Chye ML, Ciarcia R, Ciriolo MR, Clague MJ, Clark RS, Clarke PG, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronje MJ, Cuervo AM, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CA, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LM, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding WX, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht AM, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen EL, Espert L, Espina V, Fan H, Fan J, Fan QW, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farré JC, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fésüs L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao FB, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, González-Estévez C, González-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan JL, Guan KL, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S, Hazelhurst LA, He C, He YW, Hébert MJ, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Höhfeld J, Holyoake TL, Hong MH, Hood DA, Hotamisligil GS, Houwerzijl EJ, Høyer-Hansen M, Hu B, Hu CA, Hu HM, Hua Y, Huang C, Huang J, Huang S, Huang WP, Huber TB, Huh WK, Hung TH, Hupp TR, Hur GM, Hurley JB, Hussain SN, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jäättelä M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon JH, Jeung EB, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jimenez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GV, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhasz G, Juillerat-Jeanneret L, Jung CH, Jung YK, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke PY, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JA, Kiger AA, Kihara A, Kim DR, Kim DH, Kim DH, Kim EK, Kim HR, Kim JS, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim YS, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BC, Koch JC, Koga H, Koh JY, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong WJ, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovács AL, Kraft C, Krainc D, Krämer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Krüger R, Krut O, Ktistakis NT, Kuan CY, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung HJ, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, László L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJ, Lee BW, Lee GM, Lee J, Lee JH, Lee M, Lee MS, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei HY, Lei QY, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li JL, Li L, Li S, Li W, Li XJ, Li YB, Li YP, Liang C, Liang Q, Liao YF, Liberski PP, Lieberman A, Lim HJ, Lim KL, Lim K, Lin CF, Lin FC, Lin J, Lin JD, Lin K, Lin WW, Lin WC, Lin YL, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu CF, Liu K, Liu L, Liu QA, Liu W, Liu YC, Liu Y, Lockshin RA, Lok CN, Lonial S, Loos B, Lopez-Berestein G, López-Otín C, Lossi L, Lotze MT, Low P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MC, Malorni W, Man N, Mandelkow EM, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Marino G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Melendez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RF, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao CY, Miao JY, Michels PA, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE, Müller S, Muller S, Münger K, Münz C, Murphy LO, Murphy ME, Musaró A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nurnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JH, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Poggeler S, Poirot M, Poletti A, Pous C, Pozuelo-Rubio M, Praetorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian SB, Qin L, Qin ZH, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Regnier-Vigouroux A, Reichert AS, Reiners JJ, Jr., Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodríguez de Córdoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KM, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB, III, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AM, Sánchez-Alcázar JA, Sánchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AH, Scharl M, Schatzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schuller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen HM, Shen WC, Sheng ZH, Shi Y, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin EC, Simmons A, Simon AK, Simon HU, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Strålfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun SY, Sun ZJ, Sung JJ, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy LK, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takács-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KS, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tonges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai TF, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn W, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan XB, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang HG, Wang HD, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang X, Wang YJ, Wang Y, Wang Z, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen LP, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu DC, Wu WK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu DZ, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang DS, Yang E, Yang JM, Yang SY, Yang W, Yang WY, Yang Z, Yao MC, Yao TP, Yeganeh B, Yen WL, Yin JJ, Yin XM, Yoo OJ, Yoon G, Yoon SY, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu SW, Yu WH, Yuan ZM, Yue Z, Yun CH, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ, III, Zeitlin SO, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang MY, Zhang XD, Zhao M, Zhao YF, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou CZ, Zhu C, Zhu WG, Zhu XF, Zhu X, Zhu Y, Zoladek T, Zong WX, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giménez-Xavier P, Francisco R, Santidrián AF, Gil J, Ambrosio S. Effects of dopamine on LC3-II activation as a marker of autophagy in a neuroblastoma cell model. Neurotoxicology. 2009;30:658–665. doi: 10.1016/j.neuro.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Shacka JJ, Klocke BJ, Roth KA. Autophagy, bafilomycin and cell death: The “A-B-Cs” of plecomacrolide-induced neuroprotection. Autophagy. 2006;2:228–230. doi: 10.4161/auto.2703. [DOI] [PubMed] [Google Scholar]
- 43.Lin TC, Chen YR, Kensicki E, Li AY, Kong M, Li Y, Mohney RP, Shen HM, Stiles B, Mizushima N, Lin LI, Ann DK. Autophagy: Resetting glutamine-dependent metabolism and oxygen consumption. Autophagy. 2012;8:1477–1493. doi: 10.4161/auto.21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birrell JA, Hirst J. Investigation of NADH binding, hydride transfer, and NAD+ dissociation during NADH oxidation by mitochondrial complex I using modified nicotinamide nucleotides. Biochemistry. 2013;52:4048–4055. doi: 10.1021/bi3016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cecchini G. Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- 46.Ackrell BA. Progress in understanding structure–function relationships in respiratory chain complex II. FEBS Lett. 2000;466:1–5. doi: 10.1016/s0014-5793(99)01749-4. [DOI] [PubMed] [Google Scholar]
- 47.Bianchi C, Genova ML, Parenti Castelli G, Lenaz G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: Kinetic evidence using flux control analysis. J. Biol. Chem. 2004;279:36562–36569. doi: 10.1074/jbc.M405135200. [DOI] [PubMed] [Google Scholar]
- 48.Bianchi C, Fato R, Genova ML, Parenti Castelli G, Lenaz G. Structural and functional organization of Complex I in the mitochondrial respiratory chain. BioFactors. 2003;18:3–9. doi: 10.1002/biof.5520180202. [DOI] [PubMed] [Google Scholar]
- 49.Forman HJ, Kennedy JA. Role of superoxide radical in mitochondrial dehydrogenase reactions. Biochem. Biophys. Res. Commun. 1974;60:1044–1050. doi: 10.1016/0006-291x(74)90418-5. [DOI] [PubMed] [Google Scholar]
- 50.Cadenas E, Boveris A. Enhancement of hydrogen peroxide formation by protophores and ionophores in antimycin-supplemented mitochondria. Biochem. J. 1980;188:31–37. doi: 10.1042/bj1880031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, Ischiropoulos H., II Measuring reactive oxygen and nitrogen species with fluorescent probes: Challenges and limitations. Free Radic. Biol. Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2013 doi: 10.1038/onc.2013.120. 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitra K. Mitochondrial fission-fusion as an emerging key regulator of cell proliferation and differentiation. BioEssays. 2013;35:955–964. doi: 10.1002/bies.201300011. [DOI] [PubMed] [Google Scholar]
- 54.Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perkins G, Bossy-Wetzel E, Ellisman MH. New insights into mitochondrial structure during cell death. Exp. Neurol. 2009;218:183–192. doi: 10.1016/j.expneurol.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boren J, Brindle KM. Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death Differ. 2012;19:1561–1570. doi: 10.1038/cdd.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 58.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sørlie T, Dai H, He YD, van’t Veer LJ, Bartelink H, van de Rijn M, Brown PO, van de Vijver MJ. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 60.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 61.Chou TC, Martin N. CompuSyn for Drug Combinations: PC Software and User’s Guide: A Computer Program for Quantitation of Synergism and Antagonism in Drug Combinations, and the Determination of IC50 and ED50 and LD50 Values. ComboSyn Inc.; Paramus, NJ: 2005. [Google Scholar]
- 62.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 63.Chan DC. Mitochondria: Dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 64.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tolkovsky AM. Mitophagy. Biochim. Biophys. Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 67.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLOS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding WX, Guo F, Ni HM, Bockus A, Manley S, Stolz DB, Eskelinen EL, Jaeschke H, Yin XM. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J. Biol. Chem. 2012;287:42379–42388. doi: 10.1074/jbc.M112.413682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang HY, Wu WR, Wang YH, Wang JW, Fang FM, Tsai JW, Li SH, Hung HC, Yu SC, Lan J, Shiue YL, Hsing CH, Chen LT, Li CF. ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: Aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin. Cancer Res. 2013;19:2861–2872. doi: 10.1158/1078-0432.CCR-12-2641. [DOI] [PubMed] [Google Scholar]
- 74.Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N, Morioka H, Toyama Y, Hirohashi S, Kawai A, Yamada T. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol. Cancer Ther. 2010;9:535–544. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 75.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 76.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frezza C, Gottlieb E. Mitochondria in cancer: Not just innocent bystanders. Semin. Cancer Biol. 2009;19:4–11. doi: 10.1016/j.semcancer.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 78.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 79.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 80.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 81.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 82.Wang X, Liu X, Li AY, Chen L, Lai L, Lin HH, Hu S, Yao L, Peng J, Loera S, Xue L, Zhou B, Zhou L, Zheng S, Chu P, Zhang S, Ann DK, Yen Y. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin. Cancer Res. 2011;17:2570–2580. doi: 10.1158/1078-0432.CCR-10-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu FL, Liang YF, Chang YC, Yo HH, Wei MF, Shen LJ. RNA interference of argininosuccinate synthetase restores sensitivity to recombinant arginine deiminase (rADI) in resistant cancer cells. J. Biomed. Sci. 2011;18:25. doi: 10.1186/1423-0127-18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.