Abstract
Dendritic spines serve as preferential sites of excitatory synaptic connections and are pleomorphic. To address the structure–function relationship of the dendritic spines, we used two-photon uncaging of glutamate to allow mapping of functional glutamate receptors at the level of the single synapse. Our analyses of the spines of CA1 pyramidal neurons reveal that AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid)-type glutamate receptors are abundant (up to 150/spine) in mushroom spines but sparsely distributed in thin spines and filopodia. The latter may be serving as the structural substrates of the silent synapses that have been proposed to play roles in development and plasticity of synaptic transmission. Our data indicate that distribution of functional AMPA receptors is tightly correlated with spine geometry and that receptor activity is independently regulated at the level of single spines.
Most excitatory synaptic transmission in the mammalian central nervous system relies on glutamate as a neurotransmitter, and postsynaptic glutamate receptors are central in both the acquisition and maintenance of memory1, 2. Substantial biochemical evidence indicates that glutamate receptors in postsynaptic densities (PSDs) are regulated by various protein machineries that link the receptors to the cytoskeleton3, 4, 5 and that control the insertion6–10 and phosphorylation of the receptors11, 12. Individual dendritic spines have been thought to act as functional compartments of glutamate receptor expression, given that they are physically and thus metabolically separated from the body of the dendrite by the narrow spine neck13–16. Indeed, the Hebbian principle of learning as well as most theories of neuronal networks assume that the strength of synaptic connections is subject to independent control17. Moreover, spine geometry has been proposed to be a key determinant of synaptic function and memory in the brain13, 14, 18–22.
These various hypotheses have not been tested experimentally, however, because it is not possible to systematically investigate postsynaptic glutamate sensitivities at the level of the individual spine by classical electrophysiological approaches16, 23. Also, the number of functional AMPA-sensitive glutamate receptors in individual spines has not been estimated directly. Two-photon excitation of caged-glutamate compounds may overcome these difficulties24 as a result of the inherent three-dimensional resolution of neurotransmitter application associated with this technique. However, caged-glutamate compounds with a cross-section for two-photon absorption, a rate of photolysis, and a stability in aqueous solution sufficient for such studies have not previously been described25–27.
We developed a caged-glutamate compound and microscopic system for two-photon excitation that allowed systematic investigation of functional glutamate receptors at the level of the individual synapse. Our experiments with cultured hippocampal neurons have shown that it is possible to achieve the two-photon excitation-induced release of glutamate with a spatiotemporal resolution very similar to that exhibited by presynaptic terminals. Experiments with hippocampal slice preparations demonstrated that the glutamate sensitivities of individual dendritic spines are highly variable and depend on spine geometry. Our data support the notion that the fine geometry of dendrites is important in the regulation of glutamatergic synaptic transmission in the brain.
RESULTS
Two-photon uncaging of MNI-glutamate
We developed a methoxy derivative of nitroindolino-glutamate, MNI-glutamate (Fig. 1a), that possesses a two-photon cross section (0.06 GM; 1 GM = 10−50 cm4s/photon2) sufficiently large for biological application (see Methods). The efficacy of this compound was established with cultured hippocampal neurons. Under physiological conditions, MNI-glutamate was stable for 24 hours, and did not activate any current at a concentration of 10 mM. Moreover, it did not affect either the amplitude (Fig. 1b) or the time course (Fig. 1b, inset) of miniature excitatory postsynaptic currents (mEPSCs), indicating that it does not exert an antagonistic effect at AMPA receptors. Irradiation of a proximal region of the dendrites for a short period (50μs) with 720-nm-wavelength light from a mode-locked laser elicited EPSC-like currents (2pEPSC) (Fig. 1c) with a time course identical to those of spontaneous mEPSCs (Fig. 1d and e). The extremely rapid onset of 2pEPSC indicates that the uncaged glutamate acts directly on glutamate receptors on the postsynaptic membrane. The time course of 2pEPSC is well-predicted (smooth line in Fig. 1c) from rapid photolysis (10μs) of MNI-glutamate within a focal volume of two-photon excitation with lateral and axial full-width-at-half-maximal (FWHM) diameter of 0.29 and 0.89μm, respectively (black lines in Fig. f and g; see Methods). The spatial resolution of glutamate receptor activation was inferred from a small, isolated, glutamate-sensitive region of a dendrite (Fig. 2d), yielding lateral and axial FWHM diameters of 0.45 and 1.1μm, respectively (green lines in Fig. 1f and g). A slight loss of resolution was due to the diffusion of glutamate before its binding to the receptors. Nevertheless, two-photon excitation of MNI-glutamate permits the functional mapping of glutamate sensitivity with a spatiotemporal resolution similar to that achieved by activation of presynaptic terminals (~0.4μm and ~0.1 ms)28.
Figure 1. Two-photon excitation of MNI-glutamate in cultured hippocampal neurons.
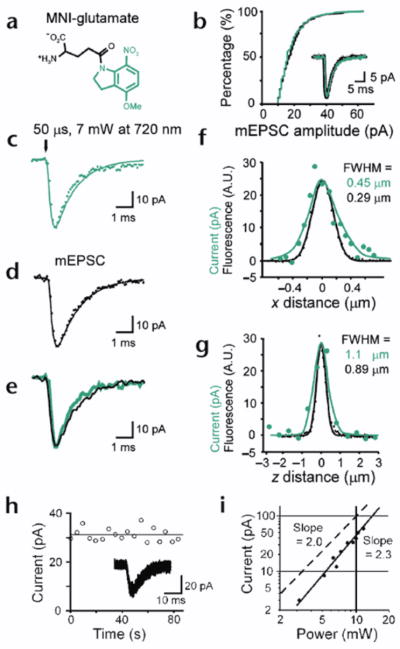
(a) Structure of MNI-glutamate. (b) Cumulative amplitude distributions of mEPSCs in the absence (black) or presence (green) of MNI-glutamate (5 mM). Traces of mEPSCs in the absence (black) and presence (green) of MNI-glutamate are overlaid. Each trace is an average of 20 mEPSCs. A representative of three experiments. (c) Average of 28 current traces obtained from a cell in response to photolysis of MNI-glutamate for 50 μs with 7 mW of 720-nm-wavelength light. (d) Average of 10 mEPSCs recorded from a cell that exhibited rapid mEPSCs. The smooth lines in (c) and (d) are drawn as described in Methods. (e) Superimposition of the 2pEPSC (green) and mEPSC (black) shown in (c) and (d), respectively. (f, g) Amplitudes of 2pEPSCs (green dots) along horizontal (f) and vertical (g) lines crossing the center of an AMPA receptor cluster, and fluorescence profiles of a 0.1-μm diameter bead (black dots) along the horizontal (f) and vertical axis (g). The smooth black lines represent Gaussian fitting of the data. The smooth green lines are predicted from a theory described in Methods. (h) Stability of 2pEPSCs. Insets, superimposed current traces. (i) Double-logarithmic plot of the dependence of 2pEPSC amplitudes on the incident power of the mode-locked laser. The duration of the irradiation was 0.6 ms. The slope of solid and dashed straight lines are 2.3 and 2, respectively.
Figure 2. Functional mapping of glutamate sensitivities in hippocampal neurons.
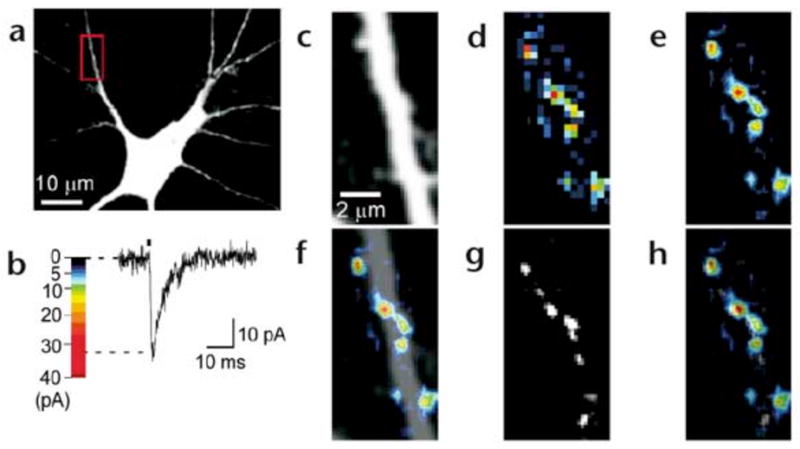
(a) Fluorescence image of a neuron in culture. The red box indicates a region of interest for functional mapping. (b) Pseudocolor coding of the amplitude of a 2pEPSC. (c) Region of interest of the neuron shown in (a) for functional mapping. (d) A pseudocolor map of peak amplitudes of 2pEPSCs (glutamate-sensitivity map). (e) A smoothed glutamate-sensitivity map by linear interpolation. (f) Overlay of (c) and (e). (g) A FM1-43 fluorescence image. (h) An overlay of (e) and (g).
In the following experiments, uncaging was effected with low peak powers and longer irradiation times compared to the above (0.6 and 2 ms). This uncaging regime produced little or no photodamage29, as shown by the stability of the amplitudes of the 2pEPSCs over 1 min (Fig. 1h). The evoked currents had a small coefficient of variation (CV) of 0.08, consistent with the post-synaptic action of glutamate23. The current amplitudes showed a supralinear (power exponent, 2.5 ± 0.6, mean ± s.d., n = 7) dependence on the radiation power, as predicted (Fig. 1i) both by the two-photon excitation30 of MNI-glutamate and by the cooperativity (1.2–1.7) of glutamate binding to AMPA receptors31 (2pEPSCs were CNQX (6-cyano-7-nitroquinoxaline)-sensitive). These data also indicate that AMPA receptors were not saturated at an incident power less than 12 mW (irradiation time, 0.6 ms).
We used a combined imaging–electrophysiology approach to record 2pEPSCs at a lateral spatial interval of 0.33μm in a region of interest (ROI; 512 points in Fig. 2a). The site of uncaging was pseudorandomized to eliminate the effects of desensitization of receptor by previous photolysis at neighboring pixels (see Methods). The mapping system simultaneously recorded the fluorescence evoked by the laser irradiation used for uncaging, to detect any structural alteration in the ROI during experiments (Fig. 2c). Maps of the amplitude of 2pEPSCs represented by pseudocolor coding (Fig. 2b) are shown in Fig. 2d and e. The regions of glutamate sensitivity were apparently clustered along the dendrites (Fig. 2f) and were adjacent to the nerve terminals stained with FM1-43 (Fig. 2g and h). Thus, our functional mapping approach was able to detect the clustering of AMPA receptors in the postsynaptic membrane23, 32, 33.
Distribution of functional AMPA receptors in vivo
We then applied our methodology to hippocampal CA1 pyramidal neurons in acute slice preparations (Fig. 3a and b). The spatial resolution of AMPA-receptor mapping was slightly reduced compared with cultured neurons, 0.6μm laterally and 1.4μm axially, because of the smaller numerical aperture (NA) of the lens used (0.9). We therefore chose to study those dendrites in which spines were relatively sparse (~1 spines/μm). The time course of 2pEPSC (Fig. 3bA–C) is similar to spontaneous mEPSCs (Fig. 3bE). To map the three-dimensional distribution of AMPA receptors on dendrites, we obtained several two-dimensional maps along the z-axis (as described for cultured neurons), with each z-axis section being separated by 0.8 μm (Fig. 3c–e). Individual spines exhibited a high degree of heterogeneity in maximal glutamate sensitivity (Fig. 3f–h), with a CV of 0.45 ± 0.11 (9 dendrites). We also detected slow NMDA-mediated currents when Mg2+ was removed from the extracellular solution; the distribution of these currents was less heterogeneous than that of AMPA receptors, and most spines expressed NMDA receptors (data not shown). Next, we determined the relationship between spine shape13 and the EPSCs evoked by two-photon uncaging of MNI-glutamate in CA1 pyramidal neurons. The expression of functional AMPA receptors was closely related to the geometry of spines. Thin spines exhibited little (Fig. 4a and b) or slight glutamate sensitivity (Fig. 4c–e), and thick mushroom spines displayed large sensitivity to uncaged glutamate (Fig. 4f–j). Regions of glutamate sensitivity tended to be asymmetrically localized to one portion of a spine (Figs. 3h and 4c–j), consistent with the predominant localization of AMPA receptors within PSDs with a diameter of 0.1 to 0.5μm13, 19, 20. The glutamate sensitivity of each PSD (as defined by the maximum glutamate sensitivity of a spine) was significantly correlated with the volume of the spine head (Fig. 5a); the correlation coefficient (r) was 0.69–0.90 (0.80 ± 0.07; n = 9) within one dendrite (0.89, Fig. 5b) and 0.77 for average of 9 dendrites (Fig. 5c). The actual correlation must be larger, because these values are reduced by the channel noise (CV ~ 0.08) and the sampling error (CV = 0.1, see Methods). The glutamate sensitivity was weakly and negatively correlated with spine length with correlation coefficient of 0.30 (Fig. 5d). Such a weak correlation may be due to the presence of several spines with long length and small spine-head volume (Fig. 5e). These data are consistent with the correlation between AMPA receptor immunoreactivity and the size of PSD19, 20, because the size of PSD is nearly proportional to the spine-head volume13.
Figure 3. Functional mapping of glutamate sensitivities in hippocampal neurons.
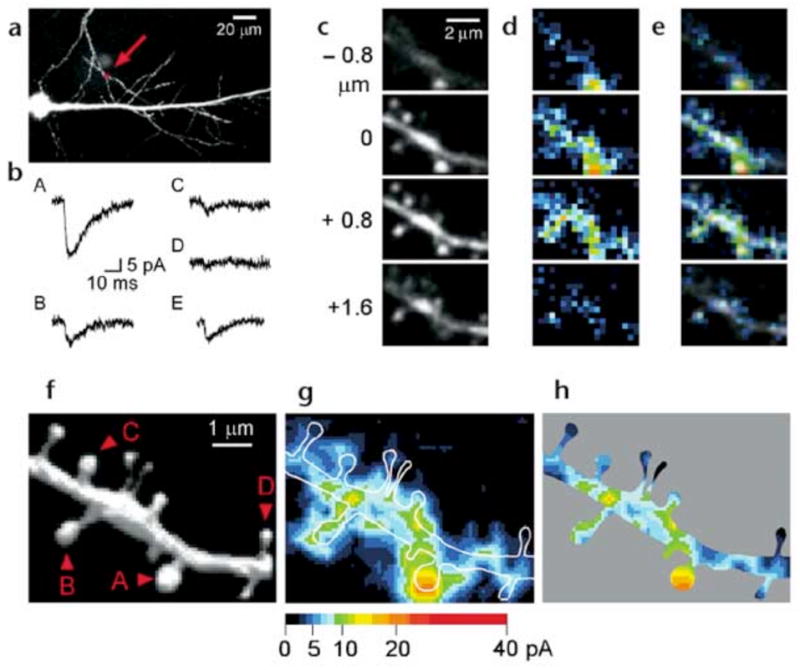
(a) Fluorescence image of a CA1 pyramidal neuron in slice. The arrow indicates the region of interest. (b) 2pEPSCs induced at four different sites of the dendrite indicated as A–D in (f). The traces were averages of data from four neighboring pixels. E shows an averaged trace of 10 mEPSCs recorded from the same neuron. (c–e) Fluorescence images at four z-axis sections as indicated (c), two-dimensional pseudocolor map of 2pEPSC amplitude (d), and overlays (e). (f) Three-dimensional reconstruction of the fluorescent images. (g) Maximal intensity plot of the glutamate sensitivity for the four sections shown in (d). The map has been smoothed by linear interpolation. White lines indicate contours of the dendritic structures. (h) Glutamate-sensitivity map within the dendritic contours.
Fgiure 4. Spine geometry and expression of functional AMPA receptors.
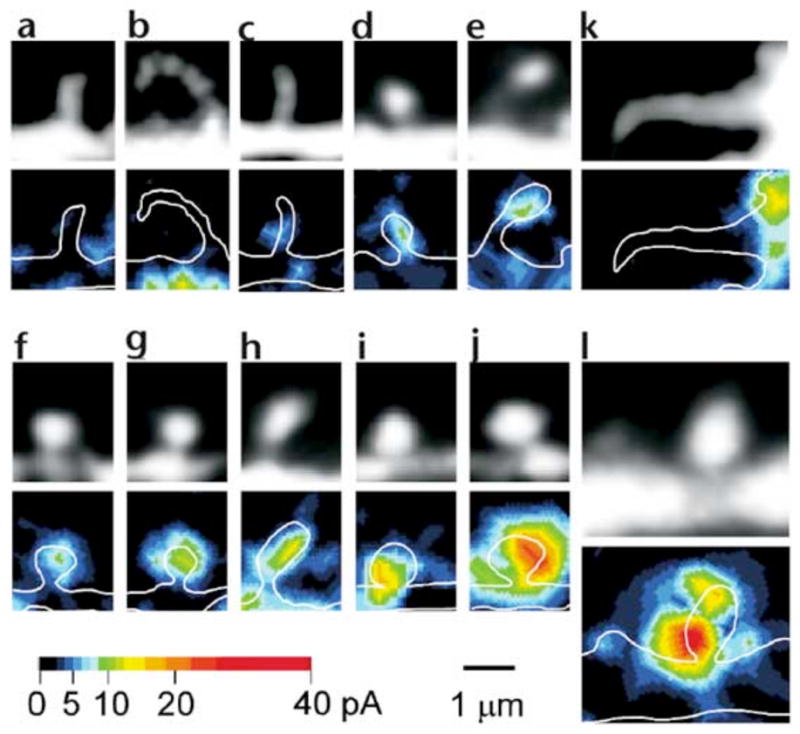
Fluorescence images (top) and glutamate-sensitivity maps (bottom) are shown for various dendritic spines of CA1 pyramidal neurons in hippocampal slices. The fluorescence images were obtained from stacked images containing the respective spine. The glutamate-sensitivity map was obtained from one x–y section in which the maximal glutamate sensitivity for the spine was detected. The maps were smoothed by linear interpolation. White lines indicate the contours of the dendritic structures. Representative data from thin spines (a–e), mushroom spines13 (f–j), filopodia (k) and a mushroom spine in a 9-day-old animal (l) are shown.
Figure 5. Correlation between spine geometry and glutamate sensitivities.
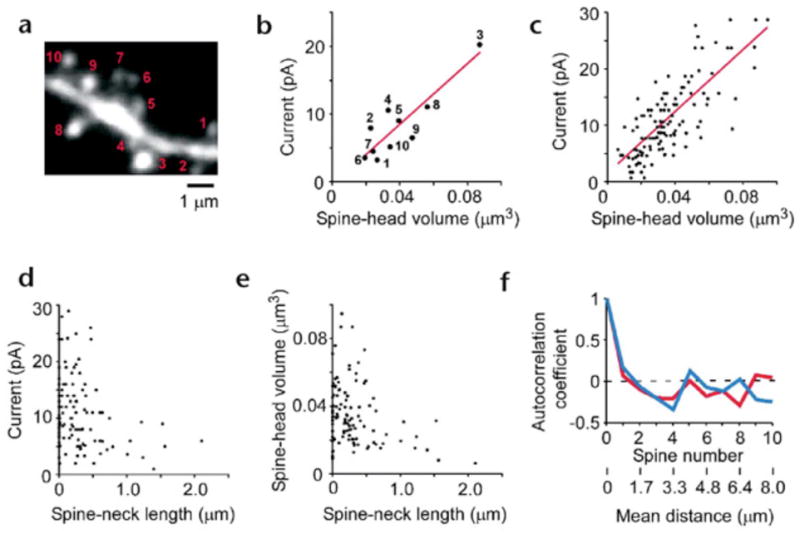
A fluorescence image of the dendrite shown in Fig. 3f. The image was generated from a stack of 40 images. (b) Correlations between the spine-head volume and glutamate sensitivities for the dendrites shown in (a). Numbered points correspond to the numbered spines in (a). (c) Correlations between the spine-head volume and glutamate sensitivities for nine different dendrites. (e) Correlation between the spine-neck length and spine-head volume for the dendrites. (f) Spatial autocorrelations of glutamate sensitivity (blue line) and of spine-head volume (red line). The mean distances between the spines obtained from the nine dendrites are also indicated.
Dendritic filopodia with a length greater than 3μm showed little glutamate sensitivity (Fig. 4kn = 7). In young animals (7 to 9 days old), dendrites were decorated only sparsely with spines34, and most of the dendritic shaft was devoid of glutamate sensitivity (Fig. 4l), although marked expression of functional AMPA receptors was already apparent in mushroom spines present (Fig. 4l). These data conform with the development of AMPA receptor immunoreactivity in CA1 pyramidal neurons35.
To examine whether expression of AMPA receptors is spatially correlated along the dendrite, we constructed an autocorrelation histogram of spine glutamate sensitivities along dendrites (Fig. 5f; 9 dendrites). Such analysis indicated the absence of a correlation, even in neighboring spines, both for glutamate sensitivities and for spine-head volumes (Fig. 5f). This observation suggests that the expression of AMPA receptors is controlled independently at a level of the individual spine, consistent with the classical notion that a single spine may behave as an independent chemical compartment to regulate synaptic transmission13, 14.
Finally, the number of functional AMPA receptors in mushroom spines was estimated by nonstationary fluctuation analysis36–38. We added the desensitization inhibitor cyclothiazide (0.1 mM, CTZ) to increase the opening probability of AMPA receptors to greater than 0.5. Photolysis was done 50 times at a site at the edge of the spines (Fig. 6a and b), and we obtained the deviations from the mean 2pEPSCs (Fig. 6c). We plotted the mean current amplitude (I) and the variance (σ2) calculated at each time point (Fig. 6d). This analysis predicted the number of AMPA receptors to be 46 to 147 (82 ± 31; mean ± s.d., n = 8) and the amplitude of single-channel currents to be 0.4 to 0.8 pA (0.6 ± 0.1). As a corollary, our data suggest that the postsynaptic AMPA receptors in the mushroom spines are not saturated by the glutamate released from a single synaptic vesicle23, 33, because the opening probability of the AMPA receptors was estimated as 0.27, given that the largest mEPSC was ~19 pA (n = 222) in our experiments, and that such mEPSC likely occurred in the mushroom spines with largest number of AMPA receptors (N = 147, i = 0.48 pA).
Figure 6. Non-stationary fluctuation analysis of AMPA receptors in a mushroom spine.
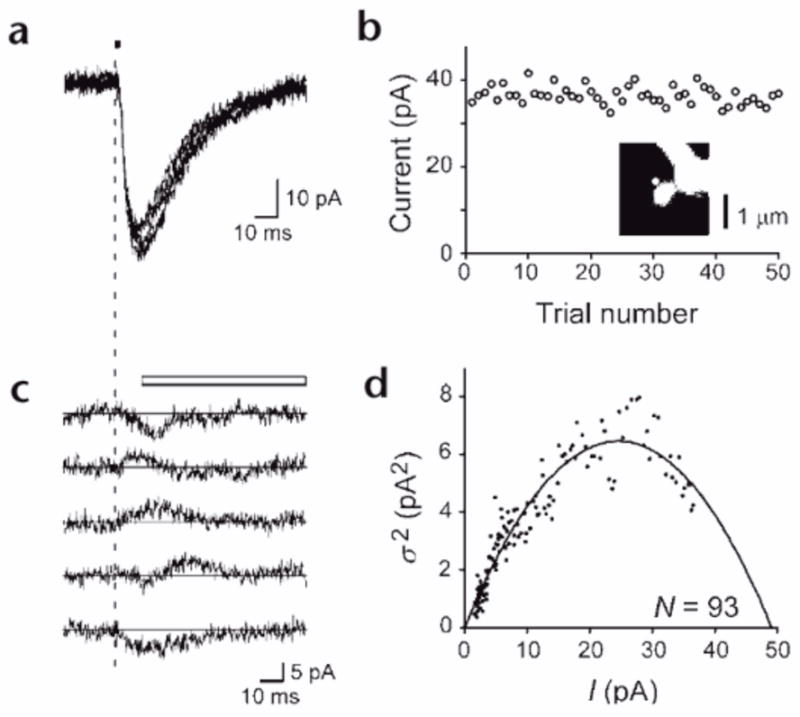
(a) Five examples of 2pEPSCs obtained during repetitive photolysis (4.5 mW, 2 ms) of the spine shown in the inset in (b). (b) Peak amplitudes of 2pEPSCs evoked 50 times in the presence of cyclothiazide. Inset, fluorescence picture of a mushroom spine. White dot indicates the site at which optical release of MNI-glutamate was made. (c) Five current traces shown in (a) after subtraction of the ensemble average of 2pEPSCs. The vertical dashed line in (a) and (c) represents the onset of uncaging. The white bar indicates the time window used for the fluctuation analysis. (d) Variance (σ2) of 2pEPSCs versus the mean amplitudes (I). Each plot represents an average of five sample points. N, calculated number of AMPA receptors.
DISCUSSION
Using two-photon excitation of a caged-glutamate compound, we showed that the expression of functional AMPA receptors among spines shows a large range (CV, 0.5), varying from the minimal (silent) to those bearing more than 140 AMPA channels, and has a strong correlation with spine-head volume. These data give strong support to previous anatomical observations13, 19, 20. Our data further suggest that the expression of glutamate receptors must be independently controlled at individual spines. Given that each spine receives only one synaptic input13, our data indicate the highest possible degree of freedom is available in synaptic connections in the dendrites, which has been assumed in the learning models of most neural network theories17.
The relationship between spine geometry and expression of functional AMPA receptors demonstrated here provides substantial support to the hypothesis that neuronal fine structures have functional correlates in the brain13, 18, 21. It has recently been suggested that electrical activity controls the size and number of spines21, 22, 39–42, and that the expression of glutamate receptors is tightly regulated by abundant cytoskeletal structures present in spines3–5. One can therefore surmise that spine structures are a key determinant of synaptic efficacy via control of the distribution of the AMPA receptors9, 10, 43. In addition, our study suggests that thin spines and filopodia may be postsynaptic components of silent synapses28, 32, 34, 44. This might result from less effective transport of AMPA receptors into thinner spines with smaller volume-to-surface ratios. The mechanism by which spine-head volume might affect trafficking of AMPA receptors remains to be determined, as does whether thin spines, filopodia and dendritic shafts eventually undergo transformation to generate thicker spines after the induction of long-term potentiation.
Two-photon uncaging provides a means to achieve unprecedented rapid, reproducible and fine three-dimensional spatial control of neurotransmitter concentration on a scale that faithfully mimics quantal release. This powerful approach thus allows the optical stimulation of dendritic processes in brain slices at the level of the individual synapse. Two-photon uncaging is a uniquely powerful tool that can be used to clarify the cellular and molecular dynamics of neurons and other cells.
METHODS
Preparations
Cultured hippocampal neurons were provided as described32. In brief, hippocampi were dissected from postnatal day 1 (P1) Wistar rats, and dispersed on a monolayer culture of primary glial cells. Neurons were cultured in DMEM (Dulbecco’s modification of Eagle’s medium)/F12 with B27 supplement. Neurons cultured for 17–25 days were used in the experiments. A cultured dish was mounted on a recording chamber that was superfused with a solution containing 120 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 30 mM glucose and 25 mM Na HEPES at pH 7.4. Hippocampal slices with a thickness of 300 μm were obtained from 15–22 day old Sprague–Dawley rats unless otherwise stated45. A slice was transferred to recording chamber that was superfused with a solution containing 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 1.25 mM NaH2PO4, 25 mM NaHCO3, 20 mM glucose, bubbled with 95% O2 and 5% CO2 gas. The bathing solutions also contained 1 μM tetrodotoxin, 50 μM picrotoxin and 200 μM Trolox (Aldrich, Milwaukee, Michigan). Whole-cell patch pipette contained a solution of 135 mM Cs-gluconate, 5 mM MgCl2, 1 mM CaCl2, 2 mM ATP, 0.3 mM GTP, 0.5 mM fura-2, 10 mM TEA, 10 mM Cs4EGTA, 10 mM Cs-HEPES at pH 7.2. All physiological experiments were performed at room temperature (23–25°C). The animal experiments were approved by the animal experiment committee of the National Institute for Physiological Sciences, Japan.
Two-photon excitation of a caged-glutamate compound
We developed MNI-glutamate (M. Matsuzaki et al., Soc. Neurosci. Abstr. 26, 425.13, 2000) using a synthetic sequence similar to that already described46. (Significant modifications are fully detailed in the Supplementary Methods, available on the supplementary information page of Nature Neuroscience online.) The compound was purified with HPLC, freeze-dried and stored at −30°C. It was applied locally from a glass pipette close to the whole-cell clamped cells at 5–10 mM. Two-photon excitation imaging of neurons was done with an inverted microscope (IX70, Olympus, Tokyo, Japan) with a water immersion objective lens (UPlanApo 60× water/IR; NA, 1.2) for cultured neurons47, and with an upright microscope (BX80, Olympus) with a water immersion objective lens (LUMPlanFI 60× water/IR; NA, 0.9). Mode-locked femtosecond-pulse Ti:sapphire laser (Tsunami, Spectra Physics, Mountain View, California) with an original pulse duration of 70 to 100 fs was attached to a laser-scanning microscope (FluoView, Olympus). The group velocity dispersion of the microscope was compensated for by chirp compensation optics. The laser power at the specimen was 3 to 7 mW, except for the experiments shown in Fig. 1i. The mode-locked laser was gated either by an electro-optical modulator (Model 350–50, Conoptics, Danbury, Connecticut) for 50 μs (Fig. 1c–g) or by a mechanical shutter at 0.6–2 ms (Uniblitz, Rochester, New York) (Figs. 2–6). Cultured neurons, stained with 10 μM FM1-43 and 40 mM KCl for 1 min and then washed for more than 5 min33, were imaged before the whole-cell clamp experiments with a cooled charged-coupled devise camera (TILL Photonics, Munich, Germany; Fig. 2g).
The two-photon action cross section of MNI-glutamate was obtained as 0.06 GM at 730 nm, using azid-1 as a standard compound with a known action cross section of 1.0 GM at 730 nm48. This was accomplished by comparing the amounts of photoproducts using a HPLC after the same dose of two-photon excitation was applied to the two compounds in an aqueous solution.
Analysis of 2pEPSCs
Neurons were whole-cell clamped at −70 mV (Axopatch 200A, Axon Instruments, Foster City, California), and the currents were low-pass filtered at 5 kHz and sampled at 10 kHz. The patch pipettes were made of borosilicate glass, and access resistances were 18–36 MΩ for culture and 13–44 MΩ for slice. The access resistance was 70% compensated for the experiments shown in Fig. 1c–g but not for others. Automatic mapping software based on LabView (National Instruments, Austin, Texas) controlled a galvano-scanner driver (General Scanning, Watertown, Massachusetts) and a shutter, and sampled the whole-cell current and the output of photomultiplier. The pseudorandom sequence of scanning the pixels in a ROI was constructed to keep the distance of two successive pixels longer than 2.5–5μm, and sampled 2pEPSCs at an interval of 100 ms. The point spread function of the focal volume of two-photon excitation were estimated using a 0.1μm fluorescent beads as 0.29 μm (FWHM) laterally and 0.89 μm axially for the inverted microscope (NA, 1.2), and 0.39μm laterally and 1.28μm axially for the upright microscope (NA, 0.9).
The glutamate concentration at a time t and at a lateral distance of x, and axial distance of z from the center of photolysis is obtained using a Gaussian approximation of the focal volume of two-photon excitation as follows49.
| (1) |
Here, M stands for the molar glutamate equivalent released by a two-photon photolysis; td, the duration of photolysis; τp, the time constant of photolysis of MNI-glutamate; Dg, the diffusion coefficient of glutamate (0.75μm2/ms); σx and σz, standard deviations of the Gaussian distribution of two-photon excitation (0.123 μm and 0.38μm, respectively); 1[s < td], the function which is 1 when the condition is satisfied, and otherwise 0. M is predicted based on the following equation30.
| (2) |
Here, CMNI represents the concentration of MNI-glutamate; δ, the two-photon cross section of MNI-glutamate (0.06 GM); g, the two-photon advantage (7.08 × 104 = 0.588/83 MHz/ 100 fs in our setup); λ, a wavelength of excitation (720 nm); n, refractive index of water (1.33); and 〈P〉, average power of incident laser (2.5 × 1016 photons/s at 7 mW). We assume τp as 10 μs, because values of τp larger than 50 μs result in significantly slower activation of 2pEPSCs than actually recorded (Fig. 1c), and those larger than 20 μs predict greater loss of spatial resolution than actually obtained (Fig. 1f) (see below). G(x, z, t) was numerically obtained using Mathematica4 (Wolfram Research, Champaign, Illinois). For instance, the peak glutamate concentration at the center of irradiation reaches 0.6 mM focally, at a laser power of 7 mW, MNI-glutamate concentration (CMNI) of 8 mM, the duration of irradiation (td) of 50 μs. The glutamate concentration during mEPSCs (Gm (x, t)) is obtained assuming a planar synaptic cleft with a width of 20 nm and a point glutamate source from which 5,000 glutamates are released according to m2t•;e−m•;t, m = 39 (ms−1)28. Using G(x, z, t) or Gm(x, t), the opening probability of AMPA receptor can be obtained by the kinetic model of AMPA receptors50. We adjusted the closing rate constant (α) from 1.1/s to 1.6/s to account for a slightly more rapid decay of mEPSCs in our preparation (Fig. 1d). In the condition listed above, the peak opening probability was 0.15 and 0.2 for 2pEPSC and mEPSC, respectively. The opening probability was then filtered at 0.83 kHz to account for uncompensated access resistance. The glutamate currents, Imf (50nm, t) or IGf (0, 0, t), were arbitrary scaled to fit the data (Fig. 1c and d), as the time course of EPSCs is not significantly dependent on the concentrations of glutamate (M) when the opening probability is below 0.2 and the duration of laser irradiation (td) is as short as 50μs. The peak amplitude of IGf (x, z, t) is plotted in Fig. 1f and g (green smooth lines). Our simulation and experiments indicated that if we keep M unchanged, a larger value of td between 50μs and 2 ms did not markedly affect the spatial resolution of glutamate sensitivity mapping.
For three-dimensional mapping of glutamate sensitivities, x–y scannings of a ROI with a pixel size of 0.33μm × 0.33μm were done at several planes along the z axis with each plane separated by 0.8μm. The error due to the sampling intervals was estimated as 0.1 (CV), given the spatial resolution of glutamate mapping as 0.6μm laterally and 1.4μm axially. For three dimensional reconstruction of fluorescence images, fine fluorescence images were acquired with a resolution of 0.083μm laterally and 0.2μm axially in the end of the experiments. The spine-head volume was estimated by total fluorescence of the spine head in the fluorescence images, which was calibrated using the large spine head whose volume (0.087μm3, spine A in Fig. 3f) could be directly estimated by a FWHM diameter. Fluorescence intensity differences between dendrites were normalized by using a dendritic shaft with a diameter of 2μm. For thin spines and filopodia, the spine head fluorescence was obtained as a total fluorescence in a0.67 × 0.67 × 0.8μm3 area that was centered at the brightest spot of a spine. The spine-neck length was estimated as the smallest distance between the edges of the head and shaft, defined as the contour where the fluorescence intensity was one-half of the fitted Gaussian maximum in z-stack images. It was defined as 0 when the fluorescence did not decay to the half-maximal values between the spine head and shaft. Autocorrelation coefficient is obtained by the following equation.
| (3) |
Here, a(i, k) represents either the maximal amplitude of the 2pEPSCs or the spine-head volume at ith spine in the kth dendrite and ak, the mean value of a(i, k) for all spines in the kth dendrite.
Nonstationary fluctuation analysis was performed based on the following equation
| (4) |
Here, σb2 stands for base line variance; i, single channel current; and N, the number of AMPA receptors37. The 2pEPSCs were filtered at 2 kHz and sampled at 10 kHz. The data between the time (3–11 ms) at the peak of 2pEPSCs and 40–80 ms after the glutamate application were used for fluctuation analysis (the open bar in Fig. 6c). We chose to analyze 33 traces (of 50) without miniature EPSCs (Fig. 6d). The estimation of N was not significantly affected by the dendritic filtering in our simulation experiments38.
We estimated the CV of the two-photon excitation to be less than 0.03, by measuring the two-photon excited fluorescence of a polar tracer, Alexa Fluor 594 hydrizide (0.2 mM, Molecular Probes, Eugene, Oregon), applied to slice preparations similarly to MNI-glutamate.
Supplementary Material
Acknowledgments
We thank S. Adams for a sample of azid-1, H. Tsubokawa for helpful suggestions on hippocampal slice experiments, R. Nichols for comments on the manuscript and R. Ijuin for technical assistance. This work was supported by CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation (JST); the Research for the Future program of the Japan Society for the Promotion of Science (JSPS); Grants-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science and Technology; and NIH (GM53395). M.M. was a JSPS research fellow.
References
- 1.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 2.Malenka RC, Nicoll RA. Long-term potentiation—a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 3.Ziff EB. Enlightening the postsynaptic density. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 4.Naisbitt S, et al. Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J Neurosci. 2000;20:4524–4534. doi: 10.1523/JNEUROSCI.20-12-04524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen L, Liang F, Walensky LD, Huganir RL. Regulation of AMPA receptor GluR1 subunit surface expression by a 4.1N-linked actin cytoskeletal association. J Neurosci. 2000;20:7932–7940. doi: 10.1523/JNEUROSCI.20-21-07932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song I, et al. Interaction of the N-ethylmaleimide-sensitive factor with AMPA receptors. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- 7.Lissin DV, et al. Activity differentially regulates the surface expression of synaptic AMPA and NMDA glutamate receptors. Proc Natl Acad Sci USA. 1998;95:7097–7102. doi: 10.1073/pnas.95.12.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luscher C, et al. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron. 1999;24:649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 9.Turrigiano GG. AMPA receptors unbound: membrane cycling and synaptic plasticity. Neuron. 2000;26:5–8. doi: 10.1016/s0896-6273(00)81131-9. [DOI] [PubMed] [Google Scholar]
- 10.Shi S, Hayashi Y, Esteban JA, Malinow R. Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell. 2001;105:331–343. doi: 10.1016/s0092-8674(01)00321-x. [DOI] [PubMed] [Google Scholar]
- 11.Benke TA, Luthi A, Isaac JT, Collingridge GL. Modulation of AMPA receptor unitary conductance by synaptic activity. Nature. 1998;393:793–797. doi: 10.1038/31709. [DOI] [PubMed] [Google Scholar]
- 12.Lee HK, Barbarosie M, Kameyama K, Bear MF, Huganir RL. Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature. 2000;405:955–959. doi: 10.1038/35016089. [DOI] [PubMed] [Google Scholar]
- 13.Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuste R, Denk W. Dendritic spines as basic functional units of neuronal integration. Nature. 1995;375:682–684. doi: 10.1038/375682a0. [DOI] [PubMed] [Google Scholar]
- 15.Svoboda K, Tank DW, Denk W. Direct measurement of coupling between dendritic spines and shafts. Science. 1996;272:716–719. doi: 10.1126/science.272.5262.716. [DOI] [PubMed] [Google Scholar]
- 16.Shepherd GM. The dendritic spine: a multifunctional integrative unit. J Neurophysiol. 1996;75:2197–2210. doi: 10.1152/jn.1996.75.6.2197. [DOI] [PubMed] [Google Scholar]
- 17.Arbib MA. The Handbook of Brain Theory and Neural Networks. MIT Press; Cambridge, Massachusetts: 1995. [Google Scholar]
- 18.Cajal Ry. Histologie du Systeme Nerveux de l’Homme et des Vertebres. Maloine, Paris: 1911. [Google Scholar]
- 19.Nusser Z, et al. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 1998;21:545–559. doi: 10.1016/s0896-6273(00)80565-6. [DOI] [PubMed] [Google Scholar]
- 20.Takumi Y, Ramirez-Leon V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 1999;2:618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- 21.Segal M, Korkotian E, Murphy DD. Dendritic spine formation and pruning: common cellular mechanisms? Trends Neurosci. 2000;23:53–57. doi: 10.1016/s0166-2236(99)01499-x. [DOI] [PubMed] [Google Scholar]
- 22.Halpain S. Actin and the agile spine: how and why do dendritic spines dance? Trends Neurosci. 2000;23:141–146. doi: 10.1016/s0166-2236(00)01576-9. [DOI] [PubMed] [Google Scholar]
- 23.Liu G, Choi S, Tsien RW. Variability of neurotransmitter concentration and nonsaturation of postsynaptic AMPA receptors at synapses in hippocampal cultures and slices. Neuron. 1999;22:395–409. doi: 10.1016/s0896-6273(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 24.Denk W. Two-photon scanning photochemical microscopy: mapping ligand-gated ion channel distributions. Proc Natl Acad Sci USA. 1994;91:6629–6633. doi: 10.1073/pnas.91.14.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettit DL, Wang SS, Gee KR, Augustine GJ. Chemical two-photon uncaging: a novel approach to mapping glutamate receptors. Neuron. 1997;19:465–471. doi: 10.1016/s0896-6273(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 26.Furuta T, et al. Brominated 7-hydroxycoumarin-4-ylmethyls: photolabile protecting groups with biologically useful cross-sections for two photon photolysis. Proc Natl Acad Sci USA. 1999;96:1193–1200. doi: 10.1073/pnas.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SS, Khiroug L, Augustine GJ. Quantification of spread of cerebellar long-term depression with chemical two-photon uncaging of glutamate. Proc Natl Acad Sci USA. 2000;97:8635–8640. doi: 10.1073/pnas.130414597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusakov DA, Kullmann DM. Extrasynaptic glutamate diffusion in the hippocampus: ultrastructural constraints, uptake, and receptor activation. J Neurosci. 1998;18:3158–3170. doi: 10.1523/JNEUROSCI.18-09-03158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopt A, Neher E. Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys J. 2001;80:2029–2036. doi: 10.1016/S0006-3495(01)76173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C, Webb WW. Measurement of two-photon excitaiton cross sections of molecular fluorophores with data from 690 to 1050 nm. J Opt Soc Am B. 1996;13:481–491. [Google Scholar]
- 31.Jonas P, Sakmann B. Glutamate receptor channels in isolated patches from CA1 and CA3 pyramidal cells of rat hippocampal slices. J Physiol (Lond) 1992;455:143–171. doi: 10.1113/jphysiol.1992.sp019294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomperts SN, Rao A, Craig AM, Malenka RC, Nicoll RA. Postsynaptically silent synapses in single neuron cultures. Neuron. 1998;21:1443–1451. doi: 10.1016/s0896-6273(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 33.McAllister AK, Stevens CF. Nonsaturation of AMPA and NMDA receptors at hippocampal synapses. Proc Natl Acad Sci USA. 2000;97:6173–6178. doi: 10.1073/pnas.100126497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durand GM, Kovalchuk Y, Konnerth A. Long-term potentiation and functional synapse induction in developing hippocampus. Nature. 1996;381:71–75. doi: 10.1038/381071a0. [DOI] [PubMed] [Google Scholar]
- 35.Petralia RS, et al. Selective acquisition of AMPA receptors over postnatal development suggests a molecular basis for silent synapses. Nat Neurosci. 1999;2:31–36. doi: 10.1038/4532. [DOI] [PubMed] [Google Scholar]
- 36.Sigworth FJ. The variance of sodium current fluctuations at the node of Ranvier. J Physiol (Lond) 1980;307:97–129. doi: 10.1113/jphysiol.1980.sp013426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spruston N, Jonas P, Sakmann B. Dendritic glutamate receptor channels in rat hippocampal CA3 and CA1 pyramidal neurons. J Physiol (Lond) 1995;482:325–352. doi: 10.1113/jphysiol.1995.sp020521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silver RA, Cull-Candy SG, Takahashi T. Non-NMDA glutamate receptor occupancy and open probability at a rat cerebellar synapse with single and multiple release sites. J Physiol (Lond) 1996;494:231–250. doi: 10.1113/jphysiol.1996.sp021487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchs PA, Muller D. Induction of long-term potentiation is associated with major ultrastructural changes of activated synapses. Proc Natl Acad Sci USA. 1996;93:8040–8045. doi: 10.1073/pnas.93.15.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 41.Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- 42.Lendvai B, Stern EA, Chen B, Svoboda K. Experience-dependent plasticity of dendritic spines in the developing rat barrel cortex in vivo. Nature. 2000;404:876–881. doi: 10.1038/35009107. [DOI] [PubMed] [Google Scholar]
- 43.Shi SH, et al. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science. 1999;284:1811–1816. doi: 10.1126/science.284.5421.1811. [DOI] [PubMed] [Google Scholar]
- 44.Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsubokawa H, Ross WN. IPSPs modulate spike backpropagation and associated [Ca2+]i changes in the dendrites of hippocampal pyramidal neurons. J Neurophysiol. 1996;76:2896–2906. doi: 10.1152/jn.1996.76.5.2896. [DOI] [PubMed] [Google Scholar]
- 46.Papageorgiou G, Corrie JET. Effects of aromatic substituents on the photocleavage of 1-acyl-7-nitroindolines. Tetrahedron. 2001;56:8197–8205. [Google Scholar]
- 47.Nemoto T, et al. Sequential-replenishment mechanism of exocytosis in pancreatic acini. Nat Cell Biol. 2001;3:253–258. doi: 10.1038/35060042. [DOI] [PubMed] [Google Scholar]
- 48.Brown EB, Shear JB, Adams SR, Tsien RY, Webb WW. Photolysis of caged calcium in femtoliter volumes using two-photon excitation. Biophys J. 1999;76:489–499. doi: 10.1016/S0006-3495(99)77217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crank J. The Mathematics of Diffusion. Oxford Univ. Press; Oxford: 1975. [Google Scholar]
- 50.Diamond JS, Jahr CE. Transporters buffer synaptically released glutamate on a submillisecond time scale. J Neurosci. 1997;17:4672–4687. doi: 10.1523/JNEUROSCI.17-12-04672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


