Abstract
A modification to the standard continuum electrostatics approach to calculate protein pKas which allows for the decoupling of histidine tautomers within a two state model is presented. Histidine with four intrinsically coupled protonation states cannot be easily incorporated into a two state formalism because the interaction between the two protonatable sites of the imidazole ring is not purely electrostatic. The presented treatment, based on a single approximation of the interrelation between histidine’s charge states, allows for a natural separation of the two protonatable sites associated with the imidazole ring as well as the inclusion of all protonation states within the calculation.
Keywords: F/FDPB, protein pKa, respiratory complex I, redox Bohr effect
INTRODUCTION
Histidine plays an essential role in protein chemistry where it is often found in the active site of enzymes. Much of histidine’s utility arises from the fact that histidine titrates near neutral pH. As such the quality of models used to describe the protonation of histidine is essential for the correct determination of pH dependent properties of proteins. The continuum electrostatics approach to protein pKa calculations is a simple and practical method that has been developed over the past two decades1–10. Work continues to improve the continuum electrostatics description while new models incorporating higher levels of detail from both the solvent and protein models have been developed11–19. Here we describe a solution of one technical issue in continuum electrostatics calculations related to the specificity of interaction between the two protonatable sites of histidine. Several applications which demonstrate the utility of the current scheme are presented.
Histidine can bind a proton at the Nδ or Nε atoms of the imidazole ring giving rise to four possible protonation states: the doubly deprotonated negative state, two neutral tautomers (Nδ and Nε) with a single bound proton, and the doubly protonated positive state. The difficulty in treating histidine within standard continuum electrostatics approaches, such as the Full Finite Difference Poisson-Boltzmann (F/FDPB) formalism, arise from the fact that the partial atomic charges of the imidazole ring depend explicitly on the location of the bound proton(s) preventing the separation of the Nδ and Nε titratable sites. The continuum electrostatics approach to pKa calculations in proteins hinges on the fact that specific groups within a covalently bonded macromolecule can be treated as separate entities that interact classically. In order to separate two ionizable sites in the protein there should not be significant through bond charge transfer between the sites (i.e. they should be separated by many single bonds). Since the imidazole ring is conjugated, a proton bound to the Nδ site will significantly alter the charges at the Nε site, and vice versa. This prevents the separation of the two imidazole sites within the standard approach.
To overcome theses problems a number of methods to treat histidine have been introduced. The simplest means to incorporate histidine into a two state model, where the protonation state of each titratable site is described by a binary variable n, is to fix one of the imidazole nitrogens in the protonated state, typically Nδ. The second imidazole nitrogen is then treated as the sole titratable site. This “single site” model1 reduces the problem’s complexity, but also limits the chemistry of the histidine side chain that gives rise to its usefulness in proteins. The “pseudo independent site” model developed by Bashford and coworkers3 allows for the titration of both the Nδ and Nε sites by essentially coupling two “single site” models for each histidine residue. In this model calculations are performed where each histidine is treated as titrating at the Nδ site. Subsequently, a complete set of calculations in which each histidine titrates at the Nε sites is then performed. Finally, calculations are performed where each histidine is treated as Nε except for one, which is treated as Nδ, in order to calculate the appropriate interactions between tautomers. These interaction energies are used to correct the free energy function to prevent sites from interacting with both the Nδ and Nε forms of the same histidine. Furthermore, an arbitrarily large interaction energy between the tautomers of the same histidine is included to prevent the simultaneous deprotonation of both sites.
Each of the described methods has drawbacks; either the chemistry of histidine is oversimplified or the complexity of the calculation is increased. Here we describe a modification of the standard two state model for histidine that alleviates these issues. A simple binary representation of the protonation state is maintained, the computational complexity is only slightly increased compared to the single site model of histidine, and all protonation/charge states of the imidazole ring are modeled. Results are presented for several small proteins, superoxide dismutase, and respiratory complex I.
METHODS
Theory of Histidine in Continuum Electrostatics calculations
Continuum electrostatics methods attempt to calculate the shift in pKa associated with transferring a model compound with known solution pKa representing the given titratable amino acid from solution to the protein environment. This pKa shift is typically considered to consist of three terms: the change in solvation energy (ΔΔGBorn), the interaction of charges in the titratable site with the fixed background charges of the protein (ΔΔGBack), and the electrostatic interaction between titratable sites i and j (Wij). Given the protonation state vector n⃑ with components ni ∈ {0,1}, where a value of one indicates the site is protonated and zero deprotonated, the free energy associated with a given protonation state can be expressed as
| (1) |
where n0 is the protonation state of the indicated site in the neutral charge (reference) state. The intrinsic pKa is the pKa associated with the given site in the protein when all other sites are in the reference state and is given by
| (2) |
where ΔΔGBorn, ΔΔGBack, and Wij can be expressed in terms of Green’s functions explicitly and have been given elsewhere4,6. These expressions involve products over charges between sites and the appropriate Green’s function yielding interaction energies.
The charges of typical ionizable residues, with a single titratable site, can be expressed in terms of the protonation state as
| (3) |
where q⃑+ and q⃑− are vectors comprised of the set of partial atomic charges for the site when protonated and deprotonated respectively. The constant term in Eq. (3) is subsumed into the protein background while the linear term determines the functional form of Eq. (1).
For histidine, we similarly define the set of charges q⃑+, q⃑δ, q⃑ε and q⃑− corresponding to the set of charges associated with the positive doubly protonated, neutral Nδ and Nε tautomers, and the negative doubly deprotonated states respectively. The charge state of a histidine residue can then be expressed as a function of its protonation state variables nδ and nε as
| (4) |
Clearly, if we substitute the values of (1, 0), (0, 1), and (1, 1) for (nδ, nε) into Eq. (4) we obtain the charge distributions q⃑δ, q⃑ε, and q⃑+. However, q⃑− does not appear in Eq. (4). When the values (0, 0) are encountered Eq. (4) yields
| (5) |
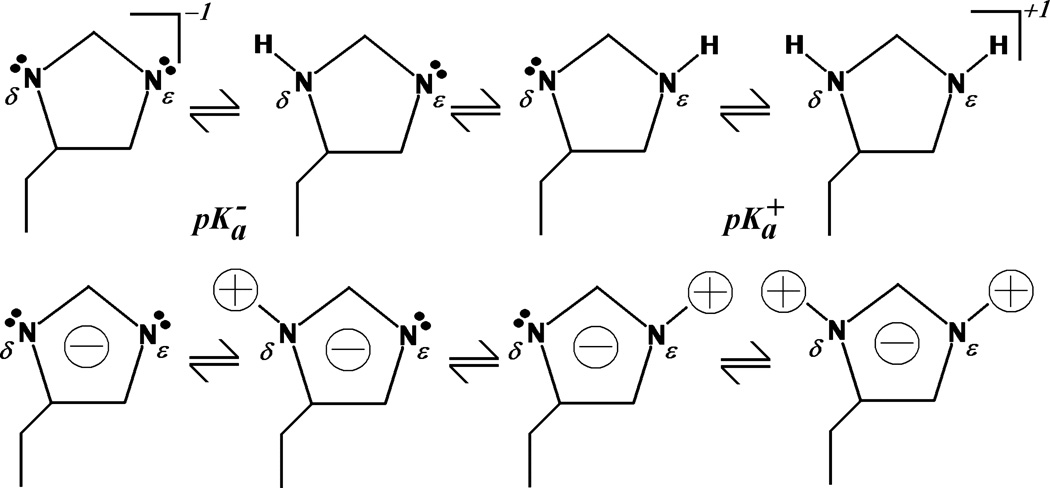
By examining Figure 1 it can be understood that Eq. (5) approximates the charge state of histidine when doubly deprotonated. In light of Eq. (5) we can rewrite Eq. (4) as
| (6) |
where Δq⃑δ = (q⃑+ − q⃑δ), etc. Eq. (6) has now essentially decoupled the Nδ and Nε titratable sites via the approximation expressed in Eq. (5), since only the linear terms in Eq. (6) are of significance.
Figure 1.
Schematic diagram of the various charge states of histidine. Top, structural formulas for the deprotonated, neutral tautomers, and protonated forms of the histidine side chain. Bottom, cartoon representation of the approximate charge distribution in each charge state. The approximation of Eq. (5) can be understood qualitatively by superimposing the neutral tautomers. The result is a structure with two peripheral positive charges and two central negative charges. Subtraction of the doubly protonated state charges leaves a single negative charge centered on the imidazole ring.
To arrive at an expression analogous to Eq. (1) we must distinguish titratable sites from titratable residues (i.e. histidine is a single titratable residue, but consists of two titratable sites). The resulting expression has the same form as Eq. (1), but the sums are now over sites rather than residues. In deriving Eq. (1) from Eq. (6) the quadratic term, , arises which can be identified as the interaction energy between the nδ and nε sites of the same histidine where and are the pKas for the negative to neutral and neutral to positive reactions shifted by the desolvation energy respectively. Assuming the solvation energies are equal for the charged states, ΔGsolv (0,0) ≈ ΔGsolv (1,1), and for the neutral states, ΔGsolv (1,0) ≈ ΔGsolv (0,1), the pKa values become
| (7) |
and the interaction energy
| (8) |
where ΔΔGBorn is determined for the positive to neutral reaction. Note that ΔΔGBorn will reduce and increase , since the lower dielectric protein environment will stabilize the neutral charge state. The difference is the quantum mechanical (i.e. electronic) interaction energy between the Nδ and Nε sites of the same histidine where and are the model pKas associated with the negative to neutral and neutral to positive protonation reactions respectively. The interaction energy, Eq. (8), then serves to adjust between the appropriate pKa value for the positive to neutral or neutral to negative reaction. In solution or if the desolvation term is neglected the interaction energy can be approximated as
| (9) |
The neutral charge state is typically chosen as the reference state, so either of the two neutral tautomers will suffice. Let us define nδ0 ≡ 1 and nε0 ≡ 0; that is the reference state is the Nδ-tautomer. From this choice we see that the Nδ site is acidic while the Nε site is basic. Model pKas are then assigned according to
| (10) |
The background term, ΔΔGBack, in Eq. (2) is then calculated with all histidines as the Nδ-tautomer. From inspection of Eq. (6) it can be seen that the charge distributions q⃑+ and q⃑ε are associated with nδ while q⃑+ and q⃑δ are associated with nε. It is important to note that the negative state charge distribution, q⃑−, is never encountered and is not utilized in the calculation explicitly. For simplicity we have assumed equivalent pKas for the Nδ and Nε sites. However, the method is not restricted to this case and the pKas in Eqn. (10) can be adjusted as desired to reflect the difference in the titration behavior of the two sites.
The described modification allows for the inclusion of the variables (nδ, nε) into a general state vector n⃑ = n1,n2…, and to treat these sites independently as with other sites. In fact any site with intrinsically linked protonatable sites, such as phosphate groups, for which an approximate charge distribution analogous to Eq. (5) can be constructed can be treated with this method.
Implementation: pKip
The method presented in the preceding section has been implemented as the standard model for histidine in the program “pKip: pKas in proteins” currently under development. pKip is a general continuum electrostatics program designed for the calculation of pKas in proteins. The program calculates all of the required terms in Eq. (1), and in turn relies on APBS20 libraries to solve the linearized Poisson-Boltzmann equation (LPBE). The program features the inclusion of membranes, an arbitrary number of user specified dielectric constants, and membrane potentials. In the calculations described in the following sections pKip was used to calculate the electrostatic terms of Eq. (1) except where otherwise stated.
The average protonation state of each enzyme was determined by Metropolis Monte Carlo (MC) simulation as described by Beroza et al.21 and performed by our in house code “Monte” also under development in this lab.
Simulation Protocols
Protonation state calculations were performed on several small proteins (1MBC3, 2LZT22, 3RN323 and 1RNZ24, 3BDC25, 1BI626, 2RN227, 4PTI28, 9RNT29) for which some or all of the protonatable groups have experimentally determined apparent pKas. Experimental pKas were taken from3 for myoglobin, from25 and30 for SNase Δ+PHS variant, and31 for all others. Protonation state calculations were also performed on Human Cu/Zn Superoxide dismutase (accession code 1L3N)32 and the hydrophilic domain of NQO-oxidoreductase (respiratory complex I, structure 2FUG) from thermus themophilus33.
In all cases a solvent dielectric of εs = 80 and ionic strength of I = 100 mM was employed. For all proteins an internal dielectric of εp = 20 was used, except for complex I and myoglobin where εp = 4 was used. CHARMM2234 charges and radii were used throughout, except for myoglobin where the Amber/Bondi charges and radii were used as in3. The model pKas were the same as those presented by Bashford and Karplus5 except for histidine which was assigned model pKas of 14 and 7 for the doubly deprotonated to Nδ and the Nε to doubly protonated reactions respectively except where otherwise noted. For each of the small test proteins and the PDB2PQR35,36 program was used to add hydrogens and assign atomic charges and radii. For Complex I the Amber737 suite was used to add hydrogens and minimize their positions. The program “loopy”38 included with the Jackal suite of programs was used to build in deleted residues and residues not resolved in the crystal structure of the hydrophilic domain of complex I. Various grid sizes and spacing were employed depending on the system. For all proteins studied the finest grid spacing was 0.2 Å or less. All MC simulations were performed at 300 K. The number of MC sweeps (MC steps per site) also varied depending on the system. For complex I 100,000 equilibration sweeps were performed followed by 50,000 production sweeps. MC simulations of the smaller proteins employed 10,000 to 20,000 production sweeps. Single and double MC moves were allowed to overcome poor sampling of highly correlated sites21. Parallel Tempering simulations performed on complex I did not show any significant differences from the single temperature simulations. Additional electrostatics calculations were also performed with the program MEAD4,39 in order to compare the presented method to the “pseudo independent site” model of3.
Hartree-Fock ab initio calculations were performed on methylimidazole in the doubly protonated, singly protonated, and deprotonated states. All calculations were performed using the Gaussian 0340 program at the 6–31G(d) level. Mulliken charges for each state were determined from the geometry optimized structures.
RESULTS AND DISCUSSION
Histidine Charges
To test the validity of the assumption expressed by Eq. (5) Mulliken partial atomic charges were calculated for the four possible protonation states of methylimidazole. The total charge for the doubly deprotonated state obtained from Eq. (5) is −1.00 e− with a precision out to five significant figures. The per atom RMSD between the approximate and calculated charge distributions for the negative charge state was determined to be 0.037 e−, with the largest deviations occurring at the Nδ and Nε protons. These protons are of course absent from the true negative protonation state, but are assigned a small charge in the approximate distribution. With per atom RMSD values below 4% this approximation should not introduce significant errors above the existing uncertainties in the F/FDPB method. Calculated charges for methylimidazole and derived charges for histidine from several force fields are provided in the Supporting Information.
Histidine Titration in Solution
To illustrate the method we first examine the titration of an isolated histidine side-chain in aqueous solution. Titration curves for the individual sites and the overall titration behavior is shown in Figure 2. Here we assume equivalent sites with intrinsic pKas of 14 and 7 with interaction element W = 7 pK units. The energies of each protonation state are calculated from Eq. (1) as a function of pH, and the average protonation was calculated exactly from the Boltzmann distribution. There are four possible states specified by (nδ, nε) with energies:
E(1,0) = (pH−14)(1−1)+(pH−7)(0−0)+7(1−1)(0−0)=0
E(0,1) = (pH−14)(0−1)+(pH−7)(1−0)+7(0−1)(1−0)=14−7−7 = 0
E(1,1) = (pH−14)(1−1)+(pH−7)(1−0)+7(1−1)(1−0)=(pH−7)
E(0,0) = (pH−14)(0−1)+(pH−7)(0−0)+7(0−1)(0−0)=−(pH−14)
For case 1 we obtain the expected result since this is the reference state, and is defined as the zero of energy. Case 2 is also correct, since we have assumed the sites are equivalent. Case 3 corresponds to the energy required to protonate a group with pKa of 7, and case 4 corresponds to deprotonating a group with pKa of 14. From the above analysis, we see that only for case 2 does the interaction energy enter the expression for the total energy (i.e. when the Nδ-tautomer is converted to the Nε-tautomer).
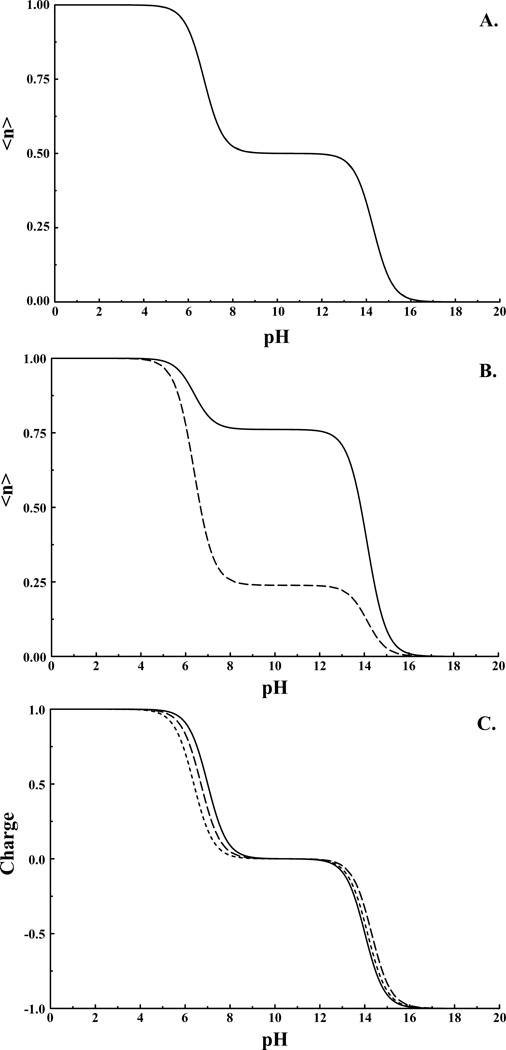
Figure 2.
Titration of histidine in solution with A) Nδ and Nε treated as equivalent with a model pKa of 7, and B) Nδ assigned model pKa of 7 and Nε assigned model pKa of 6.6 (Nδ site solid line and Nε site dashed line). Note in (A) curves are coincident. C) Overall titration behavior/charge for histidine models from (A) small dashed line, (B) large dashed line, and for two non-interacting sites with pKas of 7 and 14.
From Figure 2A & 2C we observe apparent pKas for the histidine side-chain of 6.7 and 14.3. The deviation from the expected values of 7 and 14 result from state splitting due to the interaction between states analogous to the splitting observed in coupled two level quantum systems. If we choose to differentiate the tautomers, say by assigning the Nε site an intrinsic pKa of 6.5, then cases 2 and 3 above would be adjusted to 0.5 and (pH-6.5) respectively, again as expected. Here we observe apparent pKas of 6.4 and 14.1 (Figure 2B & 2C). The splitting is less than in the previous example because the two states are no longer in resonance. The apparent pKas are very similar in both examples; however the titration behavior of the individual sites is quite different.
Histidine Titration in Proteins
Table I presents the RMSD deviation of the calculated and experimental apparent pKas (pK1/2) for a small set of eight test proteins. Table I includes comparisons between this work as well as the standard histidine treatments described in1 and3, as well as with the fully empirical approach implemented in the popular PROPKA program31. Double entries in Table I indicate the RMSD of all ionizable residues (first value) and for histidines only (second value). As can be seen from Table I there are no significant deviations from the current method and the standard approach. Furthermore the accuracy of pKas obtained from this method are comparable to those of PROPKA.
Table I.
RMSD for calculated and experimental pKas by various methods.
| Protein | PDB | RMSD (pKip) | RMSD (Standard) | RMSD (propka)c |
|---|---|---|---|---|
| Myoglobin | 1MBC | 1.66 (10), 1.20 (8) | 1.74 (10), 1.33 (8)a | / |
| HEWL | 2LZT | 0.60 (19), 0.10 (1) | 0.67 (19), 0.10 (1)a | 0.66 (19) |
| RNase A | 1RNZ | 0.94 (15), 1.27 (4) | / | 0.67 (14) |
| 3RN3 | 1.06 (16), 1.67 (4) | 1.40 (16), 1.94 (4)b | 0.94 (14) | |
| SNased | 3BDC | 1.11 (16), 0.99 (2) | / | 1.23 (16), 0.10 (2) |
| BI-VI | 1BI6 | 0.65 (22) | / | 0.68 (22) |
| RNase H | 2RN2 | 1.02 (23), 0.55 (5) | / | 0.72 (23) |
| BPTI | 4PTI | 0.46 (13) | 0.70 (10)b | 0.60 (14) |
| RNase T1 | 9RNT | 0.96 (14), 0.66 (3) | / | 1.51 (14) |
Pairs of RMSD values are for all sites and for histidines only, respectively. Values in parenthesis give the number of sites included in the calculation of the RMSD.
Myoglobin results from3, HEWL results calculated with MEAD using the “pseudo independent site” model for histidine.
Results from1 using a “single site” model of histidine.
propka results from31.
SNase Δ+PHS variant30.
HEWL contains only one histidine residue, HIS-15 (pK1/2 = 5.6)31, which is modeled equally well by both the current (pKa,calc. = 5.5) and “pseudo independent site” (pKa,calc. = 5.7) models. Figure 3 illustrates the titration behavior of HIS-15. Figure 3A illustrates the behavior when the Nδ and Nε sites are assigned the same pKa of 7 for the neutral to positive reaction. Although it is still possible for the protein environment to differentiate the two protonation sites of histidine through either the background term or through site-site interactions it is clear that both sites are nearly equally populated over the entire pH range studied. Figure 3B illustrates the titration behavior of HIS-15 when the Nδ and Nε sites are assigned model pKas of 7.0 and 6.6 respectively for the neutral to positive protonation reaction. The RMSD for all pKas is unchanged at 0.60, and the deviation for HIS-15 is 0.13 indicating no loss of accuracy. From Figure 3B it is clear that the Nδ and Nε sites have differentiated with the Nδ site being preferentially protonated. Finally, Figure 3C presents the titration behavior of HIS-15 as determined with the model of3. The curves in Figures 3B and 3C are quite similar with significant deviations arising only beyond a pH of 10 for the individual sites while the total titration behavior of HIS-15 is nearly identical in the two figures to above pH 12. Around pH 13 our model of histidine begins to deprotonate as we approach the second pKa.
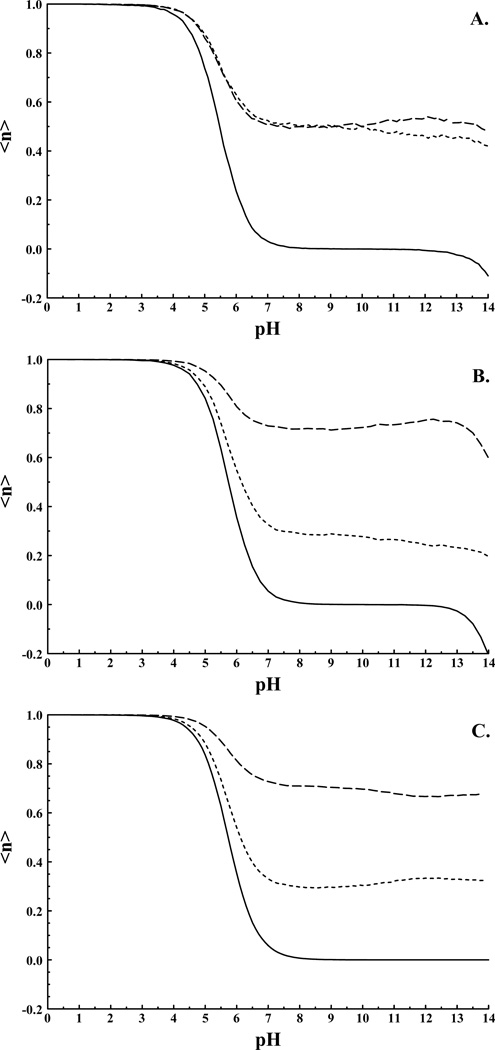
Figure 3.
HEWL HIS-15 titration curves. A) pKip result with and (Nδ and Nε equivalent). B) pKip result with and (Nδ and Nε non-equivalent). C) “pseudo independent site” result with and <n> is the average proton population of the indicated site; Large dashed line Nδ site, small dashed line Nε site. Solid curves represent the total titration behavior/charge of the histidine residue.
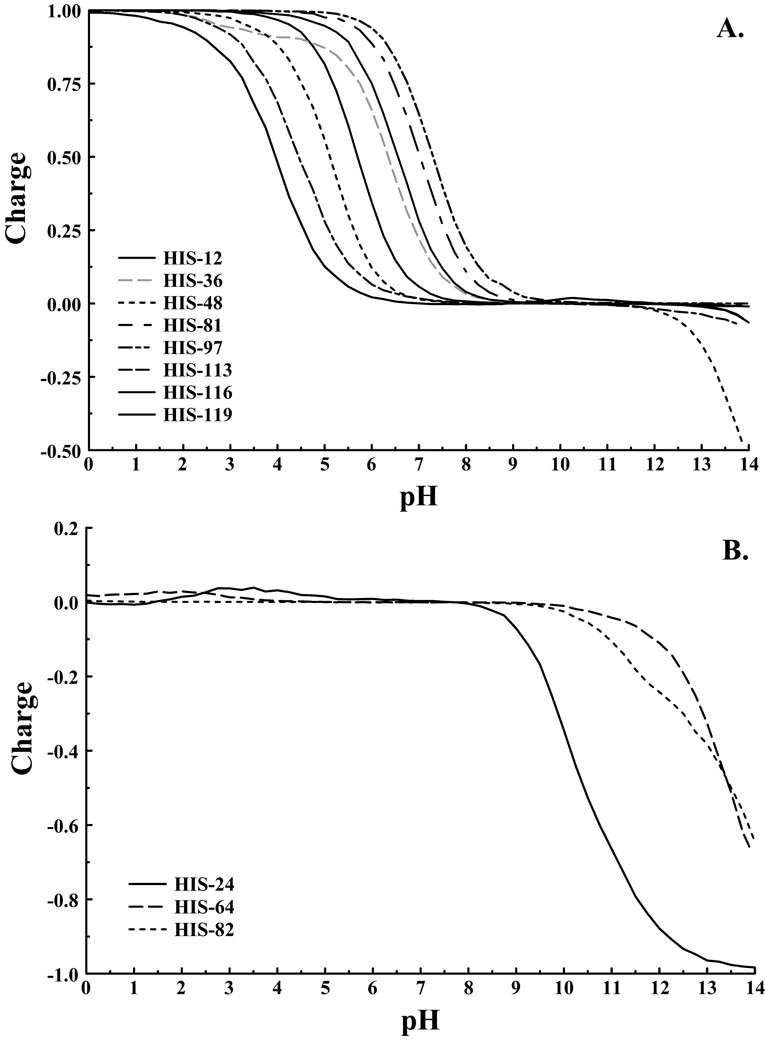
A perhaps better test case is that of myoglobin with eleven histidines exhibiting a range of behaviors. The RMSD values presented in Table I again show that the two representations of histidine both model the system with similar accuracy. The maximum deviation between experiment and the current model is 1.75 pK units for HIS-36, and is similar for both models. Figure 4 illustrates the titration behavior for the titrating and non-titrating histidines of myoglobin. Of the eight titrating histidines only HIS-48 exhibits any significant population of the fully deprotonated state. Again, deprotonation of the second proton is only observed above pH 12, well above biologically relevant pH values. For the remaining titrating histidines the second pKa is up-shifted, and even at pH 14 these residues remain primarily in the neutral charge state. Of the three non-titrating histidines our method predicts pKas below zero for the neutral to positive protonation reactions in agreement with previous calculations. Furthermore, these histidines were not observed to titrate above pH 4.7 experimentally3. Of these histidines HIS-24 is observed to deprotonate in the region between pH 9 and 13 while the remaining two histidines show apparent pKas near 14. Experiments on proteins are rarely able to exceed a pH of 9, so none of the histidines of myoglobin should be observed in the negative charge state experimentally.
Figure 4.
A) Titrating and B) Non-titrating histidines of myoglobin as determined by the pKip program.
Imidazolate Ion and Cu/Zn Superoxide Dismutase
Superoxide dismutase (SOD) facilitates the dismutation of superoxide ions into oxygen and peroxide alleviating oxidative stress due to the presence of the reactive oxygen species. Superoxide dismutase is a unique example with which to test the described approach since it contains a bridging histidine ligand which is one of the very few examples of the imidazolate ion in biology. In Cu/ZnSOD HIS-63 bridges the Cu and Zn ions in the active site. Upon reduction of the Cu center HIS-63 dissociates from Cu and exhibits an apparent pKa of 10.8 as determined from experiment41.
Six configurations out of thirty were chosen from the NMR solution structure of reduced Human Cu/ZnSOD32 to determine the pKa of HIS-63. These configurations were chosen to represent the minimum, maximum, average and ideal (as determined from density functional theory42) HIS-Cu distances. Since human Cu/ZnSOD is dimeric, a total of 12 pKa values were determined. pKas were calculated using Eqns. (8) and (9) in order to asses the significance of the desolvation term in Eqn. (8). Use of Eqn. (8), with the desolvation term fully accounted for, resulted in a pKa of 12.4 ± 1.1 while the pKa calculated from Eqn. (9) was determined to be 9.3 ± 0.8 with RMSD values of 1.9 and 1.7 respectively. It has been shown that the Born solvation model, and hence solvation energies determined from solutions of the PBE, drastically overestimate the energetic cost of burying charged groups in the low dielectric protein environment19,43,44. The above analysis indicates that the neglect of the desolvation term with respect to pK− introduces errors that are roughly equal to the inaccuracies associated with solvation energies determined from the PBE with a protein dielectric of 20. It is apparent that to obtain consistent and accurate results from continuum electrostatics calculations improved solvation models that go beyond linear response theory are required.
Complex I: The Redox Bohr Effect
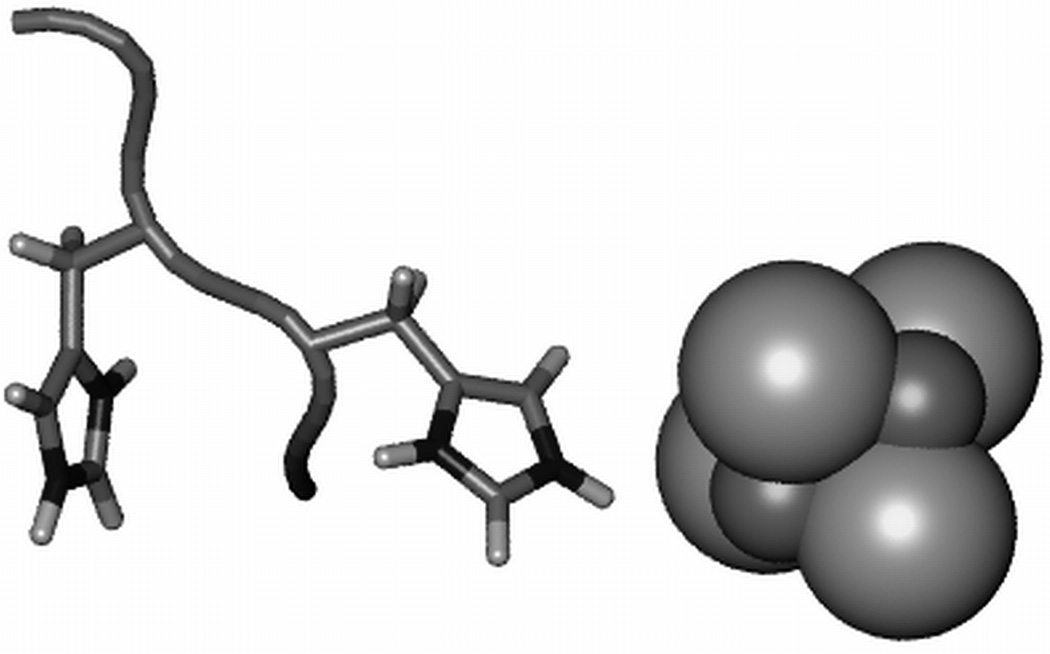
Respiratory complex I (NADH:Ubiquinone oxidoreductase) is a large (approximately 980kD in mitochondria and 550 kD in prokaryotes) membrane bound enzyme45–49. The hydrophilic domain of complex I, which protrudes into the mitochondrial matrix, contains 832 ionizable residues, 65 of which are histidines, bringing the total number of ionizable sites to 897, since Nsites = Nnon−HIS + 2NHIS. Respiratory complex I is an oxidoreductase which couples the oxidation of NADH and the reduction of quinone to proton translocation across the mitochondrial membrane. Complex I utilizes a chain of seven iron-sulfur (FeS) clusters housed in the hydrophilic domain to effect this reaction over a distance of approximately 100 Å33. Here we focus on the last FeS cluster of the redox chain, N2. It has been known for some time that N2 is a redox Bohr active group50. That is, the midpoint reduction potential of the cofactor varies with pH. It has been shown experimentally that the redox Bohr effect arises from the interaction of N2 with a nearby histidine, 4HIS-169 (Thermus numbering, 4HIS-226 in Y. lipolytica) of the 49 kDa subunit51. Figure 5 shows the configuration of FeS cluster N2, 4HIS-169, and the adjacent histidine 4HIS-170.
Figure 5.
The core of FeS cluster N2, CYS4[Fe4S4]2−/3−, shown as spheres and the adjacent histidines, 4HIS-169 (center) and 4HIS-170 (left), responsible for the redox Bohr effect.
The apparent midpoint reduction potential, , of FeS cluster N2 can be measured experimentally via EPR experiments, and is the redox analogue to the apparent pKa. is related to the shift in midpoint reduction potential, ΔEm, according to the relation,
| (11) |
where Em is the intrinsic midpoint reduction potential defined as the potential of the redox center when all other cofactors are oxidized52. Em can be defined according to a known model reduction potential if available. To calculate ΔEm we extend Eq. (1) to include redox centers in addition to protonatable sites53,54. The redox state of the enzyme is fixed during each calculation, and the linear terms over redox sites involving (the unknown self energies) Em can be dropped since they are constant. Thus the redox center, N2, only affects the titration calculations via the coulombic site-site interactions allowing us to calculate ΔEm from the average protonation state,
| (12) |
where and are the average protonation states of site I in the reduced and oxidized enzyme respectively. The subscript ν indicates the redox center.
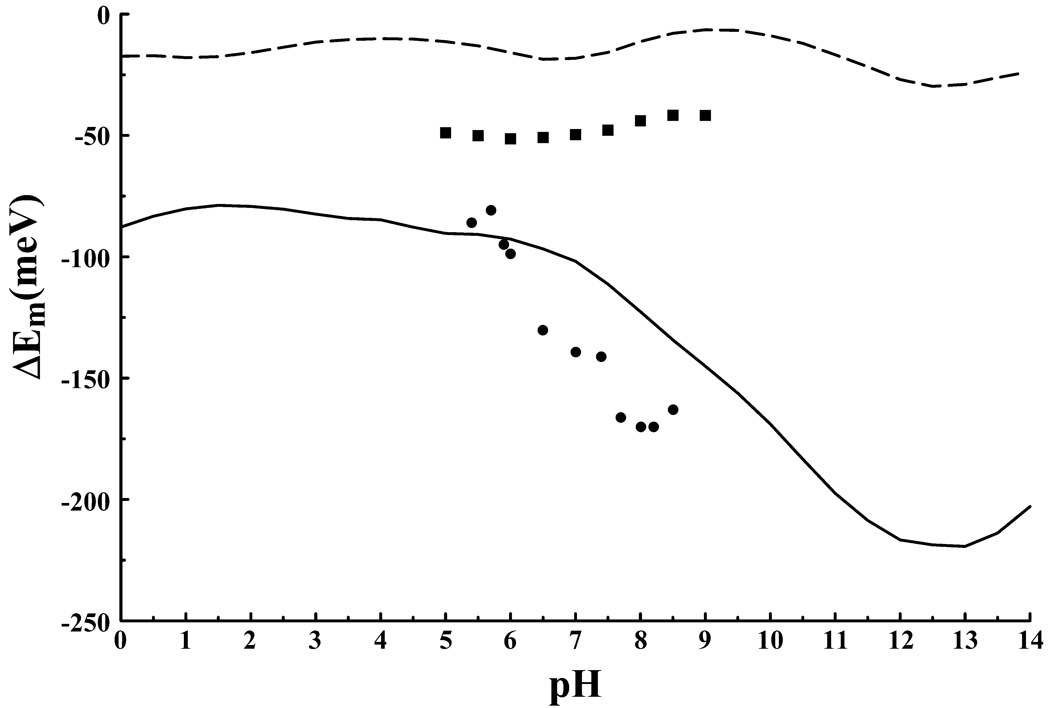
Figure 6 illustrates the results from the present calculations obtained using Eqn. (9). The solid line represents the calculated shift in midpoint reduction potential, ΔEm, resulting from the interaction of N2 with the protonation state of the enzyme. Solid circles in Figure 6 represent the experimentally observed apparent midpoint reduction potential, , of N2 from the Y. lipolytica derived enzyme51. It should be noted that the intrinsic midpoint reduction potential for the FeS cofactors of complex I varies among different species. Consequently complex I from T. thermophilus and from Y. lipolytica exhibit different values. Hence, it is coincidental that the calculated ΔEm and the measured values fall in the same range of potential. Inclusion of the intrinsic redox potential, Em, would simply shift the calculated ΔEm vertically. A precise value of for N2 from thermus is not known55,56. However, we are only interested in the variation in as a function of pH, and so a comparison between and ΔEm is sufficient. As can be seen from Figure 6 the calculated and measured variations in the reduction potential of N2 are quite similar. The calculated values are shifted up in pH by approximately 2 pH units compared to the measured values, but otherwise reproduce the effect. It has been shown via mutagenesis studies that substitution of the conserved residue 4HIS-169 by methionine eliminates the pH dependence of N2’s midpoint reduction potential in the pH range of 5 to 951. Similarly, when the protonation state of the Nδ site of 4HIS-169 is held fixed the calculated ΔEm becomes pH independent (Figure 6, solid squares). These results suggest that 4HIS-169 is the redox Bohr group associated with FeS cluster N2 in agreement with the experimental findings.
Figure 6.
The redox Bohr effect for FeS cluster N2 of complex I. The calculated shift in the midpoint reduction potential, ΔEm, of N2 calculated with pKip (solid line) and with the “pseudo independent site” model (large-dashed line). The pKip result when the protonation state of 4HIS-169 is fixed (solid squares) illustrates the loss of redox Bohr activity. Solid circles are the experimentally measured apparent midpoint reduction potentials for N2 from Y. Lipolytica21.
Of significant interest is the behavior of the adjacent histidine, 4HIS-170. Our calculations predict this residue to significantly populate the doubly deprotonated negative charge state in both the oxidized and reduced enzyme. While 4HIS-169 increases its protonation upon N2 reduction by 0.28 protons 4HIS-170 becomes completely deprotonated with an associated decrease in protonation of 0.14 protons at pH 7 indicating some level of proton transfer from 4HIS-170 to 4HIS-169.
To investigate the appearance of the unexpected negative charge state of 4HIS-170 calculations were performed with the model of3 where histidine can only populate the neutral or positive charge states. As can be seen from Figure 6 (large dashed line) this model fails to reproduce the redox Bohr effect. Furthermore fixing 4HIS-170 in a neutral protonation state also eliminates the effect (data not shown). When the protein dielectric is increased from 4 to 10 the negative charge state of histidine is no longer populated, however the redox Bohr effect is again abolished. Use of Eqn. (8), rather than Eqn. (9), produces a large up shift in the pKa of 4Hid-170 preventing deprotonation of the site and subsequently results in the observed pKa of 4HIS-169 increasing from ~9.5 to ~17.5. Redox Bohr activity is also lost.
The results are somewhat puzzling, but seem to indicate that the pKa shift of 4HIS-169 is being overestimated due to the inaccurate solvation model being used. Neglect of the desolvation term in Eqn. 8 results in an artificially lowered pKa for 4HIS-170 which in a fortuitous cancellation of errors reduces the pKa of the redox Bohr group, 4HIS-169. The fact that 4HIS-169 is not conserved among species indicates that its role in the observed effect is most likely an artifact of the present calculation. However, nearby and highly conserved 4TYR-87 which presumably suffers from the same overestimate in the desolvation contribution to the shift in pKa could serve the same function as 4HIS-170.
CONCLUSION
A convenient method to treat the two protonatable sites of histidine in protein electrostatics calculations has been presented and implemented in the program pKip. The method allows for the treatment of histidine’s protonatable sites as independent, and includes an accurate treatment of the negative charge state. We demonstrate the importance of the method in our calculations on the large protein respiratory complex I where the pH dependence of the midpoint reduction potential (redox Bohr effect) of the terminal FeS redox cofactor N2 is only reproduced when the negative charge state is included in the treatment. The primary drawbacks of the presented treatment of histidine lie within the formalism of the continuum electrostatics approach itself. The present study highlights the need for improved solvation models as suggested in43,44,57,58 and for a more consistent application of continuum electrostatics principles as suggested by Krystalik19,59.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by grants from NSF (PHY 0646273) and NIH (GM54052) to AAS.
Footnotes
Work was performed at the University of California, Davis.
REFERENCE
- 1.Antosiewicz J, Briggs JM, Elcock AH, Gilson MK, McCammon JA. Computing ionization states of proteins with a detailed charge model. J Comput Chem. 1996;17(14):1633–1644. [Google Scholar]
- 2.Antosiewicz J, McCammon JA, Gilson MK. Prediction of Ph-dependent Properties of Proteins. J Mol Biol. 1994;238(3):415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 3.Bashford D, Case DA, Dalvit C, Tennant L, Wright PE. Electrostatic Calculations of Side-Chain Pk(a) Values in Myoglobin and Comparison with NMR Data for Histidines. Biochemistry-Us. 1993;32(31):8045–8056. doi: 10.1021/bi00082a027. [DOI] [PubMed] [Google Scholar]
- 4.Bashford D, Gerwert K. Electrostatic Calculations of the pKa Values of Ionizable Groups in Bacteriorhodopsin. J Mol Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 5.Bashford D, Karplus M. Pkas of Ionizable Groups in Proteins - Atomic Detail from a Continuum Electrostatic Model. Biochemistry-Us. 1990;29(44):10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 6.Beroza P, Fredkin DR. Calculation of Amino Acid pK's in a Protein from a Continuum Electrostatic Model: Method and Sensitivity Analysis. J Comp Chem. 1996;17(10):1229–1244. [Google Scholar]
- 7.Gilson MK, Honig B. Calculation of the total electrostatic energy of a macromolecular system: Solvation energies, binding energies, and conformational analysis. Proteins. 1988;4(1):7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- 8.Gilson MK, Honig BH. Energetics of charge–charge interactions in proteins. Proteins. 1988;3(1):32–52. doi: 10.1002/prot.340030104. [DOI] [PubMed] [Google Scholar]
- 9.Warshel A, Sussman F, King G. Free energy of charges in solvated proteins: microscopic calculations using a reversible charging process. Biochemistry-Us. 1986;25(26):8368–8372. doi: 10.1021/bi00374a006. [DOI] [PubMed] [Google Scholar]
- 10.Sham YY, Muegge I, Warshel A. The effect of protein relaxation on charge-charge interactions and dielectric constants of proteins. Biophys J. 1998;74(4):1744–1753. doi: 10.1016/S0006-3495(98)77885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexov EG, Gunner MR. Incorporating protein conformational flexibility into the calculation of pH-dependent protein properties. Biophys J. 1997;72(5):2075–2093. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beroza P, Case DA. Including Side Chain Flexibility in Continuum Electrostatic Calculations of Protein Titration. J Phys Chem. 1996;100(51):20156–20163. [Google Scholar]
- 13.Georgescu RE, Alexov EG, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys J. 2002;83(4):1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieseritzky G, Knapp EW. Optimizing pKA computation in proteins with pH adapted conformations. Proteins. 2008;71(3):1335–1348. doi: 10.1002/prot.21820. [DOI] [PubMed] [Google Scholar]
- 15.Rabenstein B, Knapp E-W. Calculated pH-Dependent Population and Protonation of Carbon-Monoxy-Myoglobin Conformers. Biophys J. 2001;80(3):1141–1150. doi: 10.1016/S0006-3495(01)76091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabenstein B, Ullmann GM, Knapp EW. Calculation of protonation patterns in proteins with structural relaxation and molecular ensembles – application to the photosynthetic reaction center. Eur Biophys J. 1998;27(6):626–637. [Google Scholar]
- 17.You TJ, Bashford D. Conformation and hydrogen ion titration of proteins: a continuum electrostatic model with conformational flexibility. Biophys J. 1995;69(5):1721–1733. doi: 10.1016/S0006-3495(95)80042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunner MR, Alexov E. A pragmatic approach to structure based calculation of coupled proton and electron transfer in proteins. Bba-Bioenergetics. 2000;1458(1):63–87. doi: 10.1016/s0005-2728(00)00060-8. [DOI] [PubMed] [Google Scholar]
- 19.Krishtalik LI. Continuum electrostatics of proteins: Experimental test with model solvents and the method of the proteins pK calculations. Chem Phys. 2005;319(1–3):316–329. [Google Scholar]
- 20.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beroza P, Fredkin DR, Okamura MY, Feher G. Protonation of interacting residues in a protein by a Monte Carlo method: application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci USA. 1991;88(13):5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramanadham M, Sieker LC, Jensen LH. Refinement of triclinic lysozyme: II. The method of stereochemically restrained least squares. Acta Crystallogr B. 1990;46(Pt 1):63–69. doi: 10.1107/s0108768189009195. [DOI] [PubMed] [Google Scholar]
- 23.Howlin B, Moss DS, Harris GW. Segmented anisotropic refinement of bovine ribonuclease A by the application of the rigid-body TLS model. Acta Crystallogr A. 1989;45(Pt 12):851–861. doi: 10.1107/s0108767389009177. [DOI] [PubMed] [Google Scholar]
- 24.Fedorov AA, Joseph-McCarthy D, Fedorov E, Sirakova D, Graf I, Almo SC. Ionic Interactions in Crystalline Bovine Pancreatic Ribonuclease A†,‡. Biochemistry-Us. 1996;35(50):15962–15979. doi: 10.1021/bi961533g. [DOI] [PubMed] [Google Scholar]
- 25.Castaneda CA, Fitch CA, Majumdar A, Khangulov V, Schlessman JL, Garcia-Moreno BE. Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins. 2009;77(3):570–588. doi: 10.1002/prot.22470. [DOI] [PubMed] [Google Scholar]
- 26.Hatano K, Kojima M, Tanokura M, Takahashi K. Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean. Biochemistry-Us. 1996;35(17):5379–5384. doi: 10.1021/bi952754+. [DOI] [PubMed] [Google Scholar]
- 27.Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Nakamura H, Ikehara M, Matsuzaki T, Morikawa K. Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution. J Mol Biol. 1992;223(4):1029–1052. doi: 10.1016/0022-2836(92)90260-q. [DOI] [PubMed] [Google Scholar]
- 28.Marquart M, Walter J, Deisenhofer J, Bode W, Huber R. The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and Its Complexes with Inhibitors. Acta Crystallographica Section B-Structural Science. 1983 Aug;39:480–490. [Google Scholar]
- 29.Martinez-Oyanedel J, Choe HW, Heinemann U, Saenger W. Ribonuclease T1 with free recognition and catalytic site: crystal structure analysis at 1.5 A resolution. J Mol Biol. 1991;222(2):335–352. doi: 10.1016/0022-2836(91)90215-r. [DOI] [PubMed] [Google Scholar]
- 30.Harms MJ, Castaneda CA, Schlessman JL, Sue GR, Isom DG, Cannon BR, Garcia-Moreno EB. The pK(a) values of acidic and basic residues buried at the same internal location in a protein are governed by different factors. J Mol Biol. 2009;389(1):34–47. doi: 10.1016/j.jmb.2009.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61(4):704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 32.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. The solution structure of reduced dimeric copper zinc superoxide dismutase - The structural effects of dimerization. Eur J Biochem. 2002;269(7):1905–1915. doi: 10.1046/j.1432-1033.2002.02840.x. [DOI] [PubMed] [Google Scholar]
- 33.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311(5766):1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 34.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 35.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(suppl_2):W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32(suppl_2):W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA. Amber 7. San Fransisco, CA: University of California; 2002. [Google Scholar]
- 38.Xiang Z, Soto CS, Honig B. Evaluating conformational free energies: The colony energy and its application to the problem of loop prediction. Proc Natl Acad Sci USA. 2002;99(11):7432–7437. doi: 10.1073/pnas.102179699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bashford D. An Object-Oriented Programming Suite for Electrostatic Effects in Biological Molecules. Scientific Computing in Object-Oriented Parallel Environments. 1997;1343:233–240. [Google Scholar]
- 40.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery J, J A, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian03, Revision C.02. Wallingford CT: Gaussian, Inc.; 2004. [Google Scholar]
- 41.Azab HA, Banci L, Borsari M, Luchinat C, Sola M, Viezzoli MS. Redox Chemistry of Superoxide-Dismutase - Cyclic Voltammetry of Wild-Type Enzymes and Mutants on Functionally Relevant Residues. Inorg Chem. 1992;31(22):4649–4655. [Google Scholar]
- 42.Konecny R, Li J, Fisher CL, Dillet V, Bashford D, Noodleman L. CuZn superoxide dismutase geometry optimization, energetics, and redox potential calculations by density functional and electrostatic methods. Inorg Chem. 1999;38(5):940–950. doi: 10.1021/ic980730w. [DOI] [PubMed] [Google Scholar]
- 43.Gong HP, Hocky G, Freed KF. Influence of nonlinear electrostatics on transfer energies between liquid phases: Charge burial is far less expensive than Born model. Proc Natl Acad Sci USA. 2008;105(32):11146–11151. doi: 10.1073/pnas.0804506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koehl P, Orland H, Delarue M. Computing Ion Solvation Free Energies Using the Dipolar Poisson Model. J Phys Chem B. 2009;113(17):5694–5697. doi: 10.1021/jp9010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465(7297):441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi T, Stuchebrukhov AA. Electron tunneling in respiratory complex I. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1009181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329(5990):448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 48.Hirst J. Towards the molecular mechanism of respiratory complex I. Biochem J. 2010;425(2):327–339. doi: 10.1042/BJ20091382. [DOI] [PubMed] [Google Scholar]
- 49.Zickermann V, Kerscher S, Zwicker K, Tocilescu MA, Radermacher M, Brandt U. Architecture of complex I and its implications for electron transfer and proton pumping. Bba-Bioenergetics. 2009;1787(6):574–583. doi: 10.1016/j.bbabio.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingledew WJ, Ohnishi T. An analysis of some thermodynamic properties of iron-sulphur centres in site I of mitochondria. Biochem J. 1980;186(1):111–117. doi: 10.1042/bj1860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwicker K, Galkin A, Drose S, Grgic L, Kerscher S, Brandt U. The Redox-Bohr group associated with iron-sulfur cluster N2 of complex I. J Biol Chem. 2006;281(32):23013–23017. doi: 10.1074/jbc.M603442200. [DOI] [PubMed] [Google Scholar]
- 52.Couch VA, Medvedev ES, Stuchebrukhov AA. Electrostatics of the FeS clusters in respiratory complex I. Biochimica Et Biophysica Acta-Bioenergetics. 2009;1787(10):1266–1271. doi: 10.1016/j.bbabio.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ullmann GM. The Coupling of Protonation and Reduction in Proteins with Multiple Redox Centers: Theory, Computational Method, and Application to Cytochrome c3. J Phys Chem B. 2000;104(26):6293–6301. [Google Scholar]
- 54.Ullmann GM, Knapp E-W. Electrostatic models for computing protonation and redox equilibria in proteins. Eur Biophys J. 1999;28(7):533–551. doi: 10.1007/s002490050236. [DOI] [PubMed] [Google Scholar]
- 55.Euro L, Bloch DA, Wikström M, Verkhovsky MI, Verkhovskaya M. Electrostatic Interactions Between FeS Clusters in NADH:Ubiquinone Oxidoreductase (Complex I) from Escherichia coli. Biochemistry-Us. 2008;47:3185–3193. doi: 10.1021/bi702063t. [DOI] [PubMed] [Google Scholar]
- 56.Medvedev ES, Couch VA, Stuchebrukhov AA. Determination of the intrinsic redox potentials of FeS centers of respiratory complex I from experimental titration curves. Bba-Bioenergetics. 2010;1797(9):1665–1671. doi: 10.1016/j.bbabio.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Florian J, Warshel A. Langevin dipoles model for ab initio calculations of chemical processes in solution: Parametrization and application to phosphate ester hydrolysis and conformational analysis in aqueous solution. Abstracts of Papers of the American Chemical Society. 1997;214:85-Phys. [Google Scholar]
- 58.Sham YY, Chu ZT, Warshel A. Consistent calculations of pK(a)'s of ionizable residues in proteins: Semi-microscopic and microscopic approaches. J Phys Chem B. 1997;101(22):4458–4472. [Google Scholar]
- 59.Krishtalik LI, Kuznetsov AM, Mertz EL. Electrostatics of proteins: Description in terms of two dielectric constants simultaneously. Proteins-Structure Function and Genetics. 1997;28(2):174–182. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.