Abstract
Aeromonas salmonicida subsp. salmonicida is an important pathogen in salmonid aquaculture and is responsible for the typical furunculosis. The type-three secretion system (T3SS) is a major virulence system. In this work, we review structure and function of this highly sophisticated nanosyringe in A. salmonicida. Based on the literature as well as personal experimental observations, we document the genetic (re)organization, expression regulation, anatomy, putative functional origin and roles in the infectious process of this T3SS. We propose a model of pathogenesis where A. salmonicida induces a temporary immunosuppression state in fish in order to acquire free access to host tissues. Finally, we highlight putative important therapeutic and vaccine strategies to prevent furunculosis of salmonid fish.
Introduction
The disease furunculosis caused by Aeromonas salmonicida subsp. salmonicida (hereafter referred to as A. salmonicida) continues to be a major health problem for the growing salmonid aquaculture worldwide. The disease, known for over a century, results in significant economic losses and promotes the intensive use of antibiotics. In spite of effective commercialized oil-adjuvanted vaccines containing A. salmonicida bacterins, frequent outbreaks persist at fish farms. Different virulence factors of A. salmonicida have been described but they failed to explain the virulence phenotype of the pathogen (Ellis et al., 1988; Vipond et al., 1998). Ten years ago, our laboratory published the first descriptions of the type-three secretion system (T3SS) in the genus Aeromonas and demonstrated its role as a main virulence secretion system of A. salmonicida (Burr et al., 2002; 2003b; Stuber et al., 2003). Here, we provide the first review on this highly sophisticated nanosyringe and its putative role in a model of pathogenesis for typical furunculosis.
Genetic organization of the A. salmonicida T3SS
The structural components of A. salmonicida T3SS are encoded on a large conjugative plasmid of 140–155 kb (Stuber et al., 2003; Reith et al., 2008) by numerous genes arranged in five predicted polycistronic operons (exsADascBCDEFGHIJKL, exsCEB, aopNacr12ascXYVacrRGVHaopBD, ascNOPQRSTU and aopXsycX) (Fig. S1). The plasmid pASA5 of 155 kb has been entirely sequenced in the reference strain A. salmonicida A449. The structural genes are flanked on both sides by genes encoding for T3SS effectors and their chaperones (ati1ati2, aopHsycH, aopOsycO and putatively asa_P5G088) interspaced by various kinds of insertion elements (IS256, IS630, IS116) (Fig. S1) (Reith et al., 2008). The role of these insertion elements (ISs) in the genetic rearrangements of the T3SS loci is crucial. For example, T3SS cluster and other part of the pASA5 plasmid are lost when A. salmonicida is cultivated in stressful conditions, and it has been demonstrated that this deletion process is due to the homologous recombination between IS256 copies which are present at the extremities of the T3SS cluster (Daher et al., 2011; Tanaka et al., 2012; 2013) (Fig. S1). More complex rearrangements involving other ISs are also suspected. IS630 copies might also play a role in the genetic organization of the injectisome because they are frequently associated with T3SS loci in A. salmonicida (Studer et al., 2013) (Fig. S1) but also in other pathogenic bacteria using such secretion systems (Haneda et al., 2001; Correa et al., 2012; Stavrinides et al., 2012). The cluster of T3SS genes can also be integrated in the chromosome of some strains of certain species of Aeromonas. Such an insertion is observed in A. hydrophila strain SSU, A. veronii strain AER39 and A. diversa 2478–85 (Fig. S1, Table S1). The A. diversa 2478–85 strain is currently the only Aeromonas strain showing a second T3SS-2 similar to the one of Edwardsiella tarda (Wang et al., 2010), directly adjacent to the T3SS shared by several Aeromonas species (Fig. S1, Table S1) and associated to other T3SS virulence effectors such as ExoU described in Pseudomonas aeruginosa (Shaver and Hauser, 2004) (Fig. S1, Table S1). This observation demonstrates that T3SS gene clusters from other bacterial genera can be integrated in the Aeromonas genome.
The aopH gene has also been shown to be encoded together with its chaperone on pASA6, a smaller plasmid of 18.5 kb, which has a high identity with parts of pASA5 suggesting that pASA6 is a derivative of this latter plasmid. The effector gene aopP has been detected on pAsal1, a small plasmid of 6.4 kb (Fig. S1) which shows some identity with pASA3 (aopP negative), another small plasmids. The pAsal1 plasmid seems also to be sensitive to stressful growth conditions (Tanaka et al., 2012). Another large conjugative plasmid of 167 kb is pASA4 which is homologous to plasmid pRA1 and pRA3 of A. hydrophila and contains genes associated with resistance to antibiotics and numerous undetermined coding sequences but no known coding sequence for T3SS elements. The genes of the adenosin diphosphate (ADP) ribosylating toxin AexT and AopS (ASA_0010, homologous to VopS of Vibrio parahaemolyticus), both T3SS effectors, and their chaperones are to date the only ones detected on the chromosome of A. salmonicida A449 (Fig. S1). It has, however, to be noted that aopS is predicted to be a pseudogene because of an in-frame stop codon (Reith et al., 2008). An intact copy of aopS and the gene for its chaperone are present at the same position in the chromosome of A. salmonicida subsp. achromogenes strain AS03 (Table S1). The prevalence of intact aopS in other A. salmonicida strains is not known. Interestingly, among the different published genomes of Aeromonas sp. published, aexT and its chaperone (sycE) are always localized on the same genomic island present between glyS (asa_4261) and the transcriptional regulator asa_4278 (Table S1) which shows variability between strains from different Aeromonas species. Aeromonas salmonicida A449 and 01-B526, A. hydrophila SSU and A. veronii AER39 detain the aexTsycE cluster at this position whereas A. hydrophila ATCC7966; A. caviae Ae398; A. veronii B565, AMC34, AMC35 and AER397; and A. aquariorum AAK1 do not contain these loci (Table S1). The presence of aexTsycE at the same position in the genome of different Aeromonas species likely suggests that they would be inherited from an ancestor and lost in some strains or that they are integrated by a process specifically targeting this site.
Therefore, while the entire genetic pattern of A. salmonicida subsp. salmonicida strains is well conserved worldwide and over time (Studer et al., 2013), the cloud of virulence genes associated with the T3SS is constantly submitted to local genetic deletions in specific constraints and genetic additions through the exchanges by horizontal transfer of genes with other environmental bacteria (Burr and Frey, 2007). These processes of gene deletions might be one of the reasons for the progressive loss of virulence observed with A. salmonicida laboratory strains that are intensively cultivated under conditions that, without the selection pressure they are subjected to in the host, are unsuitable to preserve virulence. A typical example is the type strain of A. salmonicida ATCC 33658T that has lost all T3SS structural genes and is non-virulent (Burr and Frey, 2009). These genetic rearrangements highlight why it is a real challenge to work with A. salmonicida to obtain relevant data on the pathogenesis. Unless strict conditions of culture are respected, it is possible for genetic modifications to occur in the time-lapse between the isolation of the bacterium and its use for genetic characterization (molecular epidemiology) and molecular manipulations (site-directed mutagenesis). Furthermore, molecular epidemiologic studies are of concern as they take into account the presence or absence of only few T3SS genes to draw conclusions on the virulence of A. salmonicida strains. Because of the numerous mutations and genetic rearrangements affecting the A. salmonicida T3SS, epidemiological studies should include the analysis of the integrity of several genes for T3SS structural components (at least ascV and ascC) and the genes of the main effectors (aopH, aexT, ati2, aopO, aopP and aopS) (Burr and Frey, 2007; Daher et al., 2011) in order to draw conclusions regarding the presence of an intact T3SS with functional effectors.
Regulation of the A. salmonicida T3SS expression
The transcription of the T3SS genes are induced under Ca2+-limiting conditions (Burr et al., 2003a) and during the contact of A. salmonicida with the host cell (Braun et al., 2002). The transcription of the A. salmonicida T3SS genes is predicted to be controlled by a regulatory pathway similar to that observed for the T3SS of P. aeruginosa (Brutinel and Yahr, 2008; Brutinel et al., 2009). The mechanism involves four interacting regulatory proteins (ExsA, ExsD, ExsC and ExsE) (Fig. 1). ExsA is a positive activator of T3SS transcription, whereas ExsD is an anti-activator that forms a 1:1 complex with ExsA. Under non-permissive conditions for T3SS gene expression, ExsA-dependent transcription is expected to be inhibited by the formation of inhibitory ExsD-ExsA (1:1) and ExsC-ExsE (2:1) complexes. Permissive conditions would activate the T3SS secretory activity and induce the secretion and translocation of ExsE. The decrease of intracellular concentration of ExsE seems to promote the formation of ExsD-ExsC (2:2) complexes inducing the release of ExsA and the activation of T3SS gene expression. In V. parahaemolyticus, a histone-like protein (H-NS) represses T3SS gene expression by suppressing exsA gene expression (Kodama et al., 2010). This second level of transcriptional control is assumed in A. salmonicida given that several H-NS homologues are present in the chromosome and the pASA5 plasmid (ASA_P5G002 and ASA_P5G018). Another level of A. salmonicida T3SS expression control is performed by the quorum sensing (QS) pathways encoded at least by the genes asaIR and luxS, luxU, luxO (Reith et al., 2008) which produce high homoserinelactone (HSL) concentration in the extracellular medium at high cell density (Swift et al., 1997). Like for Vibrio, in growing bacterial cultures, AsaR is activated by the binding of HSL and could directly repress the expression of exsA and subsequently the expression of the T3SS (Waters et al., 2010). However, expression of A. salmonicida proteases and lipases is QS-induced at high cell density and is important to induce the virulence (Rasch et al., 2007; Schwenteit et al., 2011). These assumptions are supported by our proteomic analysis of the wild-type (wt) JF5054 strain of A. salmonicida (virulent reference strain) showing that the amount of T3SS decreases significantly from the exponential to the stationary phase of growth whereas the expression of some enzymes (proteases, lipases, chitinases, etc) increases. These observations also highlight that the T3SS activation state in the bacterial culture is a critical point to take into account for the preparation of the inoculum for experimental challenges with A. salmonicida besides the genetic variations and losses of virulence attributes. Bacterial preparations from late phases of growth with a downregulated T3SS appear to be associated with low mortality despite the genotypic presence of all the T3SS genes.
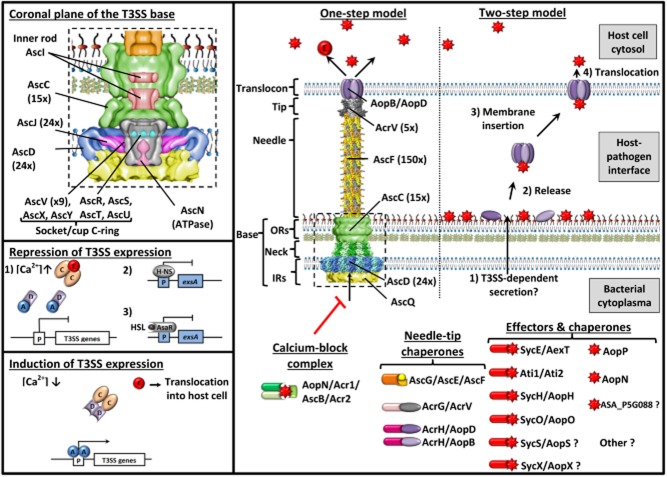
Figure 1.
A. salmonicida T3SS transcription regulation, anatomy and models of effector translocation into host cell.
Negative (ExsABDCE, Hns and quorum sensing) and positive (ExsABCDE) pathways regulating the transcriptional expression of the A. salmonicida T3SS are depicted in the two boxes at the bottom left. The structural components, chaperones, effectors and secretion regulators of the A. salmonicida T3SS and the two described models of effector translocation into fish cell are represented in the main box. The upper left box shows the structural components present inside the T3SS base.
Under non-permissive conditions (for example with high extracellular calcium concentrations), the transcription of T3SS genes induces by the positive activator ExsA is expected to be inhibited by the formation of inhibitory ExsD-ExsA (1:1) and ExsC-ExsE (2:1) complexes (Brutinel and Yahr, 2008; Brutinel et al., 2009). Permissive conditions would activate the T3SS secretory activity and induce the secretion and translocation of ExsE. Histone-like proteins (H-NS) could repress T3SS gene expression by suppressing exsA gene expression like for V. parahaemolyticus (Kodama et al., 2010). In growing bacterial cultures, AsaR from the quorum sensing is activated by the binding of homoserinelactone (HSL) and could directly repress the expression of exsA and subsequently the expression of the T3SS like for Vibrio (Waters et al., 2010).
The T3SS is a highly sophisticated nanosyringe which is mainly composed of Asc (Aeromonas secretion) proteins, the building blocks of the injectisome, and Aops (Aeromonas outer proteins) which are released outside of the bacterium (Hodgkinson et al., 2009; Diepold et al., 2011; 2012; Abrusci et al., 2013; Dewoody et al., 2013). In other bacterial species, the number of subunits in certain parts of the T3SS is known and is indicated in brackets. In the bacterial cytoplasma are represented the calcium-block complex which regulates the T3SS secretion and chaperones of the needle, the tip and effectors. Currently, two models of translocation into host cell (one-step vs two-step) are under debates (Perrett and Zhou, 2011).
Morphology of the A. salmonicida T3SS
Currently, no structural studies have specifically been carried out on the T3SS of A. salmonicida but this secretion apparatus shows conservation with the well-described T3SS of Yersinia and Salmonella (Table 1). Hence, the following information about the morphology and the functioning of the A. salmonicida T3SS is inferred from structural homologues reviewed in reference articles (Hodgkinson et al., 2009; Diepold et al., 2011; 2012; Abrusci et al., 2013; Dewoody et al., 2013). Proteins orthologue to all of the A. salmonicida T3SS components are detailed in Table 1. The T3SS structural proteins of Photorhabdus luminescens, Yersinia pestis, V. parahaemolyticus, P. aeruginosa and Photobacterium damselae present the highest conservation with the Aeromonas T3SS and constitute the ‘Ysc’ family of T3SS (Barret et al., 2013) (Fig. S1). The T3SS is a highly sophisticated nanosyringe that is mainly composed of Asc (Aeromonas secretion) proteins, the building blocks of the injectisome, and Aops (Aeromonas outer proteins) which are released outside of the bacterium (Fig. 1). Interestingly, the degree of conservation of T3SS subunits between these bacteria is the highest for the membrane components and the lowest for the translocation complex suggesting that a more important evolution of the external parts of the T3SS may be related to host adaptation (Fig. 2). Some of the T3SS effectors are present in multiple genera with a low similarity while others (for example Ati2) are shared by some genera with a high similarity suggesting a recent horizontal gene transfer (Fig. 2 and Fig. S1).
Table 1.
Proteins orthologue to A. salmonicida T3SS components
| T3SS part | Information | Ysc Family | Ssa/Esc Family | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Aeromonas | Photorhabdus | Yersinia | Vibrio | Pseudomonas | Bordetella | Salmonella | Shigella | ||
| Translocon | Pore | AopB | LopB | YopB | VopB | PopB | BopB | SipB | IpaB |
| Pore | AopD | LopD | YopD | VopD | PopD | BopD | SipC | IpaC | |
| Chaperone | AcrH | LssH | SycD/LcrH | VcrH | PcrH | BcrH1 | SicA | LpgC | |
| Needle | Tip | AcrV | LssV | LcrV | VcrV | PcrV | Bsp22 | SipD | IpaD |
| Chaperone | AcrG | LssG | LcrG | VcrG | PcrG | – | – | – | |
| Body | AscF | LscF | YscF | VscF | PscF | BscF | PrgI | MxiH | |
| Chaperone | AscG | LscG | YscG | VscG | PscG | – | – | – | |
| Chaperone | AscE | LscE | YscE | VscE | PscE | BscE | – | – | |
| Chaperone | AcrR | LssR | LcrR | VcrR | PcrR | – | – | – | |
| Polymerization control | AscH | LscH | YscH/YopR | VscH | PscH | – | – | – | |
| Length control | AscP | LscP | YscP | VscP | PscP | BscP | InvJ | Spa32 | |
| Base | OM ring | AscC | LscC | YscC | VscC | PscC | BscC | InvG | MxiD |
| Chaperone | AscW/ExsB | LscW | YscW | VscW | ExsB | – | – | – | |
| Inner rod | AscI | LscI | YscI | VscI | PscI | BscI | PrgJ | MxiI | |
| IM ring ext | AscD | LscD | YscD | VscD | PscD | BscD | PrgH | MxiG | |
| IM ring int | AscJ | LscJ | YscJ | VscJ | PscJ | BscJ | PrgK | MxiJ | |
| C-ring, socket/Cup | AscR | LscR | YscR | VscR | PscR | BscR | SpaP | Spa24 | |
| C-ring, socket/Cup | AscS | LscS | YscS | VscS | PscS | BscS | SpaQ | Spa9 | |
| C-ring, socket/Cup | AscT | LscT | YscT | VscT | PscT | – | SpaR | Spa29 | |
| C-ring, socket/Cup | AscU | LscU | YscU | VscU | PscU | BscU | SpaS | Spa40 | |
| C-ring, socket/Cup | AscV | LssD | YscV | VcrD | PcrD | BcrD | InvA | MxiA | |
| C-ring, secretion specificity | AscX | LssB | YscX | VscX | PscX/Pcr3 | – | – | – | |
| C-ring, secretion specificity | AscY | LssC | YscY | VscY | PscY/Pcr4 | Bcr4 | – | – | |
| ATPase | AscN | LscN | YscN | VscN | PscN | BscN | InvC | Spa47 | |
| Cytoplasmic | AscK | LscK | YscK | VscK | PscK | BscK | OrgA | MxiK | |
| Cytoplasmic | AscL | LscL | YscL | VscL | PscL | BscL | OrgB | MxiN | |
| Cytoplasmic | AscQ | LscQ | YscQ | VscQ | PscQ | BscQ | SpaO | Spa33 | |
| Regulators | Secretion | AopN | LopN | YopN | VopN | PopN | BopN | InvE | MxiC |
| Secretion | Acr1 | LssA | TyeA | Vcr1 | Pcr1 | – | – | – | |
| Secretion | AscB | LscB | YscB | VscB | PscB | – | – | – | |
| Secretion | Acr2 | LssN | SycN | Vcr2 | Pcr2 | – | – | – | |
| Substrate recycling? | AscO | LscO | YscO | VscO | PscO | BscO | InvI | Spa13 | |
| T3SS transcription | ExsA/AscA | LscA | LcrF | ExsA | ExsA | – | – | – | |
| T3SS transcription | ExsC | LscY | – | ExsC | ExsC | – | – | – | |
| T3SS transcription | ExsD | LscZ | – | ExsD | ExsD | – | – | – | |
| T3SS transcription | ExsE | ExsE | – | ExsE | ExsE | – | – | – | |
| Effectors | ADP-ribosylase + GAP | AexT | – | YopE | VopT | ExoT+ExoS | – | – | – |
| Chaperone | SycE | Plu3789 | SycE/YerA | – | CesT+CesT | – | – | – | |
| Phosphotyrosine phosphatase | AopH | – | YopH | – | – | – | – | – | |
| Chaperone | SycH | – | SycH | – | – | – | – | – | |
| Inositol Phosphatase | Ati2 | plu4615 | – | VPA0450 | – | – | – | – | |
| Chaperone | Ati1 | Plu4614 | – | VPA0451 | – | – | – | – | |
| Serine/threonine kinase | AopO | – | YopO/YpkA | – | – | – | – | – | |
| Chaperone | SycO | – | SycO | – | – | – | – | – | |
| NF-κB inhibition + apoptosis | AopP | – | YopJ/YopP | VopA/P | – | – | AvrA | – | |
| Lipid rafts? | AopX | Plu4750 | – | VopR | – | BteA | – | – | |
| Chaperone | SycX | – | – | – | – | – | – | – | |
| AMPylation | AopS | – | – | VopS/VepB | – | – | – | – | |
| Chaperone | SycS | – | – | VPA0451 | – | – | – | – | |
| Hydrolase | ASA_P5G088 ? | – | – | VP1677, −1678 | – | – | – | – | |
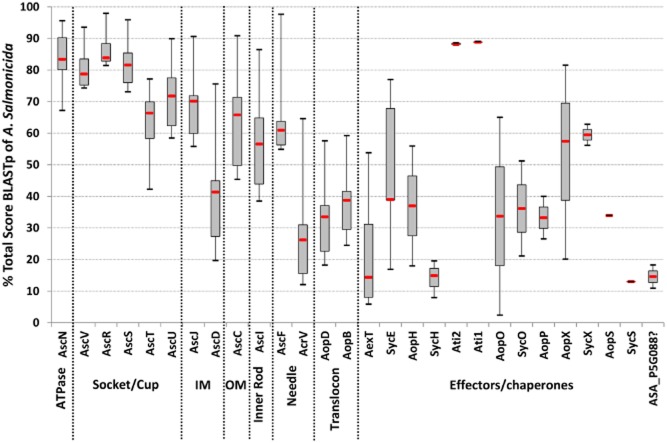
Figure 2.
T3SS homology in the Ysc family.
The diagram shows the conservation variability of T3SS structural components of Photorhabdus luminescens, Yersinia pestis, Vibrio parahaemolyticus, Pseudomonas aeruginosa and Photobacterium damselae in comparison to A. salmonicida subsp. salmonicida. For each cited T3SS component, the variability (BLASTp total score) of orthologues is represented in percentage of the A. salmonicida BLASTp total score. The homology decreases from the inner to the outer parts of the T3SS and is the lower for the translocon.
The base of the T3SS (Fig. 1) consists of a series of rings which spans the bacterial membranes and the periplasmic space (AscD and AscJ form inner membrane rings and AscC constitutes the outer membrane ring). At the centre of the inner membrane rings, a socket/cup domain (AscR, AscS, AscT, AscU and AscV) is present and is the technical platform for the secretion of T3SS components. Numerous cytoplasmic components (AscK, AscL and AscQ) are recruited to this site to orchestrate an active and orderly secretion of various T3SS substrates by an ATPase (AscN). Inside the outer membrane ring, an inner rod (AscI) connects the socket/cup to the distal part of the needle. The needle (AscF) and its tip (AcrV) make the bridge between the bacterium and the host cell, and connect the translocon (AopB/AopD) which is inserted into the host cell membrane and activates the translocation of T3SS effectors into the eukaryotic cytosol according to the ‘one-step’ model of secretion (Perrett and Zhou, 2011) (Fig. 1).
Using hybrid T3SS in Y. enterocolitica by exchanging the gene lcrV encoding the Yersinia T3SS tip structure with acrV, the analogous gene of A. salmonicida, Broz and colleagues (2007) proved the functionality of AcrV as T3SS tip structure representing the base protein of the tip. In that experiment, tip complexes formed by AcrV were larger and were diverse in shape. Using hybrid LcrV/AcrV tip structures, the authors concluded that the N-terminal domain of LcrV and AcrV form the base of the tip complex while the central globular domains of the proteins form the head of the tips.
Beside these structural elements and the effectors, the T3SS possesses numerous regulatory proteins which play different roles (Fig. 1). Chaperones stabilize T3SS proteins in the bacterial cytosol and deliver them to the ATPase AscN for the active secretion (AcrH for AopB/AopD, AcrG for AcrV, AscG/AscE/AcrR for AscF, AscW for AscC and SycE/SycH/Ati1/SycO/SycS for associated effectors). Other regulatory proteins control the polymerization (AscH) and the length (AscP) of the needle, the opening of the conduit upon host cell contact (the calcium-block complex AopN/Acr1/AscB/Acr2) and the specificity of the secretion (AscX and AscY).
The T3SS is the main virulence system of A. salmonicida
To date, several virulence factors have been characterized for A. salmonicida but the T3SS is currently the only one recognized as having a major effect on virulence. This was shown by independent studies with isogenic mutant strains for T3SS structural proteins which proved to be non-virulent both in vitro and in vivo (Burr et al., 2002; 2003b; 2005; Dacanay et al., 2006; Froquet et al., 2007; Daher et al., 2011). These results are exemplified by current results showing that the intraperitoneal (i.p.) injection of 500 cfu per fish of the fully virulent wt JF5054 strain (the JF2267 strain which was freshly reisolated from an experimentally infected dead fish) induces 70–80% of mortality whereas the isogenic T3SS-deficient mutant derivative (JF2747) deleted for ascV (protein of the socket/cup in the inner membrane) is considered to have extremely low virulence because i.p. injection of 105 cfu/fish induced no mortality (Burr et al., 2005). Furthermore, 108cfu/fish (200 000 times more than the inoculation with the wt strain), a drastic dose that does not reflect a natural infection, merely induced a weak mortality of only 20% (this work).
This phenotypic difference between the wt and the ΔascV mutant strains led us to compare the exoproteome of these bacteria by high-throughput proteomics (P. Vanden Bergh, unpublished) and established the full repertoire of in vitro T3SS effectors that are excreted in the supernatant (SN) by the wt strain of A. salmonicida. The other virulence factors described that are not part of the T3SS secretome (VapA, AerA, AerB, GCAT, Pla1, PlaC, TagA, Ahe2, GbpA and enolase) are expressed to a higher extent in the avirulent ΔascV mutant than in the wt strain and hence seem to play a secondary role in the pathogenesis.
The T3SS arsenal of A. salmonicida is mainly composed by AexT, AopH, Ati2, AopP, AopO, AopN and ExsE. AopS and AopX were not identified in our analysis but are potential T3SS effectors that could be expressed by A. salmonicida strains possessing functional copy of their genes. The occurrence of deletions in the gene of aopS and aopX effector genes suggests that A. salmonicida undergoes a pressure of selection to lose their function. Only the effect of AexT, AopP and Ati2 has been characterized in the cytotoxicity induced by A. salmonicida (Fig. 3). However, the other effectors show similarity with T3SS effectors studied in other pathogenic bacteria (Dean, 2011) and their function can thus be predicted (Fig. 3).
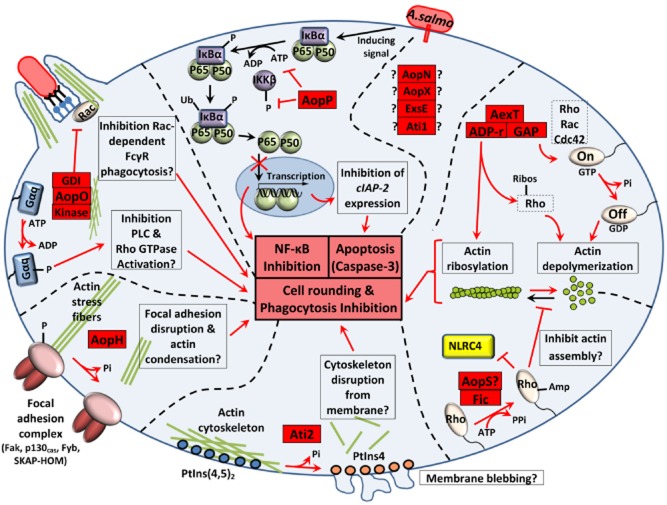
Figure 3.
Main intracellular effects of A. salmonicida T3SS effectors.
The intracellular effect of AexT, Ati2 and AopP has been characterized by previous studies while the impact of AopH, AopO and AopS is predicted from cytotoxic homologues from other bacterial species. AopN, AopX, ExsE, and Ati1 might also be translocated into the host cell but their role in virulence is not known. AopS and AopX are encoded by pseudogenes in the reference A449 strain. AexT, AopS, Ati2, AopH, and AopO would exert different negative effects on cytoskeletal dynamics. T3SS effectors are represented by red boxes with their known or predicted functional domains: ADP-ribosylating (ADP-r) and GTPase-activating (GAP) domains of AexT, filamentation induced by cAMP (Fic) domain of AopS, and the guanine-nucleotide dissociation inhibitor (GDI)-like domain of AopO. cIAP-2: cellular inhibitor of apoptosis 2; IκBα: inhibitor of the nuclear transcription factor NF-κB; IKKβ: IκB kinase β; FcγR: Fc-gamma receptor; Gαq: the heterotrimeric G protein subunit Gαq; NLRC4: Nod Like Receptor family CARD domain-containing protein 4, PLC: phospholipase C; PtIns: phosphatidylinositol; Rho, Rac, and Cdc42: GTPases of the Rho family.
The bifunctional toxin AexT possesses a GTPase-activating domain acting on small monomeric GTPases of the Rho family (Rho, Rac and Cdc42) and an ADP-ribosylating domain, which ADP-ribosylates both muscular and non-muscular actin (Braun et al., 2002; Burr et al., 2003a; Fehr et al., 2007). Both enzymatic domains play an independent role in the actin depolymerization and cell rounding (Fehr et al., 2007).
AopH is similar to Yersinia phosphotyrosine phosphatase YopH which dephosphorylates tyrosine residues of protein in focal adhesion complexes at the cellular membrane resulting in loss of focal adhesion complex, alteration of the actin cytoskeleton and blocking of phagocytosis (Cornelis, 2002; Broberg and Orth, 2010). In Yersinia, this effector inhibits also the lymphocyte proliferation and the synthesis of monocyte chemotactic protein 1 (Cornelis, 2002).
Ati2 is a phosphatidylinositol (PtIns) phosphatase that hydrolyses the D5 phosphate from PtIns(4,5)P2 and PtIns(3,4,5)P3 (Dallaire-Dufresne et al., 2013). This effector is homologous to VPA0450 of Vibrio parahaemolyticus which induces the local detachment of the actin-binding proteins from the plasma membrane, causes membrane blebbing and accelerates cytolysis by removing PtIns(4,5)P2 (Broberg et al., 2010).
AopP inhibits the NF-κB signalling pathway by preventing the translocation of the p50/p65 protein complex (NFKB1/RelA) into the nucleus of target cells (Fehr et al., 2006). AopP inhibits IκB kinase β (IKKβ) phosphorylation and attenuates IκBα phosphorylation showing that AopP-mediated inhibition of NF-κB occurs upstream of I-κB phosphorylation. This NF-κB pathway inhibition is highly proapoptotic upon concurrent tumor necrosis factor-α cellular stimulation (Jones et al., 2012). Apoptosis would be induced in part by the subsequent inhibition of the expression of anti-apoptotic factors (IAP) (Jones et al., 2012). AopP activity is unable to inhibit mitogen-activated protein kinase pathways (such as ERK, p38 and JNK) contrary to its orthologues YopJ, VopA and AvrA (Jones et al., 2012).
AopO is related to the Yersinia YopO/YpkA, a serine/threonine kinase that disrupts the normal distribution of actin in the host cell (Nejedlik et al., 2004). The kinase domain localizes the effector to the plasma membrane and is activated by actin binding. It phosphorylates and inactivates Gαq signalling pathways affecting multiple downstream targets (Broberg and Orth, 2010). Moreover, this effector inhibits phagocytosis through its guanine-nucleotide dissociation inhibitor-like domain by specifically blocking Rac-dependent Fcγ receptor internalization pathway (Groves et al., 2010).
AopN homologues in other bacteria (HrpJ and BopN) are T3SS effectors which play a role in virulence and can have a dual role: controlling the secretion of translocator proteins inside bacteria and suppressing immunity when AopN is translocated inside host cells (Nagamatsu et al., 2009; Crabill et al., 2012). In Chlamydia, translocated CopN, the analogue to AopN, is able to bind and sequester αβ-tubulin and inhibits microtubule polymerization leading to the loss of microtubule spindles and metaphase plate formation inducing mitotic arrest (Archuleta et al., 2011).
AopS is homologous to V. parahaemolyticus VopS which induces the AMPylation of Rho GTPases through its Fic domain (Yarbrough et al., 2009). This inhibition prevents the interaction of Rho GTPases with downstream effectors, thereby inhibiting actin assembly (Yarbrough et al., 2009) and preventing NLRC4 inflammasome activation (Higa et al., 2013).
AopX is related to the T3SS effector Plu4750 of P. luminescens which targets lipid raft in the cytoplasmic membrane (French et al., 2009) but its function is unknown.
We have also observed that ExsE and Ati1 were secreted by the T3SS in wt SNs (P. Vanden Bergh, unpublished). In P. aeruginosa, it was shown that the T3SS secretion in extracellular medium and the T3SS translocation into host cell of ExsE was required for transcriptional induction of the T3SS (Urbanowski et al., 2007) (Fig. 1). It is not known whether ExsE plays a role within the host cell. Ati1 is the only chaperone that is secreted by the T3SS (P. Vanden Bergh, unpublished) but it is not known whether it is also translocated into the fish cell.
Interestingly, at least five effectors (AexT, Ati2, AopH, AopP and AopS) out of the eight that have been identified so far, exert different negative effects on cytoskeletal dynamics that could explain the rapid cell-rounding within less than 1 h observed when A. salmonicida is incubated with epithelial fish cells (Movies S1 and S2). The AopS function is redundant with the actin depolymerization effect of AexT and might be the reason why mutation seen in aopS has globally no consequence on the virulence of A. salmonicida. As an essential part of the cytoskeleton, actin is involved in a multitude of cellular processes that are keys to establish structure, morphology and motility of cells and cell components (Aktories et al., 2012): epithelium barrier function (establishment and maintenance of cell junctions and cell shape), motility and cytokinesis, muscle contraction, cell division, phagocytosis, signalling of immune cells, intracellular trafficking of vesicles and organelles. This prominent role in the eukaryotic homeostasis explains why actin proteins are prime targets for bacterial virulence factors (Aktories et al., 2012). In accordance with these observations, it is likely plausible that the actin depolymerization by the T3SS of A. salmonicida contributes to the colonization of the host and to the survival in phagocytic cells (Burr et al., 2005). Their ability to destabilize the actin cytoskeleton at different critical sites in the host cell could explain why the deletion of aexT alone (Fehr et al., 2007) or a triple knock-out mutant ΔaexTΔaopOΔaopH (Fast et al., 2009) are not sufficient to prevent the cell rounding and the virulent phenotype. Hence, a quadruple ΔaexTΔaopOΔaopHΔati2 mutant would be expected to abolish the disruption of the actin cytoskeleton on the condition that the AopS gene is well inactivated. Besides the cell-rounding, another characteristic of cell death initiated by A. salmonicida is the rapid induction of nuclear morphological changes characteristic of apoptosis (nuclear shrinkage and chromatin condensation), which would be activated at least by the intracellular effect of AopP (Jones et al., 2012) (Fig. 4). At the end stage of the intoxication, the plasma membrane of completely disorganized host cells is alterated by the T3SS leading to the release of lactate dehydrogenase in the extracellular medium (P. Vanden Bergh, unpublished) (Fig. 5) as observed in V. alginolyticus (Zhao et al., 2011). This process could imply putative phospholipases (such as ASA_P5G088) thereby allowing the bacteria to damage the cell membrane and gain access to the nutrients that are released from the cytosol.
Figure 4.
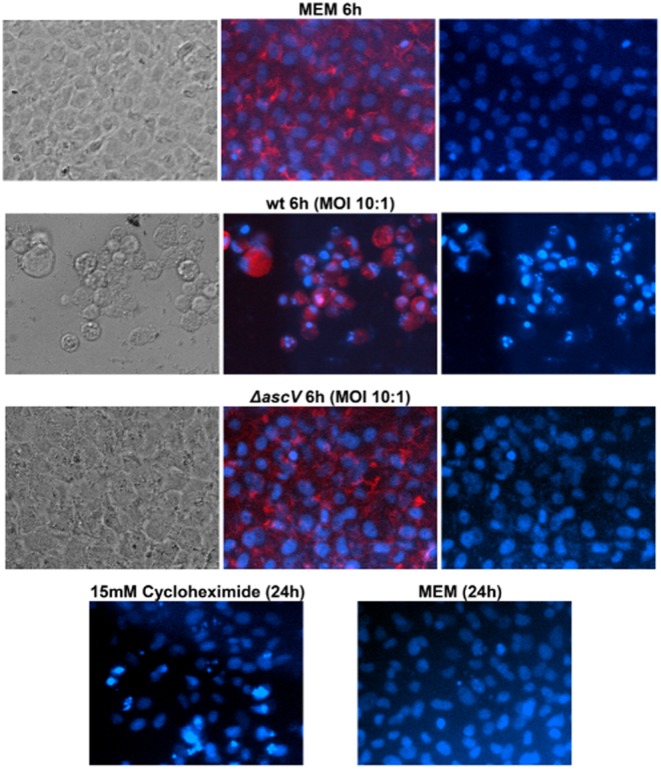
Morphological characteristics of the cytotoxicity induced by A. salmonicida.
Besides the rapid cell-rounding due to the depolymerization of actin, A. salmonicida induces also rapid nuclear morphological changes characteristic of apoptosis (nuclear shrinkage and chromatin condensation). Epithelial fish cells (epithelioma papulosum cyprinid) were incubated 6 hours with wt (JF5054), ΔascV (JF2747) A. salmonicida (MOI of 10:1) or MEM medium (negative control). As a positive control of apoptosis, cells were incubated 24h with 15 mM cycloheximide. Blue colour represents nuclei stained with DAPI and red colour, actin stained with TRITC-phalloidin.
Figure 5.
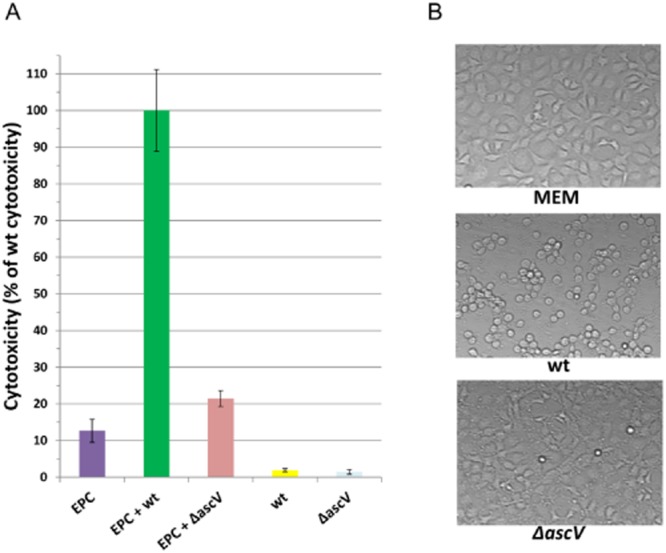
Membrane alterations induce by the A. salmonicida T3SS.
A. Virulence of A. salmonicida wt (JF2267) and ΔascV (JF2747) mutant strains for EPC cells studied by LDH release assay after 3 hours of infection at an MOI of 100:1. Cell death is expressed as the percentage of maximal LDH release measured from lysed EPCs with the wt strain.B. Microscopic analysis of A. salmonicida wt and ΔascV mutant virulence for EPC cells one hour before the LDH release assay.
Besides these classical T3SS effectors, additional proteins with predicted T3SS secretion signal were excreted to a greater extent in wt SNs (EF-G, EF-Tu, DnaK, HtpG, PNPase, MdeA, PepN and OpdA) but to a clearly lesser extent than previously described T3SS effectors (P. Vanden Bergh, unpublished). These putative effectors show a high similarity with homologues present in eukaryotic cells, where they play fundamental roles and sometimes have alternative (moonlighting) functions: EF-1α for EF-Tu (Ejiri, 2002), HSP70 and HSP90 for DnaK and HtpG (Tsan and Gao, 2009; Singhto et al., 2013), eukaryotic aminopeptidases and thimet oligopeptidase for PepN and OpdA (Kessler et al., 2011). Some of them play a role in the virulence of other pathogens and are considered to be new targets for therapy (Neckers and Tatu, 2008; Barbier et al., 2013). It is tempting to assume that they too might be injected by A. salmonicida into host cells in order to interfere with these functions.
The proteomics study that we have conducted showed that weak amounts of T3SS effectors/translocators were found in ΔascV mutant SNs (AopH, AexT, AopD, Ati2, AopP, AopN, AopB and ExsE by order of decreasing importance), and the presence of these T3SS elements in mutant SNs was not due to bacterial lysis or cross-contaminations (P. Vanden Bergh, unpublished). The mutant strain thus continues to release weak amounts of Aops in SNs, either from the resting structural T3SS components or by an alternative secretion pathway. It is accepted that the T3SS arose from an exaptation of the flagellum (Abby and Rocha, 2012), and thus, it could be possible that FlhA (ASA_1351, polar flagella) and/or LfhA (ASA_0352, lateral flagella), showing 56% and 55% of similarity with AscV, respectively, might partially supply the function of this T3SS component. Another possibility is that a second mechanism of effectors/translocators secretion, clearly less efficient than the active T3SS, operates at the same time. Further investigations are therefore necessary to clarify the mechanism leading to the presence of effectors/translocators observed in the SN of the ΔascV mutant.
The origin of the T3SS-induced virulence of A. salmonicida – A story of survival and adaptation
The adaptation of A. salmonicida T3SS effectors, shared by different environmental bacteria, to disrupt the host cytoskeleton is most intriguing, and it is tempting to hypothesize that these attack strategies are the result of a selective pressure to survive in the aquatic environment to the engulfment and killing by feral phagocytes, such as amoebas. The actin depolymerization could serve to paralyse the bacteriophagous protozoan, escape phagocytosis, survive inside the cell, spread in the environment and lyse these hosts to get an alternative source of aquatic nutriments. Hence, the well-known predator of bacteria would become a prey to be parasitized. Therefore, A. salmonicida strains virulent for fish have also been demonstrated to be virulent for amoeba (Paniagua et al., 2001; Daher et al., 2011), and it has been confirmed with our wt (JF2267) and ΔascV (JF2747) mutant strains that this cytotoxic effect was associated with the T3SS (Froquet et al., 2007). For example, in V. parahaemolyticus, it has been demonstrated that the T3SS-2 promotes survival of the bacterium in the interaction with diverse protist taxa and that the enhanced persistence is due to T3SS-2 effectors mediated cytotoxicity [some of which being homologous to A. salmonicida effectors (Table 1)] and facultative parasitism of V. parahaemolyticus on coexisting protists (Matz et al., 2011).
Because these wild phagocytes might serve as a reservoir and an amplifier for virulent A. salmonicida, it might explain why sediment is an important environmental reservoir of the bacterium as the pathogen can survive for a longer period of time and retain its pathogenicity in faecal and food waste sediment (Michel and Dubois-Darnaudpeys, 1980) at the bottom of sea cages, freshwater tanks or in pond mud (O’Brien et al., 1994). The high prevalence of protistan hosts in water with faecal contamination or plankton might serve the multiplication of virulent strains of A. salmonicida and could subsequently constitute sources of furunculosis outbreaks (King and Shotts, 1988; Nese and Enger, 1993).
The entry of A. salmonicida into the fish host
Several entry sites (skin, gills and the intestine) into fish have been described for A. salmonicida (Farto et al., 2011) but the route of infection would more likely be associated with any epithelial barrier injury (erosions/ulcerations) where the bacteria attach and penetrate into host tissues (Fig. 6). In order to initiate the disease, it could be a necessity for the bacterium to have direct access to epithelial cells and a hand-to-hand combat. Therefore, it could explain why challenges by immersion in laboratory facilities with healthy fish (intact epithelial barriers) and circulating fresh water are difficult even with high bacterial loads. This observation might support the idea that any stress perturbing mucus layer and/or epithelial integrity could be necessary to induce the furunculosis. Hence, it is frequent to read in the literature that in order to get mortality, the skin of fish is rubbed, scratched or wounded prior to an immersion challenge with A. salmonicida.
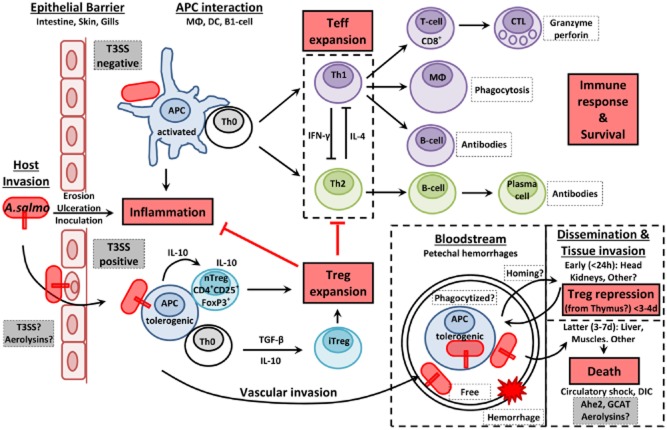
Figure 6.
A model for A. salmonicida T3SS-dependent pathogenesis.
We propose a model of pathogenesis in which T3SS+ A. salmonicida could hijack the induction of an efficient immune response by promoting tolerogenic DCs, IL-10 expression, Treg cells activation and expansion, and the inhibition of Teff (T and B cells) proliferation during the incubation period of the furunculosis.
By promoting a pseudo tolerance state like a wolf in sheep’s clothing, some A. salmonicida bacteria could rapidly disseminate through the bloodstream (free or phagocytized) into major immune organs (e.g. the head kidneys) where they attack and neutralize APCs and B lymphocytes. Some Treg cells might directly come from the thymus in the head kidneys and repress the lymphoproliferation in a T3SS-dependent manner. Later in the disease, A. salmonicida would colonize the liver, the gills and lastly cardiac and skeletal muscles freed of leukocytes. After 3–5 days post-inoculation the capacity of T and B cells to proliferate (Dautremepuits et al., 2006) and the CTL activation [especially, in the head kidney (Kumari et al., 2013)] recover their functions but would appear too late to control efficiently the infection and protect fish from death. The cause of death would be associated with a septicemic state and circulatory shock. The inoculation of T3SS-negative A. salmonicida would induce a balanced Th1/Th2 protective immune response.
At the cell contact, the actin depolymerization induced by translocated T3SS effectors of A. salmonicida might already disturb the epithelium integrity (Movies S1 and S2) (Fig. 6). Aerolysins (AerA and AerB) might also participate in the epithelial barrier injury as described in A. hydrophila (Bucker et al., 2011). In experimental challenges of naïve trout with the wt virulent strain (JF5054), it was possible to get mortality by i.p. inoculations from 50 to 500 cfu per trout showing that the infectious dose can be very low if bacteria are in a state of high virulence. This fact raises the question of definition of a threshold to distinguish virulent and non-virulent A. salmonicida inocula. If the injection of hundreds of thousands of bacteria is necessary to induce a low level of mortality, does this mean that the A. salmonicida strain is really virulent and does it represent a natural infection?
The T3SS and the escape from the immune response – The art of war
Once fish are challenged with A. salmonicida, a typical incubation period of 3–4 days occurs where bacteria rapidly disseminate (already 12 h post-challenge) in kidneys (Farto et al., 2011) (Fig. 7A). Then, A. salmonicida colonizes spleen, liver (Burr et al., 2005), and in the latest stage of the infection, the skeletal and cardiac muscles (Farto et al., 2011). In these sites, A. salmonicida gets access to its nutriments through cytolysis. The appearance of colonies in host tissues coincides with the mortality peak (Fig. 7A) (Burr et al., 2005; Farto et al., 2011). The incubation period is crucial for pathogenesis, and we hypothesize that virulent A. salmonicida strains use the T3SS and its effectors to neutralize the fish immune system along this period of the infection. In Yersinia, the inhibition of dendritic cell (DC) activation and T-cell proliferation by T3SS effectors are the mechanisms by which the bacterium evades innate and adaptative immune responses to generate the disease (Autenrieth et al., 2010). The scenario of a direct immunosuppressive effect induced by A. salmonicida was suspected by the Furunculosis Committee as early as in 1935 (Mackie et al., 1935), and thereafter, several observations have supported the hypothesis of an immunosuppressive mechanism similar to that of Yersinia:
the virulent T3SS+ JF2267 strain is able to survive phagocytosis by fish peripheral blood leukocytes in vitro (Burr et al., 2005),
the virulent T3SS+ 01-B526 strain depresses drastically B and T lymphoproliferation from head kidneys (HK) during the 3 days following the challenge and then return to the baseline at the mortality onset (Dautremepuits et al., 2006) (Fig. 7B),
there is no apparent leukocytic infiltration associated with colonies of T3SS+ A. salmonicida in fish tissues (absence of antigen-presenting cell activation and leukocyte migration) (Burr et al., 2005),
a drastic decrease in plasma antibody titres is immediately observed in the days following a challenge with virulent strains of A. salmonicida (Fig. 7B) (Bricknell et al., 1999; Romer et al., 2012). The ability of A. salmonicida to escape phagocytosis and inhibit lymphoproliferation might explain the hypoimmunoglobulinemia that could result from the combination of the inhibition of B lymphocyte differentiation/proliferation (blocking of antibody production) and the opsonization and formation of antigen/antibody complexes in the bloodstream (antibody depletion).
Figure 7.
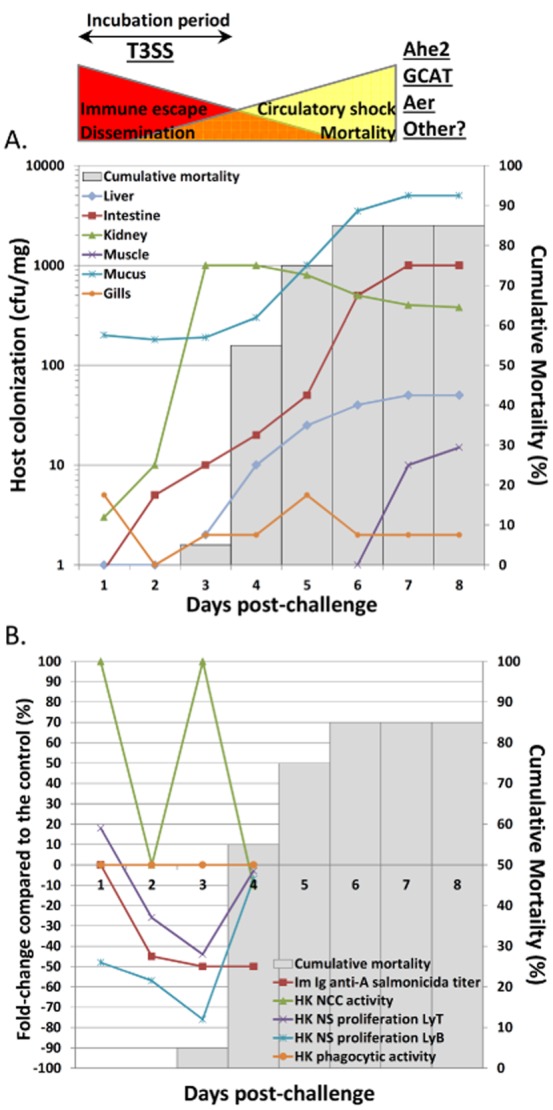
Evolution of host colonization, immune parameters and mortality during the incubation period of the furunculosis.
In these figures, the authors used results from previous publications (Dautremepuits et al., 2006; Farto et al., 2011) to put the onset of fish mortality induced by A. salmonicida in perspective with tissue colonization (A) and immune parameters from the head kidney of fish (B). The incubation period of furunculosis could be dominated by the immunosuppressive effect of the T3SS leading to the dissemination of A. salmonicida in several organs while the later stages of the disease, associated to mortality and circulatory failure, would be dominated by the high production of other virulence factors such as Ahe2 serine protease and GCAT lipase. The hypothesized importance of A. salmonicida virulence factors is represented above figures.
Diagram A represents tissue colonization of fish challenged with virulent T3SS+ A. salmonicida according to the results of Farto and collaborators (Farto et al., 2011). The kidney is the first organ massively invaded by A. salmonicida, as early as 12h post-challenge and without any symptoms, followed by the liver. Then, bacteria are isolated from muscles only in the later stages of the disease. Diagram B shows the behaviour of leukocytes from the head kidney of fish challenged with virulent T3SS+ A. salmonicida during the incubation period of furunculosis, according to the study of Dautremepuits and collaborators (Dautremepuits et al., 2006). The non-specific (NS) lymphoproliferation (B and T cells) is significantly inhibited in this organ during 3 days post-challenge and antibody titers specific of A. salmonicida are strongly depressed (Bricknell et al., 1999).
The mechanisms used by A. salmonicida to defeat the lymphocyte multiplication are currently not known but several observations tend to demonstrate that the bacterium may also induce tolerogenic DCs and activate suppressor regulatory T lymphocytes (Treg) in a T3SS-dependent manner (Fig. 6). In mammals, Treg cells in all lymphoid and intestinal organs are derived from thymus (Cebula et al., 2013). In many fish species, threads of cells directly connect the thymus with the HK, and this may be the route for lymphocyte migration from the thymus to the HK (Bowden et al., 2005). Interestingly, thymectomized fish injected with A. salmonicida show higher antibody titres than intact controls while the reconstitution of thymectomized fish with preserved thymocytes depresses the immunoglobulinemia at levels similar to those of the control group, and this suggests that A. salmonicida promotes an immunosuppressive cell population existing in this organ and acting on the B-cells (Findlay and Tatner, 1996). Moreover, IL-10 is a cytokine produced by various cell populations which downregulates the cellular immune response and contributes to Treg-mediated suppression in association with other cytokines (Yamaguchi et al., 2011). In vitro, virulent A. salmonicida elicits a significant increase in IL-10 expression by fish leukocytes from the HK whereas deletion of T3SS genes significantly decreases the expression of this cytokine (Fast et al., 2009). In mice infected with an AcrV-mutant of A. hydrophila, IL-10 levels were also significantly downregulated (Fadl et al., 2006). Furthermore, in the 3 days following a challenge with A. salmonicida, the expression of genes associated with the immunosuppressive Treg response such as Foxp3 (Zhang et al., 2011), fibroleukin Fgl2 (Millan et al., 2011; Liu et al., 2013), Es1 (HES1/KNPI) (Ostroukhova et al., 2006; Millan et al., 2011) and serum amyloid A (SAA) (Jensen et al., 1997; Nguyen et al., 2011) was enhanced in the HK of fish.
In non-infectious conditions, mammalian DCs that are present in gut, skin and lungs play a key regulatory mechanism of ‘tolerance’ to prevent excessive inflammation induced by the commensal flora. These tolerogenic DCs produce the immunomodulatory IL-10 cytokines and fail to deliver proper costimulatory signal to naïve CD4+ T-cell (Th0) for effector T (Teff) cells activation and proliferation. This results in T cell death, T cell anergy or induction and expansion of subsets of Treg cells (Kornete and Piccirillo, 2012). Specifically, Treg cells maintain order in the immune system by enforcing a dominant negative regulation on other immune cells. However, when a conventional bacterial pathogen (or T3SS negative A. salmonicida) penetrates host tissues (Fig. 6), immature DCs receive maturation signals through the pathogen-associated molecular patterns and damage associated molecular patterns receptors, and mature DCs initiate a three-step T cell activation process (Kornete and Piccirillo, 2012). In these conditions, naïve CD4+ T-cells (Th0) are then polarized towards Th1 and/or Th2 cells (depending on the stimulation and the cytokinic environment) to eliminate the pathogen.
In this view, it might be possible that A. salmonicida uses its T3SS to promote the tolerance pathways and hijack the immune response in the first days of the disease (Fig. 6). For example, LcrV (homologous to AcrV) in Yersinia induces immunosuppression by promoting the differentiation of tolerogenic DCs via the interaction with TLR2/TLR6 and CD14 receptors (DePaolo et al., 2008) and the amplified release of IL-10 from host cells (Reithmeier-Rost et al., 2004). The exacerbated release of IL-10 by Yersinia plays a crucial role in the pathogenesis since IL-10-deficient mice are resistant to the infection (Sing et al., 2002a). At least two regions of LcrV were associated with the induction of the IL-10 immunomodulatory responses (Sing et al., 2002b; Overheim et al., 2005). In A. salmonicida, AcrV shows a high conservation (66% and 93% of similarity) with these fragments. Hence, these conserved regions could play the same IL-10 dependent immunomodulatory function in AcrV.
Moreover, although virulent A. salmonicida are detected in the HK as early as 12 h post-challenge by immersion (Farto et al., 2011), the recovery of the lymphoproliferation (Dautremepuits et al., 2006) and the expression of genes associated with a Th1 response and the activation of cytotoxic T lymphocytes (CTL) (such as IFN-γ and granzyme) occur only 3–4 days after the inoculation (Millan et al., 2011; Kumari et al., 2013) (Fig. 7B) and do not appear to be sufficient to save fish from death. During this incubation period, A. salmonicida might thus trigger the migration of activated Treg cells from the thymus directly into the infected HK inhibiting the lymphocyte proliferation in this organ (Bowden et al., 2005).
The induction of the CTL response in HKs could be associated with the multiplication of A. salmonicida in this organ, the repression of the T3SS and the induction of proteases, lipases expression by the QS. It can also be associated with the observation that A. salmonicida is able to invade and survive within HK leukocytes (Fast et al., 2009), and would therefore induce an immune response specific to intracellular pathogens. The HK hosts diverse antigen-presenting cell (APC) types (melanomacrophages, B cells, reticular cells) and is thus suspected to serve as a major secondary lymphoid organ to which APCs loaded with antigens in the peripheral organs migrate (homing) (Iliev et al., 2013). This might suggest that A. salmonicida uses APCs and the homing to rapidly disseminate in the lymphoid system which explains that these organs are the first to be infected (Farto et al., 2011) (Fig. 7A). In this view, A. salmonicida has been demonstrated to be able to survive and even replicate inside fish non-phagocytic cells and macrophages, thereby supporting the notion that the bacterium can be a facultative intracellular pathogen (Garduno et al., 1993; 2000). This process would be primordial for A. salmonicida to survive in vivo soluble lytic activity present in extracellular fluids (Garduno et al., 2000).
The rapid attack and neutralization of immune functions in the early stages of the disease may explain the absence of leukocyte migration in other infected tissues in the final stage of the furunculosis. Peracute infections most often occur in fingerling fish which die from shock without showing marked clinical manifestations. Acute infections generally occur in juvenile and adult fish and are associated with haemorrhages in all organs. Aeromonas salmonicida ultimately induces a septicemic state but teleost fish are resistant to endotoxic shock (Sepulcre et al., 2009), and the cause of death in furunculosis is more likely due to a shock induced by vascular failure associated with disseminated intravascular coagulation, a consumptive coagulopathy that may rapidly be induced at least by the intravascular injection of the serine protease Ahe2 (AspA) (Salte et al., 1993) and the LPS-activated GCAT lipase (SatA) (Salte et al., 1992). Thus, the early stage of the furunculosis could be dominated by the T3SS inducing immune escape and dissemination of a few bacteria in lymphoid organs. Thereafter, the later stages of the disease would be associated with a large multiplication and bacterial dissemination in all organs, ultimately resulting in circulatory shock due to other virulence factors (Fig. 7).
The protective immune response to A. salmonicida
Various studies tend to demonstrate that the protective immune response against A. salmonicida would result from a balanced Th1/Th2 response (Fig. 6). For example, fish protection is correlated with a high level of circulating antibodies specific to A. salmonicida (Romer et al., 2012; Romstad et al., 2013). The administration to fish of specific immunostimulants such as CpG oligodeoxynucleotides or the heptanoyl tripeptide FK-565 which indirectly enhance the activation of Th1 cells (Krieg, 2002; Tada et al., 2005) increases their survival following a challenge with a virulent strain of A. salmonicida (Kitao and Yoshida, 1986; Carrington and Secombes, 2007) and suggest that promoting a Th1 polarization might be important to overcome the infection.
Like other pathogens using the T3SS for their virulence, we suspect that A. salmonicida might use certain strategies to interfere with the endogenous pathway (MHC-I) and the ultimate activation of CD8+ T-cells to avoid the destruction of its Trojan horse (APC) by CTLs. Actually, it is accepted that T3SS effectors translocated into the host cytoplasm should be, like viral proteins, perfect candidates for antigen presentation by the endogenous pathway which canonically eliminate cells containing the intracellular pathogen (Mantegazza et al., 2013). Bergman and colleagues (2009) showed that the CTL response played a critical role in eliminating Yersinia infections, and that this response was directed against Yop T3SS effectors. In Salmonella, MHC class-I-restricted CD8 T cells can play a protective role during primary Salmonella infection (Lee et al., 2012). Moreover, Salmonella T3SS effectors have been successfully used for vaccination strategy specifically through the MHC-I/CD8+ T-cell pathway (Hegazy et al., 2012). Eliciting an immune response against A. salmonicida through the endogenous pathway of antigen presentation might thus be an interesting strategy for vaccine approaches. However, it was also predicted by bioinformatics that T3SS effectors with homologues in A. salmonicida show clear escape mutations and have low epitope densities for MHC, suggesting that these bacteria, like viruses, are evolutionarily selected to ensure their survival in the presence of CD8+ T-cells (Maman et al., 2011). For example, while YopE, a homologue to AexT, was not protective in experiments using the whole protein as antigen, the N-terminal domain (YopE69-77) revealed to be a major protective antigen eliciting CD8 T-cell immunity (Zhang et al., 2012). In spite of the fact that most of the T3SS effectors are immune suppressor/modulator, this result shows that they can contain protective epitopes that might be promising candidates for vaccination. In this view, it has been demonstrated that polymorphisms in MHC-II, but also MHC-I, were significantly associated with resistance of Atlantic salmon to furunculosis (Grimholt et al., 2003; Kjoglum et al., 2008) and highlight the importance of the endogenous pathway of antigen presentation in this disease.
Vaccine and therapeutic approaches stopping the T3SS-dependent immunosuppressive strategies of A. salmonicida at the onset of the disease might thus represent the best protection of fish against furunculosis. According to this hypothesis, we recently observed that immunization of fish with bacterins of the ΔascV mutant strain of A. salmonicida (JF2747) that expresses low levels of all the T3SS components (immunosuppressors), induced a better protection (25% increase in the survival) than vaccination with the wt strain JF5054 expressing the T3SS at high levels (Vanden Bergh et al., 2013). This result was unexpected but it supports our current model of pathogenesis, and challenged the hypothesis that mounting specific antibodies against proteins of the T3SS yields better protection. At the onset of a furunculosis outbreak, therapies targeting either the inhibition of the T3SS expression (such as pro-quorum-sensing molecules) (Ng et al., 2012) or secretion (Duncan et al., 2012) by A. salmonicida, or promoting the Th1 immune response (Krieg, 2002; Tada et al., 2005), or specifically inhibiting the Treg expansion (Casares et al., 2010) are expected to help resolving the disease and constitute alternatives to classical antibiotics.
Conclusions
The typical furunculosis induced by Aeromonas salmonicida subsp. salmonicida is a disease that has been known for over a century. The mechanisms of virulence were poorly understood until the discovery of the T3SS as a major virulence system, a decade ago. In this review, we have proposed a comprehensive model of pathogenesis for A. salmonicida involving the various actions of T3SS and its effectors on the morphogenesis, the vital functions and the immune defence of the host cells. This concerted action of virulence attributes permits the pathogen to cause disease in salmonid fish by circumventing all barriers imposed by the host and the environment. Although further investigations both on the pathogen and on the host will be necessary to fully confirm its validity, this knowledge provides strategies in designing novel preventive and therapeutic approaches to optimize environmentally and economically sustainable fish farming.
Conflict of interest
Authors have no conflict of interest to declare.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Genetic comparison of T3SSs from the Ysc family.
Comparison of the organization of T3SS clusters in A. salmonicida subsp. salmonicida A449, A. hydrophila SSU, A. veronii AER39, A. diversa 2478-85, Pseudomonas aeruginosa PAO1, Photobacterium damselae subsp. damselae CIP 102761, Photorhabdus luminescens subsp. laumondii TTO1, Yersinia pestis biovar Antiqua str. E1979001, and Vibrio parahaemolyticus RIMD 2210633.
Profound analysis and comparison of published Aeromonas genomes.
Grey: conserved ORFs; light green: ORFs specific of the species; yellow: IS630; pink: other IS elements; red: putative or characterized virulence factors; mauve: ORFs for resistance to antibiotic or heavy metal; dark green: ORFs associated to pili, fimbriae or flagella; blue: ORFs associated with phage; cyan: tRNA and rRNA; orange: ORFs with homology to eukaryotic genes. Aeromonas salmonicida subsp. salmonicida A449, A. salmonicida subsp. achromogenes AS03, A. salmonicida (ATCC7966, ML09-119 and SSU), A. caviae Ae398, A. veronii (B565, AMC34, AMC35, AER39 and AER397), A. aquariorum AAK1, and A. diversa 2478-85.
Rapid cell-rounding induced by the T3SS of A. salmonicida.
Epithelioma papulosum cyprini cells (EPCs) seeded in a glass bottom well and exposed to A. salmonicida wt JF2267 (S1) and ΔascV mutant JF2747 (S2) expressing GFP (after induction with IPTG) for 120 min. Following infection at 20°C, time-lapse imaging using video microscopy was performed using a Nikon Eclipse TE2000-U inverted microscope equipped with a climate-controlled chamber. Data acquisition and image processing were performed using NIS software of Nikon Instruments. DIC and fluorescence images were acquired and assembled in movies. Image acquisition at intervals of 2 min for 120 min. MOI of 10:1 (bacteria: fish cells).
References
- Abby SS. Rocha EPC. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:1–15. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, et al. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories K, Schwan C, Papatheodorou P. Lang AE. Bidirectional attack on the actin cytoskeleton. Bacterial protein toxins causing polymerization or depolymerization of actin. Toxicon. 2012;60:572–581. doi: 10.1016/j.toxicon.2012.04.338. [DOI] [PubMed] [Google Scholar]
- Archuleta TL, Du YQ, English CA, Lory S, Lesser C, Ohi MD, et al. The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J Biol Chem. 2011;286:33992–33998. doi: 10.1074/jbc.M111.258426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autenrieth SE, Linzer TR, Hiller C, Keller B, Warnke P, Koberle M, et al. Immune evasion by Yersinia enterocolitica: differential targeting of dendritic cell subpopulations in vivo. PLoS Pathog. 2010;6:e1001212. doi: 10.1371/journal.ppat.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier M, Owings JP, Martinez-Ramos I, Damron FH, Gomila R, Blazquez J, et al. Lysine trimethylation of EF-Tu mimics platelet-activating factor to initiate Pseudomonas aeruginosa pneumonia. MBio. 2013;4:e00207–e00213. doi: 10.1128/mBio.00207-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret M, Egan F. O’Gara F. Distribution and diversity of bacterial secretion systems across metagenomic datasets. Environ Microbiol Rep. 2013;5:117–126. doi: 10.1111/j.1758-2229.2012.00394.x. [DOI] [PubMed] [Google Scholar]
- Bergman MA, Loomis WP, Mecsas J, Starnbach MN. Isberg RR. CD8(+) T cells restrict Yersinia pseudotuberculosis infection: bypass of anti-phagocytosis by targeting antigen-presenting cells. PLoS Pathog. 2009;5:1–16. doi: 10.1371/journal.ppat.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TJ, Cook P. Rombout JH. Development and function of the thymus in teleosts. Fish Shellfish Immunol. 2005;19:413–427. doi: 10.1016/j.fsi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Braun M, Stuber K, Schlatter Y, Wahli T, Kuhnert P. Frey J. Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J Bacteriol. 2002;184:1851–1858. doi: 10.1128/JB.184.7.1851-1858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricknell IR, King JA, Bowden TJ. Ellis AE. Duration of protective antibodies, and the correlation with protection in Atlantic salmon (Salmo salar L.), following vaccination with an Aeromonas salmonicida vaccine containing iron-regulated outer membrane proteins and secretory polysaccharide. Fish Shellfish Immun. 1999;9:139–151. [Google Scholar]
- Broberg CA. Orth K. Tipping the balance by manipulating post-translational modifications. Curr Opin Microbiol. 2010;13:34–40. doi: 10.1016/j.mib.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg CA, Zhang LL, Gonzalez H, Laskowski-Arce MA. Orth K. A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science. 2010;329:1660–1662. doi: 10.1126/science.1192850. [DOI] [PubMed] [Google Scholar]
- Broz P, Mueller CA, Muller SA, Philippsen A, Sorg I, Engel A. Cornelis GR. Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol. 2007;65:1311–1320. doi: 10.1111/j.1365-2958.2007.05871.x. [DOI] [PubMed] [Google Scholar]
- Brutinel ED. Yahr TL. Control of gene expression by type III secretory activity. Curr Opin Microbiol. 2008;11:128–133. doi: 10.1016/j.mib.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel ED, Vakulskas CA. Yahr TL. Functional domains of ExsA, the transcriptional activator of the Pseudomonas aeruginosa type III secretion system. J Bacteriol. 2009;191:3811–3821. doi: 10.1128/JB.00002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucker R, Krug SM, Rosenthal R, Gunzel D, Fromm A, Zeitz M, et al. Aerolysin from Aeromonas hydrophila perturbs tight junction integrity and cell lesion repair in intestinal epithelial HT-29/B6 cells. J Infect Dis. 2011;204:1283–1292. doi: 10.1093/infdis/jir504. [DOI] [PubMed] [Google Scholar]
- Burr SE. Frey J. Analysis of type III effector genes in typical and atypical Aeromonas salmonicida. J Fish Dis. 2007;30:711–714. doi: 10.1111/j.1365-2761.2007.00859.x. [DOI] [PubMed] [Google Scholar]
- Burr SE. Frey J. Aeromonas salmonicida subsp. salmonicida type strain does not possess a type III secretion system. J Clin Microbiol. 2009;47:3062–3063. doi: 10.1128/JCM.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr SE, Stuber K, Wahli T. Frey J. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J Bacteriol. 2002;184:5966–5970. doi: 10.1128/JB.184.21.5966-5970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr SE, Stuber K. Frey J. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J Bacteriol. 2003a;185:6583–6591. doi: 10.1128/JB.185.22.6583-6591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr SE, Wahli T, Segner H, Pugovkin D. Frey J. Association of type III secretion genes with virulence of Aeromonas salmonicida subsp. salmonicida. Dis Aquat Organ. 2003b;57:167–171. doi: 10.3354/dao057167. [DOI] [PubMed] [Google Scholar]
- Burr SE, Pugovkin D, Wahli T, Segner H. Frey J. Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology. 2005;151:2111–2118. doi: 10.1099/mic.0.27926-0. [DOI] [PubMed] [Google Scholar]
- Carrington AC. Secombes CJ. CpG oligodeoxynucleotides up-regulate antibacterial systems and induce protection against bacterial challenge in rainbow trout (Oncorhynchus mykiss. Fish Shellfish Immunol. 2007;23:781–792. doi: 10.1016/j.fsi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Casares N, Rudilla F, Arribillaga L, Llopiz D, Riezu-Boj JI, Lozano T, et al. A peptide inhibitor of FOXP3 impairs regulatory T cell activity and improves vaccine efficacy in mice. J Immunol. 2010;185:5150–5159. doi: 10.4049/jimmunol.1001114. [DOI] [PubMed] [Google Scholar]
- Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat Rev Mol Cell Biol. 2002;3:742–752. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- Correa VR, Majerczak DR, Ammar E, Merighi M, Pratt RC, Hogenhout SA, et al. The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector. Appl Environ Microbiol. 2012;78:6327–6336. doi: 10.1128/AEM.00892-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabill E, Karpisek A. Alfano JR. The Pseudomonas syringae HrpJ protein controls the secretion of type III translocator proteins and has a virulence role inside plant cells. Mol Microbiol. 2012;85:225–238. doi: 10.1111/j.1365-2958.2012.08097.x. [DOI] [PubMed] [Google Scholar]
- Dacanay A, Knickle L, Solanky KS, Boyd JM, Walter JA, Brown LL, et al. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology. 2006;152:1847–1856. doi: 10.1099/mic.0.28768-0. [DOI] [PubMed] [Google Scholar]
- Daher RK, Filion G, Tan SGE, Dallaire-Dufresne S, Paquet VE. Charette SJ. Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet Microbiol. 2011;152:353–360. doi: 10.1016/j.vetmic.2011.04.034. [DOI] [PubMed] [Google Scholar]
- Dallaire-Dufresne S, Barbeau X, Sarty D, Tanaka KH, Denoncourt AM, Lague P, et al. Aeromonas salmonicida Ati2 is an effector protein of the type three secretion system. Microbiology. 2013;159:1937–1945. doi: 10.1099/mic.0.067959-0. [DOI] [PubMed] [Google Scholar]
- Dautremepuits C, Fortier M, Croisetiere S, Belhumeur P. Fournier A. Modulation of juvenile brook trout (Salvelinus fontinalis) cellular immune system after Aeromonas salmonicida challenge. Vet Immunol Immunopathol. 2006;110:27–36. doi: 10.1016/j.vetimm.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev. 2011;35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- DePaolo RW, Tang F, Kim I, Han M, Levin N, Ciletti N, et al. Toll-like receptor 6 drives differentiation of tolerogenic dendritic cells and contributes to LcrV-mediated plague pathogenesis. Cell Host Microbe. 2008;4:350–361. doi: 10.1016/j.chom.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewoody RS, Merritt PM. Marketon MM. Regulation of the Yersinia type III secretion system: traffic control. Front Cell Infect Microbiol. 2013;3:4. doi: 10.3389/fcimb.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Wiesand U. Cornelis GR. The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol Microbiol. 2011;82:502–514. doi: 10.1111/j.1365-2958.2011.07830.x. [DOI] [PubMed] [Google Scholar]
- Diepold A, Wiesand U, Amstutz M. Cornelis GR. Assembly of the Yersinia injectisome: the missing pieces. Mol Microbiol. 2012;85:878–892. doi: 10.1111/j.1365-2958.2012.08146.x. [DOI] [PubMed] [Google Scholar]
- Duncan MC, Linington RG. Auerbuch V. Chemical inhibitors of the type three secretion system: disarming bacterial pathogens. Antimicrob Agents Chemother. 2012;56:5433–5441. doi: 10.1128/AAC.00975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejiri S. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci Biotechnol Biochem. 2002;66:1–21. doi: 10.1271/bbb.66.1. [DOI] [PubMed] [Google Scholar]
- Ellis AE, Burrows AS. Stapleton KJ. Lack of relationship between virulence of Aeromonas salmonicida and the putative virulence factors – A-layer, extracellular proteases and extracellular hemolysins. J Fish Dis. 1988;11:309–323. [Google Scholar]
- Fadl AA, Galindo CL, Sha J, Erova TE, Houston CW, Olano JP. Chopra AK. Deletion of the genes encoding the type III secretion system and cytotoxic enterotoxin alters host responses to Aeromonas hydrophila infection. Microb Pathog. 2006;40:198–210. doi: 10.1016/j.micpath.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Farto R, Milton DL, Bermudez MB. Nieto TP. Colonization of turbot tissues by virulent and avirulent Aeromonas salmonicida subsp. salmonicida strains during infection. Dis Aquat Organ. 2011;95:167–173. doi: 10.3354/dao02342. [DOI] [PubMed] [Google Scholar]
- Fast MD, Tse B, Boyd JM. Johnson SC. Mutations in the Aeromonas salmonicida subsp. salmonicida type III secretion system affect Atlantic salmon leucocyte activation and downstream immune responses. Fish Shellfish Immun. 2009;27:721–728. doi: 10.1016/j.fsi.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Fehr D, Casanova C, Liverman A, Blazkova H, Orth K, Dobbelaere D, et al. AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappa B signalling pathway. Microbiology. 2006;152:2809–2818. doi: 10.1099/mic.0.28889-0. [DOI] [PubMed] [Google Scholar]
- Fehr D, Burr SE, Gibert M, d’Alayer J, Frey J. Popoff MR. Aeromonas exoenzyme T of Aeromonas salmonicida is a bifunctional protein that targets the host cytoskeleton. J Biol Chem. 2007;282:28843–28852. doi: 10.1074/jbc.M704797200. [DOI] [PubMed] [Google Scholar]
- Findlay C. Tatner MF. The effect of reconstitution with cryopreserved thymocytes on the in vivo antibody response to sheep red blood cells, Aeromonas salmonicida, and DNP-KLH, in adult long term thymectomised rainbow trout (Oncorhynchus mykiss. Fish Shellfish Immunol. 1996;6:371–381. [Google Scholar]
- French CT, Panina EM, Yeh SH, Griffith N, Arambula DG. Miller JF. The Bordetella type III secretion system effector BteA contains a conserved N-terminal motif that guides bacterial virulence factors to lipid rafts. Cell Microbiol. 2009;11:1735–1749. doi: 10.1111/j.1462-5822.2009.01361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froquet R, Cherix N, Burr SE, Frey J, Vilches S, Tomas JM. Cosson P. Alternative host model to evaluate Aeromonas virulence. Appl Environ Microbiol. 2007;73:5657–5659. doi: 10.1128/AEM.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garduno R, Moore A, Olivier G, Lizama AL, Garduno E. Kay WW. Host cell invasion and intracellular residence by Aeromonas salmonicida: role of the S-layer. Can J Microbiol. 2000;46:660–668. doi: 10.1139/w00-034. [DOI] [PubMed] [Google Scholar]
- Garduno RA, Thornton JC. Kay WW. Fate of the fish pathogen Aeromonas salmonicida in the peritoneal cavity of rainbow trout. Can J Microbiol. 1993;39:1051–1058. doi: 10.1139/m93-159. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Larsen S, Nordmo R, Midtlyng P, Kjoeglum S, Storset A, et al. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- Groves E, Rittinger K, Amstutz M, Berry S, Holden DW, Cornelis GR. Caron E. Sequestering of Rac by the Yersinia effector YopO blocks Fcgamma receptor-mediated phagocytosis. J Biol Chem. 2010;285:4087–4098. doi: 10.1074/jbc.M109.071035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneda T, Okada N, Nakazawa N, Kawakami T. Danbara H. Complete DNA sequence and comparative analysis of the 50-kilobase virulence plasmid of Salmonella enterica serovar Choleraesuis. Infect Immun. 2001;69:2612–2620. doi: 10.1128/IAI.69.4.2612-2620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy WAH, Xu X, Metelitsa L. Hensel M. Evaluation of Salmonella enterica type III secretion system effector proteins as carriers for heterologous vaccine antigens. Infect Immun. 2012;80:1193–1202. doi: 10.1128/IAI.06056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa N, Toma C, Koizumi Y, Nakasone N, Nohara T, Masumoto J, et al. Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS Pathog. 2013;9:e1003142. doi: 10.1371/journal.ppat.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson JL, Horsley A, Stabat D, Simon M, Johnson S, da Fonseca PC, et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol. 2009;16:477–485. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev DB, Thim H, Lagos L, Olsen R. Jorgensen JB. Homing of antigen-presenting cells in head kidney and spleen – Salmon head kidney hosts diverse APC types. Front Immunol. 2013;4:137. doi: 10.3389/fimmu.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LE, Hiney MP, Shields DC, Uhlar CM, Lindsay AJ. Whitehead AS. Acute phase proteins in salmonids: evolutionary analyses and acute phase response. J Immunol. 1997;158:384–392. [PubMed] [Google Scholar]
- Jones RM, Luo L. Moberg KH. Aeromonas salmonicida-secreted protein AopP is a potent inducer of apoptosis in a mammalian and a Drosophila model. Cell Microbiol. 2012;14:274–285. doi: 10.1111/j.1462-5822.2011.01717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JH, Khan S, Seifert U, Le Gall S, Chow KM, Paschen A, et al. Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat Immunol. 2011;12:45–U67. doi: 10.1038/ni.1974. [DOI] [PubMed] [Google Scholar]
- King CH. Shotts EB. Enhancement of Edwardsiella tarda and Aeromonas salmonicida through ingestion by the ciliated protozoan Tetrahymena pyriformis. FEMS Microbiol Lett. 1988;51:95–99. [Google Scholar]
- Kitao T. Yoshida Y. Effect of an immunopotentiator on Aeromonas salmonicida infection in rainbow trout (Salmo gairdneri. Vet Immunol Immunopathol. 1986;12:287–296. doi: 10.1016/0165-2427(86)90132-7. [DOI] [PubMed] [Google Scholar]
- Kjoglum S, Larsen S, Bakke HG. Grimholt U. The effect of specific MHC class I and class II combinations on resistance to furunculosis in Atlantic salmon (Salmo salar. Scand J Immunol. 2008;67:160–168. doi: 10.1111/j.1365-3083.2007.02052.x. [DOI] [PubMed] [Google Scholar]
- Kodama T, Yamazaki C, Park KS, Akeda Y, Iida T. Honda T. Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol Lett. 2010;311:10–17. doi: 10.1111/j.1574-6968.2010.02066.x. [DOI] [PubMed] [Google Scholar]
- Kornete M. Piccirillo CA. Functional crosstalk between dendritic cells and Foxp3(+) regulatory T cells in the maintenance of immune tolerance. Front Immunol. 2012;3:165. doi: 10.3389/fimmu.2012.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- Kumari J, Bogwald J. Dalmo RA. Eomesodermin of atlantic salmon: an important regulator of cytolytic gene and interferon gamma expression in spleen lymphocytes. PLoS ONE. 2013;8:e55893. doi: 10.1371/journal.pone.0055893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Dunmire S. McSorley SJ. MHC class-I-restricted CD8 T cells play a protective role during primary Salmonella infection. Immunol Lett. 2012;148:138–143. doi: 10.1016/j.imlet.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yang PS, Zhu T, Manuel J, Zhang J, He W, et al. Characterization of fibrinogen-like protein 2 (FGL2): monomeric FGL2 has enhanced immunosuppressive activity in comparison to oligomeric FGL2. Int J Biochem Cell Biol. 2013;45:408–418. doi: 10.1016/j.biocel.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Mackie TJ, Arkwright JA, Pryce-Tannatt TE, Mottram JC, Johnston WD. Menzies WJM. H.M. Stationery Office. Furunculosis Committee Final Report. Edinburgh, Scotland: J. & J. Gray; 1935. pp. 1–67. [Google Scholar]
- Maman Y, Nir-Paz R. Louzoun Y. Bacteria modulate the CD8+T cell epitope repertoire of host cytosol-exposed proteins to manipulate the host immune response. PLoS Comput Biol. 2011;7:1–12. doi: 10.1371/journal.pcbi.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantegazza AR, Magalhaes JG, Amigorena S. Marks MS. Presentation of phagocytosed antigens by MHC class I and II. Traffic. 2013;14:135–152. doi: 10.1111/tra.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Nouri B, McCarter L. Martinez-Urtaza J. Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS ONE. 2011;6:e20275. doi: 10.1371/journal.pone.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel C. Dubois-Darnaudpeys A. Persistence of the virulence of Aeromonas salmonicida strains kept in river sediments. Ann Rech Vet. 1980;11:375–380. [PubMed] [Google Scholar]
- Millan A, Gomez-Tato A, Pardo BG, Fernandez C, Bouza C, Vera M, et al. Gene expression profiles of the spleen, liver, and head kidney in turbot (Scophthalmus maximus) along the infection process with Aeromonas salmonicida using an immune-enriched oligo-microarray. Mar Biotechnol (NY) 2011;13:1099–1114. doi: 10.1007/s10126-011-9374-7. [DOI] [PubMed] [Google Scholar]
- Nagamatsu K, Kuwae A, Konaka T, Nagai S, Yoshida S, Eguchi M, et al. Bordetella evades the host immune system by inducing IL-10 through a type III effector, BopN. J Exp Med. 2009;206:3073–3088. doi: 10.1084/jem.20090494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L. Tatu U. Molecular chaperones in pathogen virulence: emerging new targets for therapy. Cell Host Microbe. 2008;4:519–527. doi: 10.1016/j.chom.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejedlik L, Pierfelice T. Geiser JR. Actin distribution is disrupted upon expression of Yersinia YopO/YpkA in yeast. Yeast. 2004;21:759–768. doi: 10.1002/yea.1135. [DOI] [PubMed] [Google Scholar]
- Nese L. Enger O. Isolation of Aeromonas salmonicida from salmon lice Lepeophtheirus salmonis and marine plankton. Dis Aquat Organ. 1993;16:79–81. [Google Scholar]
- Ng WL, Perez L, Cong J, Semmelhack MF. Bassler BL. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 2012;8:e1002767. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KD, Macaubas C, Nadeau KC, Truong P, Yoon T, Lee T, et al. Serum amyloid A overrides Treg anergy via monocyte-dependent and Treg-intrinsic, SOCS3-associated pathways. Blood. 2011;117:3793–3798. doi: 10.1182/blood-2010-11-318832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien D, Mooney J, Ryan D, Powell E, Hiney M, Smith PR. Powell R. Detection of Aeromonas salmonicida, causal agent of furunculosis in salmonid fish, from the tank effluent of hatchery-reared Atlantic salmon smolts. Appl Environ Microbiol. 1994;60:3874–3877. doi: 10.1128/aem.60.10.3874-3877.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroukhova M, Qi Z, Oriss TB, Dixon-McCarthy B, Ray P. Ray A. Treg-mediated immunosuppression involves activation of the Notch-HES1 axis by membrane-bound TGF-beta. J Clin Invest. 2006;116:996–1004. doi: 10.1172/JCI26490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overheim KA, DePaolo RW, Debord KL, Morrin EM, Anderson DM, Green NM, et al. LcrV plague vaccine with altered immunomodulatory properties. Infect Immun. 2005;73:5152–5159. doi: 10.1128/IAI.73.8.5152-5159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua E, Parama A, Iglesias R, Sanmartin ML. Leiro J. Effects of bacteria on the growth of an amoeba infecting the gills of turbot. Dis Aquat Organ. 2001;45:73–76. doi: 10.3354/dao045073. [DOI] [PubMed] [Google Scholar]
- Perrett CA. Zhou D. Type three secretion system effector translocation: one step or two? Front Microbiol. 2011;2:1–2. doi: 10.3389/fmicb.2011.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasch M, Kastbjerg VG, Bruhn JB, Dalsgaard I, Givskov M. Gram L. Quorum sensing signals are produced by Aeromonas salmonicida and quorum sensing inhibitors can reduce production of a potential virulence factor. Dis Aquat Organ. 2007;78:105–113. doi: 10.3354/dao01865. [DOI] [PubMed] [Google Scholar]
- Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reithmeier-Rost D, Bierschenk S, Filippova N, Schroder-Braunstein J. Sing A. Yersinia V antigen induces both TLR homo- and heterotolerance in an IL-10-involving manner. Cell Immunol. 2004;231:63–74. doi: 10.1016/j.cellimm.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Romer VK, Dalsgaard I, Holten-Andersen L. Raida MK. Potential role of specific antibodies as important vaccine induced protective mechanism against Aeromonas salmonicida in rainbow trout. PLoS ONE. 2012;7:e46733. doi: 10.1371/journal.pone.0046733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romstad AB, Reitan LJ, Midtlyng P, Gravningen K. Evensen O. Antibody responses correlate with antigen dose and in vivo protection for oil-adjuvanted, experimental furunculosis (Aeromonas salmonicida subsp. salmonicida) vaccines in Atlantic salmon (Salmo salar L.) and can be used for batch potency testing of vaccines. Vaccine. 2013;31:791–796. doi: 10.1016/j.vaccine.2012.11.069. [DOI] [PubMed] [Google Scholar]
- Salte R, Norberg K, Arnesen JA, Odegaard OR. Eggset G. Serine protease and glycerophospholipid – cholesterol acyltransferase of Aeromonas salmonicida work in concert in thrombus formation – In vitro the process is counteracted by plasma antithrombin and alpha-2-macroglobulin. J Fish Dis. 1992;15:215–227. [Google Scholar]
- Salte R, Norberg K, Odegaard OR, Arnesen JA. Olli JJ. Exotoxin-induced consumptive coagulopathy in Atlantic salmon, Salmo salar l – Inhibitory effects of exogenous antithrombin and alpha(2)-macroglobulin on Aeromonas salmonicida serine protease. J Fish Dis. 1993;16:425–435. [Google Scholar]
- Schwenteit J, Gram L, Nielsen KF, Fridjonsson OH, Bornscheuer UT, Givskov M. Gudmundsdottir BK. Quorum sensing in Aeromonas salmonicida subsp. achromogenes and the effect of the autoinducer synthase AsaI on bacterial virulence. Vet Microbiol. 2011;147:389–397. doi: 10.1016/j.vetmic.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Sepulcre MP, Alcaraz-Perez F, Lopez-Munoz A, Roca FJ, Meseguer J, Cayuela ML. Mulero V. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- Shaver CM. Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72:6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A, Roggenkamp A, Geiger AM. Heesemann J. Yersinia enterocolitica evasion of the host innate immune response by V antigen-induced IL-10 production of macrophages is abrogated in IL-10-deficient mice. J Immunol. 2002a;168:1315–1321. doi: 10.4049/jimmunol.168.3.1315. [DOI] [PubMed] [Google Scholar]
- Sing A, Rost D, Tvardovskaia N, Roggenkamp A, Wiedemann A, Kirschning CJ, et al. Yersinia V-antigen exploits toll-like receptor 2 and CD14 for interleukin 10-mediated immunosuppression. J Exp Med. 2002b;196:1017–1024. doi: 10.1084/jem.20020908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhto N, Sintiprungrat K. Thongboonkerd V. Alterations in macrophage cellular proteome induced by calcium oxalate crystals: the association of HSP90 and F-actin is important for phagosome formation. J Proteome Res. 2013;12:3561–3572. doi: 10.1021/pr4004097. [DOI] [PubMed] [Google Scholar]
- Stavrinides J, Kirzinger MW, Beasley FC. Guttman DS. E622, a miniature, virulence-associated mobile element. J Bacteriol. 2012;194:509–517. doi: 10.1128/JB.06211-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber K, Burr SE, Braun M, Wahli T. Frey J. Type III secretion genes in Aeromonas salmonicida subsp. salmonicida are located on a large thermolabile virulence plasmid. J Clin Microbiol. 2003;41:3854–3856. doi: 10.1128/JCM.41.8.3854-3856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer N, Frey J. Vanden Bergh P. Clustering subspecies of Aeromonas salmonicida using IS630 typing. BMC Microbiol. 2013;13:36. doi: 10.1186/1471-2180-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, Karlyshev AV, Fish L, Durant EL, Winson MK, Chhabra SR, et al. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada H, Aiba S, Shibata K, Ohteki T. Takada H. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun. 2005;73:7967–7976. doi: 10.1128/IAI.73.12.7967-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KH, Dallaire-Dufresne S, Daher RK, Frenette M. Charette SJ. An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS ONE. 2012;7:1–8. doi: 10.1371/journal.pone.0033725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KH, Frenette M. Charette SJ. IS-mediated loss of virulence by Aeromonas salmonicida: a tangible piece of an evolutionary puzzle. Mob Genet Elements. 2013;3:e23498. doi: 10.4161/mge.23498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsan MF. Gao B. Heat shock proteins and immune system. J Leukoc Biol. 2009;85:905–910. doi: 10.1189/jlb.0109005. [DOI] [PubMed] [Google Scholar]
- Urbanowski ML, Brutinel ED. Yahr TL. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect Immun. 2007;75:4432–4439. doi: 10.1128/IAI.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Bergh P, Burr SE, Benedicenti O, von Siebenthal B, Frey J. Wahli T. Antigens of the type-three secretion system of Aeromonas salmonicida subsp. salmonicida prevent protective immunity in rainbow trout. Vaccine. 2013 doi: 10.1016/j.vaccine.2013.08.057. doi: 10.1016/j.vaccine.2013.08.057. [DOI] [PubMed] [Google Scholar]
- Vipond R, Bricknell IR, Durant E, Bowden TJ, Ellis AE, Smith M. MacIntyre S. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect Immun. 1998;66:1990–1998. doi: 10.1128/iai.66.5.1990-1998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Mo ZL, Xiao P, Li J, Zou YX, Hao B. Li GY. EseD, a putative T3SS translocon component of Edwardsiella tarda, contributes to virulence in fish and is a candidate for vaccine development. Mar Biotechnol. 2010;12:678–685. doi: 10.1007/s10126-009-9255-5. [DOI] [PubMed] [Google Scholar]
- Waters CM, Wu JT, Ramsey ME, Harris RC. Bassler BL. Control of the type 3 secretion system in Vibrio harveyi by quorum sensing through repression of ExsA. Appl Environ Microbiol. 2010;76:4996–5004. doi: 10.1128/AEM.00886-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Wing JB. Sakaguchi S. Two modes of immune suppression by Foxp3(+) regulatory T cells under inflammatory or non-inflammatory conditions. Semin Immunol. 2011;23:424–430. doi: 10.1016/j.smim.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL. Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mena P, Romanov G, Lin JS, Smiley ST. Bliska JB. A protective epitope in type III effector YopE is a major CD8 T cell antigen during primary infection with Yersinia pseudotuberculosis. Infect Immun. 2012;80:206–214. doi: 10.1128/IAI.05971-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chi H, Niu C, Bogwald J. Dalmo RA. Molecular cloning and characterization of Foxp3 in Atlantic salmon (Salmo salar. Fish Shellfish Immunol. 2011;30:902–909. doi: 10.1016/j.fsi.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang L, Ren C, Zhao J, Chen C, Jiang X, et al. Autophagy is induced by the type III secretion system of Vibrio alginolyticus in several mammalian cell lines. Arch Microbiol. 2011;193:53–61. doi: 10.1007/s00203-010-0646-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic comparison of T3SSs from the Ysc family.
Comparison of the organization of T3SS clusters in A. salmonicida subsp. salmonicida A449, A. hydrophila SSU, A. veronii AER39, A. diversa 2478-85, Pseudomonas aeruginosa PAO1, Photobacterium damselae subsp. damselae CIP 102761, Photorhabdus luminescens subsp. laumondii TTO1, Yersinia pestis biovar Antiqua str. E1979001, and Vibrio parahaemolyticus RIMD 2210633.
Profound analysis and comparison of published Aeromonas genomes.
Grey: conserved ORFs; light green: ORFs specific of the species; yellow: IS630; pink: other IS elements; red: putative or characterized virulence factors; mauve: ORFs for resistance to antibiotic or heavy metal; dark green: ORFs associated to pili, fimbriae or flagella; blue: ORFs associated with phage; cyan: tRNA and rRNA; orange: ORFs with homology to eukaryotic genes. Aeromonas salmonicida subsp. salmonicida A449, A. salmonicida subsp. achromogenes AS03, A. salmonicida (ATCC7966, ML09-119 and SSU), A. caviae Ae398, A. veronii (B565, AMC34, AMC35, AER39 and AER397), A. aquariorum AAK1, and A. diversa 2478-85.
Rapid cell-rounding induced by the T3SS of A. salmonicida.
Epithelioma papulosum cyprini cells (EPCs) seeded in a glass bottom well and exposed to A. salmonicida wt JF2267 (S1) and ΔascV mutant JF2747 (S2) expressing GFP (after induction with IPTG) for 120 min. Following infection at 20°C, time-lapse imaging using video microscopy was performed using a Nikon Eclipse TE2000-U inverted microscope equipped with a climate-controlled chamber. Data acquisition and image processing were performed using NIS software of Nikon Instruments. DIC and fluorescence images were acquired and assembled in movies. Image acquisition at intervals of 2 min for 120 min. MOI of 10:1 (bacteria: fish cells).