Abstract
We constructed a metabolically engineered glutamate-independent Bacillus amyloliquefaciens strain with considerable γ-PGA production. It was carried out by double-deletion of the cwlO gene and epsA-O cluster, as well as insertion of the vgb gene in the bacteria chromosome. The final generated strain NK-PV elicited the highest production of γ-PGA (5.12 g l−1), which was 63.2% higher than that of the wild-type NK-1 strain (3.14 g l−1). The γ-PGA purity also improved in the NK-PV strain of 80.4% compared with 76.8% for the control. Experiments on bacterial biofilm formation experiment showed that NK-1 and NK-c (ΔcwlO) strains can form biofilm; the epsA-O deletion NK-7 and NK-PV strains could only form an incomplete biofilm.
Introduction
Poly-γ-glutamic acid (γ-PGA) is an unusual homopolymer of D/L-glutamate acid polymerized through γ-glutamyl bonds (Ashiuchi and Misono, 2002). γ-PGA is biodegradable, edible, water-soluble and non-toxic toward humans and the environment. Therefore, it has attracted wide interests in a broad range of fields: food, medicine and water treatment (Shih and Van, 2001; Ashiuchi, 2013).
γ-PGA-producing strains are divided into two categories based on their requirement for exogenous glutamic acid: glutamate-dependent strains and glutamate-independent strains (Shih and Van, 2001). Compared with the glutamate-dependent strains, the independent strains are more preferable for the industrial production of γ-PGA with their low cost and simplified fermentation process (Cao et al., 2011). However, their lower γ-PGA productivity compared with glutamate-dependent strains limits their industrial application. Construction of a metabolically engineered glutamate-independent strain with high γ-PGA productivity is required.
Bacillus amyloliquefaciens NK-1 is a derivative of B. amyloliquefaciens LL3 (Cao et al., 2011). It is a glutamate-independent strain that produces γ-PGA comprising units of L-glutamic acid and D-glutamic acid. The endogenous plasmid pMC1 and upp gene of the NK-1 strain has been deleted, so it is suitable for genetic manipulation such as markerless deletion or insertion of genes.
The approaches employed for the enhancement of γ-PGA production has been mainly focused on two aspects: (i) optimization of the fermentation conditions (Bajaj and Singhal, 2009; Huang et al., 2011) and (ii) characterization of the genes involved in γ-PGA production and use of bioengineering methods to improve the γ-PGA yield (Ashiuchi et al., 2006; Scoffone et al., 2013; Zhang et al., 2013). Increasing numbers of studies have focused on the second approach to construct metabolically engineered microorganisms with high γ-PGA productivity. Heterologous expression of the γ-PGA synthetase complex (pgsBCA) is another strategy for γ-PGA production improvement, and it has been carried out in coryneform bacteria and Escherichia coli (Sung et al., 2005; Cao et al., 2013).
Peptidoglycan hydrolases belong to the NLPC/P60 family (DL-endopeptidase family II), such as LytE, LytF, CwlS and CwlO (Smith et al., 2000; Bisicchia et al., 2007; Vollmer et al., 2008). These enzymes can not only hydrolyse peptidoglycan, but also degrade γ-PGA. Single deletions of lytE, lytF, cwlS and cwlO were studied in glutamate-dependent Bacillus subtilis (natto), and only the cwlO deletion strain showed higher productivity of γ-PGA, and disruptions of other genes had little effect (Yamaguchi et al., 2004; Mitsui et al., 2011).
Exopolysaccharides (EPS) are synthesized primarily in microbial cells and then secreted into the extracellular environment as biofilms and capsules (Branda et al., 2001; 2005; Marvasi et al., 2010; Donot et al., 2012). Some Bacillus strains can produce abundant EPS and are essential for the formation of biofilms (Donot et al., 2012). The epsA-O cluster has been reported to be associated mainly with EPS production (Branda et al., 2004; 2006), which are the main by-products of some γ-PGA-producing strains. Thus, deletion of epsA-O can depress EPS production, and metabolic flux could be used to enhance γ-PGA productivity.
Vitreoscilla hemoglobin (VHb) is one of the most extensively studied bacterial hemoglobin molecules synthesized by the Gram-negative bacterium Vitreoscilla (Wakabayashi et al., 1986). It has been confirmed that VHb can interact directly with the terminal respiratory oxidase by delivering oxygen to it, in order to enhance oxidative phosphorylation, and thus the production of adenosine triphosphate (Park et al., 2002). It has been demonstrated that VHb expression in heterologous bacterial hosts can enhance cell density and metabolic production, especially under oxygen-limiting conditions (Kang et al., 2002; Urgun-Demirtas et al., 2003). Heterologous expression of the vgb gene (which encodes VHb) has been conducted in various microorganisms as well as plant and mammal cells (Zhang et al., 2007). During the fermentation process of γ-PGA, broth viscosity increased and dissolved oxygen in the broth reached 0% after 12–18 h of incubation, which decreased oxygen transfer to cells, inhibited cell growth and γ-PGA production. Expression of the vgb gene in a γ-PGA-producing strain will alleviate oxygen limitation at the later stage of fermentation to enhance cell density and γ-PGA production.
In the present study, we aimed to construct a high-productivity γ-PGA-producing strain based on the glutamic acid-independent B. amyloliquefaciens NK-1. We double-deleted the cwlO gene and epsA-O cluster and simultaneously inserted the vgb gene in the bacterial chromosome. The generated engineered strain NK-PV showed higher production of γ-PGA and γ-PGA purity compared with the wild-type NK-1 strain.
Results
Identification of target gene knockouts or insertion
To construct a metabolically engineered, γ-PGA high-producing strain, double-deletion of the cwlO gene and epsA-O cluster and insertion of a vgb gene were carried out in the B. amyloliquefaciens NK-1 strain. A gene marker-less knockout method was used to construct the gene deletion and insertion mutant, which was based on an upp cassette and the 5-FU selection (Keller et al., 2009).
The primers cwlO-SS/XX, AO-SS/XX and Amy-SS/XX were used to verify the gene mutants (Fig. 1). DNA sequencing was used to confirm construction of the engineered strain.
Figure 1.
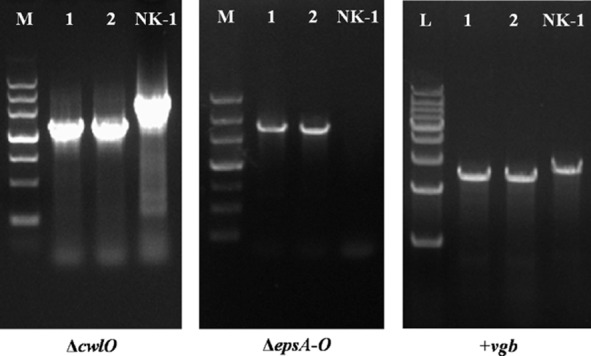
Confirmation of the gene deletion or insertion by PCR. Chromosomal DNA served as the template for amplification. Lane M, DNA marker III; Lane L, DNA ladder (1 kb); Lane 1, 2, gene deletion or insertion strains amplified with relative primers; Lane NK-1, wild-type NK-1 strains amplified with relative primers.
Confirmation of expression of the vgb gene and its effect on cell growth
It has been reported that VHb expression enhances the cell density and yield of metabolite production (Olano et al., 2008). Here, we inserted the vgb gene in the AmyA locus of the NK-7 chromosome. To identify expression of the vgb gene, RT-PCR and carbon monoxide (CO)-difference spectra were used. The vgb gene transcriptional levels of the NK-PV strain were tested: It was transcribed successfully (Fig. 2A). VHb has been reported to bind with CO and to show a characteristic absorption peak at 420 nm (Liu and Webster, 1974). CO-difference spectra of the NK-PV strain showed an absorption peak at approximately 420 nm after 36 h of induction, but this peak was absent in the NK-1 strain (Fig. 2B). These results suggested that the mutant NK-PV strain bearing the vgb gene could express functional VHb.
Figure 2.
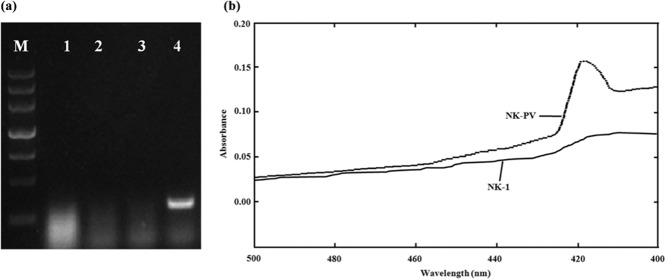
Conformation of the expression of vgb gene.
A. RT-PCR results between NK-1 and NK-PV strain. Lane M, DNA marker III; Lane 1–4, amplification results using primers VG-F and VG-R with RNA from NK-1 strain, RNA from NK-PV strain, cDNA from NK-1 strain and cDNA from NK-PV strain as template respectively.
B. CO-spectra difference analysis of NK-1 and NK-PV strain.
To confirm the effect of VHb on cell growth, the growth curve of B. amyloliquefaciens NK-1 and mutant strains were monitored by measuring OD600. The growth rate of NK-PV was higher than that of other strains without the vgb gene (Fig. 3A). The dry cell weight of the NK-PV strain was also higher than that of the other mutant strains. These results suggested that VHb expression increased cell growth and dry cell weight in our strain.
Figure 3.
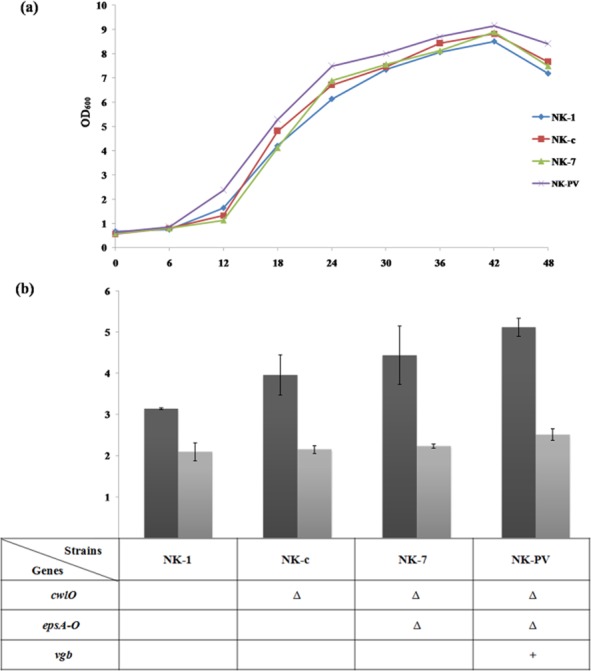
γ-PGA fermentation results between NK-1 and mutant strains.
A. Time curves of cell growth of NK-1 and mutant strains.
B. Comparison of γ-PGA production and cell dry weight between NK-1 and mutant strains after 48 h cultivation. Values represent means ± SD.
Comparison of the results of γ-PGA fermentation among B. amyloliquefaciens NK-1 and mutant strains by flask culture
To evaluate accumulation of gene-targeted B. amyloliquefaciens mutants on γ-PGA production, flask culture of B. amyloliquefaciens NK-1 and gene mutant strains NK-c, NK-7 and NK-PV was undertaken under identical conditions. The results of γ-PGA fermentation among B. amyloliquefaciens NK-1 and the other mutant strains are shown in Fig. 3B and Table 1. The γ-PGA yield from NK-c was approximately 26.2% higher than that of the NK-1 strain. The molecular weight was also higher than that of the NK-1 strain. These results were in accordance with a study carried out in the glutamate-dependent strain, in which deletion of cwlO gene increased not only the γ-PGA yield but also its molecular weight (Mitsui et al., 2011). γ-PGA yields of the NK-7 strain increased by 41.5% compared with that of the NK-1 strain, and by 12.1% compared with that of the NK-c strain. The NK-PV strain showed the highest production of γ-PGA, leading to a yield of 5.12 g l−1, which was 63.2% higher than that of the NK-1 strain (3.14 g l−1). The molecular weight of γ-PGA in NK-7 and NK-PV strains was also higher than that of the NK-1 strain (Table 1).
Table 1.
Comparison of γ-PGA fermentation results between NK-1 and mutant strains
| Strains | Yield (g l−1) | Mw (× 105) | Viscosity (cP) | CDW (g l−1) | Purity (%) |
|---|---|---|---|---|---|
| NK-1 | 3.14 ± 0.02 | 3.39 ± 0.08 | 10.05 ± 3.31 | 2.09 ± 0.22 | 76.8 ± 1.1 |
| NK-c | 3.96 ± 0.49 | 4.12 ± 0.04 | 16.70 ± 1.85 | 2.15 ± 0.09 | 75.7 ± 1.7 |
| NK-7 | 4.44 ± 0.71 | 4.01 ± 0.14 | 19.40 ± 2.40 | 2.24 ± 0.05 | 81.8 ± 0.8 |
| NK-PV | 5.12 ± 0.22 | 3.76 ± 0.49 | 31.57 ± 1.33 | 2.51 ± 0.14 | 80.4 ± 1.3 |
The viscosity of the culture broth of the NK-1 strain and other mutant strains was 10.05, 16.70, 19.40 and 31.57 (cP) respectively (Table 1). Our previous studies have shown that a higher concentration or higher molecular weight of γ-PGA leads to a more viscous broth. In the present study, the molecular weight of γ-PGA in gene mutant strains was similar whereas the γ-PGA yield changed considerably. The strain with the higher concentration of γ-PGA made the broth more viscous. Thus, even though several factors affect broth viscosity, the viscosity of the culture broth in these fours strains was mainly determined by its γ-PGA component.
We further determined the fermentation results between NK-1, NK-c, NK-PV, NK-c-HB and NK-PV-HB strains. γ-PGA yield and molecular weight of NK-1 strain were 3.4 g l−1 and 391.5 K respectively. The γ-PGA yields of cwlO gene complementary strains NK-c-HB (3.32 g l−1) and NK-PV-HB (3.73 g l−1) are all lower than their corresponding gene deletion strains NK-c (4.35 g l−1) and NK-PV (5.33 g l−1). Moreover, molecular weights of NK-c-HB and NK-PV-HB strains are 379.7 k and 401.2 k, which were all lower than NK-c (455.6 k) and NK-PV (440.1 k).
Effect of epsA-O deletion on biofilm formation
Microbes can construct structurally complex biofilms through production of an extracellular matrix. Studies have shown that a wild-type strain of the Gram-positive bacterium B. subtilis can build such a matrix. Some genetic, biochemical and cytological evidence have suggested that the matrix is composed predominantly of a protein component (TasA) and an EPS component, and that absence of TasA or the EPS results in a residual matrix, and that absence of both components leads to complete failure of biofilm formation (Branda et al., 2004; 2006). The epsA-O operon has been reported to encode biosynthetic machinery to produce EPS (Branda et al., 2004; Kearns et al., 2005). Once the epsA-O cluster is disrupted, Bacillus strains cannot secrete EPS, which greatly affects biofilm formation. In this work, the epsA-O cluster was deleted and its affect on biofilm formation determined. The NK-1 strain and NK-c strain could form a complete pellicle (Fig. 4). The NK-7 strain and NK-PV strain could only form a deficient incomplete pellicle.
Figure 4.
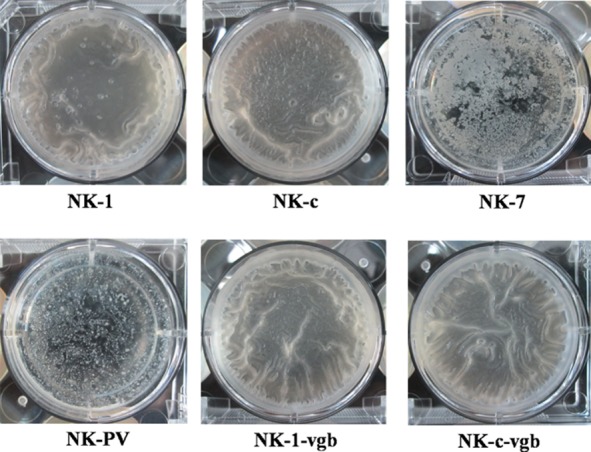
Cell pellicle formation between NK-1 and mutant strains. Cells were cultured at 30°C for 80 h in MSgg broth contained within a 6-well microtiter dish.
Effect of epsA-O deletion on γ-PGA purity and colony morphology
Some EPS can also be deposited by cold ethanol and, upon mixing with γ-PGA, decrease the purity of the γ-PGA product. To increase the purity of γ-PGA, we knocked out the epsA-O cluster. The purities of γ-PGA in the epsA-O cluster-deleted strains NK-7 and NK-PV were 81.8% and 80.4% respectively, which were higher than those of NK-1 and NK-c strains (76.8% and 75.7% respectively). The colony morphologies of NK-1 and gene mutant strains are shown in Fig. 5. The colonies of different strains were similar, only colonies of the NK-PV strain showed a slightly flatter and larger appearance than the other strains.
Figure 5.
Colony morphologies of NK-1 and mutant strains. Cells were cultured at 37°C for 36 h in LB agar plate.
Discussion
Most γ-PGA-producing Bacillus strains are unsuitable for genetic manipulation because of their low or absent genetic competence (Sung et al., 2005). Thus, few metabolic engineering strategies have been reported to improve γ-PGA production. The temperature-sensitive shuttle plasmid pKSV7 and upp counter selection-based marker-less deletion method has been used successfully for the manipulation of multiple genes (Smith and Youngman, 1992; Keller et al., 2009). In this work, we deleted the cwlO and epsA-O clusters and inserted a vgb gene in the NK-1 chromosome based on this method. The final metabolically engineered B. amyloliquefaciens NK-PV strain showed increased production of γ-PGA as well as increased purity.
Certain studies have focused on improving γ-PGA production by deletion of its degrading enzyme genes, but the ggt and pgdS gene deletion mutant strain produced similar amounts of γ-PGA to the wild-type strain (Kimura et al., 2004; Mitsui et al., 2011). This may be because GGT and PgdS are regulated appropriately in vivo and they function only if nitrogen in the medium is limited. Mitsui and colleagues (2011) investigated the effects of the cell wall lytic enzymes, LytE, LytF, CwlO and CwlS, on γ-PGA production in B. subtilis (natto). They found that disruption of the lytE, lytF and cwlS genes had little effect on γ-PGA production, whereas γ-PGA levels in the cwlO mutant were approximately twofold higher than that of the wild-type. Furthermore, they found that the cwlO deletion strain showed a higher molecular weight of γ-PGA. cwlO deletion stops γ-PGA being hydrolysed by CwlO, and its absence results in a higher yield and molecular weight of γ-PGA. Domínguez-Cuevas and colleagues (2013) reported that the cwlO gene deletion strain exhibits a shorter and wider cell size, resulting in a higher specific surface area that could increase the γ-PGA transport and thereafter the production of γ-PGA. In the present study, the cwlO gene deletion strain NK-c showed a higher yield and molecular weight of γ-PGA compared with the NK-1 strain, which was consistent with our previous work (Feng et al., 2014) and that of Mitsui and colleagues (2011); moreover, the cwlO gene complementary NK-c-HB and NK-PV-HB strains exhibited lower yield and molecular weight of γ-PGA. These results indicated that CwlO indeed related to the γ-PGA hydrolyse. No report has demonstrated that deletion of the epsA-O cluster or expression of the vgb gene increases the molecular weight of γ-PGA, so the higher molecular weights of γ-PGA in NK-c, NK-7 and NK-PV strains must be related mainly to deletion of the cwlO gene.
Bacteria can produce a wide variety of EPS such as curdlan (Shih et al., 2009), alginate (Celik et al., 2008), xanthan (Kalogiannis et al., 2003) and levan (Shih et al., 2010). The biosynthesis of EPS involves consumption of a large amount of the carbon source, which decreases the metabolic flux used for γ-PGA production. Moreover, EPS are secreted into the medium and interact with γ-PGA, thereby influencing the purity of γ-PGA. Liu and colleagues (2011) depressed EPS production in B. amyloliquefaciens C06, which caused overproduction of its γ-PGA. We deleted the epsA-O cluster based on the NK-c strain: The generated NK-7 strain showed a higher yield and purity of γ-PGA. Disruption of the epsA-O cluster blocks the pathway of EPS synthesis, which reduce the production of extracellular products as well as enables the metabolic flux to be used more efficiently for γ-PGA production. Moreover, defects in EPS production would reduce spatial competition and cell membrane permeability pressure, which would enable intracellular γ-PGA to be transported more efficiently and to accumulate outside cells. Thus, the epsA-O deletion strain showed good properties for γ-PGA production.
In Bacillus strains, EPS and TasA are two major components of the extracellular matrix that hold differentiated cell chains together to form a highly organized colony and pellicle architecture. Branda and colleagues (2006) demonstrated that single deletion of the eps cluster can result in incomplete formation of biofilms, and that double-deletion of eps and tasA ensure that the strain fails to make a pellicle. In the present study, the wild-type strain and cwlO deletion strain could form complete pellicles. In accordance with the previous study, the epsA-O deletion strains NK-7 and NK-PV could form only an incomplete pellicle (Branda et al., 2006). However, biofilm formed by NK-PV strain seems even more incomplete than NK-7 strain (Fig. 4). To test whether the expression of VHb influences bacteria biofilm formation, we determined the biofilm formation in two vgb gene expression strains NK-1-vgb and NK-c-vgb. Form Fig. 4, we can find that the two strains can all form complete pellicle like NK-1 and NK-c strains. These results indicated that VHb expression has little effect on bacteria bioflim formation, and it is not efficient to affect biofilm formation in the wild-type strains.
VHb has been used widely for the enhancement of cell density and target products, especially in highly viscous media, in which oxygen availability is limited (Frey and Kallio, 2003; Su et al., 2010). γ-PGA synthesis results in extreme viscosity of the culture medium, which seriously limit its oxygen content. p43 is a strong and well-characterized promoter in B. subtilis (Wang and Dio, 1984), which is assumed to express genes efficiently in other Bacillus genus stains. We expressed the vgb gene under the p43 promoter in the present study. However, γ-PGA in the NK-PV strain increased by only 15.3% compared with the NK-7 strain. This finding could be due to several reasons. VHb elicits its effects under oxygen-limited conditions (Zhang et al., 2007; Su et al., 2010). However, the oxygen-limited condition in NK-PV fermentation appeared after 12–18 h of incubation, which reduced the time in which VHb could elicit its effects. It has been reported that VHb is more likely to elicit its effects, for example, in a greater medium volume or at a lower rotation speed. However, the γ-PGA yields under those conditions are lower than those under optimal conditions (Zhang et al., 2007; Su et al., 2010). Therefore, under our cultivation conditions, the limitation of oxygen transfer may be less severe than that in other strains.
In summary, a metabolically engineered high γ-PGA productivity strain was created in a glutamate-independent γ-PGA-producing strain. We double-deleted the cwlO gene and epsA-O cluster and inserted the vgb gene in the NK-1 strain chromosome. The final generated NK-PV strain showed the highest γ-PGA production, which was 63.2% higher than that of the wild-type NK-1 strain; moreover, the γ-PGA purity in NK-PV strain were also higher than that of the NK-1 strain.
Experimental procedures
Strains, plasmids and growth conditions
All the strains and plasmids used in this work are listed in Table 2. Escherichia coli DH5α was used for the propagation and transformation of plasmids. Demethylation of plasmids was carried out in E. coli GM2163. The p43-vgb gene fragment was isolated from the pKSPVK plasmid (Zhang et al., 2013). The oligonucleotide primers used in this study are listed in Table 3.
Table 2.
Strains and plasmids used in this study
| Strains and plasmids | Relevant genotype and characteristics | Source |
|---|---|---|
| Strains | ||
| B. amyloliquefaciens LL3 | Poly-γ-glutamic acid (γ-PGA) producing strain | (Cao et al., 2011) |
| B. amyloliquefaciens NK-1 | LL3 derivative, ΔpMC1, Δupp | This lab |
| B. amyloliquefaciens NK-c | NK-1 derivative, ΔcwlO | This work |
| B. amyloliquefaciens NK-7 | NK-1 derivative, ΔcwlO, ΔepsA-O | This work |
| B. amyloliquefaciens NK-PV | NK-1derivative, ΔcwlO, ΔepsA-O, p43-vgb | This work |
| B. amyloliquefaciens NK-c-HB | NK-c derivative with complementary plasmid pWH1520-cwlO | (Feng et al., 2014) |
| B. amyloliquefaciens NK-PV-HB | NK-PV derivative with complementary plasmid pWH1520-cwlO | This work |
| B. amyloliquefaciens NK-1-vgb | NK-1 derivative with vgb gene expression plasmid pWHV | This work |
| B. amyloliquefaciens NK-c-vgb | NK-c derivative with vgb gene expression plasmid pWHV | This work |
| E. coli DH5α | F-, φ80dlacZΔM1, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk-, mk+), phoA, supE44, λ-thi-1, gyrA96, relA1 | This lab |
| E. coli GM2163 | F-, ara-14 leuB6 thi-1 fhuA31 lacY1 tsx-78 galK2 | This lab |
| galT22 supE44 hisG4 rpsL 136 (Strr) xyl-5 mtl-1 | ||
| dam13::Tn9 (Camr) dcm-6 mcrB1 hsdR2 mcrA | ||
| Plasmids | ||
| pKSV7 | Shuttle vector, temperature-sensitive (ts) replication origin, Ampr (gram-negative)and Cmr (gram-positive) | (Smith and Youngman, 1992) |
| pMD19-T | T easy vector for gene cloning; Ampr | Takara |
| pKSPVK | pKSV7 derivative consists of LL3 16S rRNA gene, promoter p43, vgb, and Kanr for selection | (Zhang et al., 2013) |
| p-upp | pKSV7-derivation with upp gene | This lab |
| p-upp-ΔAO | pKSV7-derivation with upp gene and deletion fragment of epsA-O | This work |
| p-upp-ΔcwlO | pKSV7-derivation with upp gene and the deletion of fragment cwlO | (Feng et al., 2014) |
| p-upp-spvx | pKSV7-derivation with upp gene and insertion fragment p43-vgb | This work |
| pWHV | pWH1520 derivative, vgb expression vector coding for VHb | (Zhang et al., 2013) |
| pWH1520-cwlO | pWH1520 derivative carrying the structural gene cwlO | (Feng et al., 2014) |
Table 3.
Primers used in this study
| Primers | Sequence(5′-3′) |
|---|---|
| AO-SF | GCCGGATCCAGTCGGCATTTTTTACGCCGTCC |
| AO-SR | CAGTCTCGATCAGACGTGTCATGATAAAAATCAGTAA |
| AO-XF | GATTTTTATCATGACACGTCTGATCGAGACTGCAGGCA |
| AO-XR | CCCGTCGACACGTAAAAACCCGGTTCCTCAT |
| AO-SS | GAAAAATGCGCCGCCATGAATCCATAC |
| AO-ZZ | CGTCTGGTCATCATCAATAAAAGCCACAGG |
| AO-XX | CGCGACAGATAATCTTTCGTGTCACGG |
| cwlO-SS | GACTGACGTCATGAGCTGCTGGGTTTTT |
| cwlO-XX | CCAAGTTCTTTTTCACCGGGAACGCC |
| Amy-SF | CCCCGTCGACAATGTTGCATTAAGAAGGCTGAAAACG |
| Amy-SR | TGCATGCACGAAGCTCTTTCGTTTTTGAATCATTTTTCT |
| Amy-XF | GCGGTTGAATAATGAATATGTACGGGACAAAAGGGAC |
| Amy-XR | CCCCGGATCCTTATTTTTCCAAGGCGAAAGATT |
| PV-F | ATTCAAAAACGAAAGAGCTTCGTGCATGCAGGCCG |
| PV-R | TTGTCCCGTACATATTCATTATTCAACCGCTTGAG |
| Amy-SS | AGGTTTTCACCCGCATATTAAGCAG |
| Amy-XX | AGGACAGAAAAAACAGAAACAGCACG |
| VG-F | TCCTGTATTGAAGGAGCATGGCGTTAC |
| VG-R | CGCCTGCTTGACAATGTTTGACTGC |
The underlined is the restriction enzyme cleavage site.
B. amyloliquefaciens and E. coli strains were grown at 37°C in lysogeny broth (LB) medium for the routine construction and maintenance of strains. For γ-PGA production, B. amyloliquefaciens was cultured at 37°C at 180 r.p.m. for 48 h in γ-PGA fermentation medium (Cao et al., 2011). If required, antibiotics were used (100 μg ml−1 ampicillin, 5 μg ml−1 chloramphenicol). 5-fluorouracil (5-FU) was added to the medium at a final concentration of 100 μg ml−1.
DNA manipulation and construction of mutant strains
The temperature-sensitive plasmid p-upp was used to construct the epsA-O deletion vector. The upstream and downstream fragments of epsA-O were amplified by PrimeSTAR HS DNA Polymerase (Takara Bio, Tokyo, Japan) using primers AO-SF/AO-SR and AO-XF/AO-XR respectively. The two regions of DNA were joined by overlap-polymerase chain reaction (PCR). The generated fragment was restricted by SalI and BamHI, and cloned into the p-upp vector using the same enzymes to create the deletion plasmid p-upp-ΔAO. The vgb gene combined with the p43 promoter was inserted into the amylase gene AmyA region of the chromosome. To construct the vgb insertion vector, the p43-vgb fragment was obtained from the plasmid pKSPVK (Zhang et al., 2013) by primers PV-F/PV-R, and the upstream and downstream regions were amplified using primers Amy-SF/Amy-SR and Amy-XF/Amy-XR respectively. These three regions of DNA fragments were joined by overlap-PCR, and the generated fragment cloned into the pMD19-T simple vector for sequencing. The generated plasmid T-SPVX was digested with BamHI and SalI, and the fragment cloned into the p-upp vector to generate the insertion vector p-upp-spvx.
Gene knockout mutant strains were constructed by a gene marker-less deletion method based on the upp cassette (Keller et al., 2009). The cwlO deletion strain NK-c has been constructed in our previous work (Feng et al., 2014). p-upp-ΔAO and p-upp-spvx plasmids were first introduced into E. coli GM2163 for demethylation. Plasmids isolated from E. coli GM2163 were treated with BamHI methyltransferase before being transformed into target strains (Feng et al., 2013). Cells were incubated on LB agar plates with 5 μg ml−1 chloramphenicol. Single colonies were picked, and primers AO-SS/AO-ZZ and Amy-SS/Amy-XX used to determine single-cross clones by PCR. Selected single-cross clones were then incubated in LB medium, supplemented with 100 μg ml−1 5-FU, for 24 h at 42°C. Cells were diluted 105 times and spread on LB agar plates with 5-FU. Single colonies were picked, and primers AO-SS/AO-XX and Amy-SS/Amy-XX used to determine the gene mutant clones by PCR. Double deletion of cwlO and epsA-O strains was designated ‘B. amyloliquefaciens NK-7’. Double deletion of cwlO and epsA-O combined with the vgb gene insertion strain was designated ‘B. amyloliquefaciens NK-PV’.
Production of γ-PGA by flask culture
B. amyloliquefaciens NK-1, B. amyloliquefaciens NK-c, B. amyloliquefaciens NK-7, B. amyloliquefaciens NK-PV, B. amyloliquefaciens NK-c-HB and B. amyloliquefaciens NK-PV-HB strains were first incubated in 50 ml LB medium respectively, in a 250-ml flask, and cultured aerobically for 16 h at 37°C with shaking at 180 r.p.m. One milliliter seed culture was then transferred into a 500-ml flask containing 100 ml γ-PGA fermentation medium. Flask cultures were incubated at 180 r.p.m. for 48 h at 37°C. For inducible expression, 0.5% xylose was added to the gene complementary strains medium after 3 h incubation. All cultivations were repeated at least five times.
Reverse transcription-polymerase chain reaction (RT-PCR) analyses
RT-PCR was undertaken to test expression of the vgb gene between NK-1 and NK-PV strains. Cells were harvested for RNA extraction after 16 h of cultivation in LB medium. The commercial RNApure Bacteria kit (Cwbio, Beijing, China) was used to extract total RNA. cDNA was obtained by reverse transcriptional synthesis with a HiFi-MMLV cDNA kit (Cwbio). Primers VG-F and VG-R were used to determine gene transcription.
Carbon monoxide-difference spectral analysis
VHb activity was detected by CO-difference spectra. Hemoglobin can react with CO. The complex comprising CO and reduced hemoglobin has a characteristic peak at 420 nm (Choi et al., 2003; Zhang et al., 2013). B. amyloliquefaciens NK-1 and B. amyloliquefaciens NK-PV were harvested by centrifugation at 8000 r.p.m. for 20 min at 4°C after 36-h cultivation in γ-PGA fermentation medium. Cell pellets were washed with 0.1 M potassium phosphate buffer (pH 7.2) thrice. Cells were re-suspended in 20 ml of the same buffer, and were broken with a sonicator on ice (600 W for 20 min with cycles of sonication of 30 s each and 30 s pause). The crude extract was centrifuged at 8000 r.p.m. for 20 min at 4°C to remove cell debris. Samples were divided into two aliquots. One was exposed to CO for 2 min and the other to air. Then, hemoglobin levels were obtained by CO-difference spectra using a UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan).
Analyses of formation of colonies and biofilms
Bacteria were grown on LB agar for 24 h and then incubated in LB medium to an optical density at 600 nm (OD600) of 1.0. For analyses of colony architecture, 2 μl of starting culture were spotted onto LB agar plates for 24 h at 37°C. For analyses of pellicle formation, 10 μl of starting culture were added to 10 ml of MSgg broth (Branda et al., 2006) contained within a six-well microtiter dish, and the dish incubated for 80 h at 30°C without agitation. For inducible expression of vgb gene, 0.5% xylose was added to MSgg medium. All colonies and pellicles were photographed using a digital camera equipped with a close-up lens (Canon, Tokyo, Japan).
Analytical procedures
γ-PGA was purified by a method described previously (Kubota et al., 1993). The OD of the fermentation broth was measured by the UV-1800 spectrophotometer (Shimadzu). The molecular weight of γ-PGA was measured by a gel permeation chromatography system (Cao et al., 2010). Relative viscosity was measured by a DV-I Digital Rheometer (Brookfield, Middleborough, MA, USA) fitted with a spindle S00 code at the shear rate of 10 r.p.m. (25°C). Whole polysaccharide content (%) in the γ-PGA product was measured by the phenol-sulfuric acid method (DuBois et al., 1951). The purity of γ-PGA (%) was defined as 1-polysaccharide content (%).
Conflict of interest
None declared.
References
- Ashiuchi M. Microbial production and chemical transformation of poly-γ-glutamate. Microb Biotechnol. 2013;6:664–674. doi: 10.1111/1751-7915.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiuchi M. Misono H. Biochemistry and molecular genetics of poly-γ-glutamate synthesis. Appl Biochem Biotechnol. 2002;59:9–14. doi: 10.1007/s00253-002-0984-x. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Shimanouchi K, Horiuchi T, Kamei T. Misono H. Genetically engineered poly-γ-glutamate producer from Bacillus subtilis ISW1214. Biosci Biotechnol Biochem. 2006;70:1794–1797. doi: 10.1271/bbb.60082. [DOI] [PubMed] [Google Scholar]
- Bajaj IB. Singhal RS. Enhanced production of poly (γ-glutamic acid) from Bacillus licheniformis NCIM 2324 by using metabolic precursors. Appl Biochem Biotechnol. 2009;159:133–141. doi: 10.1007/s12010-008-8427-5. [DOI] [PubMed] [Google Scholar]
- Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, et al. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol. 2007;65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R. Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R. Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L. Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Loslck R. Kolter RA. Major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Cao MF, Song CJ, Jin YH, Liu L, Liu J, Xie H, et al. Synthesis of poly(γ-glutamic acid) and heterologous expression of pgsBCA genes. BJ Mol Catal B. 2010;67:111–116. [Google Scholar]
- Cao MF, Geng WT, Liu L, Song CJ, Xie H, Guo WB, et al. Glutamic acid independent production of poly-γ-glutamic acid by Bacillus amyloliquefaciens LL3 and cloning of pgsBCA genes. Bioresour Technol. 2011;102:4251–4257. doi: 10.1016/j.biortech.2010.12.065. [DOI] [PubMed] [Google Scholar]
- Cao MF, Geng WT, Zhang W, Sun JB, Wang SF, Feng J, et al. Engineering of recombinant Escherichia coli cells co-expressing poly-γ-glutamic acid (γ-PGA) synthetase and glutamate racemase for differential yielding of γ-PGA. Microb Biotechnol. 2013;6:675–684. doi: 10.1111/1751-7915.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik GY, Aslim B. Beyatli Y. Characterization and production of the exopolysaccharide (EPS) from Pseudomonas aeruginosa G1 and Pseudomonas putida G12 strains. Carbohydr Polym. 2008;73:178–182. [Google Scholar]
- Choi SJ, Kim M, Kim SI. Jeon JK. Microplate assay measurement of cytochrome p450-carbon monoxide complexes. J Biochem Mol Biol. 2003;36:332–335. doi: 10.5483/bmbrep.2003.36.3.332. [DOI] [PubMed] [Google Scholar]
- Domínguez-Cuevas P, Porcelli I, Daniel RA. Errington J. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol. 2013;89:1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donot F, Fontana A, Baccou JC. Schorr-Galindo S. Microbial exopolysaccharides: main examples of synthesis, excretion, genetics and extraction. Carbohydr Polym. 2012;87:951–962. [Google Scholar]
- DuBois M, Gilles KA, Hamilton JK, Rebers PA. Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1951;28:350–356. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- Feng J, Gu YY, Wang JQ, Song CJ, Yang C, Xie H, et al. Curing the plasmid pMC1 from the poly (γ-glutamic acid) producing Bacillus amyloliquefaciens LL3 strain using plasmid incompatibility. Appl Biochem Biotechnol. 2013;171:532–542. doi: 10.1007/s12010-013-0382-0. [DOI] [PubMed] [Google Scholar]
- Feng J, Gao WX, Gu YY, Zhang W, Cao MF, Song CJ, et al. Functions of poly-gamma-glutamic acid (γ-PGA) degradation genes in γ-PGA synthesis and cell morphology maintenance. Appl Microbiol Biotechnol. 2014 doi: 10.1007/s00253-014-5729-0. doi: 10.1007/s00253-014-5729-0. [DOI] [PubMed] [Google Scholar]
- Frey AD. Kallio PT. Bacterial hemoglobins and flavohemoglobins: versatile proteins and their impact on microbiology and biotechnology. FEMS Microbiol Rev. 2003;27:525–545. doi: 10.1016/S0168-6445(03)00056-1. [DOI] [PubMed] [Google Scholar]
- Huang B, Qin P, Xu Z, Zhu R. Meng Y. Effects of CaCl2 on viscosity of culture broth, and on activities of enzymes around the 2-oxoglutarate branch, in Bacillus subtilis CGMCC 2108 producing poly-(γ-glutamic acid) Bioresour Technol. 2011;102:3595–3598. doi: 10.1016/j.biortech.2010.10.073. [DOI] [PubMed] [Google Scholar]
- Kalogiannis S, Iakovidou G, Liakopoulou-Kyriakides M, Kyriakidis DA. Skaracis GN. Optimization of xanthan gum production by Xanthomonas campestris grown in molasses. Process Biochem. 2003;39:249–256. [Google Scholar]
- Kang DG, Kim JYH. Cha HJ. Enhanced detoxification of organophosphates using recombinant Escherichia coli with co-expression of organophosphorus hydrolase and bacterial hemoglobin. Biotechnol Lett. 2002;24:879–883. [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R. Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Keller KL, Bender KS. Wall JD. Development of a markerless genetic exchange system for Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Appl Environ Microbiol. 2009;75:7682–7691. doi: 10.1128/AEM.01839-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Tran LS, Uchida I. Itoh Y. Characterization of Bacillus subtilis gamma-glutamyltransferase and its involvement in the degradation of capsule poly-gamma-glutamate. Microbiology. 2004;150:4115–4123. doi: 10.1099/mic.0.27467-0. [DOI] [PubMed] [Google Scholar]
- Kubota H, Matsunobu T, Uotani K, Takebe H, Satoh A, Tanaka T, et al. Production of poly(γ-glutamic acid) by Bacillus subtilis F-2-01. Biosci Biotech Biochem. 1993;57:1212–1213. doi: 10.1271/bbb.57.1212. [DOI] [PubMed] [Google Scholar]
- Liu CY. Webster DA. Spectral characteristics and interconversions of the reduced, oxidized, and oxygenated forms of purified cytochrome o. J Biol Chem. 1974;249:4261–4266. [PubMed] [Google Scholar]
- Liu J, Ma X, Wang Y, Liu F, Qia JQ, Li XZ, et al. Depressed biofilm production in Bacillus amyloliquefaciens C06 causes γ-polyglutamic acid (γ-PGA) overproduction. Curr Microbiol. 2011;62:235–241. doi: 10.1007/s00284-010-9696-0. [DOI] [PubMed] [Google Scholar]
- Marvasi M, Visscher PT. Martinez LC. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett. 2010;313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- Mitsui N, Murasawa H. Sekiguchi J. Disruption of the cell wall lytic enzyme CwlO affects the amount and molecular size of poly-γ-glutamic acid produced by Bacillus subtilis (natto) J Gen Appl Microbiol. 2011;57:35–43. doi: 10.2323/jgam.57.35. [DOI] [PubMed] [Google Scholar]
- Olano C, Lombo F, Mendez C. Salas JA. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng. 2008;10:281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Park KW, Kim KJ, Howard AJ, Stark BC. Webster DA. Vitreoscilla hemoglobin binds to subunit I of cytochrome bo ubiquinol oxidases. J Biol Chem. 2002;277:33334–33337. doi: 10.1074/jbc.M203820200. [DOI] [PubMed] [Google Scholar]
- Scoffone V, Dondi D, Biino G, Borghese G, Pasini D, Galizzi A. Calvio C. Knockout of pgdS and ggt genes improves γ-PGA yield in B. subtilis. Biotechnol Bioeng. 2013;110:2006–2012. doi: 10.1002/bit.24846. [DOI] [PubMed] [Google Scholar]
- Shih IL. Van YT. The production of poly-(γ-glutamic acid) from microorganisms and its various applications. Bioresour Technol. 2001;79:207–225. doi: 10.1016/s0960-8524(01)00074-8. [DOI] [PubMed] [Google Scholar]
- Shih IL, Yu JY, Hsieh C. Wu JY. Production and characterization of curdlan by Agrobacterium sp. Biochem Eng J. 2009;43:33–40. [Google Scholar]
- Shih IL, Chen LD. Wu JY. Levan production using Bacillus subtilis natto cells immobilized on alginate. Carbohydr Polym. 2010;82:111–117. [Google Scholar]
- Smith K. Youngman P. Use a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie. 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Blackman SA. Foster SJ. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 2000;146:249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- Su Y, Li X, Liu Q, Hou Z, Zhu X, Guo X. Ling P. Improved poly-γ-glutamic acid production by chromosomal integration of the Vitreoscilla hemoglobin gene (vgb) in Bacillus subtilis. Bioresour Technol. 2010;101:473–476. doi: 10.1016/j.biortech.2010.01.128. [DOI] [PubMed] [Google Scholar]
- Sung MH, Park C, Kim CJ, Poo H, Soda K. Ashiuchi M. Natural and edible biopolymer poly-γ-glutamic acid: synthesis, production, and applications. Chem Rec. 2005;5:352–366. doi: 10.1002/tcr.20061. [DOI] [PubMed] [Google Scholar]
- Urgun-Demirtas M, Pagilla KR, Stark BC. Webster D. Biodegradation of 2-chlorobenzoate by recombinant Burkholderia cepacia expressing Vitreoscilla hemoglobin under variable levels of oxygen availability. Biodegradation. 2003;14:357–365. doi: 10.1023/a:1025672528291. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P. Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Wakabayashi S, Matsubara H. Webster DA. Primary sequence of a dimeric bacterial hemoglobin from Vitreoscilla. Nature. 1986;322:481–483. doi: 10.1038/322481a0. [DOI] [PubMed] [Google Scholar]
- Wang PZ. Dio RH. Overlapping promoters transcribed by Bacillus subtilis σ55 and σ37 RNA polymerase holoenzymes during growth and stationary phase. J Biol Chem. 1984;259:8619–8625. [PubMed] [Google Scholar]
- Yamaguchi H, Furuhata K, Fukushima T, Yamamoto H. Sekiguchi J. Characterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named cwlO), and the enzymatic properties of its encoded protein. J Biosci Bioeng. 2004;98:174–181. doi: 10.1016/S1389-1723(04)00262-2. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Y, Wang Z, Xia Y, Chen W. Tang K. Recent developments and future prospects of Vitreoscilla hemoglobin application in metabolic engineering. Biotechnol Adv. 2007;25:123–136. doi: 10.1016/j.biotechadv.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Zhang W, Xie H, He Y, Feng J, Gao WX, Gu YY, et al. Chromosome integration of the Vitreoscilla hemoglobin gene (vgb) mediated by temperature-sensitive plasmid enhances γ-PGA production in Bacillus amyloliquefaciens. FEMS Microbiol Lett. 2013;343:127–134. doi: 10.1111/1574-6968.12139. [DOI] [PubMed] [Google Scholar]


